Abstract
Tumor necrosis factor (TNF) is critical for the control of hepatitis B virus (HBV) in the clinical setting and in model systems. TNF induces noncytopathic suppression and clearance of HBV in animal models, possibly through reduction of viral nucleocapsids, but the mechanism is not well described. Here, we demonstrate the molecular mechanism and broad host range for TNF action against HBV. We show that TNF rapidly blocks HBV replication by promoting destabilization of preexisting cytoplasmic viral nucleocapsids containing viral RNA and DNA, as well as empty nucleocapsids. TNF destabilized human HBV nucleocapsids in a variety of human hepatocytic cell lines and in primary rat hepatocytes and also destabilized duck HBV (DHBV) nucleocapsids in chicken hepatocytic cells. Lysates from TNF-treated uninfected cells also destabilized HBV nucleocapsids in vitro. Moreover, inhibition of DHBV DNA replication by TNF blocks nuclear accumulation of the viral transcription template, maintenance of which is essential for the establishment and maintenance of chronic infection. We show that TNF destabilization of HBV nucleocapsids does not involve ubiquitination or methylation of the viral core protein and is not mediated by the nitric oxide free radical arm of the TNF pathway. These results define a novel antiviral mechanism mediated by TNF against multiple types of HBVs in different species.
Despite recent medical advances, chronic infection of the liver by hepatitis B virus (HBV) continues to be a major international health problem. The prevalence of HBV chronic infection ranges from 0.1 to 20% worldwide, and it is estimated that over 1 million deaths annually are attributable to HBV infection (14). Most morbidity from HBV arises as a consequence of chronic infection, which occurs in 5 to 10% of adult infections and as many as 90 to 95% of neonatal infections (14). The consequences of chronic HBV infection include hepatitis, cirrhosis, and a significantly increased risk for development of hepatocellular carcinoma (14). Current antiviral therapies, including nucleoside reverse transcriptase inhibitors, can be effective in suppressing viral load, but resistance to these drugs commonly evolves (44, 69). Improving therapy for HBV infection remains a global health priority.
HBV is a member of the viral family Hepadnaviridae, which includes woodchuck HBV (WHV), ground squirrel HBV, and duck HBV (DHBV) (14). Hepadnaviruses are distinguished by a unique replication cycle which involves the generation of an RNA genomic template, known as pregenomic RNA (pgRNA), that is incorporated into viral nucleocapsids in the cytoplasm, where it undergoes reverse transcription into an infectious cDNA form (14). Nucleocapsids are comprised entirely of HBV core protein. Encapsidation of the pgRNA into nucleocapsids is required for viral replication to occur. HBV DNA also resides in the nucleus as a nonreplicating epichromosomal covalently closed circular DNA (cccDNA) that functions as the viral transcription template and is essential for maintenance of chronic infection and latency (14). Levels of cccDNA in the nucleus appear to be maintained by constant shunting of newly formed DNA-containing nucleocapsids from the cytoplasm to the nucleus (35, 45). Although the mechanism by which shunting of cytoplasmic nucleocapsids to the nucleus occurs is unclear, studies have shown that interruption of nuclear accumulation of HBV nucleocapsids is important for curing chronic infection (11).
The majority of adults infected by HBV resolve the infection within a few months and do not progress to having a chronically infected liver. Resolution of acute HBV infection is critically dependent on cellular (adaptive) immunity, in which hepatocytes bearing HBV antigens are targeted for killing by cytotoxic T lymphocytes (CTLs) (2, 26, 32). In general, adaptive immune responses take 7 to 10 days to become apparent (14) but can maintain surveillance against HBV for years following an acute hepatitis infection (56). It is clear that an anti-HBV response mediated by natural killer (NK) cells and NK T cells precedes the adaptive CTL response and is important for controlling HBV during the early phase of host infection (3). Many studies also indicate that an innate immune response is crucial for early clearance of HBV from the liver of infected individuals. For instance, in chimpanzees infected with HBV, viral levels begin to decline before CTL killing of infected hepatocytes becomes evident (24). Early noncytopathic mechanisms are therefore also important to remove intrahepatic HBV. Moreover, DHBV, a more distantly related hepadnavirus, can be efficiently cleared from ducks without massive hepatic necrosis, even when virtually all of the hepatocytes are infected (31). Thus, innate immune responses, which can be noncytolytic, precede and complement adaptive immune responses to HBV and likely play a key role in a successful antiviral defense against HBV.
Secretion of antiviral cytokines by intrahepatic immune cells and infected hepatocytes is thought to play an important role in suppression or clearance of HBV from the infected liver (3, 33). Best characterized are the type I interferons (alpha/beta interferon [IFN-α/β]), which inhibit replication of viruses through a variety of mechanisms, including inhibition of protein synthesis, viral transcription, and nuclear trafficking of viral DNA (57). Specifically in the case of HBV, it was shown that type I interferons suppress HBV replication primarily by downregulating transcription of viral mRNAs (22). Other supporting studies for clearance of DHBV and WHV infection also suggest that these hepadnaviruses are cleared, in part, through noncytolytic inflammatory cytokine-mediated mechanisms (29, 30, 54). Recovery of WHV infection is preceded by increased hepatic expression of IFN-γ and tumor necrosis factor (TNF) (15, 29, 54).
TNF is suspected to have a direct antihepadnaviral activity (5, 9, 20). During HBV infection in humans, increased TNF levels are associated with viral clearance (1, 8, 12). Increased hepatic levels of both TNF and IFN-γ are each individually associated with suppression of HBV replication in transgenic mice that express HBV in the liver (18, 19, 52). TNF can directly inhibit HBV DNA replication in a noncytopathic manner (5, 22, 25). We previously showed that TNF inhibition of HBV is dependent on NF-κB activation and resulted in decreased levels of viral nucleocapsids (5). Here we characterize the TNF response to HBV and provide a mechanistic understanding for TNF inhibition of HBV replication.
MATERIALS AND METHODS
Cell culture.
HepG2 cells are a differentiated hepatoblastoma cell line (ATCC). HepG2.215 cells are derived from HepG2 cells (ATCC) and contain multiple copies of an integrated human HBV genome. Cells were propagated in modified Eagle's medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 1 mM sodium pyruvate, nonessential amino acids (Cellgro Mediatec Inc.), and 25 μg/ml gentamicin sulfate (Sigma-Aldrich). Huh7 cells are a transformed human hepatoblastoma cell line (ATCC) and were grown in Dulbecco's MEM supplemented with 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, and 25 μg/ml gentamicin sulfate. LMH cells, a chicken hepatoma cell line, were propagated in Dulbecco's MEM-F-12 medium without glutamine supplemented with 10% FBS, nonessential amino acids, and 25 μg/ml gentamicin sulfate. Human TNF (Calbiochem) was added to the culture media at a concentration of 5 ng/ml unless otherwise indicated.
Transfection and plasmids.
Cells were transfected with a 3:1 ratio of Fugene reagent (Roche) to DNA. Prior to the addition of TNF, cells were washed three times with phosphate-buffered saline. Plasmid pGem7Z(+)ayw contains a 120% DNA copy of the human HBV strain ayw strain genome and is referred to here as the HBV replicon. This plasmid, when transfected into hepatocytic cell lines, supports viral gene expression and replication (7). All mutant HBV plasmids were produced by site-directed mutagenesis and constructed into the pGem7Z(+)ayw HBV replicon. Plasmid pGem7Z(+)aywΔɛ encodes a G-to-A point mutation in the third amino acid of the epsilon (ɛ) loop region, creating a pgRNA encapsidation mutant (59), referred to here as ΔɛHBV. Plasmid pGem7Z(+)ayw env− contains two premature stop codons, one in the pre-S1 gene (codon 51) and the other in the S gene (codon 15) preventing HBsAg envelopment of nucleocapsids. Plasmid pGEMayw-Cp149 encodes a truncated core protein, produced by truncating the core open reading frame at position 149 by insertion of a premature stop codon, which is then constructed into the HBV replicon. Plasmid pSG5-pgRNA encodes pgRNA under a T7 promoter for in vitro transcription. Plasmid pcDNA-DHBV contains three end-to-end copies of the DHBV genome, and like the HBV replicon, directs viral replication. Adenoviral (Ad) vector Ad-HBV was constructed by insertion of the 120% HBV replicon DNA into the pAd-Easy Ad system in Ad region E1 following the manufacturer's protocol (Stratagene). Ad-GFP was similarly produced. Both Ad vectors were propagated in 293 human kidney epithelial cells and titers obtained before use. Plasmids pRelA, pIKKα, pIKKβ, and pIκB-SR were the gift of M. Karin (USCD). The NF-κB reporter plasmid (gift of E. Skolnik, NYU) contains four NF-κB binding sites adjacent to a minimal promoter and firefly luciferase coding region.
Determination of capsid levels and replication assays.
Capsid levels and replication assays were performed as described previously (6). Briefly, cells were transfected with either human HBV or DHBV plasmids. Cytoplasmic extracts were prepared by lysis in Tris lysis buffer (50 mM Tris-HCl [pH 7.4], 100 mM EDTA, 10 mM magnesium acetate, 1% NP-40) followed by centrifugation to remove nuclei and debris. Cellular DNA and RNA were digested with 150 μg/ml DNase I and 100 μg/ml RNase A. Intact cytoplasmic core particles were resolved by electrophoresis through a 1.2% agarose gel in 15 mM sodium phosphate buffer (pH 7.5) prior to capillary transfer onto a nylon membrane. Membrane-immobilized core particles were visualized by immunoblotting with anticore antibody (DAKO) and enhanced chemiluminescence. For replication assays, nucleocapsids were precipitated with 35% polyethylene glycol-1.5 M NaCl and digested with proteinase K, and the encapsidated DNA was isolated and analyzed by Southern blot DNA hybridization as described previously (7).
