Abstract
MobilomeFINDER (http://mml.sjtu.edu.cn/MobilomeFINDER) is an interactive online tool that facilitates bacterial genomic island or ‘mobile genome’ (mobilome) discovery; it integrates the ArrayOme and tRNAcc software packages. ArrayOme utilizes a microarray-derived comparative genomic hybridization input data set to generate ‘inferred contigs’ produced by merging adjacent genes classified as ‘present’. Collectively these ‘fragments’ represent a hypothetical ‘microarray-visualized genome (MVG)’. ArrayOme permits recognition of discordances between physical genome and MVG sizes, thereby enabling identification of strains rich in microarray-elusive novel genes. Individual tRNAcc tools facilitate automated identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites and other integration hotspots in closely related sequenced genomes. Accessory tools facilitate design of hotspot-flanking primers for in silico and/or wet-science-based interrogation of cognate loci in unsequenced strains and analysis of islands for features suggestive of foreign origins; island-specific and genome-contextual features are tabulated and represented in schematic and graphical forms. To date we have used MobilomeFINDER to analyse several Enterobacteriaceae, Pseudomonas aeruginosa and Streptococcus suis genomes. MobilomeFINDER enables high-throughput island identification and characterization through increased exploitation of emerging sequence data and PCR-based profiling of unsequenced test strains; subsequent targeted yeast recombination-based capture permits full-length sequencing and detailed functional studies of novel genomic islands.
INTRODUCTION
Comparative analyses of multiple bacterial genomes have revealed that some bacterial species possess an extremely plastic genome (1,2). Horizontal gene transfer events have led to the integration of foreign DNA segments into species-specific syntenic backbones, often within tRNA and tmRNA gene sites (3,4). This ‘optional’ genomic repertoire, termed ‘mobilome’ (mobile genome) (5,6), which can vary considerably between members of the same bacterial species, includes episomal plasmids, transposons, integrons, prophages and a growing list of pathogenicity islands (PAIs) or genomic islands (GIs) (2,7). Many in silico approaches for detecting mobile genetic elements in sequenced bacterial genomes have been developed recently. These include methods based on anomalous codon usage, G+C content, dinucleotide bias, and amino acid usage patterns (8–11), identification of archetypal GI-specific features (12) and comparative genomics (3,13); for excellent reviews see (1,5,14).
The main barrier to high-throughput prospecting of the mobilome has been a paucity of bacterial genome sequence information, and so it has become a major challenge to develop rapid and cost-effective approaches to discover strain-specific DNA that is dispersed amongst hundreds of members of bacterial species of principal interest to man (1). Recently we have developed a high-throughput strategy, dubbed MobilomeFINDER, for experimental and in-silico discovery of bacterial GIs (Figure 1). This approach combines the newly proposed ‘MAmP’ (6), ‘tRNAcc’ (3) and ‘tRIP’ (3) comparative genomics-based approaches with an experimental island capture step facilitated by island probing (15) and/or a yeast-based homologous recombination system (16). MAmP (Microarray-Assisted mobilome Prospecting) is underpinned by comparative genomic hybridization (CGH), ArrayOme (6) and pulsed-field gel electrophoresis (PFGE) genome sizing (Figure 1A) and is used to screen large numbers of isolates to identify strains that are particularly rich in mobilome DNA sequences to which the species meta-array would have been ‘blind’. tRNAcc (tRNA gene contents and contexts analysis), complemented by an in silico PCR approach (Figure 1B), is used to identify putative GIs in closely related complete and near-complete genomes. Finally, the tRIP (tRNA site interrogation for pathogenicity islands, prophages and other GIs) (Figure 1C) strategy permits high-throughput experimental identification and characterization of new GIs through PCR-based profiling of MAmP-selected or otherwise chosen test strains, followed by large-scale targeted capture and full-length sequencing of GIs. We have now incorporated and improved the previously reported ArrayOme (6) and tRNAcc standalone tools (3) into a user-friendly MobilomeFINDER web-server as a public resource: http://mml.sjtu.edu.cn/MobilomeFINDER/.
Figure 1.
