Abstract
This article presents a web server iPDA, which aims at identifying the disordered regions of a query protein. Automatic prediction of disordered regions from protein sequences is an important problem in the study of structural biology. The proposed classifier DisPSSMP2 is different from several existing disorder predictors by its employment of position-specific scoring matrices with respect to physicochemical properties (PSSMP), where the physicochemical properties adopted here especially take the disorder propensity of amino acids into account. The web server iPDA integrates DisPSSMP2 with several other sequence predictors in order to investigate the functional role of the detected disordered region. The predicted information includes sequence conservation, secondary structure, sequence complexity and hydrophobic clusters. According to the proportion of the secondary structure elements predicted, iPDA dynamically adjusts the cutting threshold of determining protein disorder. Furthermore, a pattern mining package for detecting sequence conservation is embedded in iPDA for discovering potential binding regions of the query protein, which is really helpful to uncovering the relationship between protein function and its primary sequence. The web service is available at http://biominer.bime.ntu.edu.tw/ipda and mirrored at http://biominer.cse.yzu.edu.tw/ipda.
INTRODUCTION
Intrinsically disordered proteins or protein regions exhibit unstable and changeable three-dimensional structures under physiological conditions (1). Although lacking fixed structures, protein disorder has been identified to carry out important functions in many biological processes (1,2). In addition, it is observed that the absence of a rigid structure allows disordered binding regions to interact with several different targets (3,4). These regions, sometimes called ‘molecular recognition elements’, usually undergo a disorder-to-order transition when binding to their targets (5,6). In this regard, predicting protein disorder and investigating its potential of induced folding is a necessary preliminary to understanding protein structure and function (7).
The proposed web server iPDA aims at providing an integrated environment for detecting disordered regions and exploring their functional roles. In our recent work DisPSSMP (8), it is demonstrated that the accuracy of protein disorder prediction can be greatly improved if the disorder propensity of amino acids is considered when generating the condensed position-specific scoring matrix (PSSM) features. For iPDA, we implement a two-stage classifier of Radial Basis Function Networks (RBFN) to further enhance the predicting power of DisPSSMP. As unbalanced datasets, a large amount of ordered residues over disordered residues, are employed when training the new classifier DisPSSMP2, an alternative decision function is recently adopted and the cutting threshold is dynamically determined by the proportion of predicted secondary structure in the query protein.
iPDA takes an amino acid sequence as the input and reports the prediction of disordered residues with graphical plots, along with various sequence characteristics which are believed to be important when investigating the so-called induced folding behavior (6). The provided information includes sequence conservation from multiple sequence alignment (ClustalW) (9), concurrent sequence conservation from pattern mining (WildSpan) (10,11), secondary structure prediction (Jnet and PSIPRED) (12,13), low-complexity regions (CARD) (14) and hydrophobic clusters. Romero et al. stated that low-complexity regions are usually located in the long disordered regions (15), where the sequence complexity is measured by Shannon's entropy. In addition, Ferron et al. mentioned in their recent study that hydrophobic clusters and secondary structures can provide distinct clues for investigating induced folding (6). Meanwhile, we observe that sequence conservation is essential for disordered regions to maintain their functionality. Therefore, iPDA further provides a pattern mining utility to detect motifs in the specified disordered and/or ordered regions in order to predict potential intra- and inter-molecular interactions.
METHODS
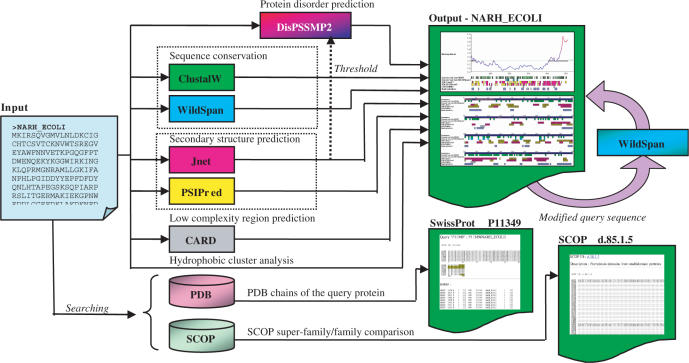
The architecture of iPDA is shown in Figure 1. Given an amino acid sequence, iPDA performs various sequence analyses by invoking several well-established predictors. In addition, iPDA provides two utilities for examining missing residues in PDB structures. The details of how each predicting package is incorporated into iPDA are discussed in the following subsections.