Capsid stability assay.
Stability of nucleocapsids was determined as follows. HepG2 cells were transfected with HBV replicons and then labeled 12 h later with [35S]methionine-cysteine protein labeling mix (Perkin Elmer) for 6 h to allow for efficient labeling of core protein (core protein contains two methionines and four cysteines). The labeled core protein was “chased” into nucleocapsids by the addition of unlabeled medium. Cells were then mock treated or treated with 5 ng/ml TNF for the times shown. Following lysis, intact core particles were separated from free core protein by centrifugation through a Centricon filter with a retention cutoff of 100 kDa. Free core protein and nucleocapsids were immunoprecipitated and transferred to polyvinylidene difluoride membrane for immunoblot analysis, or labeled core protein was visualized using a PhosphorImager.
In vitro capsid stability assay.
HepG2 cells were transfected with HBV replicons, a crude extract was prepared by Dounce homogenization (40 strokes in 50 mM Tris-HCl [pH. 8.0], 1 mM EDTA, 100 mM NaCl) and clarified by centrifugation at 10,000 × g, and an S10 supernatant was reserved as a source of nucleocapsids. Uninfected HepG2 cells were treated for 0, 1, or 4 h with 5 ng/ml TNF and cytoplasmic extracts prepared by Dounce homogenization in TB buffer as described above. The nucleocapsid extracts were incubated with equal amounts of the uninfected extracts prepared from mock-treated or TNF-treated cells for up to 24 h at 30°C. Nucleocapsid levels were determined by native agarose gel electrophoresis and immunoblot analysis using an anti-HBcAg antibody. Total core protein levels were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis.
Isolation of primary rat hepatocytes.
For each isolation, a Sprague-Dawley male rat of between 6 and 8 weeks of age was anesthetized by intraperitoneal injection with 75 mg ketamine/kg of body weight and 5 mg xylazine/kg. The liver was perfused via the portal vein with 100 ml of 0.05 mM EGTA phosphate buffer (pH 7.4) at a flow rate of 10 ml/min. Perfusion was continued at the same flow rate with 100 ml phosphate-buffered saline containing 5 mM CaCl2 and 0.2% collagenase type I. All solutions were kept at 37°C during liver perfusion. The liver was removed and the capsule teased apart to release the hepatocytes. The cells were gently filtered through a nylon mesh and centrifuged for 2 min at 700 rpm. The cell pellet was washed twice, resuspended in 10 ml Williams E medium, and applied to a 30% Percol cushion to separate live from dead cells. Hepatocytes were plated on collagen-coated plates and used the following day for infection with recombinant Ads (described above) at a multiplicity of infection of 50 particles/cell. Twenty-four hours after infection, cells were treated with 5 ng/ml TNF. Two days postplating, the cells were covered with Matrigel (BD Biosciences). At days 5 and 10, the cells were harvested, and isolation of HBV nucleocapsids was performed as described above.
Generation of pgRNA and transfection of poly(A) mRNA.
pgRNA was generated from plasmid pSG5-pgRNA by use of a mMessage machine kit (Ambion) and subsequently poly(A) tailed following the manufacturer's protocol. Poly(A) mRNA (2 μg) was transfected into HepG2 cells using DMRIE-C reagent (Invitrogen) following the manufacturer's protocol.
Analysis of cccDNA levels.
Isolation of cccDNA was performed as described previously (45). Briefly, cells were lysed with cccDNA lysis buffer (1% SDS, 10 mM Tris-HCl [pH 7.5], 10 mM EDTA) for 10 min at 37°C. After addition to 0.6 M in potassium chloride, the lysates were briefly vortexed and incubated on ice for 7 min. Lysates were centrifuged for 10 min at 13,000 rpm and the pellet was discarded. The DNA was precipitated, digested with exonuclease III to remove DNA contaminants and with DpnI to remove chromosomal and methylated (transfected) plasmid DNA, and then analyzed by electrophoresis and Southern blot DNA hybridization. The authenticity of cccDNA was established by restriction enzyme digestion with EcoRI followed by Southern blot hybridization.
Real-time PCR.
Real-time PCR was carried out using a SYBR green real-time PCR kit (Sigma-Aldrich) in a Roche Lightcycler. Reverse transcription was conducted at 60°C for 30 min, denaturation at 95°C for 30 s, amplification at 95°C for 5 s, 58°C for 8 s, 72°C for 7 s (40 cycles), determination of melting curve at 60°C for 15 s, and cooling 40°C for 30 s. The following primers were generated for RT-PCR: 5′-GAAGCGTCACGGACTTGATAACTA and 3′-CCCATCGGTGTTACGGTTTG (chicken lysozyme) and 5′-AACGACCCCTTCATTGAC and 3′-TCCACGACATACTCAGCAC (chicken GAPDH).
Detection of ROS and nitrogen species.
HepG2 cells were transfected with pGEMayw1.2 and then 24 h later pretreated with 10 mM N-acetylcysteine (NAC) for 2 h. Following pretreatment, cells were incubated with either 1 ng/ml lipopolysaccharide, 5 ng/ml TNF, or 0.1% H2O2. Following the addition of lucigenin or luminol at a concentration of 30 μM, reactive oxygen species (ROS) and nitric oxide synthetase (NOS) were detected using a luminometer over a 2-h period.
Luciferase assay.
Luciferase activity was determined using a luciferase assay kit (Promega). Briefly, cells transfected with luciferase reporter genes were lysed in 1× passive lysis buffer (Promega), 10 μl of cell lysate was placed into a 96-well opaque dish, and luciferase activity was determined in triplicate.
Data analysis.
All figures present typical results of no fewer than three independent analyses. Autoradiograms were quantified by densitometry and the means of at least three experiments calculated; the ratio of nucleocapsid levels to total core protein levels is presented below each figure. In all cases, the untreated wild-type HBV samples were normalized to 100%.
RESULTS
TNF downregulates HBV nucleocapsid levels in hepatocytic cells of multiple species.
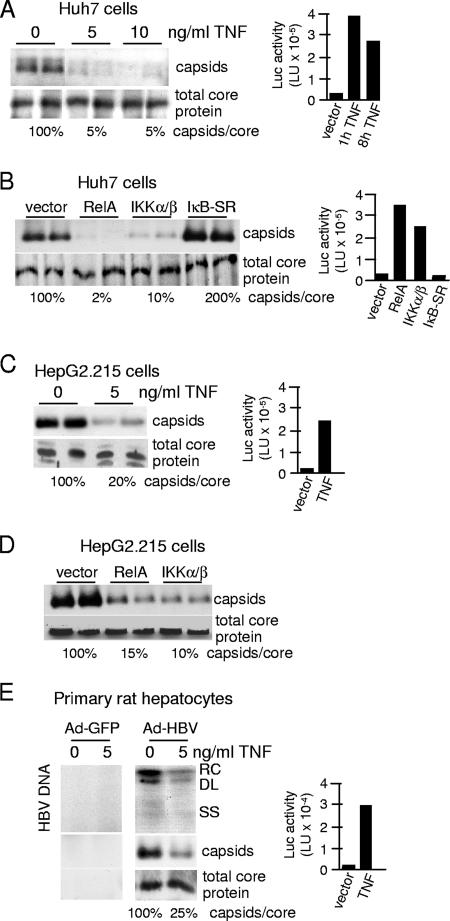
We previously showed that TNF noncytopathically downregulates human HBV DNA replication and cytoplasmic nucleocapsid levels in HepG2 cells through the action of NF-κB (5). Additionally, overexpression of NF-κB-activating kinases IKKα/β or the RelA component of NF-κB in the absence of TNF treatment is sufficient to reduce nucleocapsid levels in HepG2 cells (5). To determine whether the anti-HBV effect is cell type dependent or widespread, a variety of hepatocytic cell lines and primary hepatocytes containing replicating HBV were treated with TNF. Huh7 cells, a human differentiated hepatoma cell line, were transfected with vectors expressing the HBV replicon and then treated with 5 ng/ml TNF for 24 h (Fig. 1A). Nucleocapsid levels were determined by agarose gel electrophoresis and immunoblot analysis. Nucleocapsid levels were markedly decreased in Huh7 cells treated with TNF to 5% of those for mock-treated cells (Fig. 1A). There was no evidence of cell death, as determined by release of lactate dehydrogenase (data not shown), at the lowest dose of TNF (5 ng/ml), similar to what was observed previously for TNF-treated HepG2 cells (5). TNF at 5 ng/ml strongly activated a cotransfected NF-κB-dependent luciferase reporter (Fig. 1A, right panel), demonstrating the strong responsiveness of these cells to TNF. It should also be noted that NF-κB activation by TNF is rapid, as shown here, occurring within the first hour of treatment and to high levels. Additionally, overexpression of either RelA or IKKβ in Huh7 cells, which strongly activates NF-κB (Fig. 1B), reduced nucleocapsid levels to 2 to 10% that of the vector control. Overexpression of the NF-κB inhibitor IκB-SR in transfected Huh7 cells blocked NF-κB activation (Fig. 1B, right panel) and increased the ratio of nucleocapsids to core protein by twofold. As suggested previously (5), since the HBV replicons express HBx protein, a weak activator of NF-κB, suppression of NF-κB activation by expression of the IκB superrepressor modestly restored nucleocapsid levels. In all cases, the reduction in nucleocapsid levels was not a result of reduced total core protein levels, suggesting that nucleocapsid assembly or stability may be compromised and that core protein is not degraded by TNF treatment. A similar effect of 5 ng/ml of TNF on nucleocapsid levels and NF-κB activity was observed for HepG2.215 cells, a HepG2 cell line that expresses chromosomally integrated HBV (Fig. 1C), without a change in total core protein levels. Weak costaining of cellular proteins with core protein was often apparent in these blots. Although we previously found that HepG2.215 cells were not sensitive to TNF downregulation of HBV nucleocapsids (5), extensive washing with serum-free medium prior to the addition of TNF rendered these cells sensitive, possibly by removal of secreted soluble TNF receptor (unpublished results). In HepG2.215 cells, nucleocapsid levels were strongly decreased (to 10 to 15%) in cells that overexpressed either RelA or IKK-β, in contrast to cells that were transfected with vector alone, demonstrating that nucleocapsid reduction is mediated by NF-κB (Fig. 1D). Cells were transfected by lipophilic reagent at >90% efficiency, accounting for the almost quantitative inhibition of NF-κB activation (data not shown).
