Abstract
Viral recombination has been postulated to play two roles in the development of human immunodeficiency virus (HIV) resistance to antiretroviral drugs. First, recombination has the capacity to associate resistance mutations expressed by distinct viruses, thereby contributing to the development of viruses with improved drug resistance. In addition, recombination could preserve diversity in regions outside those subject to strong selective pressure. In this study, we sought direct evidence for the occurrence of these processes in vivo by evaluating clonal virus populations obtained from the same patient before and after a treatment change that, while unsuccessful in controlling viral replication, led to the emergence of viruses expressing a different profile of resistance mutations. Phylogenetic studies supported the conclusion that the genotype arising after the treatment change resulted from the emergence of recombinant viruses carrying previously existing resistance mutations in novel combinations, whereas alternative explanations, including convergent evolution, were not consistent with observed genotypic changes. Despite evidence for a strong loss of genetic diversity in genomic regions coding for the protease and reverse transcriptase, diversity in regions coding for Gag and envelope was considerably higher, and recombination between the emerging viruses expressing the new pattern of resistance mutations and viral quasispecies in the previously dominant population contributed to this preservation of diversity in the envelope gene. These findings emphasize that recombination can participate in the adaptation of HIV to changing selective pressure, both by generating novel combinations of resistance mutations and by maintaining diversity in genomic regions outside those implicated in a selective sweep.
Human immunodeficiency virus type 1 (HIV-1) has a high capacity to adapt to changes in selective pressure, such as changes in immune pressure or in pharmacological pressure. Although error-prone reverse transcription (34), in the context of rapid viral turnover (22, 56), is known to play an important role in generating the viral diversity necessary for the selection of variants with improved fitness, viral recombination may also contribute to this process (5). Two copies of viral RNA are encapsidated in HIV-1 virions, and the two genomes can be genetically distinct if the cell producing the virus has been infected with two or more different viruses. During the ensuing replicative cycle, the alternate use of the two templates by reverse transcriptase (RT) produces a recombinant DNA genome that is different from that of either parental strain. Extensive studies performed in vitro have shown that recombination occurs frequently throughout the genome during HIV replication (2 to 20 events/genome/replicative cycle) and can efficiently shuffle closely linked genetic markers (10, 11, 18, 23, 25, 32, 38, 39, 45, 59). In vivo, recombination between viral variants also occurs at a high rate (8, 49), reflecting the frequent occurrence of cells dually infected by genetically distinct HIV variants (9, 12, 28).
Following changes in antiretroviral treatment, recombination could influence the viral evolution in two distinct ways. First, recombination may contribute to the development of drug resistance. Consistent with this idea, several groups have shown that when cell lines are infected in vitro with two viruses carrying distinct resistance mutations and cultured in the presence of antiretroviral drugs, double mutants produced by recombination that express an improved drug resistance profile can readily be selected (21, 29, 36, 57). However, this process has not been clearly documented in vivo.
Recombination also has the potential to influence viral diversity during the development of drug resistance. Following a change in antiviral therapy, the emergence of viruses expressing mutations required for resistance to the new regimen has been shown to be associated with a decrease in viral diversity (14, 24, 30, 37, 47, 58). Consistent with the occurrence of a genetic sweep, this loss of diversity can affect both the region where novel resistance mutations have emerged and regions not directly subjected to the change in selective pressure, such as the env gene. Although a decrease in viral diversity in env has been described for this setting, such reductions in diversity may be transient (14, 30, 58) and have not been identified for all patients (4, 16, 24, 30, 47). A potential explanation for the preservation of genetic diversity outside the region under selection following a genetic sweep is the occurrence of recombination between the newly emerging virus and the preexisting majority viral population (30, 37).
Despite its potential importance, the direct demonstration that recombination either contributes to the evolution of drug resistance or influences viral diversity in individual patients has been hampered by technical problems inherent in the evaluation of viral sequences amplified by RT-PCR from clinical samples. First, it is difficult to establish whether putative recombination events observed in sequences amplified from clinical samples reflect events occurring in vivo or were introduced during the amplification reaction (27, 35). Second, sequences from widely separated regions must be amplified in separate reactions. Thus, it is impossible to directly compare diversity in such widely separated genomic regions from the same viruses. Recently we described an approach based on the analysis of contemporaneous clonal viral populations that overcomes these limitations (8). The analysis of such clones from several patients demonstrated that extensive recombination is occurring in vivo, confirming studies performed using limiting-dilution PCR (49).
Using this approach, we have evaluated clonal virus populations obtained from the same patient before and after a treatment change that, while unsuccessful in controlling viral replication, led to the emergence of viruses expressing an alternative pattern of resistance mutations. We observed that (i) the pol gene of the viruses that emerged after the treatment change was created through recombination; (ii) the emergence of the recombinant viruses was associated with a loss of diversity in the regions of the pol gene that were under selective pressure, but diversity was preserved in other genomic regions; and (iii) recombination events between the emerging recombinant virus and the prior majority viral population made a contribution to this preservation of diversity in regions not under selective pressure by antiviral treatment.
MATERIALS AND METHODS
Study subject.