Figure 1.
iPDA employs seven sequence analyzers and provides utilities for discovering sequence motifs and extracting missing residues in PDB structures.
Protein disorder prediction
In our recent work DisPSSMP (8), a condensed PSSM with respect to physicochemical properties (PSSMP) was considered when generating feature profiles to build the classifier, where the PSSMP merges several amino acid columns of a PSSM that belong to a certain property into a single column. Besides, DisPSSMP decomposed each conventional physicochemical property of amino acids into two disjoint groups which have a propensity for order and disorder, respectively. The experimental results revealed that the PSSMP features with disorder propensity considered perform better than both the PSSMP features from traditional physicochemical properties and the original PSSM features on this problem.
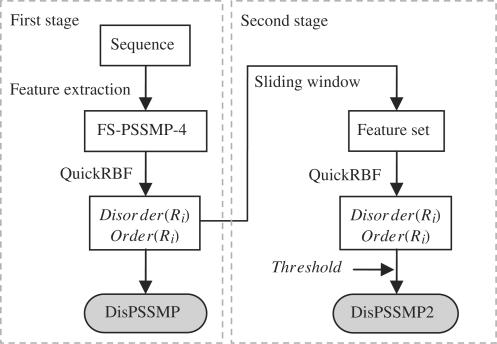
The web server iPDA implements a two-stage classifier of RBFN, named DisPSSMP2, to further enhance the predicting power of DisPSSMP. Figure 2 shows the system flow, in which the procedure of generating the feature set FS-PSSMP-4 from a protein sequence was described in (8). In the first stage, the RBFN outputs the probabilities of being disordered and ordered for a given residue Ri, named Disorder(Ri) and Order(Ri), respectively. While our previous predictor DisPSSMP takes the larger probability as the prediction, in this article the values of Disorder(Ri) and Order(Ri) are collected with a sliding window to generate the feature set used in the second stage. In DisPSSMP2, the size of the sliding window and the cutting threshold of predicting disorder used in the second stage have to be determined through cross-validation.
Figure 2.
The system flow of the proposed two-stage classifier DisPSSMP2. In both stages, an efficient package for constructing RBFN, named QuickRBF (16), is employed to build the classifier.
For training and validation processes, six datasets have been extracted from different databases, as summarized in Table 1. The datasets PDB693 and D184 were first collected when developing DisPSSMP (8). However, only about 30% ordered residues in the training set were included by DisPSSMP when constructing the classifier. In order to completely exploit the knowledge present in the training datasets, DisPSSMP2 recruits all ordered residues in the datasets PDB652 [PDB652 excludes the sequences of PDB693 used in (8) that have similarity identity of more than 70% against any protein sequence in the other training sets by running CD-HIT (21), resulting 652 proteins.] and D184, and further combines another globular protein set named G200 (17) to enhance the accuracy of predicting ordered residues.
Table 1.
Summary of the datasets employed in this study
| Training data | Testing data | |||||
|---|---|---|---|---|---|---|
| PDB652 | D184 | G200 | R80 | U79 | P80 | |
| Number of chains | 652 | 184 | 200 | 80 | 79 | 80 |
| Number of ordered regions | 1281 | 257 | 200 | 151 | 0 | 80 |
| Number of disordered regions | 1613 | 274 | 0 | 183 | 79 | 0 |
| Number of ordered residues | 190936 | 55164 | 37959 | 29909 | 0 | 16568 |
| Number of disordered residues | 49365 | 27116 | 0 | 3649 | 14462 | 0 |
| Total residues | 240301 | 82280 | 37959 | 33558 | 14462 | 16568 |
As unbalanced datasets, 284 059 ordered and 76 481 disordered residues, are employed when training the RBFN classifier, an alternative decision function is newly adopted to avoid the problem of under-prediction (17). A residue is predicted as disorder if [Disorder(Ri) − Order(Ri) + 1]/2 is greater than a cutting threshold. The 936 protein chains for training are partitioned into five groups. According to the 5-fold cross-validation, the performance of DisPSSMP2 is about the same when the cutting threshold is set in between 0.3 and 0.4. It has been observed that unstructured proteins in average contain fewer secondary structure elements elements than globular proteins (17). In this regard, we propose setting the cutting threshold of DisPSSMP2 dynamically by the estimated proportion of secondary structure in the query protein. Since we expect DisPSSMP2 to predict more disorder for the practical use of iPDA, the cutting threshold is set by ‘0.4 − the proportion of coils × 0.2’, resulting in a cutting threshold lower than 0.3 if the proportion of coils is greater than 50%. The window size used in the second stage is also determined through cross-validation. Window sizes in between 35 and 59 perform similarly. Thus, a window size of 47 is adopted. To evaluate the performance of DisPSSMP2, the benchmark proposed by Yang et al. (18) is employed as the blind testing data, as listed in Table 1.