FIG. 1.
TNF downregulates human HBV nucleocapsid levels in different hepatocytic cells lines and primary hepatocytes. (A) Huh7 cells were transfected with an HBV genomic replicon plasmid and an NF-κB-dependent luciferase reporter and then treated 2 days later with 5 ng/ml TNF for up to 24 h. Cytoplasmic nucleocapsids were resolved by native agarose gel electrophoresis and subjected to immunoblot analysis with antibodies to HBcAg. Total core protein levels were quantified by SDS-PAGE and immunoblot analysis with antibodies to HBcAg. Luciferase activity was determined at the times shown using a luminometer, and the results of three experiments were averaged for presentation. (B) Huh7 cells were transfected with an HBV genomic replicon plasmid and vector alone or expression vectors for RelA, IKKα/β or IκB-SR, and an NF-κB-dependent luciferase reporter. Nucleocapsids and total core protein levels were assessed at 48 h as described above, in duplicate. Luciferase activities were determined at the same time and averaged from three independent experiments. (C) HepG2.215 cells were transfected with the NF-κB luciferase reporter and mock treated or treated with 5 ng/ml TNF for 24 h, and nucleocapsids/total core protein levels and luciferase activity were determined as described above. (D) HepG2.215 cells were transfected with vector alone or expression vectors for RelA or IKK-α/β, and nucleocapsids/total core protein levels were examined as described above. (E) Primary rat hepatocytes were purified, transfected with an NF-κB luciferase reporter plasmid, and transduced with an Ad vector at 50 particles per cell expressing a replicon of HBV (Ad-HBV) or control Ad-GFP for 3 days and then treated with 5 ng/ml human TNF for 3 days. Cytoplasmic viral nucleocapsids were purified from equal numbers of cells, and associated HBV DNA was detected by Southern blot analysis. Nucleocapsids and total core protein levels were assessed as described above. Luciferase activity was determined as described above. Results shown are representative of three independent experiments performed in duplicate. Abbreviations: RC, relaxed circular form; DL, double-strand linear form; SS, single-strand form; Luc, luciferase.
Studies were carried out to determine whether TNF downregulates HBV nucleocapsid levels and viral DNA replication in primary hepatocytes or is restricted to hepatocytic cell lines. Isolated primary rat hepatocytes were transduced with an Ad-HBV recombinant Ad vector containing the HBV replicon or an Ad vector expressing green fluorescent protein (Ad-GFP) as a control and transfected with an NF-κB-dependent luciferase reporter. When treatment was done with 5 ng/ml TNF for 24 h, activation of NF-κB was apparent, but at a level roughly 10-fold lower than that for human hepatoma cell lines (Fig. 1E, right panel). The weak activation of NF-κB in primary rat hepatocytes compared to that in continuous human hepatocytic cell lines likely reflects both poorer reporter plasmid transfection of these cells and only partial cross-species activity of human TNF. Nevertheless, both HBV replication and nucleocapsid levels were decreased by TNF action by about 75% compared to what was seen for the untreated control (Fig. 1E). There was no detectable HBV DNA or nucleocapsids in the Ad-GFP control. These results therefore demonstrate that TNF inhibition of human HBV replication occurs in a variety of human and rat hepatocytic cells via an NF-κB-dependent decrease in nucleocapsid abundance.
NF-κB decreases DHBV nucleocapsid levels.
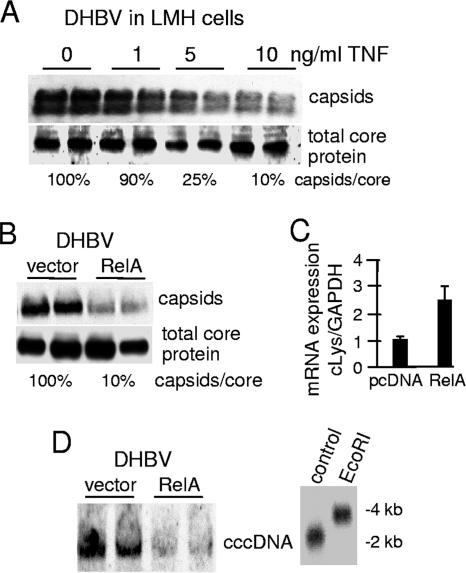
Other hepadnaviruses have been observed to be inhibited by the antiviral activity of cytokines (15, 30, 50, 63, 64). For instance, WHV, which causes a disease in woodchucks similar to that caused by HBV in humans, has been found to be suppressed by type I and type II interferons in addition to TNF (15, 30, 50). Moreover, DHBV shares 65% homology with HBV and is also inhibited by type I and type II interferons (51, 63, 64). However, the role of TNF and NF-κB on DHBV replication has not been well studied. To determine whether the antiviral effect of TNF on hepadnaviruses is conserved across different viral and host species, we investigated the effect of TNF on DHBV replication by use of a chicken hepatocytic cell line. A DHBV replicon was transfected into LMH cells and incubated with increasing concentrations of TNF (Fig. 2A) or transfected with a vector control plasmid and a plasmid overexpressing RelA to activate NF-κB (Fig. 2B). DHBV capsid levels were decreased 90% by treatment with 10 ng/ml of human TNF and 80% by coexpression of RelA (Fig. 2A and B). All data are expressed as the means of three independent studies. Although the RelA utilized for these experiments was the human homolog, it is sufficiently conserved to upregulate known avian effectors of TNF signaling, such as lysozyme (Fig. 2C) (58). The somewhat lower response of DHBV nucleocapsid reduction in LMH cells is probably a result of restricted action of human TNF on chicken hepatocytes. Nevertheless, these data indicate that TNF-mediated signaling downregulates transspecies hepadnaviral replication of both human and duck viruses in a variety of hepatocytes.
FIG. 2.
TNF and NF-κB downregulate levels of DHBV cytoplasmic nucleocapsids and nuclear cccDNA. (A) LMH cells (chicken hepatocytic cell line) were transfected with a DHBV replicon consisting of a trimer of head-to-tail genomic DNA and treated with 0, 1, 5, or 10 ng/ml TNF for 24 h. Cytoplasmic nucleocapsids were resolved by native agarose gel electrophoresis and subjected to immunoblot analysis with DHBcAg antibody. Total core protein levels were determined by SDS-PAGE followed by immunoblot analysis. All studies were performed in duplicate. (B) LMH cells were transfected with the DHBV replicon and either vector alone or a plasmid expressing RelA. Capsid and total core protein levels were determined as described above and values are presented as the means of three studies. (C) mRNA was isolated from LMH cells cotransfected with the DHBV replicon and vector alone or RelA plasmid, and expression levels of chicken lysozyme were quantified and analyzed by quantitative real-time PCR. (D) LMH cells were cotransfected with the DHBV replicon and vector alone or with RelA plasmid. cccDNA was isolated from equal numbers of cell nuclei, and levels were analyzed by Southern hybridization analysis as described in Materials and Methods. The authenticity of cccDNA was established by restriction enzyme digestion and an electrophoretic mobility shift change from 2 kb (cccDNA) to 4 kb (linear DNA) with EcoRI, which linearizes the cccDNA. Results shown are representative of at least three independent experiments performed in duplicate.
Activation of NF-κB prevents accumulation of nuclear cccDNA.
The maintenance of cccDNA within the nucleus of the infected hepatocyte is essential for the establishment and maintenance of hepadnavirus persistent infection and is widely thought to require shunting of cytoplasmic DNA containing nucleocapsids to the nucleus (14). Destruction of cccDNA or its clearance from infected hepatocytes, or destruction of the infected hepatocyte itself, is thought to be critical for complete viral clearance and to prevent recurrent infection (11). Schultz and Chisari demonstrated that IFN-γ profoundly reduces the rate of cccDNA nuclear accumulation in primary duck hepatocytes (63). To investigate whether TNF similarly decreases levels of cccDNA as a result of disruption of cytoplasmic nucleocapsids, LMH chicken hepatocytic cells were transfected with a DHBV replicon. DHBV was utilized for these experiments because cccDNA is readily detectable in this system but not with human HBV in cultured cells. RelA was used to activate NF-κB, as there is no validated source of chicken TNF. Four days posttransfection, cccDNA levels were assessed by Southern blot DNA hybridization analysis. cccDNA levels were found to be strongly reduced in cells that were cotransfected with RelA compared to vector control (Fig. 2D). Restriction enzyme digestion and Southern blot analysis of the vector control cccDNA sample demonstrated its authenticity as viral cccDNA (Fig. 2D, right panel). These data indicate that NF-κB activation prevents accumulation of DHBV nuclear cccDNA pools.
TNF and NF-κB downregulate HBV nucleocapsids and replication posttranscriptionally and rapidly.
