Abstract
Phosphatase and tensin homolog (PTEN), deleted on chromosome 10, is a potent tumor suppressor. PTEN expression is reduced in advanced bladder cancer and reduction correlates with disease stage. To gain insights into the function of PTEN in human bladder cancer by identifying its binding partners, we developed a novel IPTG inducible PTEN expression system and evaluated this system in the PTEN null UMUC-3 human bladder cancer xenograft model. In this model, induction of PTEN in vivo resulted in reduced tumor growth. We used mass spectrometry to identify PTEN interaction partners in these cells, which identified known interaction partners Major Vault Protein (MVP) and Paxillin as well as a novel interaction partner, TRK Fused Gene (TFG). In conclusion, using a biologically relevant model system to dissect PTEN tumor suppressor function in human bladder cancer, we identified three molecules important for many cellular functions in complex with PTEN.
Keywords: PTEN, Major Vault Protein, AKT, EGFR
INTRODUCTION
The potent tumor suppressor Phosphatase and Tensin Homolog Deleted on Chromosome 10 (PTEN) acts as a moderator of the Phosphoinositol 3 Kinase / AKT signaling cascade (PI3K/AKT pathway) [1]. The relevance for PTEN in human bladder cancer has recently been reported. When expression levels in the cytoplasm as well as in the nucleus were assessed, 53% of tumors showed a lower PTEN protein expression in one or both of the assessed cellular compartments. This loss of expression was as high as 94% in patients with advanced tumor stages. The same study also showed the importance of PTEN in tumor formation as urothelial specific PTEN suppression by a Cre-loxP system increased spontaneous as well as chemically induced transitional cell carcinomas. In summary this study emphasizes the multifaceted role PTEN plays in tumor formation and progression [2].
To date there have been few studies reporting on PTEN expression systems in bladder cancers where the mechanism of action of PTEN can be studied at a physiologically relevant expression level. To address this gap in the literature and further expand our knowledge of PTEN function, we sought to develop a novel inducible PTEN system in human bladder cancer. Here a system was created where PTEN expression is controlled by IPTG, in the human bladder cancer cell line UMUC-3, enabling reconstitution studies of PTEN at low physiological protein expression levels. This inducible system would prevent supra physiological expression of the transgene, causing potentially erroneous experimental endpoints. We show this model has biological significance by its effect on tumor xenograft establishment and growth in bladder cancer. Further we demonstrate utility of this model in a pilot study by identifying both known and novel PTEN interaction partners by a mass spectroscopy (MS) based approach.
MATERIALS AND METHODS
Recombinant DNA techniques
PTEN Inducible system
Humanized LacI and the inducible SV40 promoter have been described elsewhere [3]. The LacI cDNA was cloned into pIRESpuro2 (Clonetech, Mountain View, CA, USA) at the Eco RI site. The CMV promoter in pcDNA3.1zeo (Invitrogen, Carlsbad, CA, USA) was cut out with Nhe I and Nru I and substituted with the LacI suppressed SV40 promoter. PTEN with a C-terminal hemagglutinin (HA) tag [4] was cloned at the Eco RI site in this vector.
Cloning Putative PTEN binding partners
All tested proteins were cloned by RT-PCR with primers provided in Supplementary Data Section and subsequently inserted in pENTRdTOPO. The Gateway rfb-cloning cassette was put into pIRESpuro3 vector at the Bst BI site, enabling it to accept constructs from pENTRdTOPO by recombination according to manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Furthermore, a 3xFLAG tag was inserted between Eco RI and Bam HI, such that a C-terminal 3xFLAG fusion protein is created after recombination. Clones were sequenced with the M13 forward (-20) and M13 reverse primers.