Sequence conservation
It has been observed that residues within structural domains usually have higher conservation scores than in domain linkers (19). For deriving conservation information, the homologues of a query protein are collected by invoking PSI-BLAST (20) against Swiss-Prot database with e-value cutting threshold of 0.01. After that, redundant sequences are removed by executing CD-HIT (21) with threshold set to 70%. Using these homologues, iPDA provides two levels of sequence conservation to investigate the functional regions of the query protein. The sequence conservation with respect to a single position is calculated based on the multiple sequence alignment generated by ClustalW (9). The conservation of a given position is defined by the proportion of the particular amino acid type observed in the query protein. Only the top 10% conserved residues are highlighted by iPDA. Next, a second level of sequence conservation, called concurrent conservation, is derived by employing sequential pattern mining. The employed algorithm is named WildSpan for its ability of generating patterns across large wildcard regions (11). WildSpan has been recruited in the web server MAGIIC-PRO (10) in detecting functional signatures directly from unaligned sequences. A pattern generated by WildSpan contains the residues that are simultaneously conserved but largely separated in the protein sequence. Hsu et al. (22) observed that 90% of the concurrent conserved blocks discovered by WildSpan interact with at least one of the other blocks in space. Since disordered fragments of a protein might undergo a disorder-to-order transition to interact with each other when binding ligands or other proteins, it is expected that their conservation propensity would be revealed by ClustalW and the concurrent conservation can be discovered by WildSpan.
Iterative pattern mining
As more and more the disordered regions of proteins are found to be functionally significant, mining conserved patterns in the disordered regions is essential for understanding protein function (23). However, Brown et al. (24) observed that disordered regions usually have higher evolutionary rates than ordered regions, which makes it difficult to detect the conserved patterns of the disordered regions when using the entire protein as a query. Therefore, iPDA provides users an iterative mining strategy. The users can select regions of interest or mask unwanted segments of the query protein, and then invoke WildSpan iteratively to find conserved fragments other than the most highly conserved positions. Two parameters are requested upon calling WildSpan: (1) b stands for the minimum number of conserved regions (also called blocks) in a pattern and (2) k is the minimum number of such patterns wanted. The default setting for the first call of WildSpan is b = 3 and k = 1 in iPDA.
Secondary structure
Since induced folding regions are shown to have secondary structure propensity, iPDA provides predicting results from two secondary structure predictors, Jnet (v0.1) and PSIPred (v2.5) (12,13). These predictors are selected to complement each other, because it was observed in our recent study that each available package of secondary structure prediction behaves divergently, especially in short secondary structure elements (17). More confidence arises when two predictors concur, while users should be aware of risks when the results are inconsistent. The predicted information by Jnet is also recruited by DisPSSMP2 in determining the cutting threshold of the classifier.
Low-complexity regions
Low-complexity regions of a protein are usually disordered, but disordered regions are not always with low-complexity property (15). iPDA adopts CARD (14) to perform prediction of low-complexity regions. This information helps to strengthen the confidence of the disordered regions predicted by DisPSSMP2.
Hydrophobic clusters
It has been shown in many studies that disordered regions of proteins are comprised of a category of amino acids distinct from that of ordered ones (25). For example, amino acids of aromatic hydrophobic groups are known to be favored in the ordered regions, and thus are less found in the disordered regions (26). Callebaut et al. categorized 20 amino acids into three groups for hydrophobic cluster analysis and identified ‘VILFMYW’ as hydrophobic residues (27). It has been shown that structured segments have more hydrophobic clusters than the linker ones. Thus, the information of hydrophobic clusters is provided to validate the prediction of ordered regions. Here, iPDA assigns a position as a hydrophobic cluster if more than 5 of itself and its 10 neighbors (five from its left and five from its right) belong to the hydrophobic group ‘VILFMYW’.