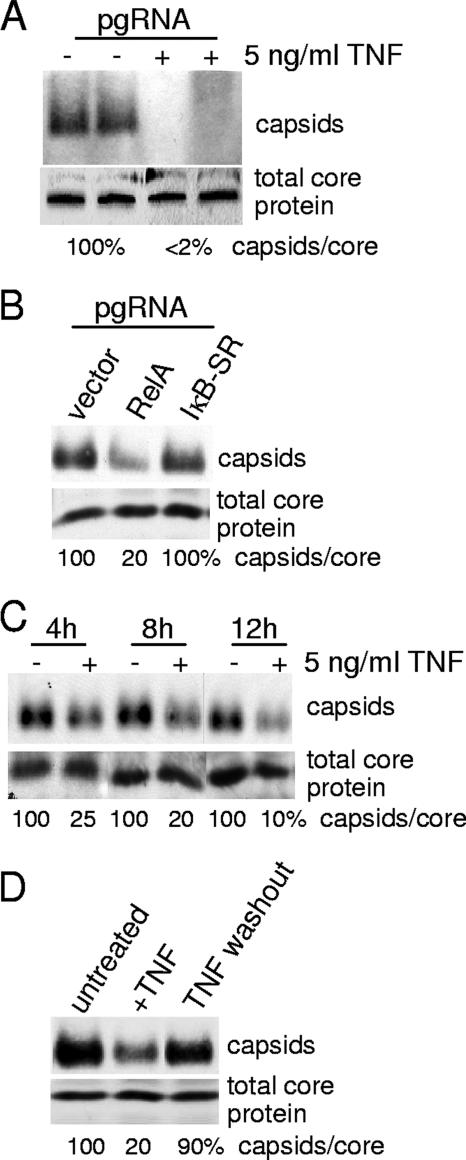
We previously reported that TNF only slightly decreases viral mRNA levels over the course of 1 to 2 days of treatment (5). The decrease in viral RNA with TNF treatment is therefore an unlikely explanation for the much larger decrease in viral DNA replication and capsid levels observed. To further determine whether TNF/NF-κB inhibition of HBV replication acts posttranscriptionally, HepG2 cells were transfected with in vitro-transcribed HBV full-length pgRNA. Transfected pgRNA expresses core and polymerase proteins and can be encapsidated in nucleocapsids and reverse transcribed but does not give rise to cccDNA and therefore cannot act as a transcriptional template, thus removing any potential effect of viral transcription on TNF antiviral action. HepG2 cells were transfected with pgRNA and then treated 1 day later with 5 ng/ml TNF for 24 h (Fig. 3A). TNF decreased the abundance of nucleocapsids derived from pgRNA by ∼98% (they were virtually undetectable) without affecting the total level of core protein. HepG2 cells were also transfected with expression plasmids expressing RelA, IκB-SR, or vector alone and then transfected with in vitro-transcribed pgRNA (Fig. 3B). Compared to what was seen for untreated vector control samples, expression of RelA reduced the abundance of nucleocapsids derived from transfected pgRNA by 80% but did not decrease overall core protein levels. Thus, these data demonstrate that TNF mediates the downregulation of nucleocapsid levels posttranscriptionally. Expression of IκB-SR had no effect on nucleocapsid levels, unlike that of transfected HBV replicons. Interestingly, the pgRNA does not express HBV HBx protein, and there was no evidence for the increased abundance of nucleocapsids observed with NF-κB inhibition in HBx-expressing HBV replicon-transfected cells.
FIG. 3.
TNF acts posttranscriptionally and rapidly to downregulate HBV nucleocapsid levels. (A) HepG2 cells were transfected with in vitro-transcribed pgRNA (see Materials and Methods for details). Following transfection, cells were treated with 5 ng/ml TNF for 24 h in duplicate studies as shown or (B) secondarily transfected with a vector control or expression plasmids for RelA or IκB-SR. Nucleocapsids were isolated and analyzed by native agarose gel electrophoresis and immunoblotting, and total core protein was analyzed by SDS-PAGE and immunoblotting using an anti-HBcAg antibody. (C) HepG2 cells were transfected with an HBV replicon plasmid for 2 days and then treated with 5 ng/ml TNF for 4, 8, or 12 h. Cytoplasmic nucleocapsids and total core protein levels were determined as described above. (D) HepG2 cells were transfected with an HBV replicon as described above, treated with 5 ng/ml of TNF for 24 h, washed, and cultured for another 24 h. Total core protein and nucleocapsid levels were analyzed by electrophoresis and immunoblotting. Results shown are representative of at least three independent experiments.
It was previously established that HBV nucleocapsids are disrupted when cells are treated with 5 ng/ml recombinant human TNF for 4 days (5). To determine the kinetics for response to treatment with TNF, HepG2 cells transfected with the HBV replicon were treated with 5 ng/ml of TNF for 4, 8, 12, or 24 h. Surprisingly, within 4 h of treatment, TNF significantly decreased viral nucleocapsid levels (75%) with no effect on total core protein levels (Fig. 3C). Between 8 and 12 h of TNF treatment, TNF reduction of nucleocapsid levels was maximally achieved (∼90%). The destabilization of viral nucleocapsids by TNF was also found to be transient in the absence of continuous cytokine exposure. Whereas HepG2 cells treated with 5 ng/ml TNF for 24 h downregulated nucleocapsid abundance by 80%, 24 h after TNF washout the abundance of nucleocapsids recovered to 90% of the level of untreated cells (Fig. 3D). Thus, TNF acts rapidly, transiently, and through NF-κB as an antiviral mediator of viral replication.
TNF destabilizes preexisting HBV nucleocapsids.
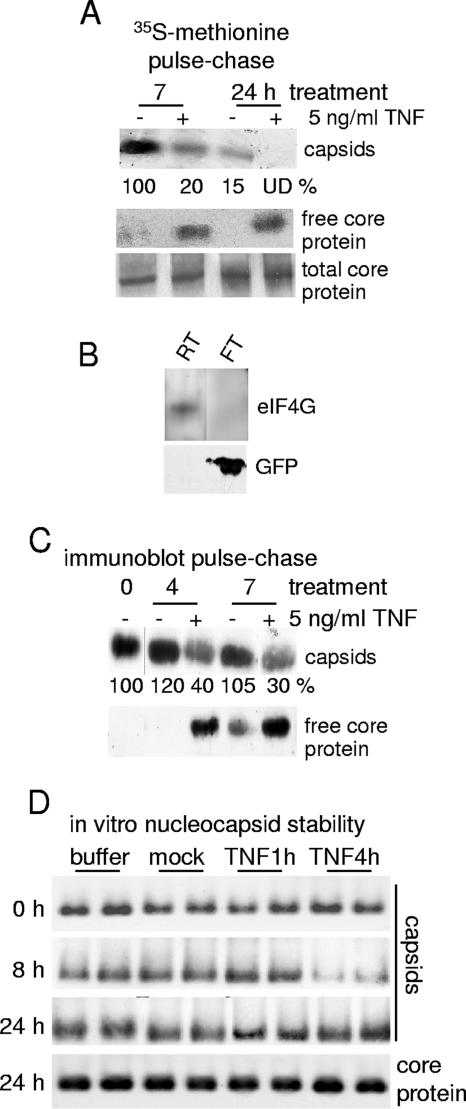
TNF could act by inhibiting assembly of new nucleocapsids and/or by promoting disassembly of existing nucleocapsids. To determine whether TNF treatment destabilizes existing nucleocapsids, HepG2.215 cells were labeled with [35S]methionine-cysteine for 6 h. Following labeling, the medium was changed and chased with nonradiolabeled medium for 6 h. Almost all of the labeled core protein assembles into nucleocapsids during this period because the concentration for spontaneous assembly is very low (0.8 μM) (65). In addition, nucleocapsids are extremely stable and were not observed to undergo degradation or disruption in the absence of TNF treatment (data not shown). Cells were treated with 5 ng/ml TNF (or mock treated) for up to 24 h, and the ratio of free core protein to assembled nucleocapsids was determined by size fractionation of cytoplasmic extracts into a <100-kDa fraction which retains 25 kDa of free core protein and a >100-kDa fraction that contains nucleocapsids (see Materials and Methods for details). Labeled intact nucleocapsids and free core protein were resolved by SDS-PAGE and quantified by autoradiography. There was very little detectable free core protein in the untreated control samples (Fig. 4A). In contrast, nucleocapsids decreased significantly with TNF treatment, reciprocally with an increase in the abundance of labeled free core protein (Fig. 4A). The steady-state levels of unlabeled core protein in mock- and TNF-treated cells were similar, as measured by SDS-PAGE and immunoblot analysis. The total abundance of labeled nucleocapsids decreased by 24 h of cold chasing, as expected due to envelopment and secretion, whereas with TNF disruption, total core protein levels remained unchanged. The altered migration of core protein is an electrophoretic artifact in this particular experiment. Studies in which unlabeled free core protein and nucleocapsids from HepG2.215 cells were fractionated with and without TNF treatment were also performed (Fig. 4C). As observed for the pulse-chase analysis, in the absence of TNF treatment almost all core protein was found in nucleocapsids, which was reciprocally reduced as free core protein levels increased at 4 h and 7 h of TNF treatment.
FIG. 4.
TNF mediates disassembly of preexisting nucleocapsids. (A) HepG2.215 cells were subjected to pulse-chase analysis with [35S]methionine-cysteine (see Materials and Methods for details). Nucleocapsids were separated from “free” core protein (nonnucleocapsid protein) using Centricon-100 filtration with a >100-kDa molecular size exclusion limit, and nucleocapsids, free core proteins, and total core protein in lysates were immunoprecipitated using an anti-HBcAg antibody. Capsid data were quantified by densitometry and expressed as normalized to the untreated 7-h mock control lane. UD, undetectable. (B) Centricon-100 analysis control data are shown, demonstrating the retention of eukaryotic initiation factor 4 subunit G (eIF4G), a 220-kDa protein, in the retentate fraction (RT) and the passage of 25-kDa green fluorescent protein (GFP) in the flowthrough fraction (FT). (C) HepG2.215 cells were unlabeled and analyzed as described above. Nucleocapsids and free core protein levels were determined by Centricon-100 filtration as described above, and proteins were detected by immunoblot analysis. Capsid data were quantified as described above and normalized to the untreated (0-h) capsid level. (D) HepG2 cells transfected with HBV replicons and untransfected HepG2 cells were left untreated or were treated with 5 ng/ml TNF for 1 h or 4 h. Cells were harvested and lysed by Dounce homogenization, and crude extracts from HBV-transfected cells as a source of viral nucleocapsids were incubated at 30°C with lysates obtained from the untransfected HepG2 cells. Nucleocapsid integrity was assessed over a 24-h period by native agarose gel electrophoresis and immunoblot analysis, in duplicate samples. Results shown are representative of at least three independent experiments.
