Abstract
Understanding interactions between proteins and RNA is key to deciphering the mechanisms of many important biological processes. Here we describe RNABindR, a web-based server that identifies and displays RNA-binding residues in known protein–RNA complexes and predicts RNA-binding residues in proteins of unknown structure. RNABindR uses a distance cutoff to identify which amino acids contact RNA in solved complex structures (from the Protein Data Bank) and provides a labeled amino acid sequence and a Jmol graphical viewer in which RNA-binding residues are displayed in the context of the three-dimensional structure. Alternatively, RNABindR can use a Naive Bayes classifier trained on a non-redundant set of protein–RNA complexes from the PDB to predict which amino acids in a protein sequence of unknown structure are most likely to bind RNA. RNABindR automatically displays ‘high specificity’ and ‘high sensitivity’ predictions of RNA-binding residues. RNABindR is freely available at http://bindr.gdcb.iastate.edu/RNABindR.
INTRODUCTION
Protein–RNA interactions are vital to a wide range of biological processes, including regulation of gene expression, protein synthesis and replication and assembly of many viruses (1–4). A more detailed understanding of protein–RNA interactions is especially important for understanding how miRNA and other non-coding RNAs regulate gene expression. The ability to computationally predict which residues of a protein directly participate in RNA-binding has already contributed to the design of wet-lab experiments to decipher mechanisms of protein–RNA recognition (5,6) and has the potential to enhance our fundamental understanding of how proteins recognize RNA.
Here we describe RNABindR, a web-based server that uses machine learning approaches to identify amino acids in a protein that are most likely to participate in RNA-binding. In previous work, we demonstrated that RNABindR can predict RNA-binding residues with high accuracy, using only the amino acid sequence of a query protein (and no information about the bound RNA) as input (7). In the current web-based implementation, RNABindR allows users to: (i) identify actual binding residues for a given protein–RNA complex in the Protein Data Bank (PDB) (8) and (ii) predict RNA-binding residues in a protein sequence whose RNA-bound structure is not available in the PDB. When calculating actual binding residues for a known structure, the only required input is the PDB ID of a protein–RNA complex and an interface distance cutoff in angstroms (Å). The RNABindR server calculates which amino acids in the protein have atoms within the defined cutoff distance of atoms in the bound RNA. It returns a display of the labeled amino acid sequence and a Jmol (www.jmol.org) graphical viewer in which RNA-binding residues are highlighted within the three-dimensional structure of the complex. To predict RNA-binding residues for a protein of unknown structure, the user must provide the amino acid sequence of a protein of interest. The RNABindR server returns the amino acid sequence with the predicted RNA-binding status (+ or −) for each residue. Three different prediction results, reflecting different expected specificity values, are provided for each query sequence, allowing users to compare results with high ‘specificity’ versus high ‘sensitivity’ for RNA-binding residues. RNABindR is designed to be fast and easy to use; results are typically returned within a few seconds. Output can be displayed as described above, or can be downloaded as a file to facilitate transfer into other programs.
MATERIALS AND METHODS
Dataset of protein–RNA interactions
A training dataset of protein–RNA interactions was extracted from structures of known protein–RNA complexes in the PDB solved by X-ray crystallography. Proteins with >30% sequence identity or structures with resolution worse than 3.5 Å were removed using PISCES (9). This resulted in a dataset, RB147, containing 147 non-redundant protein chains and a total of 32 324 amino acids. This dataset is larger than the RB109 dataset used in our previously published work (5,7), where a different method was used to define RNA-binding residues. Previously, we used the ENTANGLE program (10) to identify amino acids in contact with RNA. For the dataset used in the current implementation of RNABindR, RNA-binding residues were identified according to a distance-based cutoff definition: an RNA-binding residue is an amino acid containing at least one atom within 5 Å of any atom in the bound RNA. According to this definition, RB147 contains a total of 6157 RNA-binding residues and 26 167 non-binding residues.