Cell culture and transfection, Western Blots, In vitro and in vivo bladder cancer cell growth and Statistical Analysis procedures
These approaches and procedures are standard and have been described in detail in Supplementary Data Section
Immunoprecipitations and Mass Spectrometry (MS) analysis
18μg of DNA and 56μl FuGene was used to transfect a 70% confluent 150mm plate and incubated for 48 hours with 1mM IPTG before harvested in 500μl lysis buffer (see above) including protein and phosphatase inhibitors (Sigma, St. Louis, MO, USA)). The lysates were incubated for 2 hours with covalently conjugated anti-HA agarose beads (Sigma, St. Louis, MO, USA) and washed in lysis buffer four times. For MVP control precipitations covalently linked ProteinG sepharose or IgG-linked agarose nonspecific control, was utilized. LRP56 mouse monoclonal antibody was used for precipitating endogenous MVP (Abcam, Cambridge, MA, USA). For MS analysis, co-precipitated proteins were eluted with 500mM LiCl for 30 minutes at 4°C. MS/MS analysis was performed using a Finnigan LCQ Deca ion trap mass spectrometer (ThermoElectron, San Jose, CA) as previously described [5], with the following changes. The samples were reduced with DTT at 52°C for one hour and then alkylated with iodoacetamide for 1 hour at room temperature in the dark. A subsequent trypsin digest was performed in ammonium bicarbonate buffer (100 mM pH 8-8.5) prior to analysis.
RESULTS
IPTG induces PTEN protein expression at physiological levels and alters AKT phosphorylation
To facilitate biologically relevant PTEN binding partner discovery, an inducible expression system was constructed (Figure 1A) in the PTEN null human bladder cancer cell line UMUC-3. PTEN-HA fusion protein was expressed under the control of an IPTG inducible SV40 promoter and single cell clones were created through limiting dilutions. These were screened for PTEN-HA induction by western blot analysis (Figure 1B). Maximum induction was reached at 1mM IPTG and further increased concentration did not lead to higher PTEN-HA expression (Figure 1C).
Figure 1.
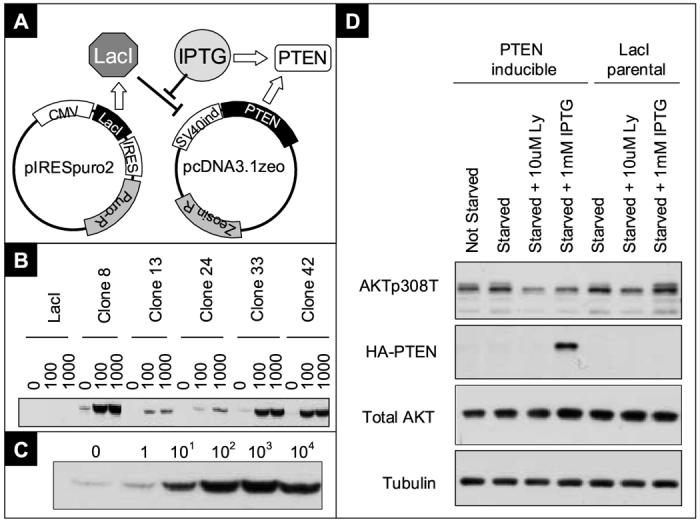
Development of the PTEN inducible bladder cancer cell system
A) A novel PTEN inducible vector system was created by cloning the humanized LacI suppressor gene in the mammalian bicistronic expression vector pIRESpuro2. A SV40 promoter with a short lac operator sequence was inserted in pcDNA3.1zeo to control the expression of PTEN-HA. Expression of LacI suppresses expression of PTEN, which can be induced by incubation with 1mM IPTG. B) Western blot showing PTEN-HA expression of a representative subset of clones utilizing HA.11 mouse monoclonal antibody at the indicated IPTG concentrations (mM). C) Western blotting of clone 42 showing the range of expression after overnight incubation with indicated concentrations of IPTG (μM). D) Western blot of clone 42 showing downstream PTEN signaling effect. PTEN-HA inducible cells, or Lac I parental, were induced with IPTG for 48 hours.
The downstream effects of PTEN induction was evaluated by incubating the cells for 24 hours in complete media with 1mM IPTG, and transferred to serum free media for 24 hours before harvest. The phosphorylation levels of AKT308T was compared between parental and PTEN induced cells. AKT phosphorylation was reduced after PTEN induction in cells expressing PTEN-HA, but not in LacI parental cells, indicating that IPTG in itself has no effect on the PI3K / AKT pathway and that PTEN-HA is functional in UMUC-3. Further, serum withdrawal has no effect on AKT phosphorylation levels if PTEN is not expressed (Figure 1D). Clones with both low baseline PTEN-HA expression and high PTEN expression with IPTG induction, were selected for further investigation.