Studies were conducted to determine whether preformed HBV nucleocapsids could also be destabilized in vitro in extracts prepared from TNF-treated uninfected cells. HepG2 cells were transfected with HBV replicons, and crude cytoplasmic extracts were prepared. Cytoplasmic extracts were also prepared from nontransfected cells that were either mock treated or treated with 5 ng/ml TNF for 1 h or 4 h. Following high-speed centrifugation to remove nuclear and membrane debris, the HBV vector capsid-containing extracts were incubated with the extracts prepared from mock-treated or TNF-treated uninfected cells for up to 24 h at 30°C. The stability of nucleocapsids was assessed by native agarose gel electrophoresis and immunoblot analysis. Nucleocapsid levels were markedly decreased when extracts were incubated with lysates prepared from cells treated with TNF for 4 h but not with those from cells treated with TNF for only 1 h (Fig. 4D). Interestingly, nucleocapsids were found to reassemble by 24 h of incubation, suggesting that the nucleocapsid-destabilizing activity or factor is labile or becomes depleted. Total core protein levels are shown for the 24-h time point and did not vary regardless of the treated extract. These results further confirm that TNF destabilizes existing nucleocapsids and suggest that TNF induces a cellular antinucleocapsid activity. Reformed capsids were not examined for replicative function or presence of pgRNA due to the small amount of capsids present. These data therefore indicate that TNF destabilizes preformed nucleocapsids without inducing the degradation of core protein.
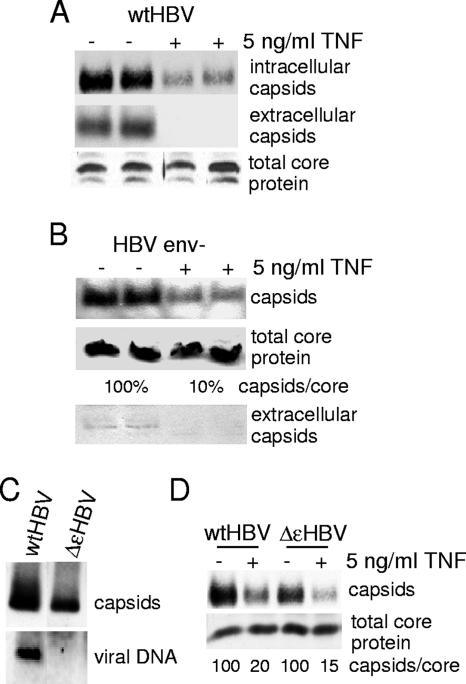
We asked whether the decrease in cytoplasmic capsid levels might in part be a result of TNF induction of premature secretion of nucleocapsids and/or mature virions. To investigate this possibility, we first assessed the capsid and mature virion levels in the extracellular medium. After transfection with wild-type HBV replicon DNA, cells were either mock treated or treated with 5 ng/ml TNF, viral particles were recovered from the medium by centrifugation, and cytoplasmic extracts were also prepared for analysis of cytoplasmic nucleocapsid and total core protein levels. Immunoblot detection of nucleocapsids was carried out on both the medium and the cytoplasmic extracts to determine whether TNF promoted shunting of both immature and mature nucleocapsids into the extracellular space (Fig. 5A). TNF treatment significantly reduced the levels of intracellular cytoplasmic nucleocapsids, as expected, but also caused a disproportionately larger reduction in the levels of nucleocapsids or viral particles released into the medium (Fig. 5A). There was no change in total core protein levels, as expected. To develop a second line of evidence, HepG2 cells were transfected with a plasmid encoding an HBV replicon containing an envelope mutation [pGem7Z(+)ayw env−] that is unable to produce enveloped nucleocapsids for secretion. The mutation consists of the insertion of two stop codons in the pres1 and S open reading frames. Both cytoplasm and medium were probed for nucleocapsid and total core protein levels (Fig. 5B). Treatment with TNF did not promote the secretion of nonenveloped (immature) nucleocapsids, as the level of extracellular capsids was almost undetectable, as expected for a nucleocapsid secretion-defective phenotype, and there was no change in total core protein levels. Collectively, these data indicate that TNF inhibits HBV replication by promoting the disassembly of existing nucleocapsids in the cytoplasm, thereby decreasing the secretion of infectious virus rather than acting to inhibit secretion itself.
FIG. 5.
TNF promotes destabilization and not premature secretion of mature, empty, and nonenveloped nucleocapsids. (A) HepG2 cells were transfected with a wild-type HBV replicon (wtHBV) plasmid and then treated with 5 ng/ml TNF for 24 h. Extracellular and intracellular nucleocapsids were collected and analyzed by native agarose gel electrophoresis and immunoblot analysis in duplicate. Total core protein levels were determined by SDS-PAGE and immunoblot analysis. (B) HepG2 cells were transfected with an env− HBV replicon (HBV env-) that cannot produce HBsAgs or secrete mature enveloped nucleocapsids and then treated with 5 ng/ml TNF for 24 h. Cytoplasmic and extracellular nucleocapsids and total core protein levels were analyzed as described above. (C) Cytoplasmic nucleocapsids were purified from HepG2 cells transfected with wild-type HBV or ΔɛHBV replicons and nucleocapsid and encapsidated viral DNA were examined by immunoblotting and Southern blot DNA analysis. (D) HepG2 cells were transfected with either a wild-type HBV replicon or ΔɛHBV, a replicon mutated in the epsilon region that cannot encapsidate pgRNA into nucleocapsids. Cells were treated with 5 ng/ml TNF, and nucleocapsids and total core protein were analyzed as described above. Results shown are representative of at least three independent experiments.
TNF downregulates nucleocapsids comprised of core protein alone.
Of particular interest was whether TNF destabilization of nucleocapsids differentiates between genome-containing and empty nucleocapsids. Approximately 90% of HBV nucleocapsids in the cytoplasm contain nucleic acids (DNA or RNA) which form a T = 4 conformation (55). The other 10% of nucleocapsids adopt a T = 3 conformation and do not appear to contain nucleic acid (55). To determine whether the TNF response distinguishes DNA-containing from empty nucleocapsid species, HepG2 cells were transfected with the plasmid ΔɛHBV, which encodes a 120% replicon containing a mutation in the loop region of the epsilon hairpin of the viral pgRNA. The ɛ mutation prevents formation of the Pol-pregenome ribonucleoprotein complex and therefore precludes incorporation of pgRNA into nucleocapsids (59), as shown for isolated wild-type and ΔɛHBV nucleocapsids (Fig. 5C). All nucleocapsids produced in cells expressing the ɛ mutation are therefore comprised of core protein alone. TNF treatment of wild-type and ΔɛHBV replicons containing HepG2 cells reduced nucleocapsid levels to roughly the same extent (∼80 to 85%), with no effect on the levels of total core protein (Fig. 5D). These data therefore demonstrate that in addition to RNA- and DNA-containing nucleocapsids, empty nucleocapsids are also targeted for destabilization by TNF.
Nucleocapsids comprised of C-terminally truncated core protein are disrupted by TNF.
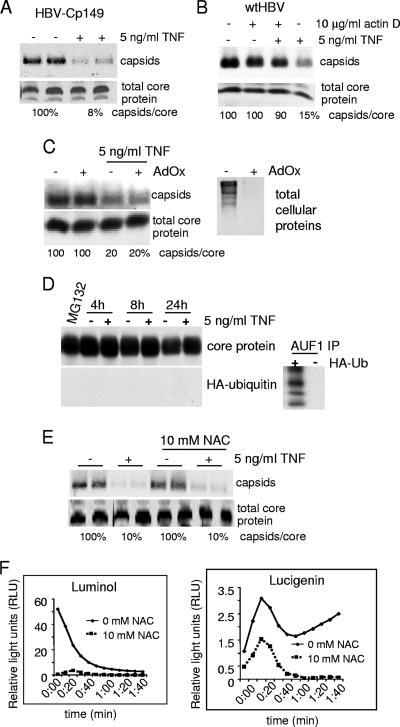
The C-terminal region of core protein has been previously shown to be essential for several steps in the maturation process of the nucleocapsid (17, 36, 38, 41, 47, 53). In particular, phosphorylation of serines 155, 162, and 170 has been shown to be important for encapsidation of pgRNA, replication of the viral genome, and localization of nucleocapsids (17, 37, 53). Analysis of serine-to-alanine mutations at positions 155, 162, and 170 determined that the phosphorylations of these serines were not essential for TNF disassembly of nucleocapsid levels (data not shown). To determine if there were other C-terminal elements that might be essential for TNF-mediated suppression of viral nucleocapsids, a truncated core protein (Cp149) was developed within the viral replicon. Cp149 is capable of forming nucleocapsids similar to full-length core protein both in vitro and in vivo (65). Treatment of cells containing Cp149 nucleocapsids with 5 ng/ml TNF resulted in their reduction by ∼90% (Fig. 6A), a sensitivity similar to that of wild-type nucleocapsids. We can conclude that TNF acts independently of the C-terminal regulatory region of core protein and therefore must target a region of core protein within the first 149 amino acids to destabilize nucleocapsids.