Naive Bayes classifier
RNABindR uses a Naive Bayes classifier (11) as implemented in the Weka software package (12) for all predictions. A detailed description of the algorithm and evaluation of its performance on several different datasets of RNA-binding proteins has been published (7). Briefly, the Naive Bayes classifier assumes the independence of attributes. This assumption greatly reduces the complexity of the classifier. In RNABindR, the input to a Naive Bayes classifier consists of a window x = (x- n, x- n + 1,…, xT - 1, xT, xT + 1, … xn - 1, xn) of 2n + 1 contiguous amino acid identities, with n amino acid sequence residues on either side of the target residue xT. The output is an instance c ∈ { +, -} where ‘+’ indicates that the target residue xT at the center of the window is an RNA-binding residue and ‘−’ indicates xT is not an RNA-binding residue. The Naive Bayes classifier assigns the class label ‘+’ to input x if:
and the class label ‘−’ otherwise. The desired trade-off of sensitivity versus specificity can be achieved by varying θ, which is the classification threshold. Specificity is the fraction of residues predicted to be RNA-binding residues that are in fact RNA-binding residues. Sensitivity is the fraction of actual RNA-binding residues that are predicted to be RNA-binding residues by RNABindR (7,13).
Reliability of RNABindR predictions
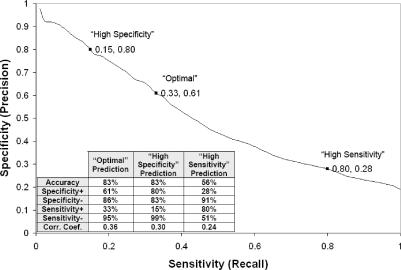
RNABindR has been evaluated using leave-one-out cross validation experiments with several different datasets of RNA-binding proteins (7). For the Naive Bayes classifier implemented in the current web-based version of RNABindR, one protein sequence was used as the test set and the other 146 sequences in the RB147 dataset were used as the training set for each round of training. This process was repeated until each protein had been used as the test set. Figure 1 depicts RNABindR performance over all values of θ and the inset table provides a summary of the average classification performance of RNABindR on the RB147 dataset, using three different values of the classification threshold, θ. The results illustrate that, as with other machine learning methods, in the RNABindR predictions there is a trade-off between the specificity (or ‘precision’) and sensitivity (or ‘recall’). Changing the value of θ changes the number of predicted RNA-binding residues and the ‘confidence’ with which binding residues are predicted. In classification tasks that involve unbalanced training sets (i.e. unequal numbers of positive and negative examples), as is the case here, the correlation coefficient (CC) is perhaps the best single parameter for comparing the ‘overall’ performance of different machine learning algorithms (13; also see 7 for further discussion and precise definitions of performance parameters used in our work.).
Figure 1.
Summary of RNABindR performance in predicting RNA-binding residues. Specificity versus sensitivity trade-off and the average performance statistics for RNABindR in leave-one-out cross-validation experiments on the RB147 dataset are shown. The plot shows the specificity and sensitivity values across the entire range of the classification threshold θ, with the ‘Optimal,’ ‘High Specificity,’ and ‘High Sensitivity’ points marked. The columns in the table show results obtained using the three different classification thresholds employed by RNABindR. The ‘Optimal Prediction’ uses the threshold value that maximizes the correlation coefficient on the training dataset; this prediction represents a balance between the competing goals of identifying as many RNA-binding residues as possible and minimizing the number of false positives. The ‘High Specificity Prediction’ identifies fewer RNA-binding residues, but with higher confidence in the positive predictions. The ‘High Sensitivity Prediction’ identifies more RNA-binding residues, but at the cost of an increased false positive rate. Definitions of performance measures are according to Baldi et al. (2001) (13). Specificity ‘+’ and ‘−’refer to specificity on the positive class (RNA-binding residues) and negative class (non-RNA-binding residues), respectively.
As shown in Figure 1, using the ‘high specificity’ classification threshold, RNABindR predicts a smaller number of RNA-binding residues, with higher confidence: 80% of the RNA-binding residues predicted for the RB147 dataset are, in fact RNA-binding residues. In contrast, using the ‘high sensitivity’ classification threshold, RNABindR predicts a larger number of RNA-binding residues, but with lower confidence: only 28% of the RNA-binding residues predicted for the RB147 dataset are actually RNA-binding. Using this high sensitivity threshold, however, a much higher fraction (∼80%) of the actual binding residues is identified. The third prediction provided by RNABindR, referred to as the ‘optimal’ prediction, uses a threshold corresponding to the value of θ that maximizes the correlation coefficient for predictions on the RB147 dataset. The ‘optimal’ prediction is not guaranteed to be the best prediction. Instead, it is a prediction in which the trade-off between specificity and sensitivity has been optimized on the training dataset.
