Abstract
RNA Movies is a simple, yet powerful visualization tool in likeness to a media player application, which enables to browse sequential paths through RNA secondary structure landscapes. It can be used to visualize structural rearrangement processes of RNA, such as folding pathways and conformational switches, or to browse lists of alternative structure candidates. Besides extending the feature set, retaining and improving usability and availability in the web is the main aim of this new version. RNA Movies now supports the DCSE and RNAStructML input formats besides its own RNM format. Pseudoknots and ‘entangled helices’ can be superimposed on the RNA secondary structure layout. Publication quality output is provided through the Scalable Vector Graphics output format understood by most current drawing programs. The software has been completely re-implemented in Java to enable pure client-side operation as applet and web-start application available at the Bielefeld Bioinformatics Server http://bibiserv.techfak.uni-bielefeld.de/rnamovies
INTRODUCTION
It is well known that RNA secondary structure prediction via energy minimization is limited in accuracy due to efficiency constraints imposed on the models and missing or imprecise thermodynamic data (1). Nonetheless, simulations of RNA folding pathways (2–5) can provide interesting insights into RNA folding dynamics and structural rearrangements. Since complete RNA secondary structure landscapes have become tractable (6), the limitations of predictions may be overcome by sampling and browsing candidate structures sorted by free energy or other properties. To facilitate this new approach of interrogation of an RNA secondary structure landscape a dedicated visualization tool was developed.
RNA Movies (7) uses as input a script—essentially a list of secondary structures—that it then draws at an adjustable rate using the NAVIEW RNA secondary structure layout algorithm (8), while smoothing the transitions between the nucleotide positions in subsequent structures linearly. The main aim was to create an intuitively usable web application. Therefore, the user interface matches the layout of a typical media player with play, pause, forward, backward and rewind buttons below the display area. Structures in the central viewing area can be scaled, moved and rotated by mouse controls. Multiple configuration options, such as animation speed and nucleotide colouring options are provided in a menu system.
To provide dynamic extensibility of the application the menu paths are determined by an XML framework at the program start-up. This enables the adaptation of the menu structure and its associated actions to match the requirements of a given application domain.
MATERIALS AND METHODS
Input formats
An RNA Movies script consists of an RNA sequence succeeded by a list of compatible secondary structures. The order in which the structures are displayed is determined by the order in which they appear in the script. A script may contain the complete secondary structure space generated by a folding algorithm or any subspace thereof. It is up to the folding program to arrange the structures in a meaningful order.
Currently, RNA Movies supports three input formats. The RNM-format makes use of the Vienna (dot-bracket) format for secondary structures. Pseudoknots may be modelled using additional brace characters {}, [], and <>. Furthermore, RNA Movies now supports the DCSE format (9) and the RNAStructML format (10). The DCSE format has been extended to include pseudoknots and entangled helices. RNAStructML is an XML-based and XML Schema language defined format for all kinds of RNA secondary structure information including pseudoknots and entangled helices.
Key features
The well-known NAVIEW algorithm (8) was chosen because of its many favourable properties. The layout is nearly free of overlaps, consistent and intuitively interpretable. Consistency is a very important property with regard to animation, as common parts of consecutive structures remain fixed while the rest of the structure moves about changing its shape. Therefore, a linear interpolation of coordinates associated with every base suffices as transition between different structures and works very well in practice. Moreover, co-transcriptional folding can be visualized by consecutively longer secondary structures.
Control over the program is provided by so-called actions that are available through drop down menus at the top of the application window. Actions provide all functionality for loading data, image export and operations concerning animation control. In some cases it might be necessary to adapt the set of actions to match the requirements of a certain application (e.g. as a visualization front-end for an RNA folding web service). As the structure of the menu is declared in XML this can easily be done by exchanging or editing the appropriate file. Removing the exit button, which does not make sense in the context of an applet, serves as a simple example. Combined with the ability to dynamically load new modules declared in the XML framework, adding additional features, such as new layout algorithms, becomes easy, because there is no need to change existing code.