MobilomeFINDER island discovery strategy: (A) MAmP facilitates identification of strains rich in microarray-elusive novel DNA (6), (B) tRNAcc identifies putative GIs in complete and near-complete closely related genomes (3), (C) tRIP PCR permits high-throughput identification and characterization of new GIs in test strains which can then be systematically targeted for capture (15,16) and full-length sequencing. ‘Island probing’ refers to a technique of tagging GIs with a dual positive–negative selectable cassette to allow for subsequent marker rescue and/or GI deletion experiments to size and further characterize islands (15). The yeast recombinational method is based on an E. coli–yeast shuttle vector that is constructed to carry targeting sequence homologues to conserved sequences flanking the putative island (16, 28). See text for further details.
ANALYSIS TOOLS
The MobilomeFINDER web-server comprises a flexible suite of individual interactive tools that greatly facilitate high-throughput experimental and in-silico discovery of bacterial GIs in closely related bacteria (Table 1; Figure 1).
Table 1.
Interactive tools available at the MobilomeFINDER web-server that facilitate the process of bacterial genomic island or mobile genome (mobilome) discovery
| Tool | Description |
|---|---|
| Microarray-assisted mobilome prospecting | Identify strains that are particularly rich in novel mobilome DNA; underpinned by CGH, ArrayOme and PFGE (Figure 1A) |
| ArrayOme | Estimate the cumulative size of parts of the genome that harbour genes identified as ‘present’ by microarray-based CGH; the size of the novel mobilome is thus predicted by comparing the microarray-visualized genome size with the PFGE-measured chromosome size |
| Island identificationa | Identify GIs by tRNAcc analysis of closely related bacteria (Figure 1B) |
| IdentifyIsland | Identify putative islands based on conserved flanking blocks recognized by the multiple aligner Mauve 1.2.2 (18) |
| TabulateIsland | Tabulate islands identified by Identify- Island following analysis of different subsets of genomes |
| LocateHotspots | Locate proposed hotspots in non-annotated chromosomal sequences using BLASTN-based searches |
| IslandScreen | Identify putative islands by single-step crude analysis with combination of IdentifyIsland, LocateHotspots and DNAnalyser. |
| Primer designa | |
| ExtractFlank | Generate ClustalW-derived MSA files that contain upstream or downstream flanking regions of identified islands to serve as inputs for Primaclade-facilitated (21) design of conserved PCR primers |
| insilicotRIP | Interrogate identified hotspots for the presence or absence of an integrated element using a locally installed version of Electronic PCR (22); generates 0–500 kb virtual fragments |
| Island analysisa | |
| DNAnalyser | Calculate the GC content and dinucleotide bias of identified islands, and plot the negative cumulative GC profile of genomes |
| GenomeSubtrator | High throughput BLASTN-based comparison of CDS sequences against test genomes to identify strain-specific CDS based on the level of nucleotide similarity |
aThese tools can also be used for the generic identification and preliminary characterization of putative genomic islands located at other user-specified hotspots and for the analysis of cognate flanking sequences.
GI, genomic island; CGH, comparative genomic hybridization; PFGE, pulsed-field gel electrophoresis; MSA, multiple sequence alignment; CDS, protein coding sequence.
ArrayOme web-interface: a tool for identification of strains rich in novel mobilome DNA
Microarray-derived CGH data identifies genes common to both the microarray used and the genome of the strain under investigation. This complement of genes represents an entity we previously defined as the microarray-visualized genome (MVG) (6). Briefly, ArrayOme produces an accurate estimate of MVG sizes based on microarray-CGH data alone (Figure 1A; Table 1). The sizes of novel mobilomes borne by individual strains can be estimated by comparing MVG sizes with PFGE-measured chromosome sizes to identify isolates rich in novel or highly divergent genetic material that are likely to carry unique GIs or prophages (Figure 1A).