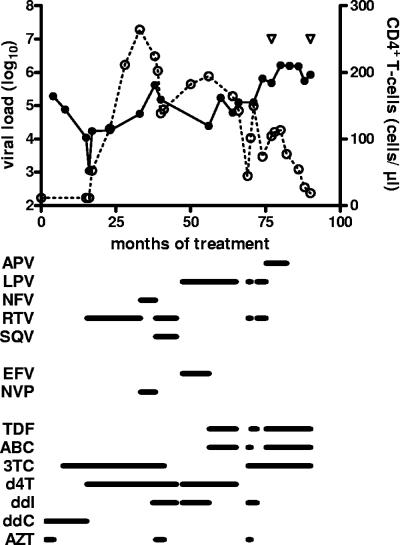
The HIV-1-infected patient evaluated in this study had been followed at the Hôpital Bichat - Claude Bernard; informed consent was obtained for participation in the study. A summary of the patient's treatment and the evolution of viral load and CD4+ T-lymphocytes are shown in Fig. 1. The patient had been treated with antiretroviral agents since May 1996 but had a long history of treatment failure; genotypic resistance testing had been performed on six occasions during the emergence of drug-resistant viruses. At month 82 of antiretroviral therapy (M82), the patient underwent a change in treatment regimen that was unsuccessful in controlling viral replication but led to the emergence of viruses expressing a different pattern of resistance mutations. Clonal viral populations were obtained prior to (M77) and after (M90) this change in therapy.
FIG. 1.
Treatment history and evolution of viral load and CD4+ T-cell counts in the study subject. The log10 viral load (solid symbols) and CD4+ T-cell counts (open symbols) are shown as a function of time after initiating treatment with antiretroviral agents. The agents received by the patient are indicated below the graph. Abbreviations: ABC, abacavir; APV, amprenavir; ddI, didanosine; EFV, efavirenz; 3TC, lamivudine; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; RTV, ritonavir; SQV, saquinavir; d4T, stavudine; TDF, tenofovir; ddC, zalcitabine; AZT, zidovudine. The open triangles indicate the times at which clonal viral populations were obtained (left and right, M77 and M90, respectively).
Clonal viral populations.
Clonal viral populations were obtained as previously described (8). Briefly, MT4 cells expressing CCR5 and CXCR4 receptors were resuspended at 2 × 106 cells/ml in complete medium containing 1% (vol/vol) dimethyl sulfoxide, and 0.25-ml aliquots were distributed in 24-well plates. An equal volume of plasma, diluted in complete medium containing (final concentration) 1% dimethyl sulfoxide and 2 μg/ml DEAE-dextran, was added to each well. The plates were centrifuged (860 × g; 2 h; 22°C) and cultured for 4 h at 37°C to permit viral entry. Cells were recovered and washed once, and 200-μl aliquots containing 2 × 104 cells were distributed into 96-well plates. The cultures were maintained at 37°C in 5% CO2 and were passaged with a 1:10 dilution every 7 days for up to 40 days. Cultures were inspected by light microscopy, and when patent cytopathic changes were observed, the culture supernatant and the cell pellet from infected wells were recovered separately and frozen at −80°C. If viral replication was observed in >20% of the wells, the experiment was repeated after further dilution of the plasma.
Sequencing of viral genomes.
DNA was extracted from infected cell pellets using a QIAmp viral DNA mini kit (QIAGEN, Valencia, CA) and used to amplify proviral DNA corresponding to the following nucleotide regions: (i) HBX2 nucleotides 1147 to 2549, coding for the C-terminal portion of Gag and all of protease; (ii) nucleotides 2670 to 3308, coding for amino acids 41 to 253 of RT; and (iii) nucleotides 6246 to 7568, coding for the C1, V1/V2, C2, V3, C3, and a portion of the V4 regions of envelope. Amplification products were directly sequenced in both directions using an ABI automated sequencing platform (Applied Biosystems, Foster City, CA). Sequences were aligned using CLUSTAL X (version 1.81) (53), and alignments in regions with insertions were verified manually. All chromatograms were visually inspected, and none contained sequences with ambiguous or polymorphic bases.
Standard genotypic resistance testing was performed on bulk plasma viral RNA using the consensus technique of the ANRS Resistance study group (42) or the TruGene HIV-1 genotyping kit (Bayer Healthcare, Eragny, France).
Analysis of nucleotide sequences.
All viral nucleotide positions correspond to the HXB2 reference strain. For the sake of this study, a resistance mutation is defined as mutation that satisfied either or both of the following two criteria: (i) a resistance mutation identified as such by the expert panel of the International AIDS Society-USA (26) and/or (ii) a nonpolymorphic treatment-selected mutation identified in the Stanford HIV drug resistance database (44). The following antiretroviral drugs were used: abacavir, amprenavir, didanosine, efavirenz, lamivudine, lopinavir, nelfinavir, nevirapine, ritonavir, saquinavir, stavudine, tenofovir, zalcitabine, and zidovudine.
Neighbor-joining phylogenetic trees and bootstrap scores (1,000 replicates) were obtained using the Mega package (version 3.1) (31). Maximum-likelihood (M-L) phylogenies were obtained using the HKY85 model; nucleotide substitution and gamma distribution were estimated from the original data using PAUP* (version 4.0b10) (50). Topological incongruence of the M-L topologies was assessed using the incongruence length difference test as implemented in PAUP*.
S-H tests.
To evaluate the relatedness of different portions of the pol sequence of clonal viruses obtained at 90 months to viral sequences obtained earlier in the patient's course, the following approach was used. For each region of the pol sequence considered, an unconstrained M-L phylogeny was generated and compared to tree topologies in which the M90 sequences were constrained to be monophyletic with each of the earlier pol sequences. These comparisons were made using the Shimodaira-Hasegawa (S-H) test (20, 48) as implemented in PAUP*. A significant result in this test (P < 0.05) indicates that the constrained topology is significantly worse and therefore incongruent with the unconstrained topology.
Nucleotide diversity and spectrum analysis.