The supported input formats include the RNM format that is akin to the output generated by most folding routines and the forward-looking XML formats provided by BioDOM. These formats allow the notation of pseudoknots within RNA secondary structures. Pseudoknots are displayed through highlighting the region of the backbone and connecting the first and last bases with lines. For image export of one or more structure graphs RNA Movies provides JPEG and PNG output as well as vector graphics through SVG. Moreover, whole movies can be exported as animated GIFs.
Usage example
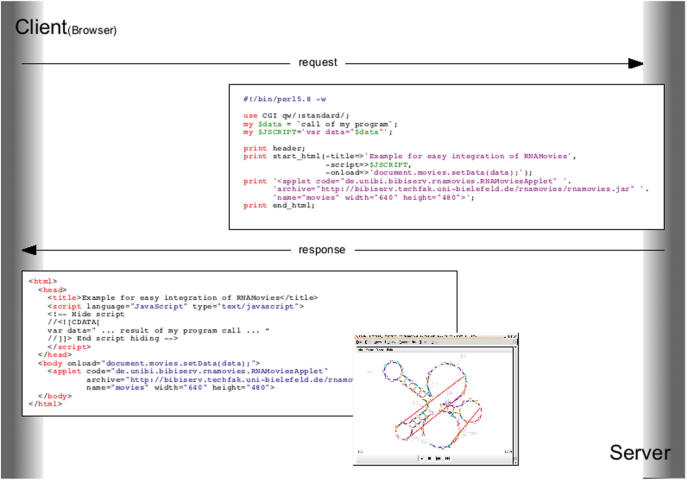
A great benefit of RNA Movies is the easy integration of the program into application workflows on other web servers. Figure 1 shows the integration of the RNA Movies applet in a typical web browser/web server environment. For simplicity the source code was reduced to a minimum, using Perl as server side language and HTML and Java as client side languages. Note the use of JavaScript in the HTML page to set input parameters of the Java applet.
Figure 1.
Easy Integration of RNA Movies in your own application using a Client-/Server architecture.
APPLICATIONS
RNA Movies is available online as applet. Moreover, it can also be run as fully fledged application, either locally or from a web page using the Java WebStart technology.
Currently, two tools on the Bielefeld Bioinformatics Server use the applet as visualization front-end.
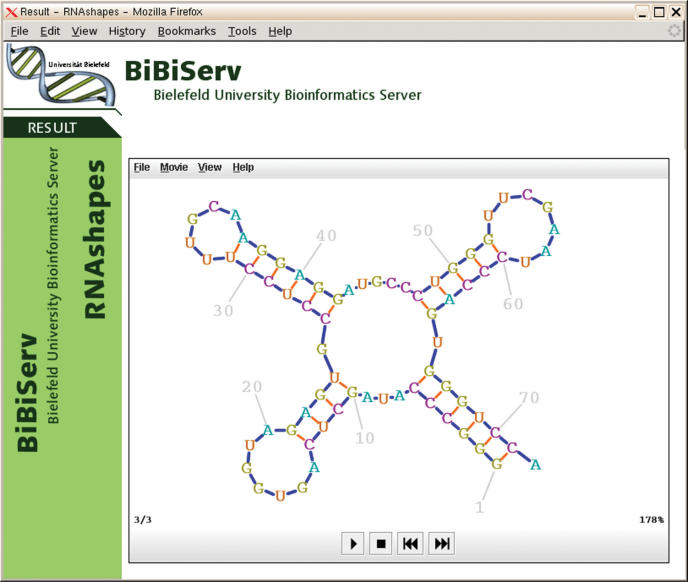
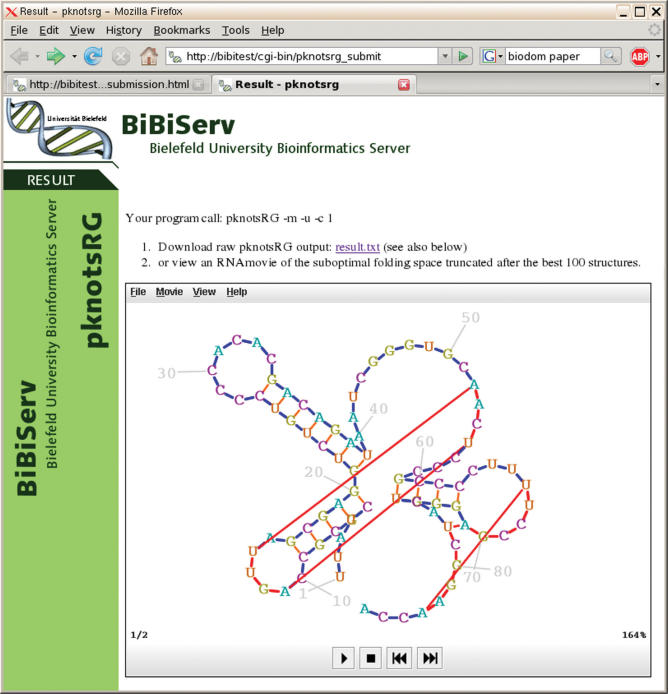
Figure 2 shows a screen shot of the third RNA secondary structure of the Alanine tRNA of Natronobacterium paharoanis calculated by RNAshapes (6) using the shape folding mode with the most abstract shape class. PknotsRG is a fast algorithm for pseudoknot computation (11). Since the online version supports suboptimal folding of RNA secondary structures with pseudoknots, RNA Movies is the preferred visualization front-end. Figure 3 shows a pknotsRG folding of the tRNA-like structure from turnip yellow mosaic virus.
Figure 2.
Third structure of ‘Alanine tRNA of Natronobacterium phiranis’ calculated by RNAShapes.
Figure 3.
First structure of tRNA/like structure from turnip yellow mosaic virus folded by pknotsRG.
FUTURE WORK