IPTG induced PTEN expression does not suppress in vitro cell growth but is sufficient for xenograft tumor suppression
Clone 42, met the criteria outlined above and was named UM42wt. We further evaluated this clone for in vitro and in vivo consequences of PTEN reconstitution. Clonal cell lines shown in Figure 1B were analyzed for proliferation rate after PTEN induction in 10 % and 2 % serum. No growth retardation was observed after IPTG induction in any of the analyzed cell lines (data not shown). Data for UM42wt and LacI parental is shown in Figure 2A. To evaluate whether PTEN re-expression would be associated with a biologically relevant vivo phenotype, UM42wt cells were injected subcutaneously in nude mice after overnight induction by 1mM IPTG (Figure 2B). As seen in Figure 2C, all tumor-bearing mice without PTEN induction had rapidly increasing tumor burdens. One mouse in IPTG fed animals also showed rapid tumor growth, whereas two mice showed a slower increase. In addition, two IPTG fed mice showed tumors initially, which later became not palpable. These two mice were not given more IPTG after day 60, and kept for an additional 180 days, without any signs of tumor reoccurrence (data not shown). The difference in tumor growth in vivo between the first and second group, receiving water alone or water with 10mM IPTG, respectively, is statistically significant (p-value=0.01). From these data, we conclude that the described LacI/IPTG controlled PTEN inducible system in UM42wt is sufficient for suppression of AKT phosphorylation in vitro and tumor suppression in vivo and thus constitutes an excellent system for the discovery of functionally important PTEN binding partners.
Figure 2.
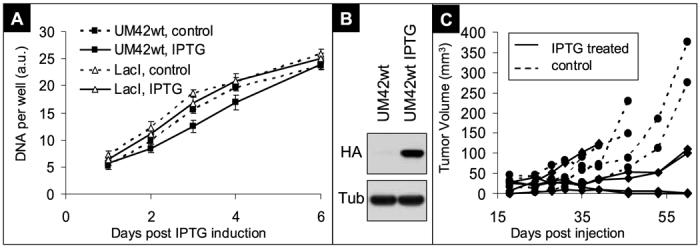
Biochemical and in vivo functional validation of the PTEN inducible bladder cancer cell system
A) In vitro growth curves. DNA was measured as surrogate measure of cell numbers, each data point represents the average of two wells measured in duplicates, and error bars are standard deviations of these four measurements. Similar data was observed in two separate experiments. A.U.: arbitrary fluorescent units from CyQuant analysis. B) Western blot showing the induction of PTEN-HA at the time of injection. Tubulin (Tub) shown to indicate equal loading. C) Tumor burden per mouse expressed as tumor volume. Ten mice divided into two groups were injected with non-treated UM42wt cells (control, dashed lines) or IPTG treated UM42wt cells that expressed high levels of PTEN-HA at the time of injection (data not shown). The mice receiving the IPTG treated cells (IPTG treated, solid lines) were given 10mM IPTG in their drinking water throughout the duration of the experiment.
Mass Spectroscopy reveals a large number of potential binding partners
To identify new interaction partners to the tumor suppressor PTEN, we induced PTEN-HA expression in UM42wt and immunoprecipitated PTEN-HA. Although a small amount of PTEN was eluted by this method, coprecipitated proteins were readily eluted as shown in Figure 3. Cells not expressing PTEN (LacI parental) were used as negative controls for the immunoprecipitation, and eluates from these HAdirected precipitates were also analyzed by mass spectroscopy. Four separate PTEN-HA precipitations were analyzed, as well as two negative pull downs from LacI parental cells. A list of over 400 proteins was generated and prioritized for further investigation as described in Supplementary Data Section.
Figure 3.