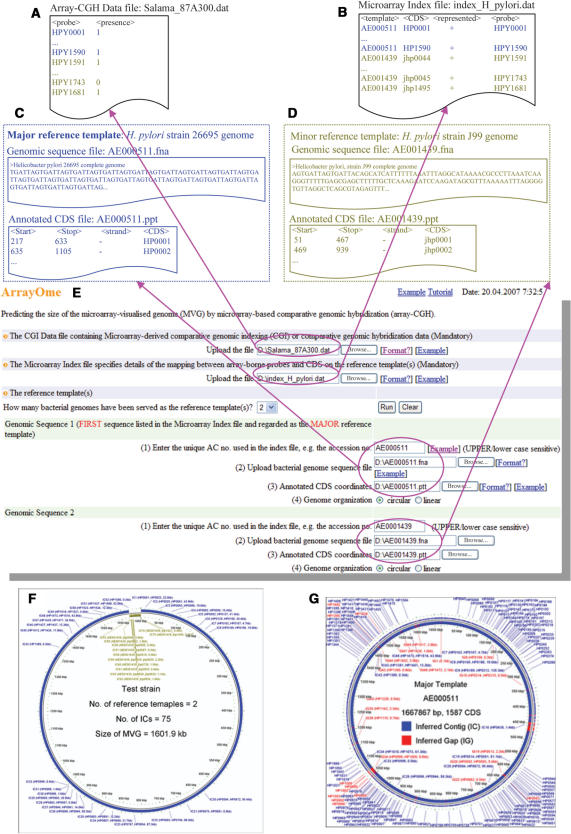
The ArrayOme web-interface that we have now developed provides a universally accessible biologist-friendly tool that is complemented by a broad repertoire of DNA-prospecting accessories within the wider MobilomeFINDER web-based resource. To highlight the ease of use and benefits of this new tool we present the example of MVG size determination for Helicobacter pylori strain 87A300 using a previously published two-strain array (26695 and J99) CGH data set (17). The ArrayOme web-interface (Figure 2E) utilizes input data comprising: (i) a microarray-derived CGH dataset (Figure 2A), (ii) a microarray-specific index file in which array probes are mapped to CDS on either the major or minor reference templates (Figure 2B), (iii) major DNA template (H. pylori 26695 genome) files that provide location and size information for all CDS detectable with the particular microarray used (Figure 2C), and (iv) minor template (H. pylori J99 genome) files (Figure 2D). Guidance on creation and/or sourcing of files is readily accessible via ‘Format’ and ‘Example’ links. Similarly, the selection of options to maximize the utility and adaptability of ArrayOme for individual user applications is facilitated through push buttons, file-browsers and tick-boxes and explanatory notes. A major enhancement is the hyperlink-embedded graphical virtual genome map constructed by stringing together inferred contigs (ICs) that have been produced by merging adjacent genes classified as ‘present’. The ICs are ordered based on major template coordinates followed by a tail made up of ICs derived from each of the minor templates in turn. The hypothetical MVG size and the contribution of each template to the virtual genome of a test strain is displayed (Figure 2F). The hyperlinks allow visualization of individual ICs using the NCBI Sequence Viewer v2.0 utility; subsequent selection of the NCBI ‘Protein coding genes’ link opens up the wider knowledge repository specific to genes within the selected IC. ArrayOme web-server also generates, a second graphical output comprising a circular map of the major template only that highlights the locations and sizes of identified ICs and inferred gaps (IGs) within this template (Figure 2G); IGs refer to contiguous regions defined as ‘absent’ following ArrayOme analysis. Hyperlinks within this output also allow access to individual NCBI Entrez Gene entries. High-quality image files in a PNG format, text files listing coordinates and CDS contents of ICs/IGs and a detailed ArrayOme result file are also generated.
Figure 2.
The ArrayOme web-tool. (A) Test strain CGH data file (17); (B) microarray index file that maps array probes to template CDS; (C–D) genome sequence and the annotated CDS files for major and minor reference templates, respectively; (E) input screenshot; (F) virtual MVG map of test strain; (G) major reference template map showing inferred contigs and gaps. See text for details.
MobilomeFINDER web-tools for genomic island identification by comparative analysis
The MobilomeFINDER web-server streamlines identification of GIs by comparative analysis of tRNAcc and other integration hotspots (Figure 1B). Briefly, IdentifyIsland (Table 1) exploits the multiple sequence aligner Mauve 1.2.2 (18) to investigate whether tRNA sites across multiple genomes are occupied by anomalous strain-specific DNA segments lying between the 3’-ends of tRNA genes and downstream conserved flanks. In addition to the original multi-step, manually curated procedure (Figure 3), MobilomeFINDER supports single-step, entirely automated crude analysis using IslandScreen, a newly available tool that has been developed by integrating LocateHotspots, IdentifyIsland and DNAnalyser. We have described the tRNAcc strategy and individual tools in detail previously (3). Table 1 outlines key features of the tools. The web-interface varies with each tool but typically consists of an input and run parameter page and status and results pages. For example, with IdentifyIsland the input page permits uploading of the genome sequence data and details of tRNA sites to be interrogated (Figure 3A), whilst with DNAnalyser results pages display feature tables and graphical representations of islands (Figure 3G–H). In the example shown, the complete and near-complete genome sequences of Klebsiella pneumoniae MGH 78578 and Kp342, respectively (Figure 3A–C), were submitted. In addition, the 87 tRNA/tmRNA genes to be interrogated and their locations in the two genomes were also entered into the interface. After automated IdentifyIsland analysis (Figure 3D), potentially occupied sites were manually examined to exclude misassignments [see Ou et al. (3) for details].