Retrieving PDB missing residues
In the study of protein disorder, it is of interest to examine missing coordinates of backbone atoms in PDB structures. Residues present in the SEQRES records but not in the ATOM records are called missing residues (18). iPDA provides an utility to find missing residues from PDB database. All PDB chains are preprocessed to construct protein-structure mapping according to their SwissProt entry names or AC numbers. By marking the missing residues on the protein chains aligned by ClustalW, iPDA provides a clear view about which segments might be unstable. Disordered proteins usually activate their biological functions when undergoing disorder-to-order transitions. Therefore, the protein segments which are disordered in some PDBs but ordered in some others attract more attention for further analyses. Additionally, iPDA provides a similar utility of finding missing residues among all PDB chains belonging to a SCOP super-family/family.
RESULTS AND DISCUSSIONS
In this section, we first evaluate how DisPSSMP2 performs in comparison with DisPSSMP and other existing packages for disorder prediction. After that, several interesting examples are provided to illustrate how iPDA helps users to explore the functional roles of the detected disorder regions.
Many measures have been introduced to evaluate the performance of protein disorder predictors (8,18,28,29). Since sensitivity, specificity and accuracy listed in Table 2 are seriously affected by the relative frequency of the target classes (29), two more appropriate measures are included in Table 2 to reveal the properties of different packages. The first one, Matthews’ correlation coefficient, is widely used in many bioinformatics problems (30,31). The other evaluation measure, named probability excess, was recommended by CASP (28,29) and Yang et al. (18) for this problem.
Table 2.
Definition of measures employed in this study
| Measure | Abbr. | Equation |
|---|---|---|
| Sensitivity | Sens. | TP/(TP + FN) |
| Specificity | Spec. | TN/(TN + FP) |
| Accuracy | Accu. | (TP + TN)/(TP + FP + TN + FN) |
| Matthews' correlation coefficient | MCC | (TP × TN − FP × FN)/sqrt[(TP + FP) × (TN + FN) × (TP + FN) × (TN + FP)] |
| Probability excess | Prob. Excess | (TP × TN − FP × FN)/[(TP + FN) × (TN + FP)] |
Note: The definition of the abbreviations used: TP and TN is the number of correctly classified disordered and ordered residues, respectively; FP is the number of ordered residues incorrectly classified as disordered; FN is the number of disordered residues incorrectly classified as ordered.
To evaluate the performance of DisPSSMP2, we use a benchmark proposed by Yang et al. (18), comprising 239 proteins. When preparing the training data of DisPSSMP2, the redundancy between the training and testing data has been avoided using the same criterion adopted in Yang's paper (18). This benchmark also helps to judge whether a predictor tends to over-predict or under-predict disorder (8,17,18,32). As listed in Table 3, DisPSSMP2 has a better performance than DisPSSMP no matter which evaluation measure is used. The improvement of DisPSSMP2 is mainly from its including more ordered residues as training samples and the two-stage architecture employed. In addition, the performance of the existing packages for predicting protein disorder is ranked by its Prob. Excess in Table 3. It should be aware that these packages were trained with different databases and some of them have different definitions for protein disorder from ours. Although many methods achieve specificity in excess of 90%, they usually result in low sensitivity. Since iPDA expects to discover potential disorder-to-order transitions, it is expected that employed predictor should deliver a high sensitivity rate of disordered regions without an explicit drop on specificity.
Table 3.