Nucleotide distance was determined using the Tajima-Nei method (52), as implemented in MEGA; gaps, when present, were handled by pairwise deletion. Statistical comparisons of diversity were performed by one-way analysis of variance; posttest comparisons, performed only if P values were <0.05, were made using Bonferroni's multiple comparisons test. Tajima's D statistic was calculated without an outgroup as described previously (51) using DNA-SP (46). Fay and Wu's H statistic for different genomic regions of the month 90 clones was determined using the consensus sequence for month 77 clones as an outgroup (17). Coalescent simulations to determine the statistical significance of the H statistic were performed using DNA-SP.
RESULTS
Evolution of resistance between 77 and 90 months of antiretroviral treatment.
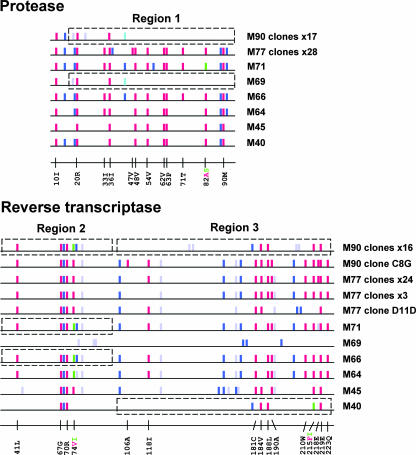
At M77, and 6 weeks after starting a regimen with abacavir, tenofovir, lamivudine, and boosted amprenavir, the patient was experiencing virologic failure. All clonal viruses obtained at M77 (n = 28) expressed the same protease resistance mutations, with the exception of a single clone that expressed the V82I instead of the V82A substitution (Fig. 2). Most clonal viruses also shared the same 14 RT resistance mutations, although one clone without K70R (clone D3E), three clones without VII8I and D218E mutations, and a clone with a distinct pattern of resistance mutations in the C-terminal portion of the RT (clone D11D) were also present.
FIG. 2.
Graphical representation of the protease (top) and RT (bottom) genotypes obtained for the study subject. Each bar represents a codon that was polymorphic in at least one of the sequences obtained from the patient. The bars are color coded as follows: red, resistance mutation as defined in Materials and Methods; green, alternative resistance mutation found at the same position; dark blue, other nonsynonymous polymorphism; light blue, alternative nonsynonymous polymorphism found at the same position; violet, synonymous polymorphism. For the genotypes obtained by sequencing bulk cDNA obtained from plasma by RT-PCR (M40, M45, M64, M66, M69, and M71), the consensus sequence is shown. For sequences representing a single clonal virus (M77 clone D11D and M90 clone C8G), all polymorphisms are shown. For sequences representing groups of clones (the group of 17 M90 clones [M90 clones ×17], M90 clones ×16, and M77 clones ×28, etc.), only polymorphisms expressed by the majority of the sequences in the given group are shown, although most polymorphisms were present in all clones in a given group. Resistance mutations were expressed by all the clones in a given group with the following exceptions: for protease, M77 clones ×28, one clone expressed the V82I resistance mutation, not the V82A mutation, and for RT, M77 clones ×24, one clone did not express the K70R mutation. Boxes surround genotypes obtained at earlier times that are most similar to the sequences of the M90 clones in the regions coding for amino acids 15 to 99 of protease (region 1), 41 to 100 of RT (region 2), and 101 to 236 of RT (region 3). The positions of all resistance mutations encountered in these sequences are also identified.
At M82, amprenavir was discontinued, but the same RT inhibitors were continued. Eight months later (M90), the amino acid sequence of the Pol region of the clonal viruses had changed substantially. Three resistance mutations in the N-terminal portion of the protease were still present (L10I, K20R, M36I), but nine resistance mutations and four other polymorphisms identified at M77 were no longer present at M90. For 16/17 clonal viruses, notable changes in the RT were also observed, including the loss of six resistance mutations and four other polymorphisms present at M77 and the replacement of the L74V mutation by the L74I allele. For the remaining M90 clone (C8G), the sequence coding for the RT was very similar to that expressed by clones obtained at M77 (Fig. 2). The pattern of resistance mutations observed for the M90 clones was identical to that of a bulk resistance genotype (data not shown).
The evaluation of nucleotide sequences obtained at earlier times in the patient's treatment history indicated that viruses expressing the combination of resistance mutations in protease and RT identified at M90 had not previously been identified. We noticed, however, that a pattern of resistance-associated mutations very similar to that observed at M90 could be produced by associating the C-terminal portion of the protease observed at M69 (a time when the patient was undergoing a structured treatment interruption), the N-terminal portion of the RT observed at M66 or M71, and the C-terminal portion of the RT observed at M40 (boxed regions in Fig. 2). The pattern of resistance mutations of this putative recombinant virus would be identical to that observed at M90, with the exception of the T215I mutation in lieu of the T215F mutation. The T215F resistance mutation requires two nucleotide changes and is often preceded by the T215I mutation. In addition, all neutral and silent polymorphisms present in the putative M69+M66/M71+M40 recombinant virus are also present in the M90 sequence.
To further evaluate the possibility that the pol region of M90 viruses was produced through recombination events, the sequences were divided into the three regions indicated in Fig. 2, and phylogenetic studies were performed. As shown in Fig. 3, all M90 clones were monophyletic with the M69 nucleotide sequence in the neighbor-joining tree covering region 1 (C-terminal portion of protease), and all M90 clones except for clone C8G were monophyletic, respectively, with the M66/M71 sequences for region 2 (N-terminal portion of the RT) and the M40 sequence for region 3 (C-terminal portion of RT). As expected, the M90 clone C8G clustered with the M77 clones in regions 2 and 3 but not region 1. Compatible with the occurrence of recombination events between these regions, the phylogeny for each of these three regions was incongruent with those of the other two regions according to the incongruence length difference test (P ≤ 0.02 for each comparison).