Figure 3.
Web-based tRNAcc analysis of two Klebsiella pneumoniae genomes. (A) IdentifyIsland inputs specifying the K. pneumoniae genome sequences and the coordinates of tRNA genes. (B) MGH 78578 chromosomal map with tRNA genes marked. (C) Kp342 contigs were compared with the MGH 78578 genome using PipMaker (31) and ordered by coordinates of best alignment to produce a ‘virtual’ Kp342 genome. (D) IdentifyIsland output listing boundary coordinates of putative islands. (E) DNAnalyser input interface. (F–H) DNAnalyser output data for the two K. pneumoniae genomes analysed. See text for details.
MobilomeFINDER web-tools for detection of foreign DNA signatures
The manually edited IdentifyIsland output file is then fed into DNAnalyser (Figure 3E), which generates a tabulated output of key island features including location, size, GC content and dinucleotide frequency distribution (10) (Figure 3F). Furthermore, the web-based DNAnalyser is an intuitive tool that: (i) draws a circular genome map with the locations of identified GIs marked (Figure 3G) and (ii) plots the negative cumulative GC profile, in which a sharp upward spike indicates a relatively sharp increase in GC content whereas an abrupt fall indicates a relatively sharp decrease in GC content (11) (Figure 3H), thus highlighting the wider genomic context of islands. Hyperlinks to the NCBI database in output tables and schematics permit visualization of islands and access to annotation data. In the example presented, amongst other entities we identified a 33 kb prophage-like mobile element inserted into a met tRNA gene site (56_Met) in the K. pneumoniae MGH 78578 chromosome that possessed an integrase gene, recognizable flanking direct repeats and exhibited distinct GC content and dinucleotide usage; all features typical of archetypal integrative elements. The web-based GenomeSubtractor performs in silico ‘subtractive hybridization’ and outputs data in a tabulated form showing the locations of single and clustered strain-specific CDS based on the length of match and degree of identity (3,19).
Web-tools for high-throughput design and validation of tRIP primers
The newly developed ExtractFlank (Table 1) web-interface facilitates one-step extraction and ClustalW (20) mediated alignment of upstream (UF) and downstream (DF) hotspot flanking regions for the set of genomes being investigated. The multiple sequence alignment files are then fed into Primaclade (http://www.umsl.edu/services/kellogg/primaclade.html) (21) to facilitate the design of tRIP PCR primers specific for each flank. The newly developed insilicotRIP tool, supported by Electronic PCR (22), allows multiple primer pairs to be checked simultaneously for specificity versus numerous hotspots in multiple genomes (Table 1). With ready browser access to primer set and genome template files, a 500 kb cap on virtual amplicon size and an output comprising amplicon sizes and FASTA-formatted sequences, the insilicotRIP tool serves as a generic multiplex-PCR tool that is ideal for large-scale primer validation and/or in silico PCR-based genomic exploration.
IMPLEMENTATION
MobilomeFINDER runs on a Linux platform and has integrated and improved the ArrayOme (6) and tRNAcc standalone packages (3) that we reported previously. Specific enhancements include: (i) a newly developed insilicotRIP tool, (ii) an intuitive DNAnalyser tool that generates additional outputs comprising a circular genome map with the locations/sizes of GIs marked and a plot of the negative cumulative GC profile of the genome, (iii) a schematic ArrayOme output, (iv) hyperlinks to visualize DNA fragment details using NCBI Sequence Viewer, (v) an ExtractFlank tool that automatically generates ClustalW multiple sequence alignment files and (vi) ‘example’ and ‘format’ prompts, ready access to tutorial files and enhanced warnings re file formatting errors aid file construction and entry. In addition, the following freely available components were employed: Mauve 1.2.2 (18); NCBI Blast 2.2.9 (23); ClustalW (20); Primaclade (21); Electronic PCR (22); CGview (24); gnuplot (http://www.gnuplot.info) and Bioperl (25). Each run is assigned a job-id and the output files are kept on the server for 7 days allowing the user to inspect the results at any given time. The server web site includes a step-by-step tutorial for general users as well as detailed technical documentation and the open source codes of tRNAcc and ArrayOme for software developers. In addition, users can download the standalone versions of tRNAcc and ArrayOme to run locally.