Server description
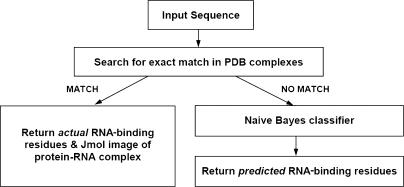
RNABindR provides two main services: (i) identification of RNA-binding residues, given the structure of a protein–RNA complex and (ii) prediction of RNA-binding residues given a protein sequence. An overview of RNABindR is provided in Figure 2.
Figure 2.
RNABindR flowchart. The query sequence is first compared with every protein sequence in every protein–RNA complex structure in the PDB to search for an exact match. If a match is found, the prediction program is not run and the actual RNA-binding residues are calculated using a distance cutoff and returned, along with an interactive Jmol image highlighting interface residues within the protein–RNA complex structure. If an exact sequence match is not identified, the Naive Bayes classifier is run and the predicted RNA-binding and non-binding residues are returned (using three different classification threshold values, see text).
Calculation of RNA-binding residues in protein–RNA complexes of known structure
Input—To identify RNA-binding residues (i.e. amino acid residues that lie in the interface between protein and bound RNA) in a known protein–RNA complex, the only required input is the PDB ID of the complex. RNABindR parses the PDB file to determine which chains in the complex are protein and which are RNA. Interactions are calculated for each protein chain with every RNA chain in the complex. For example, for a protein–RNA complex with two protein chains (A and B) and two RNA chains, (C and D), interactions will be calculated between the following pairs of chains: A and C, A and D, B and C and B and D. If desired, the user can enter a single protein chain identifier to restrict the output to only those interactions between the specified protein chain and the RNA chain(s) in the complex.
By default, RNABindR uses a distance cutoff of 5 Å between any atom of the amino acid and any atom of the RNA to determine which residues interact with the RNA. However, RNABindR allows the user to change this parameter to any desired value (between 0 and 100 Å) to make the definition of RNA-binding more or less stringent.
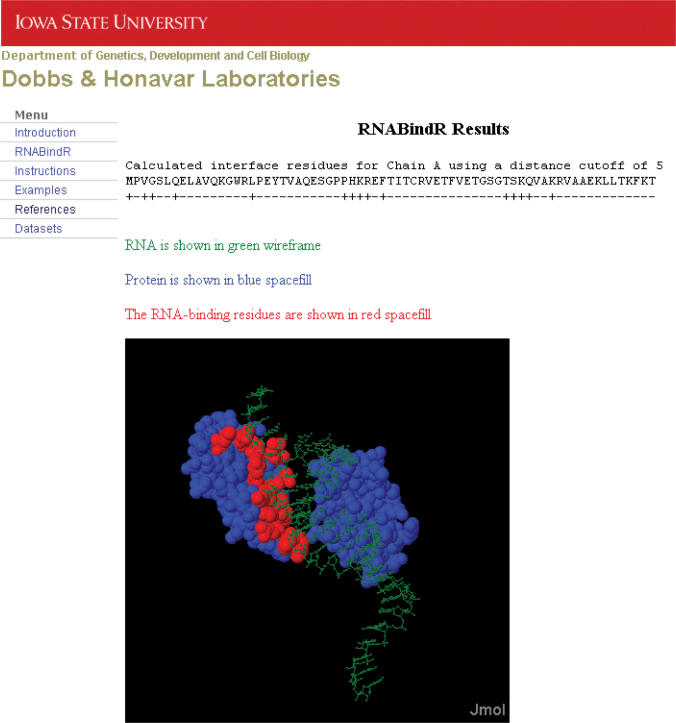
Output—Figure 3 shows an example of RNABindR output to identify RNA-binding residues in a known protein–RNA complex. The output is a display of the sequence of each chain in the complex, with a label for each residue; ‘+’ for residues that are within the specified distance cutoff and ‘−’ for residues that do not have any atoms within the distance cutoff. The calculated RNA-binding residues are also displayed on the PDB structure of the protein–RNA complex using Jmol (www.jmol.org). By default, the RNA-binding residues are displayed in red space-fill representation, the rest of the protein is displayed in blue space-fill and the bound RNA is displayed in green wireframe. Users can also print or download the text output to facilitate further analysis of the calculated RNA-binding residues.