FIG. 3.
Neighbor-joining phylogenetic trees for the three genomic regions identified in Fig. 2 and covering the sequences coding for amino acids 15 to 99 of protease (A), 41 to 100 of RT (B), and 101 to 236 of RT (C). The genotypes obtained by sequencing bulk cDNA obtained from plasma by RT-PCR (Geno) and sequences from individual clones obtained at M77 and M90 are shown. The names of the bulk genotypes that grouped most closely in each region with the clones obtained at M90 have been boxed. Selected bootstrap scores are also shown. The scale line at the bottom of each tree represents 0.5% divergence.
In the case where the M90 pol sequence was created through recombination, one would anticipate that each of the donor sequences would be monophyletic with the M90 sequence in the region that it contributed through recombination, but that the donor sequences would not cluster closely with the M90 sequence upstream and downstream of the putative recombination breakpoints. To test these points, unconstrained M-L tree topologies were generated for each of the three regions and were compared using the S-H test to tree topologies in which M90 sequences (excluding clone C8G for regions 2 and 3) were constrained to be monophyletic with each of the sequences from earlier times. The null hypothesis of the S-H test (P > 0.05) indicates no significant difference in the likelihoods of the unconstrained and constrained topologies. The results of this analysis are shown in Table 1. For region 1, only the topology of the tree in which the M90 sequences were constrained to be monophyletic with the M69 sequence was not significantly different from the unconstrained M-L tree topology. In regions 2 and 3, however, the topologies of the trees in which the M90 sequences were constrained to be monophyletic with the M69 sequence were incongruent with the unconstrained topology. Similarly, for region 3, only the topology of the tree in which the M90 sequences were constrained to be monophyletic with the M40 sequence was not significantly different from the unconstrained M-L tree topology, but for regions 1 and 2, the topologies of the trees in which the M90 sequences were constrained to be monophyletic with the M40 sequence were incongruent with the unconstrained topology. For region 2, the topologies of trees in which the M90 sequences were constrained to be monophyletic with the M64, M66, and M71 sequences were not significantly different from the unconstrained M-L tree topology. This finding was not surprising, because the M66 and M71 sequences are identical to each other and identical to the consensus M90 sequence in this region, and the M64 sequence differs by only two polymorphisms (Fig. 2). Thus, viruses from any of these times would have been good candidates to have participated in recombination. Importantly, however, for regions 1 and 3, the topologies of the trees in which the M90 sequences were constrained to be monophyletic with the M64, M66, and M71 sequences were phylogenetically incongruent with the corresponding unconstrained topologies. Thus, the sequences that clustered closely with the M90 sequences within a given region failed to cluster closely with the M90 sequences upstream and/or downstream of the putative recombination breakpoints, an observation that further supports a recombinant origin for the M90 sequences.
TABLE 1.
S-H tests of topological incongruence for regions of the pol genea
| Sequence | Likelihood (ln L) scores (P value) for:
|
||
|---|---|---|---|
| Region 1 | Region 2 | Region 3 | |
| Best tree sequence | −540.69 | −414.62 | −921.32 |
| M90 sequence constrained to group with sequence from: | |||
| M40 | −575.75 (<0.001) | −432.56 (<0.05) | −924.37 (0.22) |
| M45 | −574.56 (<0.001) | −425.96 (0.05) | −933.93 (<0.05) |
| M64 | −579.88 (<0.001) | −418.41 (0.16) | −955.33 (<0.001) |
| M66 | −586.02 (<0.001) | −414.62 (1.0) | −955.33 (<0.001) |
| M69 | −540.69 (1.0) | −426.94 (<0.05) | −939.69 (<0.05) |
| M71 | −581.86 (<0.001) | −414.62 (1.0) | −955.33 (<0.001) |
| M77 | −576.71 (<0.001) | −429.50 (<0.05) | −974.48 (<0.001) |
For each region, the best-tree topology was compared using the S-H test to the topologies in which the M90 sequences were constrained to be monophyletic with each of the genotypes obtained after starting antiretroviral treatment (months 40 to 71) or with the sequences of clonal viruses obtained at M77. Results for comparisons that did not show topological incongruence (P > 0.05) are shown in boldface. Nucleotide sequences coding for amino acids 15 to 99 of protease (region 1), 41 to 100 of RT (region 2), and 101 to 236 of RT (region 3) were evaluated.
Although these results suggest that the pol region of M90 clones arose through recombination between viruses similar to those present at M69 (protease), M40 (C-terminal portion of RT), and M64, M66, or M71 (N-terminal portion of RT), they do not indicate when such a recombinant was formed or the order in which the fragments were assembled. As indicated above, for one M90 clone (clone C8G), the sequence of the protease was similar to those of the other M90 clones, but the RT sequence was similar to that observed in many of the M77 viruses. This should not be taken as evidence that the C-terminal portion of the RT was the last segment recovered, because this clone could have arisen by recombination between a virus carrying the pol region seen for the other M90 clones and a virus from the previously majority population present at M77. Consistent with this possibility, the evaluation of the diversity of the env region presented below supports the conclusion that viruses carrying the pol region characteristic of M90 viruses underwent recombination with the previously majority population present at M77.
Impact of the emergence of viruses expressing a novel pol sequence on viral diversity.