Because sequence alignment algorithms, such as the multigenome comparison tool Mauve (18) and the pairwise alignment tool BLAST (23), are computationally intensive, it may not be possible to return results to users immediately when the input is large. With the current hardware configuration using two Dual-Core Intel Xeon 2.8GHz processors and 8GB RAM, the MAUVE-facilitated tool IdentifyIsland takes about 1 h to discover islands by comparative analysis of the contents and contexts of ∼80 tRNA sites across three closely related bacterial genomes. Three tools, LocateHotspots and GenomeSubtractor that use BLASTN and IdentifyIsland that uses MAUVE, display a URL for subsequent retrieval of results if the job cannot be completed promptly. Alternatively, if users supply their e-mail address, results will be emailed automatically upon completion of the job.
APPLICATIONS
MobilomeFINDER and the related experimental methodologies are applicable to a wide range of bacterial species. To date the web-server has been used to perform comparative bacterial genomic analyses for several species including nine Escherichia coli genomes, four Salmonella enterica genomes, two K. pneumoniae genomes (Figure 3), two Pseudomonas aeruginosa genomes and two Streptococcus suis genomes; the resulting data are shown at http://mml.sjtu.edu.cn/MobilomeFINDER/database.htm.
We have used the MobilomeFINDER web-server to characterize the GI contents of blood culture-derived E. coli isolates obtained from patients with no laboratory evidence of concurrent urinary tract infections. CGH analyses using the ShE.coli metagenome microarray (http://www.ifr.ac.uk/safety/microarrays/) (6), together with PFGE-based genome sizing has been used to identify mobilome-rich strains by MAmP (Figure 1A). In addition, PCR-based tRNA site interrogation (tRIP) (Figure 1C) coupled with chromosome walking and sequencing has been used to investigate sixteen tRNA loci in ten selected E. coli isolates. Approximately half of the 85 GIs identified were related to UPEC strain CFT073 islands, with an equal number resembling elements in Shigella and EAEC, EHEC, EPEC pathotypes of E. coli. Based on a limited preview data, at least seven GIs contained sequences novel to E. coli, with six possessing stretches of sequence without any counterparts in the entire DNA database (K. Rajakumar, unpublished data). In addition to the 95 E. coli GIs we identified within sequenced genomes in our recent study (3), we have also discovered by insilicotRIP analysis a large leuX tRNA gene-associated GI that contains a likely DNA modifying dnd gene cluster (26,27) within the unfinished genome of enterotoxigenic (ETEC) E. coli B7A (RefSeq accession no. NZ_AAJT00000000).
YEAST RECOMBINATIONAL SYSTEM-BASED CAPTURE OF GENOMIC ISLANDS
The yeast recombinational capture system, originally described by Raymond et al. (28) and subsequently modified by Wolfgang et al. (16), is a laboratory complement of the MobilomeFINDER tool. A single capture vector is constructed carrying targeting sequence homologues to conserved sequences flanking the putative island insertion site (Figure 1C). It can subsequently be used to capture and characterize in detail chromosomal intervals from any number of strains (16). A set of plasmids has been engineered and these are available on request from Stephen Lory (Email: stephen_lory@hms.harvard.edu). Additional information about the system is provided online at http://mml.sjtu.edu.cn/MobilomeFINDER/ycv.htm.
CONCLUSION
The MobilomeFINDER web-server has been developed to facilitate high-throughput experimental and in-silico discovery of bacterial GIs by combining MAmP, tRNAcc, tRIP and other related approaches. We present it as a comprehensive, comparative-genomics-based mobilome discovery platform dedicated to biologists. It is clear that even with current high-throughput genomic sequencing facilities (29) it will not be feasible to sequence hundreds of isolates to identify and decode the novel gene pool accessible to each and every bacterial species. Furthermore, microarray CGH data alone is limited to genes represented on the array and provides no insight as to the extent of novel DNA in a test strain. We propose that a strategy such as MobilomeFINDER will help address this challenge by facilitating the identification of key mobilome-rich strains and the rapid, high-throughput discovery and characterization of GIs, thereby focussing increased research effort on understanding the role of the bacterial pan-genome. In combination with shotgun sample pyrosequencing (29,30), the MobilomeFINDER strategy could even aid prioritization of strains for full-length genome characterization, thus maximizing the ‘genetic return’ per genome sequenced.