Figure 3.
Example of RNABindR results: identifying actual RNA-binding residues in a known protein–RNA complex. RNABindR output includes the amino acid sequence of the identified protein chain(s) in the complex, with a ‘+’ label for each interacting residue (those having atoms within the selected RNA contact cutoff distance) and ‘−’ for non-binding residues. Below, a Jmol applet displays the structure of the protein–RNA complex. RNA-binding residues are displayed in red space-fill, non-binding residues in blue space-fill and the RNA in green wireframe. Users can manipulate the image using the Jmol applet.
Prediction of RNA-binding residues in proteins of unknown structure
Input—To predict RNA-binding residues in a protein of interest, the only required input is the amino acid sequence of the protein. RNABindR accepts FASTA-formatted protein sequences in the single-letter amino acid representation, but is able to read any standard amino acid sequence format; any characters (e.g. sequence numbering or blank spaces) that are not part of the standard 20-letter amino acid alphabet are ignored. After processing the sequence to remove any extra characters, RNABindR determines whether the query sequence has an exact match in any protein–RNA complexes available in the PDB. If an exact match to the query sequence is identified, the prediction program is not run. Instead, RNABindR returns the actual RNA-binding residues from the PDB complex and a Jmol image of its structure, in which the RNA-binding residues are highlighted as described above. If no exact match is found, RNABindR predicts RNA-binding residues in the query protein sequence. In the current implementation, RNABindR predictions are made using a Naive Bayes classifier trained on all 147 protein chains in the RB147 dataset; the input query sequence is used as the test case.
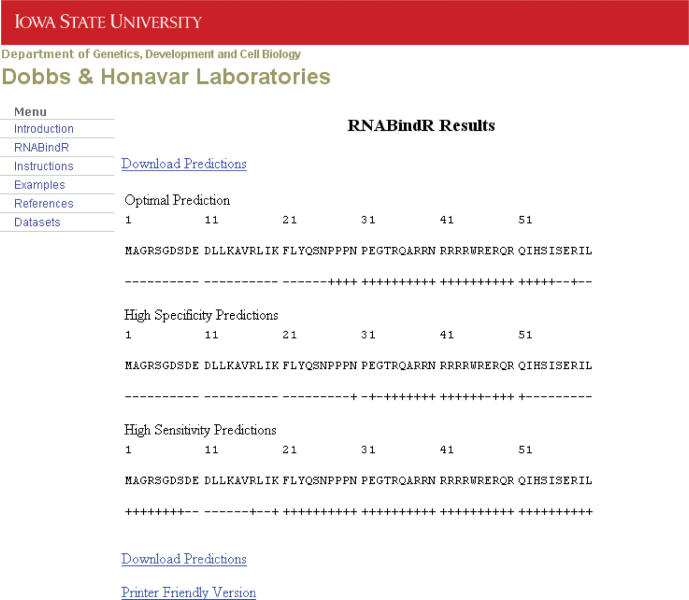
Output—Figure 4 shows an example of RNABindR output obtained for predicting RNA-binding residues in a protein of unknown structure. The input amino acid sequence is shown at the top, and labels ‘+’ and ‘−’ for predicted RNA-binding and non-binding residues, respectively, are shown immediately below the sequence. Users can also print or download the text output to facilitate further analysis of the predicted RNA-binding residues.
Figure 4.
Example of RNABindR results: predicting RNA-binding residues in a protein of unknown structure. RNABindR output includes the query sequence and three predictions obtained using three different classification thresholds. Residues predicted to bind RNA are indicated by ‘+’ and non-binding residues by ‘−’ on the line below the sequence. The ‘optimal prediction’ uses the threshold value that maximizes the correlation coefficient on the RB147 dataset. The ‘high specificity prediction’ provides fewer predicted RNA-binding residues, with higher confidence, and the ‘high sensitivity prediction’ provides more predicted RNA-binding residues, but with lower confidence. Links are provided for downloading the predictions in a text-only format or a printer friendly format.