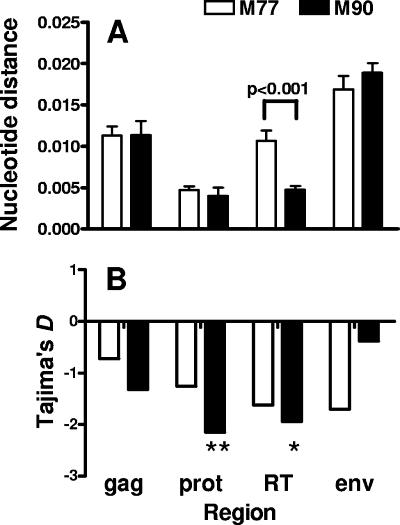
Nucleotide diversity in gag, pol, and env at M77 and M90 is shown in Fig. 4A. The diversity of the viruses expressing a new pattern of resistance mutations that emerged at M90 was low in both the protease and RT regions, and nucleotide diversity in the RT region was significantly lower than that observed at M77. In contrast, diversity in the gag and env regions was significantly higher than that observed for protease and RT (P < 0.01 for all comparisons), and no significant difference in viral diversity was observed for gag and env when sequences obtained at M77 and M90 were compared. If the low nucleotide diversity seen for protease and RT and the reduction in diversity observed for RT between M77 and M90 resulted from the occurrence of a selective sweep, a significant excess of low-frequency allelic variants would be expected, producing negative values for Tajima's D statistic. Indeed, in both the protease and RT, D was significantly negative at M90 and had decreased compared to the value observed at M77 (Fig. 4B). In the gag region, however, D was not significantly negative, and for the env region, the value had increased compared to the value obtained at M77.
FIG. 4.
Comparison of nucleotide diversities and Tajima's D statistic values for different genomic regions of clones obtained at months 77 (open bars) and 90 (solid bars). (A) Nucleotide distance was calculated pairwise for all clones using the method of Tajima and Nei (52), and results are expressed as the mean ± standard error of the mean. (B) Tajima's D statistic was determined as described previously (51). Significant departure from neutrality is indicated by asterisks (*, P < 0.05; **, P < 0.01).
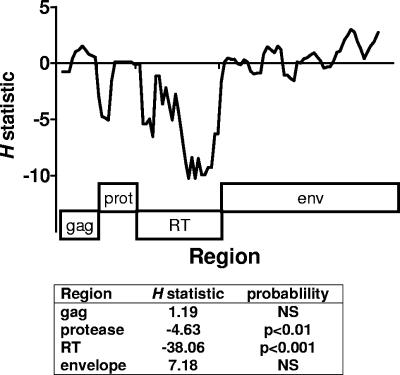
These findings are consistent with a reduction of diversity in protease and RT resulting from a selective sweep, but a preservation of diversity in regions not under selection (gag and env), through recombination. This conclusion was supported by evaluation of Fay and Wu's H statistic, which detects an excess of high-frequency variants from a precursor state, a characteristic of selective sweeps. As presented in Fig. 5, both as a sliding window analysis and as summary statistic for each region, the H statistic was strongly negative in regions coding for the protease and RT, consistent with a selective sweep focused on these regions. In contrast, no evidence for the extension of this sweep into the neighboring gag region or the more distant envelope region was observed, consistent with the preservation of diversity in these regions through recombination.
FIG. 5.
Fay and Wu's H statistic values for different genomic regions of clonal viruses obtained at month 90. The H statistic was determined using the consensus sequence of M77 clones as an outgroup. In the upper panel, a sliding window representation of the H statistic is shown (window length, 100; step size, 25); the corresponding regions of the genome are shown in boxes below. The H statistic was also determined for each of the genomic regions, and the significance of the value compared to neutral expectations was determined by coalescent simulation, as implemented in DNA-SP.
Viral species that are dominant at the time that viruses carrying the new pattern of resistance mutations begin to emerge are likely partners for such recombination events. If the envelope sequences observed after the genetic sweep were derived, at least in part, from the preexisting population through recombination, two results would be expected. First, some of the sites that were polymorphic in the envelope prior to the selective sweep in protease and RT would also be polymorphic after the sweep. Indeed, env polymorphisms present in more than one of the 17 sequences obtained at M90 were observed at 56 positions. In 41/56 cases, the same polymorphism was present in the M77 viral population.
In addition, if polymorphisms were recovered by recombination with viruses in the preexisting majority population, polymorphisms with a higher allele frequency in the preexisting viral population should have a higher probability of being present following the evolutionary sweep. Polymorphisms were identified at 161 positions in envelope sequences obtained at M77. For polymorphisms present in only a single sequence, 26/107 (24%) were also polymorphic at M90, whereas 28/54 (52%) of polymorphisms present in two or more sequences at M77 were also present at M90 (P < 0.001 using Fisher's exact test).
Consistent with extensive recombination, the comparison of the amino acid sequences for individual envelope regions identified numerous examples where two or more alternative haplotypes present in the viral population before the evolutionary sweep had been preserved in viruses obtained after the sweep. For example, of the five alternative V4 region haplotypes present at M77, three were identified at M90 (Table 2).
TABLE 2.
Preservation of alternative haplotypes in the envelope V4 region after an evolutionary sweep in pola
| Haplotype sequence | No. of clones with indicated haplotype at:
|
|
|---|---|---|
| M77 | M90 | |
| T A Q N --- A P | ||
| . . . . . . . | 17 | 12 |
| N . P . . . . | 6 | 2 |
| . . P . . . . | 2 | 0 |
| . V . . . . . | 2 | 0 |
| K . . D DME S L | 1 | 3 |
The consensus sequence for all amino acids that were polymorphic in the indicated region of envelope is shown on the first row. For each haplotype, only the amino acids differing from the consensus sequence are shown, and the numbers of clones expressing that haplotype before and after the evolutionary sweep are indicated.