Comparison with other existing packages
| Method | Sens. | Spec. | Accu. | MCC | Prob. Excess |
|---|---|---|---|---|---|
| DisPSSMP2 | 0.848 | 0.867 | 0.862 | 0.681 | 0.715 |
| VSL2 (33) | 0.821 | 0.815 | 0.817 | 0.594 | 0.636 |
| DisPSSMP (8) | 0.814 | 0.818 | 0.817 | 0.592 | 0.632 |
| IUPred[long] (34,35) | 0.629 | 0.954 | 0.863 | 0.644 | 0.583 |
| RONN (18) | 0.661 | 0.882 | 0.820 | 0.549 | 0.542 |
| FoldIndex (36,37) | 0.675 | 0.812 | 0.774 | 0.467 | 0.487 |
| IUPred[short] (34,35) | 0.549 | 0.934 | 0.826 | 0.541 | 0.483 |
| SPRITZ[long] (38) | 0.543 | 0.917 | 0.812 | 0.506 | 0.460 |
| DISOPRED2 (39) | 0.455 | 0.976 | 0.834 | 0.550 | 0.430 |
| PONDR (15) | 0.617 | 0.804 | 0.751 | 0.407 | 0.420 |
| DISpro (40) | 0.390 | 0.989 | 0.821 | 0.530 | 0.379 |
| FoldUnfold (41) | 0.631 | 0.737 | 0.707 | 0.343 | 0.368 |
| DisEMBL[465] (7) | 0.345 | 0.980 | 0.802 | 0.465 | 0.326 |
| DisEMBL[hot] (7) | 0.500 | 0.807 | 0.721 | 0.308 | 0.308 |
| PreLink (42) | 0.302 | 0.963 | 0.777 | 0.378 | 0.265 |
| SPRITZ [short] (38) | 0.290 | 0.893 | 0.724 | 0.226 | 0.183 |
| DisEMBL[coils] (7) | 0.723 | 0.432 | 0.514 | 0.143 | 0.155 |
| GlobPlot (43) | 0.321 | 0.814 | 0.676 | 0.146 | 0.136 |
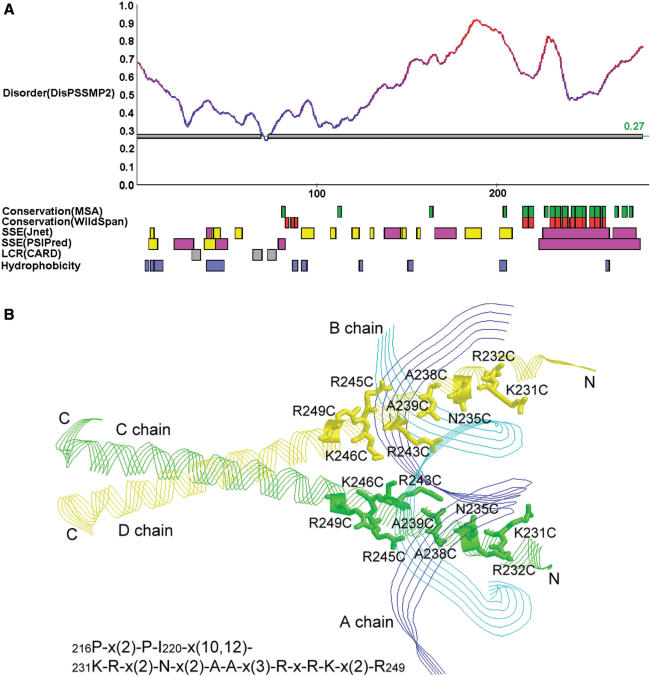
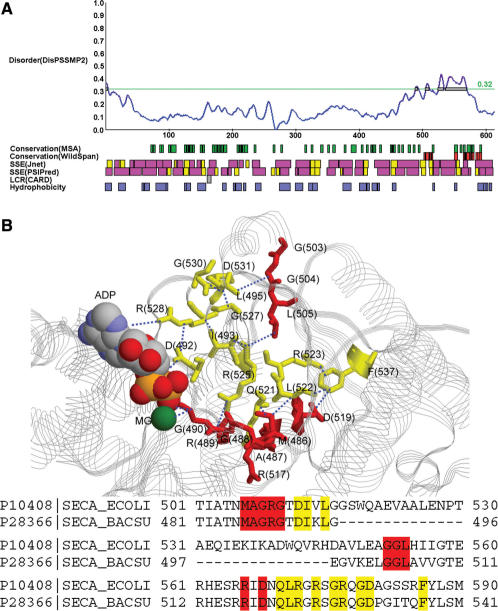
Next we provide some examples discussed in (1) to illustrate how iPDA facilitates the study of protein disorder and induced folding. The first example used is a DNA-binding protein GCN4. According to the prediction shown in Figure 3A, this protein might be largely unstructured. Meanwhile, it is observed that the region 225–281 is provided with large helical components. WildSpan also indicates high concurrent conservation in this area. Figure 3B shows that one pattern found by WildSpan identifies the important residues with respect to the DNA-binding region. Similar discoveries are observed on the proteins NFATC1 and RXR discussed in (1). In many cases, we observed that the regions undergoing disorder-to-order transitions when binding DNA usually possess both high disorder and secondary structure propensity, and additionally at least one pattern is found within this region to indicate potential intra- and/or inter-molecular interactions. This observation can be again justified by another protein, SecA, which undergoes locally disorder-to-order transition upon ADP binding in high temperature (44). The partial result of analyzing SecA is shown in Figure 4A. It shows that the range of 500–600 exhibits both disorder and concurrent conservation property. In this region, WildSpan detects 26 residues, and 10 of them are predicted as disorder. We highlight these 10 residues as red sticks in Figure 4B to examine their positions with respect to the molecule ADP.