Typical users of RNABindR may have different goals in mind when predicting RNA-binding residues: some may wish to identify a relatively small number of amino acids predicted to bind RNA with ‘high confidence,’ while others may wish to identify as many potential RNA-binding residues as possible, with more potentially ‘false positive’ predictions. To accommodate these different uses, RNABindR displays three different predictions for each query sequence: an ‘optimal’ prediction, a ‘high specificity’ prediction and a ‘high sensitivity’ prediction. As discussed above, the high specificity prediction uses a more stringent classification threshold to identify the most likely RNA-binding residues, whereas the high sensitivity prediction uses a less stringent threshold to identify more potential RNA-binding residues. Because the reliability of RNABindR predictions for any particular protein depends on the extent to which the query protein shares features that are ‘captured’ by the Naive Bayes classifier (during training on the RB147 dataset), prediction performance for any particular query sequence cannot be guaranteed. The three types of predictions are supplied as a guide to help the user make best use of RNABindR predictions.
Related servers
Predicting RNA-binding residues has proven to be an important and difficult computational task (7,14–16). Since RNABindR was developed, two other web-based servers for RNA-binding site predictions have become available, BindN (14) and KYG (15). BindN (http://bioinformatics.ksu.edu/bindn) uses a support vector machine (SVM) to predict both RNA-binding and DNA-binding residues in a protein sequence. BindN is a sequence-based server, requiring only the amino acid sequence of a query protein. The feature vector used as input to the SVM classifier consists of the side chain pKa value, hydrophobicity index and molecular mass for each amino acid in a window of 11 residues. The BindN server requires users to choose an estimated specificity or sensitivity, which is used to determine the classification threshold (14). KYG (http://yayoi.kansai.jaea.go.jp/qbg/kyg/index.php) provides several methods for statistically analyzing a protein structure and predicting RNA-binding residues. KYG is a structure-based server and relies on estimating the interface propensity for single amino acids and pairs of amino acids. KYG also utilizes evolutionary information in the form of a multiple sequence alignment profile, which must be supplied by the user. Users are allowed to choose among nine different predictions, each of which is based on a different combination of residue propensities and profile scores. The KYG server can predict RNA-binding residues only for those proteins whose structures are known.
RNABindR offers some potential advantages over BindN and KYG. RNABindR has been designed to be user-friendly and widely applicable. Like BindN, RNABindR requires only a protein sequence as input, so researchers can obtain predictions for any protein sequence of interest. RNABindR does not require users to specify any parameters or choose between different methods. Also, RNABindR provides a quick and easy way to visualize RNA-binding residues and examine the protein–RNA interface(s) within the three-dimensional structure of any known protein–RNA complex.
RNABindR, BindN and KYG each use different methods, are trained on different datasets and often provide different predictions of RNA-binding residues for the same query protein sequence. Users may use all three servers and apply their biological expertise regarding their protein of interest to determine which predictions warrant further investigation.
SIGNIFICANCE AND FUTURE DIRECTIONS
Over the last decade, there has been a dramatic increase in the number of available structures of protein–nucleic acid complexes: the Protein Data Bank (PDB) included only 198 protein–nucleic acid complexes in 1996, but by April 2007, this number had grown to 1734, of which 529 were protein–RNA complexes (PDB, accessed April 3, 2007, http://www.pdb.org).The resulting availability of larger and more diverse training sets can be expected to significantly improve the performance of RNABindR. RNABindR will be updated periodically to take advantage of the latest data available in the PDB. A beta-version with three types of enhancements is under development. In recent work, we have generated a comprehensive database that includes every protein–RNA interface for which structural information is available in the PDB. The next version of RNABindR will incorporate this complete database. Users will have the option of choosing a classifier that is trained on the comprehensive dataset or on one of several ‘non-redundant’ datasets (e.g. RB 147). Alternatively, users will be able to train a new classifier using a ‘customized’ training dataset (e.g. any subset of known protein–RNA complexes, chosen based on similarities in sequence or biological function). Recent unpublished and earlier published results (7) indicate that using such training datasets can provide a significant increase in the reliability of RNA-binding site predictions. A second enhancement will be to allow users to choose among several machine learning algorithms (e.g. SVMs) or statistical methods that have been shown to be effective for RNA-binding site prediction by our group and by others (5,7,14–16). Third, RNABindR will allow users to take advantage of structural and/or evolutionary information, when available. If the structure of a query protein is available in the PDB (but the structure of the query protein in complex with RNA is not), predicted RNA- binding residues will be identified and displayed on the three-dimensional structure of the protein, as is done for calculated RNA-binding residues in known protein–RNA complexes in the current implementation (see Figure 3). In the longer term, structural predictions will also be included for such RNA-binding sites, based on structure fragment libraries and other homology modeling approaches.