FIG. 6.
TNF disruption of nucleocapsids does not require the regulatory C-terminal region of core protein but does require cellular transcription. (A) HepG2 cells were transfected with a Cp149 HBV replicon (HBV-Cp149) that lacks the C terminus of core protein from amino acid 149 and then treated with 5 ng/ml TNF for 24 h. Nucleocapsids and total core protein were analyzed as described earlier. wtHBV, wild-type HBV replicon. (B) HepG2 cells were transfected with a wild-type HBV replicon for 2 days and pretreated with actinomycin D (actin D) for 30 min, followed by the addition of 5 ng/ml TNF for 4 h. Nucleocapsids and total core protein were analyzed as described earlier. Results shown are representative of at least three independent experiments. (C) HepG2 cells were transfected with the HBV replicon plasmid, and then 24 h later cells were pretreated with 100 μM for 24 h followed by the addition of 5 ng/ml TNF for 6 h. Nucleocapsids and total core protein were analyzed as described above. The inhibition of protein methylation by AdOx was examined by labeling cells with l-[methyl-3H]-methionine in the presence or absence of AdOx followed by SDS-PAGE and fluorography. (D) HepG2 cells were cotransfected with the HBV replicon for 48 h and with an HA-ubiquitin expression plasmid and then treated with 5 ng/ml TNF for 4, 8, or 24 h or incubated with 10 μM of proteasome inhibitor MG132, and AUF1 or HA was immunoprecipitated and analyzed by SDS-PAGE and immunoblot analysis with anti-HA or anti-HBcAg antibodies. (E) HepG2 cells were transfected with HBV replicon DNA, treated 24 h later with 10 mM NAC for 2 h, and then incubated with 5 ng/ml TNF for 6 h (in the presence of NAC). Nucleocapsid and total core protein levels were assayed in duplicate as described above. (F) HepG2 cells were transfected with HBV replicons, and then at 24 h cells were left untreated or were treated with 10 mM NAC to block ROS and incubated with luminol or lucigenin. Cells were then exposed to 0.1% H2O2, and free radical production was measured by luminol or lucigenin fluorescence. Data are expressed in relative light units.
TNF destabilization of nucleocapsids requires active cellular transcription and is not mediated by reactive oxygen species, ubiquitination, or methylation of core protein.
TNF stimulates the transcription of a variety of cellular genes, many through the activation of NF-κB (10). Moreover, we showed earlier that TNF destabilizes nucleocapsids without a need for HBV gene transcription. We therefore investigated whether TNF destabilization of HBV nucleocapsids requires new cellular gene transcription. HBV replicon-transfected cells were treated with actinomycin D to prevent transcription beginning 30 min prior to addition of 5 ng/ml TNF for 4 h. Resolution of nucleocapsids by nondenaturing agarose gel electrophoresis followed by immunoblot analysis demonstrated that actinomycin D almost fully blocked TNF reduction of nucleocapsid levels (∼10%), whereas TNF alone decreased nucleocapsid levels by 85% compared to what was seen for mock-treated cells (Fig. 6B). There was no change in total core protein levels. These data indicate that TNF stimulates the transcription of cellular genes that are essential for the destabilization of nucleocapsids.
Arginines within the core protein potentially could be targets of methylation by methyltransferases, which would be destabilizing to nucleocapsid integrity. Inflammatory cytokines such as IL-1β and possibly TNF, as well as nitric oxide produced by inducible NOS (iNOS) activation (which is induced by TNF), have been shown to stimulate the methylation of arginine-rich proteins such as RNA-binding protein HuR by arginine methyltransferase (46). We therefore investigated whether core protein is also a target of TNF-induced methyltransferase. HBV replicon-transfected cells were pretreated for 24 h with oxidized adenosine (AdOx) to block the methylation of proteins prior to the addition of 5 ng/ml TNF for 24 h. Resolution of nucleocapsids by nondenaturing gel electrophoresis followed by immunoblot analysis demonstrated that in the presence of AdOx, TNF samples decrease nucleocapsid levels equally as well as do untreated samples (Fig. 6C). As a positive control for AdOx action, the methylation of bulk proteins was determined from cells labeled for 3 h with l-[methyl-3H]-methionine and subjected to fluorography as described previously (62). AdOx efficiently blocked methylation of cell proteins (Fig. 6C, right panel).
It was reported that downregulation of HBV replication by IFN-γ requires proteasomal activity (60), and TNF is known to cause ubiquitination of proteins such as IκBα. Although there is no decrease in overall core protein levels in the presence of TNF, it is plausible that TNF might mediate the monoubiquitination of core protein and thus the destabilization of nucleocapsids without core protein degradation. To determine whether TNF induces the monoubiquitination of core protein, HepG2 cells were cotransfected with HBV genomic DNA and a construct encoding a hemagglutinin (HA)-tagged octameric ubiquitin. Following treatment with 5 ng/ml TNF for 24 h, core protein from isolated nucleocapsids and free core protein were immunoprecipitated, resolved by SDS-PAGE, and subjected to immunoblot analysis with HBcAg and anti-HA antibodies. As a control, AUF1 protein known to be ubiquitinated (43) was immunoprecipitated. AUF1 was ubiquitinated when cotransfected with HA-Ub8 (Fig. 6B), but there was no evidence for HA-Ub8-associated core protein.
Another possible mediator of capsid destabilization could be the formation of reactive oxygen and nitrogen intermediates. Studies with a human HBV-transgenic mouse demonstrated that iNOS is critical for IFN-γ suppression of HBV replication (23), and in immune cells, TNF mediates the release of free radicals (10). However, hepatocytes are reported to have only a weak free radical burst (16). To determine whether free radical production is responsible for the TNF-mediated downregulation of HBV, HepG2 cells were transfected with the HBV replicon, pretreated with 10 mM NAC, a free radical scavenger, and then treated with 5 ng/ml TNF. There was no detectable impact of NAC on TNF destabilization of nucleocapsids or total core protein levels (Fig. 6E). To demonstrate that NAC scavenges free radicals in hepatocytes, HepG2 cells were pretreated with 10 mM NAC and then treated with 1% H2O2. Lucigenin or luminol was added to cells to detect the production of free radicals. Luminol also emits light when oxidized by hydrogen peroxide, perioxynitrate, or hydroxyl radicals. Lucigenin emits light when oxidized by hydrogen peroxide or hydroxyl radicals. NAC completely scavenged H2O2 and free radicals produced as a result of H2O2 exposure and measured by either luminol or lucigenin (Fig. 6F), demonstrating that NAC was effective in this system at inhibiting ROS. Thus, TNF suppresses nucleocapsid levels without requiring the production of reactive oxygen or nitrogen species.
DISCUSSION
In this report, we describe a novel antiviral mechanism utilized by TNF to inhibit HBV replication in a noncytopathic manner. TNF activates NF-κB, which serves to induce new transcription of cellular gene products that lead to the rapid destabilization of HBV nucleocapsids. The consequence of TNF action is a profound decrease in secretion of viral particles, which in vivo may slow the early progression of HBV infection in the liver. Furthermore, TNF-mediated disruption of cytoplasmic nucleocapsids leads to a decline in nuclear cccDNA levels, probably by preventing the formation of nucleocapsids that deliver cccDNA to the nucleus. Maintenance of the cccDNA pool is thought to be critical for HBV persistence in infected hepatocytes. These data highlight that destabilization of HBV nucleocapsids and reduction of cccDNA could ultimately limit chronic infection. TNF destabilization of nucleocapsids does not require the presence of any other HBV genes except for core protein. TNF disrupts all types of nucleocapsids, including empty nucleocapsids. Importantly, TNF appears to be as effective against DHBV as against HBV and acts to destabilize nucleocapsids and block viral replication in a panspecies manner and in hepatocytic cells from a variety of different species. Thus, these data indicate that TNF action is phylogenetically conserved across the Hepadnaviridae family and different species of hepatocytes. This conservation argues that TNF antiviral action represents an important and possibly ancient innate defense mechanism against HBV in vivo.
TNF inhibition of HBV differs from that of other described cytokine inhibitors in that it targets the stability of nascent nucleocapsids. Type I IFNs likely suppress HBV mRNA transcription (22) and type II IFN-γ (27) might regulate the activity of La proteins, which may play a putative role in HBV mRNA stability (27). IFN-γ might also require both proteasome activity and iNOS activity (23, 60), which we show does not play a role in TNF inhibition of HBV. Other cytokines implicated in regulation of HBV infection include interleukin-1 (IL-1), IL-2, IL-6, IL-12, and IL-18 (9, 15, 21, 25, 40). Of note is that IL-2 was shown to decrease HBV replication, in part by inducing the production of TNF (22, 25). It is likely that these and other cytokines work in concert to simultaneously inhibit multiple steps in the HBV life cycle, thereby laying the groundwork for resolution of infection.
It should be noted that TNF is associated with the clearance of a variety of mammalian viruses in addition to HBV. For example, herpes simplex virus is prevented from infecting mice that are pretreated with TNF (61). TNF has also been shown to noncytopathically decrease Sin Nombre virus nucleocapsid protein (39). In addition, treatment of peripheral blood monocytes with TNF reduces the reverse transcriptase activity of human immunodeficiency virus type 1 and thus inhibits its replication in experimental systems (42). Other viruses reportedly inhibited by TNF include vesicular stomatitis virus, encephalomyocarditis virus, Ad2, and herpes simplex virus type II (28). Suppression of these viruses involves a variety of mechanisms, including induction of chemokine and cytokine production, downregulation of receptors required for viral infection, and apoptosis of infected cells (28). This raises the question of how one innate immune molecule could employ a variety of antiviral mechanisms. However, as known from interferons, this is in fact the pattern of many innate immune molecules that have antiviral effects (57). In this regard, we showed that TNF activity against HBV critically requires new transcription of cellular genes mediated by NF-κB. It is known that transcriptional activation by NF-κB can result in the upregulation of as many as 150 genes, at least in some contexts such as in immune cells. The pleiotropic induction of multiple gene products may explain the many different antiviral responses of TNF. The initial invasion by a microbe must be responded to rapidly to prevent the spread of infection. The early host response, unlike the later adaptive response, is therefore unlikely to enact a specific biochemical response to a pathogen. Consequently, a diverse approach in which panels of antiviral effectors are upregulated simultaneously can provide the best early host defense.