Figure 3.
(A) The result page of iPDA on protein GCN4_YEAST (P03069). (B) One pattern derived by WildSpan identifies nine important residues for DNA binding (PDB structure used: 1YSA).
Figure 4.
(A) The result of P10408 (SECA_ECOLI). (B) The 10 residues predicted by both WildSpan and DisPSSMP2 are plotted as red sticks on the structure of a homologous protein, SECA_BACSU (P28366). Other 12 WildSpan predicted residues that either provides inter-molecular interactions to ADP or intra-molecular interactions with each other and/or the previous 10 residues are plotted as yellow sticks. Distances smaller than 5 Å are shown in blue dotted lines (PDB structure used: 1M74).
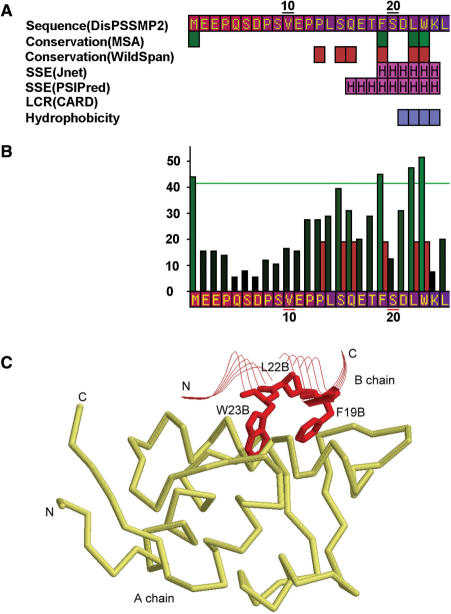
Another example of disordered regions containing functional motifs is the protein p53. The iPDA result is shown in Figure 5. In the disordered N-terminal domain (NTD) of p53, a short motif ‘FxxLW’, called the MDM2 functional motif, is discussed by Dawson's et al. (45). The key residues are detected by ClustalW, as well as the second run of WildSpan (b = 3 and k = 1). Those residues were not found in the first run of WildSpan, because the DNA-binding domain of p53 is more conserved than the MDM2 binding domain. If only the first disordered region (1–117) predicted by DisPSSMP2 are selected, the motif will be detected, as shown in Figure 5A and B. In Figure 5C, an available PDB structure shows the interaction of this polypeptide with the protein MDM2.
Figure 5.
(A) The partial result of iPDA on protein P53_HUMAN (P04637). (B) The partial conservation plot of ClustalW and WildSpan, after invoking the second run of pattern mining on the first predicted disordered region. (C) The discovered conserved segment ‘FxxLW’ contains three important residues for MDM2 binding, where chain B is a short polypeptide of p53 and chain A is the protein MDM2 (PDB structure used: 1YCQ).
CONCLUSION
iPDA provides comprehensive information for annotating the disordered regions of a query sequence. The integrated resource recognizes intrinsically unstructured proteins and helps to tell whether a disordered protein or protein fragment is with tendency toward being folded upon binding other molecules. According to the experiments conducted in this study, the disorder predictor DisPSSMP2 achieves a higher sensitivity rate than other existing packages performing the similar task without sacrificing the specificity rate. Besides, iPDA employs sequential pattern mining to identify concurrent conservation iteratively, from highly conserved regions to lightly conserved regions one at a time. It is observed in many cases that the disordered regions undergoing disorder-to-order transitions upon binding usually exhibit high concurrent conservation and clear secondary structure propensity. This association deserves further studies in the near future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank National Science Council of Republic of China, Taiwan, for the financial support under the contracts NSC 95-2221-E-002-274-MY2 and 95-3114-P-002-005-Y. Funding to pay the Open Access publication charges for this article was provided by the National Science Council of the Republic of China, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Fink AL. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53(Suppl. 6):573–578. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- 4.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac. Symp. Biocomput. 1998:473–484. [PubMed] [Google Scholar]
- 5.Romero P, Obradovic Z, Dunker AK. Sequence data analysis for long disordered regions prediction in the Calcineurin family. Genome Inform Ser. Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 6.Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 7.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Su CT, Chen CY, Ou YY. Protein disorder prediction by condensed PSSM considering propensity for order or disorder. BMC Bioinformatics. 2006;7:319. doi: 10.1186/1471-2105-7-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu CM, Chen CY, Liu BJ. MAGIIC-PRO: detecting functional signatures by efficient discovery of long patterns in protein sequences. Nucleic Acids Res. 2006;34:W356–W361. doi: 10.1093/nar/gkl309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WildSpan: Efficient Discovery of Motifs Spanning Large Wildcard Regions. http://biominer.bime.ntu.edu.tw/wildspan/
- 12.Cuff JA, Barton GJ. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 13.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 14.Shin SW, Kim SM. A new algorithm for detecting low-complexity regions in protein sequences. Bioinformatics. 2005;21:160–170. doi: 10.1093/bioinformatics/bth497. [DOI] [PubMed] [Google Scholar]
- 15.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 16.QuickRBF: an efficient RBFN Package. http://csie.org/~yien/quickrbf/quickstart.php.