DISCUSSION
In this study, we have compared the sequences of viral clones obtained from the same patient before and after a change in antiretroviral treatment that, while unsuccessful in suppressing viral replication, resulted in the emergence of viruses expressing a different pattern of resistance mutations in protease and RT. We present evidence that recombination played an important role in shaping the viral evolutionary changes in two distinct ways. First, our analysis indicates that the pol sequence observed for viruses emerging after the treatment change was generated through recombination, producing viruses carrying previously existing resistance mutations in a novel combination. Second, despite evidence for a strong genetic sweep in regions coding for the protease and RT, viral diversity in gag and env was considerably higher, and recombination between the emerging viruses expressing the new resistance mutation profile and viral quasispecies in the previously dominant population contributed to this preservation of diversity. These findings emphasize that recombination can influence the adaptation of HIV to changing selective pressure, both by generating viruses expressing novel combinations of resistance mutations and by preserving diversity in genomic regions outside those implicated in a selective sweep.
Recombination and drug resistance.
Studies performed in vitro have clearly shown that following simultaneous infection of cells with two viral strains expressing distinct drug resistance mutations, recombinant viruses that express both mutations and exhibit improved drug resistance can be selected (21, 29, 36, 57). The extent to which this process contributes to the development of drug resistance in vivo is less clear. Mathematical modeling indicates that whether recombination would accelerate or retard the appearance of such double mutants in vivo is dependent on numerous factors, including viral population size, the rates of mutation and recombination, and epistatic effects (1), none of which are accurately defined. Thus, although recombination events have been demonstrated to occur in vivo (8, 33, 42, 49), the contribution of this process to the evolution of drug resistance has not previously been clearly documented.
For the patient evaluated here, however, a treatment change was associated with the emergence of recombinant viruses carrying gene segments derived from distinct viral populations previously identified in the patient, indicating that recombination can participate in the evolution of drug resistance. The sequence of segments forming the protease and the N-terminal portion of the RT of the viruses identified at M90 were essentially identical to the sequences of these segments in viruses identified at M69 and at M66/M71. The segment most similar to the C-terminal portion of the RT was identified at M40, but viruses present at this time expressed the T215I resistance mutation, not T215F. Thus, the actual donor sequence contributing the C-terminal portion of the RT, although closely related to the M40 sequence, may have already undergone the additional mutational event required to produce T215F. Alternatively, this event could have occurred after the formation of a recombinant virus. It is also noteworthy that M90 viruses carried five synonymous base changes that had not been identified in the consensus sequences of viral populations previously identified in the patient and were not present in the M77 clonal viruses. The fixation of silent mutations is to be expected during a genetic sweep, such as would occur during the emergence of a recombinant virus.
Several factors helped us identify these viruses as true recombinants. First, the evaluation of biological clones, instead of viral sequences obtained by RT-PCR, assured that the sequences obtained were colinear in the same virus. Thus, artifacts due to recombination occurring during PCR were not a concern. Second, nucleotide sequences of the pol region had been obtained at multiple times during the development of resistance in this patient, allowing us to identify potential donor fragments involved in recombination. Third, the magnitude and nature of the changes in the pattern of resistance mutations observed was helpful in excluding the possibility that the M90 viruses arose through convergent evolution. In particular, only 8 months after discontinuation of treatment with protease inhibitors, many but not all of the resistance mutations in the protease were no longer detected at month 90. Studies evaluating patients primarily infected with multidrug-resistant viruses have repeatedly found that resistance mutations are very slowly lost through back-mutation (2, 3, 6, 13, 19, 41), excluding the possibility that the virus identified at M90 could have arisen through convergent evolution from a virus expressing resistance to protease inhibitors. Conversely, the persistence of protease resistance mutations in the N-terminal portion of the protease, as well as the pattern of resistance mutations and other polymorphisms present in the RT, was not consistent with convergent evolution from a virus not previously expressing drug resistance. Taken together, these arguments support the idea that recombination was responsible for creating the novel pattern of resistance mutations observed for the viruses that emerged at M90 following the change in treatment. Interestingly, a number of mutations associated with resistance to RT inhibitors have been shown to directly increase the rate of viral recombination (38). Thus, the development of drug resistance may promote the emergence of such recombinant viruses.
Impact of recombination on viral diversity.
The emergence of viral variants carrying a novel pattern of resistance mutations following changes in antiretroviral therapy, such as the recombinant viruses described here, can lead to a loss of diversity throughout the viral genome, even when selective pressure is exerted only upon the pol region. Indeed, reductions in viral diversity in env associated with the emergence of drug-resistant viruses have been seen in some studies (14, 24, 30, 37, 47, 58), but such changes can be transient and are not detected for all patients (4, 16, 24, 30, 47). The recent study by Kitrinos et al. (30) using heteroduplex analysis found that losses in diversity in env were most marked when treatment changes produced an initial large reduction in viral load, whereas little or no loss of heterogeneity in envelope regions was seen for patients whose viral load did not initially decrease. Based on these findings, these authors suggested that an initial drop in viral load reduces the likelihood of recombination between the emerging virus and the prior majority population, which otherwise preserved diversity in env. Our results provide strong genetic support for this model. In the patient evaluated here, a marked fall in viral load following the treatment change did not occur. The continued replication of quasispecies in the prior majority population permitted recombination events to occur between this population and viruses expressing the new pattern of resistance mutations in pol that subsequently became dominant. Consistent with this interpretation, the envelope sequences present after the selective sweep in pol appeared to be derived through recombination from those expressed by viruses present before the sweep, as indicated by the extensive sharing of the same polymorphisms in the two populations and the preferential retention of polymorphisms present at higher frequency before the genetic sweep in the viral population emerging after the sweep.