We also showed that core protein ubiquitination and methylation are not critical for TNF-mediated destabilization of nucleocapsids. In our studies we found that the TNF anti-HBV activity, unlike that of IFN-γ, is not mediated by an iNOS-dependent mechanism. Furthermore, we found that TNF does not destabilize nucleocapsids by the production of free radicals, as their inhibition has no effect. Although we have not observed gross modifications of core protein, such as proteolysis, ubiquitination, or methylation, we cannot eliminate the possibility that minor changes, undetectable by traditional methods, are sufficient to destabilize nucleocapsids. For instance, Barrasa et al. (4) demonstrated that mutation of core protein threonine 174 to alanine disrupts the formation of nucleocapsids even when wild-type core protein is present, suggesting that the modification of only a small number of core proteins is sufficient to block or destabilize capsid formation. In addition, it was recently shown that HBV core protein assembly into nucleocapsids requires the phosphorylation of serine 87 (34). Further studies are required to determine whether nucleocapsids undergo more-subtle modifications mediated by TNF that may contribute to their instability.
To begin to understand the mechanism by which TNF destabilizes nucleocapsids, it is helpful to review the mechanism and kinetics of HBV capsid assembly, which have been well studied in vitro. Core protein is typically found as a dimer, which assembles into a 35-nm icosahedral capsid (T = 4) composed of 120 core dimers once a critical concentration of approximately 0.8 μM has been reached (65). Assembly of HBV nucleocapsids in vitro is nucleated by a trimer of core protein dimers (70). Further assembly into nucleocapsids proceeds without accumulation of observable populations of intermediates (70). The core dimer association energy becomes progressively stronger in increasing concentrations of NaCl (68) and is facilitated by the local ion concentration, particularly Zn2+ ions (68). Thus, Stray et al. have hypothesized that the effect of high salt concentrations and Zn2+ ions is to induce the nucleocapsid protein to adopt an assembly active conformation (68). At a 17-Å resolution, T = 4 HBV nucleocapsids contain holes at each of the quasi-sixfold axes (71). The presence of these holes likely aids HBV nucleocapsid maturation, particularly during reverse transcription of the viral genome, when access to cytoplasmic nucleotides is critical. It may also play a role in the egress of digested RNA (71). It has been proposed that the presence of nucleocapsid pores as well as flexible associations between core dimers within the nucleocapsid may be attributed to nucleocapsid “breathing” (13, 70, 71). This theory is further supported by the ability of several small molecules to misdirect and disassemble already formed nucleocapsids (13, 70). For example, heteroaryldihydropyrimidines have been shown to bind near a cluster of histidine residues at the base of the intradimer surface of HBV nucleocapsids (13, 70). Access to these histidines requires that the capsid be flexible, not structurally inert. In addition, heteroaryldihydropyrimidines likely extract dimers from the capsid and thus lower the energy barrier to nucleocapsid dissociation (70). This interaction and extraction is likely to occur only if the nucleocapsid “breathes.” This is consistent with the hypothesis proposed for the mechanism of HBV nucleocapsid dissociation, where contacts between core protein dimers are continuously made and broken, resulting in metastable nucleocapsids (66). The stochastic nature of core dimer interactions could therefore be a target of innate immune inhibition by TNF.
Most of what is known about HBV nucleocapsid assembly has been derived from in vitro models. In vivo, it is believed that the encapsidation of pgRNA acts as a nucleating event that lowers the core protein concentration threshold for nucleocapsid assembly. In addition, several host proteins have been found to play important roles in HBV nucleocapsid assembly (14, 48, 49, 67). For example, several heat shock and heat shock-related proteins have been found to be associated with HBV nucleocapsids. The encapsidation of pgRNA requires that HBV Pol, heat shock protein 90 (hsp90) chaperones, and hsp60 associate with the 5′-end region of pgRNA (48). A chaperonin t-complex polypeptide I (TCP-1)-related protein has also been hypothesized to be involved in core protein multimerization (48). In addition, it was suggested that hsp40 might bind core polypeptides and intermediate forms of nucleocapsids in the early stages of virus assembly and thereby regulate HBV nucleocapsid levels (67). Thus, TNF might act on heat shock or host cell proteins involved in the regulation of viral nucleocapsid assembly, promoting disassembly. This is currently under investigation.
In summary, we have described a phylogenetically conserved TNF-mediated innate antiviral mechanism that targets the integrity of HBV nucleocapsids. Our results demonstrate a novel antiviral mechanism induced by TNF. The elucidation of this antiviral strategy may lead to the development of new therapeutics for acute and chronic HBV infection.
Acknowledgments
This work was supported by NIH grant CA056533 to R.J.S. R.P. was supported partially by a T32 training grant from NIAID.
We thank Sarah Fabes for critical review of the manuscript.
Footnotes
Published ahead of print on 2 May 2007.
REFERENCES
- 1.al-Wabel, A., M. al-Janadi, and S. Raziuddin. 1993. Cytokine profile of viral and autoimmune chronic active hepatitis. J. Allergy Clin. Immunol. 92:902-908. [DOI] [PubMed] [Google Scholar]
- 2.Ando, K., L. G. Guidotti, S. Wirth, T. Ishikawa, G. Missale, T. Moriyama, R. D. Schreiber, H. J. Schlicht, S. N. Huang, and F. V. Chisari. 1994. Class I-restricted cytotoxic T lymphocytes are directly cytopathic for their target cells in vivo. J. Immunol. 152:3245-3253. [PubMed] [Google Scholar]
- 3.Baron, J. L., L. Gardiner, S. Nishimura, K. Shinkai, R. Locksley, and D. Ganem. 2002. Activation of a nonclassical NKT cell subset in a transgenic mouse model of hepatitis B virus infection. Immunity 16:583-594. [DOI] [PubMed] [Google Scholar]
- 4.Barrasa, M. I., J. T. Guo, J. Saputelli, W. S. Mason, and C. Seeger. 2001. Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids? J. Virol. 75:2024-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biermer, M., R. Puro, and R. J. Schneider. 2003. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid integrity through activation of NF-κB. J. Virol. 77:4033-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, M., S. Giannakopoulos, E. Wang, N. Tanese, and R. J. Schneider. 2001. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J. Virol. 75:4247-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 8.Bozkaya, H., M. Bozdayi, R. Turkyilmaz, M. Sarioglu, H. Cetinkaya, K. Cinar, K. Kose, C. Yurdaydin, and O. Uzunalimoglu. 2000. Circulating IL-2, IL-10 and TNF-alpha in chronic hepatitis B: their relations to HBeAg status and the activity of liver disease. Hepatogastroenterology 47:1675-1679. [PubMed] [Google Scholar]
- 9.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1997. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J. Virol. 71:3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, G., and D. V. Goeddel. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634-1635. [DOI] [PubMed] [Google Scholar]
- 11.Dandri, M., and J. Petersen. 2005. Hepatitis B virus cccDNA clearance: killing for curing? Hepatology 42:1453-1455. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, H., A. Meager, A. W. F. L. Eddleston, G. J. M. Alexander, and R. Williams. 1990. Spontaneous TNF-alpha and IL-1beta production during successful interferon therapy in chronic HBV infection. Lancet 335:875-877. [DOI] [PubMed] [Google Scholar]
- 13.Deres, K., C. H. Schröder, A. Paessens, S. Goldmann, H. J. Hacker, O. Weber, T. Krämer, U. Niewöhner, U. Pleiss, J. Stoltefuss, E. Graef, D. Koletzki, R. N. Masantschek, A. Reimann, R. Jaeger, R. Grob, B. Beckermann, K. Schlemmer, D. Haebich, and H. Rübsamen-Waigmann. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893-896. [DOI] [PubMed] [Google Scholar]
- 14.Ganem, D., and R. J. Schneider. 2001. The molecular biology of the hepatitis B viruses, p. 2923-2970. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott, New York, NY. [Google Scholar]
- 15.Garcia-Navarro, R., B. Blanco-Urgoiti, P. Berraondo, R. Sanchez de la Rosa, A. Vales, S. Hervas-Stubbs, J. J. Lasarte, F. Borras, J. Ruiz, and J. Prieto. 2001. Protection against woodchuck hepatitis virus (WHV) infection by gene gun coimmunization with WHV core and interleukin-12. J. Virol. 75:9068-9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Ruiz, C., and J. C. Fernandez-Checa. 2006. Mitochondrial glutathione: hepatocellular survival-death switch. J. Gastroenterol. Hepatol. 21:S3-S6. [DOI] [PubMed] [Google Scholar]
- 17.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilles, P. N., G. Fey, and F. V. Chisari. 1992. Tumor necrosis factor alpha negatively regulates hepatitis B virus gene expression in transgenic mice. J. Virol. 66:3955-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., K. Ando, M. V. Hobbs, T. Ishikawa, L. Runkel, R. D. Schreiber, and F. V. Chisari. 1994. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. USA 91:3764-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidotti, L. G., and F. V. Chisari. 2000. Cytokine-mediated control of viral infections. Virology 273:221-227. [DOI] [PubMed] [Google Scholar]
- 21.Guidotti, L. G., and F. V. Chisari. 1996. To kill or to cure: options in host defense against viral infection. Curr. Opin. Immunol. 8:478-483. [DOI] [PubMed] [Google Scholar]