We intend to augment RNA Movies with new layout algorithms for better visualization of pseudoknot structures, as currently available algorithms do not fulfil the property of layout consistency to a degree needed for this application domain. Command line functionality is desirable for batch processing and web services applications to allow integration into existing workflow engines. Furthermore, the output of movie file formats would enable end users to easily publish files generated with RNA Movies.
ACKNOWLEDGEMENTS
This work was partly supported by the German Research Council (DFG) Initiative for Bioinformatics and the NRW Graduate School Program of the Ministry of Innovation, Science, Research and Technology of the state of North Rhine-Westphalia (MIWFT-NRW). Funding to pay the Open Access publication charge was provided by MIWFT-NRW.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Flamm C, Hofacker IL, Maurer-Stroh S, Stadler PF, Zehl M. Design of multistable RNA molecules. RNA. 2001;7:254–265. doi: 10.1017/s1355838201000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gultyaev AP. The computer simulation of RNA folding involving pseudoknot formation. Nucleic Acids Res. 1991;19:2489–2494. doi: 10.1093/nar/19.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isambert H, Siggia E. Modeling RNA folding paths with pseudoknots: application to hepatitis delta virus ribozyme. Proc. Natl Acad. Sci. USA. 2000;97:6515–6520. doi: 10.1073/pnas.110533697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mironov AA, Dyakonovo LP, Kister AE. A kinetic approach to the prediction of RNA secondary structures. J. Biomol. Struct. Dynam. 1985;2:953–962. doi: 10.1080/07391102.1985.10507611. [DOI] [PubMed] [Google Scholar]
- 6.Steffen P, Voß B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- 7.Evers DJ, Giegerich R. RNA Movies: visualizing RNA secondary structure spaces. Bioinformatics. 1999;15:32–37. doi: 10.1093/bioinformatics/15.1.32. [DOI] [PubMed] [Google Scholar]
- 8.Bruccoleri RE, Heinrich G. An improved algorithm for nucleic acid secondary structure display. Comput. Appl. Biosci. 1988;4:167–173. doi: 10.1093/bioinformatics/4.1.167. [DOI] [PubMed] [Google Scholar]
- 9.De Rijk P, De Wachter R. DCSE, an interactive tool for sequence alignment and secondary structure search. Comput. Appl. Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- 10.Seibel PN, Krüger J, Hartmeier S, Schwarzer K, Loewenthal K, Mersch H, Dandekar T, Giegerich R. XML schemas for common bioinformatic data types and their application in workflow systems. BMC Bioinformatics. 2006;7:490. doi: 10.1186/1471-2105-7-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]