Previous studies have found that viral populations expressing different resistance profiles, including those with suboptimal drug resistance, can continue to replicate in “sanctuary sites” (7, 15, 40, 43, 54, 55). It is noteworthy that treatment changes may influence the repertoire of resistant viruses available to participate in recombination events. In our patient, the discontinuation of protease inhibitors was associated with striking changes in the pattern of RT resistance mutations (including the loss of resistance mutations V118I, Y181C, G190A, L210W, and 218E), despite continued treatment with the same RT inhibitors. These mutations, which had accumulated during treatment with other RT inhibitors, including nonnucleoside RT inhibitors, were not required for optimal resistance to the currently used regimen. The replication of viruses not expressing these RT mutations (e.g., the viruses present at M40) may have been suppressed by treatment with amprenavir, because protease resistance was less developed in these viruses. Following the discontinuation of protease inhibitors, the improved replication of these viruses would favor their participation in recombination events, thereby permitting the elimination of RT resistance mutations not required for resistance that exerted a fitness cost. These findings raise the possibility that when treatment changes are anticipated for patients failing a current regimen, a reinforced regimen including drugs effective against viruses expressing patterns of resistance mutations previously detected in the patient might be useful until viral load has fallen to low levels. Even if these drugs are ineffective against the current majority viral population, the enhanced suppression of the replication of viruses expressing alternative genotypes might reduce the risk that preexisting resistance mutations can be recovered through recombination, thereby limiting one avenue for rapid viral evolution.
Acknowledgments
This work was supported in part by the Agence Nationale de Recherches sur le SIDA (ANRS) grant 2005/002.
We gratefully acknowledge the help of Florence Damond and Diane Descamps in performing these studies.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Althaus, C. L., and S. Bonhoeffer. 2005. Stochastic interplay between mutation and recombination during the acquisition of drug resistance mutations in human immunodeficiency virus type 1. J. Virol. 79:13572-13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, J. D., F. M. Hecht, T. Wrin, T. J. Liegler, C. A. Ramstead, M. P. Busch, M. R. Segal, C. J. Petropoulos, and R. M. Grant. 2004. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS 18:1683-1689. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, B., J. P. Routy, Y. Quan, D. Moisi, M. Oliveira, D. Turner, and M. A. Wainberg. 2004. Persistence of multidrug-resistant HIV-1 in primary infection leading to superinfection. AIDS 18:1653-1660. [DOI] [PubMed] [Google Scholar]
- 4.Brown, A. J., and A. Cleland. 1996. Independent evolution of the env and pol genes of HIV-1 during zidovudine therapy. AIDS 10:1067-1073. [PubMed] [Google Scholar]
- 5.Burke, D. S. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg. Infect. Dis. 3:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cane, P. A. 2005. Stability of transmitted drug-resistant HIV-1 species. Curr. Opin. Infect. Dis. 18:537-542. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier, C., D. E. Dwyer, F. Mammano, D. Lecossier, F. Clavel, and A. J. Hance. 2004. Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors. J. Virol. 78:4234-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier, C., T. Nora, O. Tenaillon, F. Clavel, and A. J. Hance. 2006. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 80:2472-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., Q. Dang, D. Unutmaz, V. K. Pathak, F. Maldarelli, D. Powell, and W. S. Hu. 2005. Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor. J. Virol. 79:4140-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., T. D. Rhodes, and W. S. Hu. 2005. Comparison of the genetic recombination rates of human immunodeficiency virus type 1 in macrophages and T cells. J. Virol. 79:9337-9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavel, F., M. D. Hoggan, R. L. Willey, K. Strebel, M. A. Martin, and R. Repaske. 1989. Genetic recombination of human immunodeficiency virus. J. Virol. 63:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang, Q., J. Chen, D. Unutmaz, J. M. Coffin, V. K. Pathak, D. Powell, V. N. KewalRamani, F. Maldarelli, and W. S. Hu. 2004. Nonrandom HIV-1 infection and double infection via direct and cell-mediated pathways. Proc. Natl. Acad. Sci. USA 101:632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaugerre, C., L. Morand-Joubert, M. L. Chaix, O. Picard, A. G. Marcelin, V. Schneider, A. Krivine, A. Compagnucci, C. Katlama, P. M. Girard, and V. Calvez. 2004. Persistence of multidrug-resistant HIV-1 without antiretroviral treatment 2 years after sexual transmission. Antivir. Ther. 9:415-421. [PubMed] [Google Scholar]
- 14.Delwart, E. L., H. Pan, A. Neumann, and M. Markowitz. 1998. Rapid, transient changes at the env locus of plasma human immunodeficiency virus type 1 populations during the emergence of protease inhibitor resistance. J. Virol. 72:2416-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Pasquale, M. P., A. J. Leigh Brown, S. C. Uvin, J. Allega-Ingersoll, A. M. Caliendo, L. Sutton, S. Donahue, and R. T. D'Aquila. 2003. Differences in HIV-1 pol sequences from female genital tract and blood during antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 34:37-44. [DOI] [PubMed] [Google Scholar]
- 16.Dykes, C., P. Mootsikapun, A. Dexter, L. Berrios, M. Chiulli, R. C. Reichman, and L. M. Demeter. 2000. Analysis of env sequence evolution in human immunodeficiency virus-infected patients receiving therapy with nonnucleoside reverse-transcriptase inhibitors. J. Infect. Dis. 182:316-320. [DOI] [PubMed] [Google Scholar]