- 22.Guidotti, L. G., S. Guilhot, and F. V. Chisari. 1994. Interleukin-2 and alpha/beta interferon down-regulate hepatitis B virus gene expression in vivo by tumor necrosis factor-dependent and -independent pathways. J. Virol. 68:1265-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guidotti, L. G., H. McClary, J. M. Loudis, and F. V. Chisari. 2000. Nitric oxide inhibits hepatitis B virus replication in the livers of transgenic mice. J. Exp. Med. 191:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 25.Guilhot, S., L. G. Guidotti, and F. V. Chisari. 1993. Interleukin-2 downregulates hepatitis B virus gene expression in transgenic mice by a posttranscriptional mechanism. J. Virol. 67:7444-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo, J. T., H. Zhou, C. Liu, C. Aldrich, J. Saputelli, T. Whitaker, M. I. Barrasa, W. S. Mason, and C. Seeger. 2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J. Virol. 74:1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise, T., L. G. Guidotti, and F. V. Chisari. 1999. La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA. J. Virol. 73:5767-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbein, G., and W. A. O'Brien. 2000. Tumor necrosis factor (TNF)-alpha and TNF receptors in viral pathogenesis. Proc. Soc. Exp. Biol. Med. 223:241-257. [DOI] [PubMed] [Google Scholar]
- 29.Hodgson, P. D., and T. I. Michalak. 2001. Augmented hepatic interferon gamma expression and T-cell influx characterize acute hepatitis progressing to recovery and residual lifelong virus persistence in experimental adult woodchuck hepatitis virus infection. Hepatology 34:1049-1059. [DOI] [PubMed] [Google Scholar]
- 30.Jilbert, A. R., and I. Kotlarski. 2000. Immune responses to duck hepatitis B virus infection. Dev. Comp. Immunol. 24:285-302. [DOI] [PubMed] [Google Scholar]
- 31.Jilbert, A. R., T. T. Wu, J. M. England, P. M. Hall, N. Z. Carp, and A. P. O'Connell. 1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J. Virol. 66:1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajino, K., A. R. Jilbert, J. Saputelli, C. E. Aldrich, J. Cullen, and W. Mason. 1994. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J. Virol. 68:5792-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakimi, K., T. E. Lane, S. Wieland, V. C. Asensio, I. L. Campbell, F. V. Chisari, and L. G. Guidotti. 2001. Blocking chemokine responsive to gamma-2/interferon (IFN)-gamma inducible protein and monokine induced by IFN-gamma activity in vivo reduces the pathogenetic but not the antiviral potential of hepatitis B virus-specific cytotoxic T lymphocytes. J. Exp. Med. 194:1755-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang, H. Y., S. Lee, S. G. Park, J. Yu, Y. Kim, and G. Jung. 2006. Phosphorylation of hepatitis B virus Cp at Ser87 facilitates core assembly. Biochem. J. 398:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kann, M., A. Bischof, and W. H. Gerlich. 1997. In vitro model for the nuclear transport of the hepadnavirus genome. J. Virol. 71:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kann, M., and W. H. Gerlich. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68:7993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kau, J.-H., and L.-P. Ting. 1998. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J. Virol. 72:3796-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khaiboullina, S. F., D. M. Netski, P. Krumpe, and S. C. St Jeor. 2000. Effects of tumor necrosis factor alpha on Sin Nombre virus infection in vitro. J. Virol. 74:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura, K., K. Kakimi, S. Wieland, L. G. Guidotti, and F. V. Chisari. 2002. Interleukin-18 inhibits hepatitis B virus replication in the livers of transgenic mice. J. Virol. 76:10702-10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan, Y. T., J. Li, W. Liao, and J. Ou. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 42.Lane, B. R., D. M. Markovitz, N. L. Woodford, R. Rochford, R. M. Strieter, and M. J. Coffey. 1999. TNF inhibits HIV-1 replication in peripheral blood monocytes and alveolar macrophages by inducing the production of RANTES and decreasing C-C chemokine receptor 5 (CCR5) expression. J. Immunol. 163:3653-3661. [PubMed] [Google Scholar]
- 43.Laroia, G., and R. J. Schneider. 2002. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 30:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavanchy, D. 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J. Viral Hepat. 11:97-107. [DOI] [PubMed] [Google Scholar]
- 45.Lenhoff, R. J., and J. Summers. 1994. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J. Virol. 68:5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, H., S. Park, B. Kilburn, M. A. Jelinek, A. Henschen-Edman, D. W. Aswad, M. R. Stallcup, and I. A. Laird-Offringa. 2002. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 277:44623-44630. [DOI] [PubMed] [Google Scholar]
- 47.Liao, W., and J. H. Ou. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 69:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lingappa, J. R., R. L. Martin, M. L. Wong, D. Ganem, W. J. Welch, and V. R. Lingappa. 1994. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J. Cell Biol. 125:99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lingappa, J. R., M. A. Newman, K. C. Klein, and J. E. Dooher. 2005. Comparing capsid assembly of primate lentiviruses and hepatitis B virus using cell-free systems. Virology 333:114-123. [DOI] [PubMed] [Google Scholar]
- 50.Lu, M., B. Lohrengel, G. Hilken, T. Kemper, and M. Roggendorf. 2002. Woodchuck gamma interferon upregulates major histocompatibility complex class I transcription but is unable to deplete woodchuck hepatitis virus replication intermediates and RNAs in persistently infected woodchuck primary hepatocytes. J. Virol. 76:58-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandart, E., A. Kay, and F. Galibert. 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J. Virol. 49:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melegari, M., S. K. Wolf, and R. J. Schneider. 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. Virol. 79:9810-9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura, I., J. T. Nupp, M. Cowlen, W. C. Hall, B. C. Tennant, J. L. Casey, J. L. Gerin, and P. J. Cote. 2001. Pathogenesis of experimental neonatal woodchuck hepatitis virus infection: chronicity as an outcome of infection is associated with a diminished acute hepatitis that is temporally deficient for the expression of interferon gamma and tumor necrosis factor-alpha messenger RNAs. Hepatology 33:439-447. [DOI] [PubMed] [Google Scholar]
- 55.Newman, M., F. M. Suk, M. Cajimat, P. K. Chua, and C. Shih. 2003. Stability and morphology comparisons of self-assembled virus-like particles from wild-type and mutant human hepatitis B virus capsid proteins. J. Virol. 77:12950-12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Penna, A., M. Artini, A. Cavalli, M. Levrero, A. Bertoletti, M. Pilli, F. V. Chisari, B. Rehermann, G. Del Prete, F. Fiaccadori, and C. Ferrari. 1996. Long-lasting memory T cell responses following self-limited acute hepatitis B. J. Clin. Investig. 98:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry, A. K., G. Chen, D. Zheng, H. Tang, and G. Cheng. 2005. The host type I interferon response to viral and bacterial infections. Cell Res. 15:407-422. [DOI] [PubMed] [Google Scholar]
- 58.Phi Van, L. 1996. Transcriptional regulation of chicken lysozyme gene by NF-kB. Biochem. J. 313:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pollack, J. R., and D. Ganem. 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol. 67:3254-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robek, M. D., S. F. Wieland, and F. V. Chisari. 2002. Inhibition of hepatitis B virus replication by interferon requires proteasome activity. J. Virol. 76:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossol-Voth, R., S. Rossol, K. H. Schutt, S. Corridori, W. de Cian, and D. Falke. 1991. In vivo protective effect of tumour necrosis factor alpha against experimental infection with herpes simplex virus type 1. J. Gen. Virol. 72:143-147. [DOI] [PubMed] [Google Scholar]
- 62.Sarkar, B., J.-Y. Liu, and R. J. Schneider. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 278:20700-20707. [DOI] [PubMed] [Google Scholar]
- 63.Schultz, U., and F. V. Chisari. 1999. Recombinant duck interferon gamma inhibits duck hepatitis B virus replication in primary hepatocytes. J. Virol. 73:3162-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schultz, U., J. Summers, P. Staeheli, and F. V. Chisari. 1999. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J. Virol. 73:5459-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seifer, M., and D. N. Standring. 1995. Assembly and antigenicity of hepatitis B virus core particles. Intervirology 38:47-62. [DOI] [PubMed] [Google Scholar]
- 66.Singh, S., and A. Zlotnick. 2003. Observed hysteresis of virus capsid diassembly is implicit in kinetic models of assembly. J. Biol. Chem. 278:18249-18255. [DOI] [PubMed] [Google Scholar]
- 67.Sohn, S.-Y., S.-B. Kim, J. Kim, and B.-Y. Ahn. 2006. Negative regulation of hepatitis B virus replication by cellular Hsp40/DnaJ proteins through destabilization of viral core and X proteins. J. Gen. Virol. 87:1883-1891. [DOI] [PubMed] [Google Scholar]
- 68.Stray, S. J., P. Ceres, and A. Zlotnick. 2004. Zinc ions trigger conformational change and oligomerization of hepatitis B virus capsid protein. Biochemistry 43:9989-9998. [DOI] [PubMed] [Google Scholar]
- 69.Tipples, G. A., M. M. Ma, K. P. Fischer, V. G. Bain, N. M. Kneteman, and D. L. Tyrrell. 1996. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology 24:714-717. [DOI] [PubMed] [Google Scholar]
- 70.Zlotnick, A., P. Ceres, S. Singh, and J. M. Johnson. 2002. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J. Virol. 76:4848-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zlotnick, A., N. Cheng, J. F. Conway, F. P. Booy, A. C. Steven, S. J. Stahl, and P. T. Wingfield. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35:7412-7421. [DOI] [PubMed] [Google Scholar]