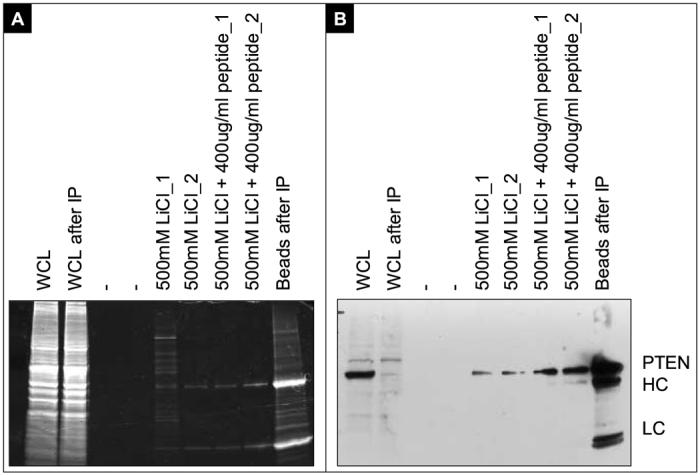
Total protein and HA-western blotting on HA immunoprecipitations and elutions for mass spectrometric analysis
A) SyproRuby stained gel. Precipitations were performed with HA-antibodies covalently linked to Sepharose beads. The whole cell lysate (WCL) is compared before and after IP. The beads were washed four times after incubation in the WCL, and eluted as described. After elutions the beads were boiled in SDS loading buffer and loaded for comparison. B) Western blot with HA-antibodies. The heavy and light chains from the precipitating antibodies are indicated with HC and LC respectively.
The most predominant protein identified of these 21 was the Major Vault Protein (MVP). There were 34 peptides identified in three of the four precipitations, where 12 were separate peptides, covering 20% of the protein (Figure 4A). It is evident from these data that the use of negative control experiments and the generation of independent datasets are of high value to prioritize the large number of proteins that are identified by the mass spectroscopy technique, and that manual verification of this data is also of utmost importance.
Figure 4.
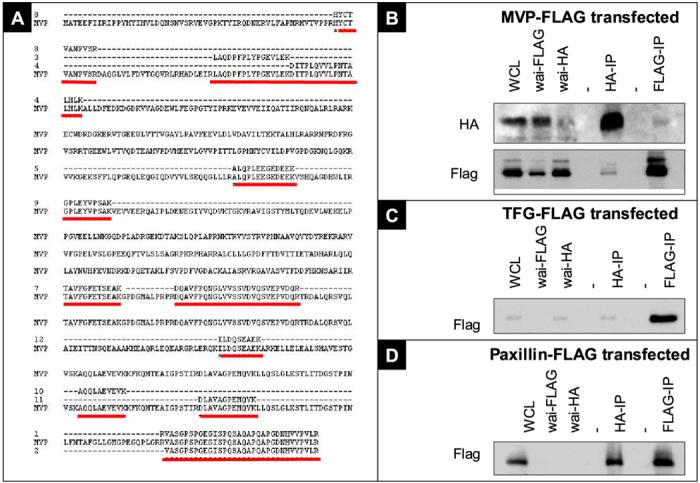
Mass spectrometric identification of Major Vault Protein (MVP), Paxillin (PXN) and TRK-fused gene (TFG) as PTEN binding partners
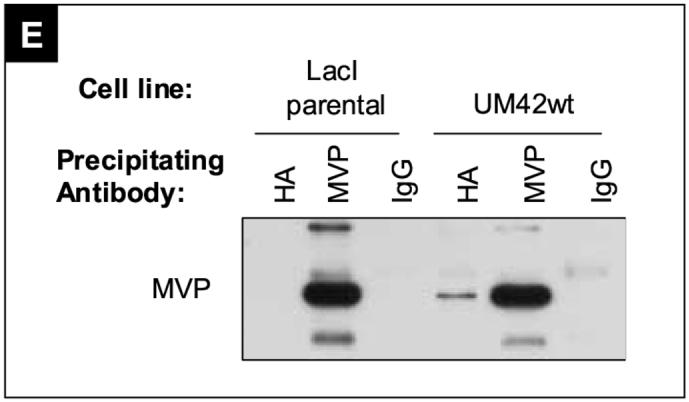
A) The amino acid sequence of MVP is aligned with the different peptides (red lines) identified in the MS-based screen for PTEN interacting proteins. 12 distinct peptides, with 100% homology to MVP primary amino acid sequence were found, covering 20.2% of the protein. B) Western blot showing HA and FLAG immunoprecipitation results. UM42wt cells were transfected with MVP-FLAG fusion protein, induced with 1mM IPTG to express PTEN for 48 hours, and subjected to immunoprecipitations with antibodies against the HA and FLAG tags. A partial clearance of HA and FLAG tagged proteins were observed after comparing the whole cell lysates (WCL) with the supernatant after immunoprecipitation (WAI). PTEN-HA was detected in the MVP-FLAG precipitation, and MVP-FLAG was detected in the PTEN-HA precipitations. (-) indicates empty lanes. C and D) Precipitations as described in B) transfected with indicated constructs. E) Cells were induced with 1mM IPTG for 48 hours prior to harvest, and precipitated with anti HA, anti MVP or nonspecific mouse IgG antibodies. Precipitates were western blotted against the MVP protein.