- 17.Su CT, Chen CY, Hsu TM. 1st International Conference on Bioinformatics Research and Development (BIRD) 2007. Enhancing protein disorder detection by refined secondary structure prediction. [Google Scholar]
- 18.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 19.Udwary DW, Merski M, Townsend CA. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J. Mol. Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large proteins databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 22.Hsu CM, Chen CY, Liu BJ, Huang CC, Laio MH, Lin CC, Wu TL. Identification of hot regions in protein-protein interactions by sequential pattern mining. BMC Bioinformatics. 2007;8(Suppl. 5):S8. doi: 10.1186/1471-2105-8-S5-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lise S, Jones DT. Sequence patterns associated with disordered regions in proteins. Proteins. 2005;58:144–150. doi: 10.1002/prot.20279. [DOI] [PubMed] [Google Scholar]
- 24.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 25.Garner E, Cannon P, Romero P, Obradovic Z, Dunker AK. Predicting disordered regions from amino acid sequence: common themes despite differing structural characterization. Genome Inform Ser. Workshop Genome Inform. 1998;9:201–213. [PubMed] [Google Scholar]
- 26.Romero P, Obradovic Z, Kissinger C, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequence. Proc. IEEE Int. Conf. Neural Networks. 1997;1:90–95. [Google Scholar]
- 27.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon JP. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melamud E, Moult J. Evaluation of disorder predictions in CASP5. Proteins. 2003;53(Suppl. 6):561–565. doi: 10.1002/prot.10533. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y, Dunbrack RL., Jr Assessment of disorder predictions in CASP6. Proteins. 2005;61(Suppl. 7):167–175. doi: 10.1002/prot.20734. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Yoon S, Welsh WJ. Improved method for predicting beta-turn using support vector machine. Bioinformatics. 2005;21:2370–2374. doi: 10.1093/bioinformatics/bti358. [DOI] [PubMed] [Google Scholar]
- 31.Natt NK, Kaur H, Raghava GP. Prediction of transmembrane regions of beta-barrel proteins using ANN- and SVM-based methods. Proteins. 2004;56:11–18. doi: 10.1002/prot.20092. [DOI] [PubMed] [Google Scholar]
- 32.Su C-T, Chen C-Y. International Symposium on Biomedical Engineering (ISOBME) 2006. A two-stage RBFN classifier for protein disorder prediction. [Google Scholar]
- 33.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 35.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 36.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 37.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 38.Vullo A, Bortolami O, Pollastri G, Tosatto SC. Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006;34:W164–W168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Cheng J, Sweredoski MJ, Baldi P. Accurate prediction of protein disordered regions by mining protein structure data. Data Mining Knowledge Discov. 2005;11:213–222. [Google Scholar]
- 41.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- 42.Coeytaux K, Poupon A. Prediction of unfolded segments in a protein sequence based on amino acid composition. Bioinformatics. 2005;21:1891–1900. doi: 10.1093/bioinformatics/bti266. [DOI] [PubMed] [Google Scholar]
- 43.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keramisanou D, Biris N, Gelis I, Sianidis G, Karamanou S, Economou A, Kalodimos CG. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat. Struct. Mol. Biol. 2006;13:594–602. doi: 10.1038/nsmb1108. [DOI] [PubMed] [Google Scholar]
- 45.Dawson R, Muller L, Dehner A, Klein C, Kessler H, Buchner J. The N-terminal domain of p53 is natively unfolded. J. Mol. Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]