Protein–RNA interactions play many essential and diverse roles in biological regulation, ranging from structural and catalytic roles in ribosomes and spliceosomes, to regulatory roles in microRNA-mediated gene regulation and cellular signaling, to storage and propagation of genetic information (17–20). Despite their obvious functional importance, the details of the molecular mechanisms of protein–RNA recognition are still poorly understood. The impressive diversity of structures and functions of protein–RNA complexes makes understanding what dictates specificity in protein–RNA interaction an especially challenging problem (18). Hence, computational tools for analyzing protein–RNA interfaces and for predicting RNA-binding sites in proteins are becoming increasingly important for deciphering the amino acid sequence and structural underpinnings of protein–RNA interactions (7,14–16,21–25). RNABindR predictions have already helped guide the experimental investigation of the RNA-binding domains in proteins (5,6). Approaches that combine computational prediction and experimental validation of RNA-binding sites in proteins will increase our understanding of the mechanisms of protein–RNA recognition.
ACKNOWLEDGEMENTS
We are grateful to members of our groups and colleagues who have tested RNABindR and provided valuable feedback. This work was supported in part by NIH grant GM066387 and by graduate research assistantships provided by USDA MGET grant 2001-52100-11506, the ISU Center for Integrated Animal Genomics (CIAG) and the ISU Center for Computational Intelligence, Learning, and Discovery (CCILD). Funding to pay the Open Access publication charges for this article was provided by NIH grant GM066387.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 4.Freed EO, Mouland AJ. The cell biology of HIV-1 and other retroviruses. Retrovirology. 2006;3:77. doi: 10.1186/1742-4690-3-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terribilini M, Lee JH, Yan C, Jernigan RL, Carpenter S, Honavar V, Dobbs D. Identifying interaction sites in “recalcitrant” proteins: predicted protein and RNA binding sites in rev proteins of HIV-1 and EIAV agree with experimental data. Pac. Symp. Biocomput. 2006:415–426. [PMC free article] [PubMed] [Google Scholar]
- 6.Bechara E, Davidovic L, Melko M, Bensaid M, Tremblay S, Grosgeorge J, Khandjian EW, Lalli E, Bardoni B. Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Res. 2007;35:299–306. doi: 10.1093/nar/gkl1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terribilini M, Lee JH, Yan C, Jernigan RL, Honavar V, and Dobbs D. Prediction of RNA binding sites in proteins from amino acid sequence. RNA. 2006;12:1450–1462. doi: 10.1261/rna.2197306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Dunbrack RL., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 10.Allers J, Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:2746–2752. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell T. Machine Learning. 1997 McGraw-Hill, Boston, MA. [Google Scholar]
- 12.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 13.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34(Web Server issue):W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim OT, Yura K, Go N. Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Res. 2006;34:6450–6460. doi: 10.1093/nar/gkl819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong E, Miyano S. A weighted profile method for protein-RNA interacting residue prediction. Trans. On Comput. Syst. Biol. 2006;IV:123–139. [Google Scholar]
- 17.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272:2088–2097. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 20.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 21.Auweter SD, Oberstrass FC, Allain FHT. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draper DE. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 23.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein–RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treger M, Westhof E. Statistical analysis of atomic contacts at RNA–protein interfaces. J. Mol. Recognit. 2001;14:199–214. doi: 10.1002/jmr.534. [DOI] [PubMed] [Google Scholar]
- 25.Kim H, Jeong E, Lee SW, Han K. Computational analysis of hydrogen bonds in protein–RNA complexes for interaction patterns. FEBS Lett. 2003;552:231–239. doi: 10.1016/s0014-5793(03)00930-x. [DOI] [PubMed] [Google Scholar]