ACKNOWLEDGEMENTS
We are grateful to James Lonnen, Mansi Mukesh Patel and Jon van Aartsen for support in developing this resource, and to many other colleagues for testing and suggesting enhancements to MobilomeFINDER. We are also grateful to Dr Ling-Ling Chen at Shandong University of Technology, China for critical reading of the manuscript and many valuable comments. We thank the Institute for Genomic Research (TIGR) and the Genome Sequencing Centre at Washington University in St Louis for their policy of making preliminary sequence data publicly available and acknowledge the use in this study of unpublished genome data corresponding to K. pneumoniae strain Kp342 and MGH 78578, respectively. This study was supported by grants from the 863 program, Ministry of Science and Technology of China (Grant No. 2006AA02Z328) to H.Y.O.; National Science Foundation of China (30500285/c0110) to X.H.; and a mediSearch grant from The Leicestershire Medical Research Foundation to K.R. and M.R.B. Work in the Hinton lab was supported by a Core Strategic Grant from the BBSRC. E.H. was supported by a MRC/University of Leicester PhD studentship and A.B.T. by the University of Bahrain. Work in S.L.'s laboratory was supported by the NIH grant (GM068516). Funding to pay the Open Access publication charges for this article was provided by the 863 program from the Ministry of Science and Technology, China (2006AA02Z328).
Conflict of interest statement. None declared.
REFERENCES
- 1.Binnewies TT, Motro Y, Hallin PF, Lund O, Dunn D, La T, Hampson DJ, Bellgard M, Wassenaar TM, et al. Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct. Integr. Genomics. 2006;6:165–185. doi: 10.1007/s10142-006-0027-2. [DOI] [PubMed] [Google Scholar]
- 2.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 3.Ou HY, Chen LL, Lonnen J, Chaudhuri RR, Thani AB, Smith R, Garton NJ, Hinton J, Pallen M, Barer MR, et al. A novel strategy for the identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites in closely related bacteria. Nucleic Acids Res. 2006;34:e3. doi: 10.1093/nar/gnj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 6.Ou HY, Smith R, Lucchini S, Hinton J, Chaudhuri RR, Pallen M, Barer MR, Rajakumar K. ArrayOme: a program for estimating the sizes of microarray-visualized bacterial genomes. Nucleic Acids Res. 2005;33:e3. doi: 10.1093/nar/gni005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiao W, Wan I, Jones SJ, Brinkman FS. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics. 2003;19:418–420. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- 10.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Zhang CT. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics. 2004;20:612–622. doi: 10.1093/bioinformatics/btg453. [DOI] [PubMed] [Google Scholar]
- 12.Mantri Y, Williams KP. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 2004;32:D55–D58. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiapello H, Bourgait I, Sourivong F, Heuclin G, Gendrault-Jacquemard A, Petit MA, El Karoui M. Systematic determination of the mosaic structure of bacterial genomes: species backbone versus strain-specific loops. BMC Bioinformatics. 2005;6:171. doi: 10.1186/1471-2105-6-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, Miyada CG, Lory S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salama N, Guillemin K, McDaniel TK, Sherlock G, Tompkins L, Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl Acad. Sci. USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukiya S, Mizoguchi H, Tobe T, Mori H. Extensive genomic diversity in pathogenic Escherichia coli and Shigella Strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 2004;186:3911–3921. doi: 10.1128/JB.186.12.3911-3921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gadberry MD, Malcomber ST, Doust AN, Kellogg EA. Primaclade–a flexible tool to find conserved PCR primers across multiple species. Bioinformatics. 2005;21:1263–1264. doi: 10.1093/bioinformatics/bti134. [DOI] [PubMed] [Google Scholar]
- 22.Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7:541–550. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 25.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. A novel DNA modification by sulphur. Mol. Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X, He X, Li A, Lei F, Kieser T, Deng Z. Streptomyces coelicolor A3(2) lacks a genomic island present in the chromosome of Streptomyces lividans 66. Appl. Environ. Microbiol. 2004;70:7110–7118. doi: 10.1128/AEM.70.12.7110-7118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, Zhou Y, Kaul R, Clendenning JB, et al. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 2002;184:3614–3622. doi: 10.1128/JB.184.13.3614-3622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spencer DH, Kas A, Smith EE, Raymond CK, Sims EH, Hastings M, Burns JL, Kaul R, Olson MV. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 2003;185:1316–1325. doi: 10.1128/JB.185.4.1316-1325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]