- 17.Fay, J. C., and C. I. Wu. 2000. Hitchhiking under positive Darwinian selection. Genetics 155:1405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galetto, R., V. Giacomoni, M. Veron, and M. Negroni. 2006. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J. Biol. Chem. 281:2711-2720. [DOI] [PubMed] [Google Scholar]
- 19.Gandhi, R. T., A. Wurcel, E. S. Rosenberg, M. N. Johnston, N. Hellmann, M. Bates, M. S. Hirsch, and B. D. Walker. 2003. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin. Infect. Dis. 37:1693-1698. [DOI] [PubMed] [Google Scholar]
- 20.Goldman, N., J. P. Anderson, and A. G. Rodrigo. 2000. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 49:652-670. [DOI] [PubMed] [Google Scholar]
- 21.Gu, Z., Q. Gao, E. A. Faust, and M. A. Wainberg. 1995. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J. Gen. Virol. 76:2601-2605. [DOI] [PubMed] [Google Scholar]
- 22.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 23.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibanez, A., B. Clotet, and M. A. Martinez. 2000. Human immunodeficiency virus type 1 population bottleneck during indinavir therapy causes a genetic drift in the env quasispecies. J. Gen. Virol. 81:85-95. [DOI] [PubMed] [Google Scholar]
- 25.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, D. Pillay, J. Schapiro, A. Telenti, and D. Richman. 2005. Update of the drug resistance mutations in HIV-1: 2005. Top. HIV Med. 13:51-57. [PubMed] [Google Scholar]
- 27.Judo, M. S., A. B. Wedel, and C. Wilson. 1998. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 26:1819-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 29.Kellam, P., and B. A. Larder. 1995. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J. Virol. 69:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitrinos, K. M., J. A. Nelson, W. Resch, and R. Swanstrom. 2005. Effect of a protease inhibitor-induced genetic bottleneck on human immunodeficiency virus type 1 env gene populations. J. Virol. 79:10627-10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 32.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. USA 101:4204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyerhans, A., J. P. Vartanian, and S. Wain-Hobson. 1990. DNA recombination during PCR. Nucleic Acids Res. 18:1687-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moutouh, L., J. Corbeil, and D. D. Richman. 1996. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. USA 93:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nijhuis, M., C. A. Boucher, P. Schipper, T. Leitner, R. Schuurman, and J. Albert. 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 95:14441-14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolenko, G. N., E. S. Svarovskaia, K. A. Delviks, and V. K. Pathak. 2004. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78:8761-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onafuwa, A., W. An, N. D. Robson, and A. Telesnitsky. 2003. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 77:4577-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, Jr., R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pao, D., U. Andrady, J. Clarke, G. Dean, S. Drake, M. Fisher, T. Green, S. Kumar, M. Murphy, A. Tang, S. Taylor, D. White, G. Underhill, D. Pillay, and P. Cane. 2004. Long-term persistence of primary genotypic resistance after HIV-1 seroconversion. J. Acquir. Immune Defic. Syndr. 37:1570-1573. [DOI] [PubMed] [Google Scholar]
- 42.Pasquier, C., N. Millot, R. Njouom, K. Sandres, M. Cazabat, J. Puel, and J. Izopet. 2001. HIV-1 subtyping using phylogenetic analysis of pol gene sequences. J. Virol. Methods 94:45-54. [DOI] [PubMed] [Google Scholar]
- 43.Philpott, S., H. Burger, C. Tsoukas, B. Foley, K. Anastos, C. Kitchen, and B. Weiser. 2005. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J. Virol. 79:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee, S. Y., J. Taylor, G. Wadhera, A. Ben-Hur, D. L. Brutlag, and R. W. Shafer. 2006. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. USA 103:17355-17360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhodes, T. D., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Palomino, S., I. Olivares, E. Yuste, D. D. Richman, and C. Lopez-Galindez. 1996. Random important alterations in HIV-1 viral quasispecies after antiviral treatment. Antivir. Ther. 1:225-236. [PubMed] [Google Scholar]
- 48.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 49.Shriner, D., A. G. Rodrigo, D. C. Nickle, and J. I. Mullins. 2004. Pervasive genomic recombination of HIV-1 in vivo. Genetics 167:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4b10. Sinauer Associates, Sunderland, MA.
- 51.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tajima, F., and M. Nei. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1:269-285. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirado, G., G. Jove, R. Kumar, R. J. Noel, E. Reyes, G. Sepulveda, Y. Yamamura, and A. Kumar. 2004. Differential virus evolution in blood and genital tract of HIV-infected females: evidence for the involvement of drug and non-drug resistance-associated mutations. Virology 324:577-586. [DOI] [PubMed] [Google Scholar]
- 55.Venturi, G., M. Catucci, L. Romano, P. Corsi, F. Leoncini, P. E. Valensin, and M. Zazzi. 2000. Antiretroviral resistance mutations in human immunodeficiency virus type 1 reverse transcriptase and protease from paired cerebrospinal fluid and plasma samples. J. Infect. Dis. 181:740-745. [DOI] [PubMed] [Google Scholar]
- 56.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 57.Yusa, K., M. F. Kavlick, P. Kosalaraksa, and H. Mitsuya. 1997. HIV-1 acquires resistance to two classes of antiviral drugs through homologous recombination. Antivir. Res. 36:179-189. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y. M., S. C. Dawson, D. Landsman, H. C. Lane, and N. P. Salzman. 1994. Persistence of four related human immunodeficiency virus subtypes during the course of zidovudine therapy: relationship between virion RNA and proviral DNA. J. Virol. 68:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuang, J., A. E. Jetzt, G. Sun, H. Yu, G. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]