Three interaction partners are verified by immunoprecipitations
The 21 potential interaction partners shown in Table 1 (Supplementary Data Section) were selected for further analysis and fourteen were successfully cloned and FLAG-tagged as described in the Materials and Methods section (Table 2, Supplementary Data Section). Briefly, primers were designed to the 21 target cDNAs, and used in RT-PCR reactions utilizing cDNA from UMUC-3 cells. 17 of these PCR reactions produced a product at the right size. Primers that had not produced a PCR product after the third attempt were excluded from subsequent analysis. These PCR products were cloned into pENTR vector and sequenced. Fourteen clones were verified in this manner as the correctly cloned cDNA (Table 2, Supplementary Data Section). When three independent clones from a RT-PCR reaction had failed to produce the correct insert, they were excluded from the study. Immunoprecipitations with anti-HA antibodies were made from UM42wt cells transiently transfected with expression clones of the potential interaction partners as well as cells not expressing PTEN-HA as a negative control. FLAG expression in these precipitations was detected by western blotting.
Out of the fourteen cloned interaction partners two did not express any protein detectable under western conditions (HNRPH3 and C1QBP) utilizing the anti FLAG antibody. Another five candidates (SFRS1; CFL1; G3BP; SYNCRIP; NPM and TUBB2C) exhibited significant pull downs in cells lacking PTEN expression, indicating that they were false positive candidates. Three candidates (EWS, TNRC6B and ACTG2) were not detected in the HA-PTEN pulldowns and the results from the Mass Spectometry detections could hence not be confirmed. Three proteins, the major vault protein (MVP) (Figure 4B) TRK-fused gene (TFG) (Figure 4C) and Paxillin (PXN) (Figure 4D) were not detected in the negative control blot and were detected in the pulldowns where PTEN were present and were hence confirmed as real direct or indirect interaction partners to PTEN.
To further verify these interactions reciprocal precipitations with FLAG were also performed for these 3 proteins and HA detected. MVP-FLAG was the only interaction partner that pulled down PTEN-HA in detectable levels (Figure 4E). We also investigated whether endogenous levels of MVP expression were enough to detect in PTEN pulldowns. To verify that MVP was detected in the PTEN-HA pull-down due to a specific (although not necessarily direct) interaction with PTEN we also included a control antibody precipitation. In this experiment, the endogenous MVP was detected only in cells expressing PTEN-HA, and not in cells that were negative for PTEN, showing that PTEN-HA expression is necessary for the MVP pull down to occur, but the FLAG tag is not, indicating that the interaction is specific (Figure 4E) We conclude from these experiments that PTEN and MVP are part of the same protein complex in UM42wt cells and that our screen for interaction partners indeed identified verifiable candidates.
DISCUSSION
The study of tumor suppressors is complicated by the fact that their exogenous introduction in a cancer cell population in vitro is susceptible for the direct selection against transfected cells by the very nature of the gene product. One way of circumventing this problem is to utilize an inducible system regulating the expression of the transgene. Here we modified the inducible SV40 / humanized LacI system previously developed for transgenic mouse models [3]. The system has advantages for this application such as low background expression, high inducibility above background and an inexpensive inducing agent that is amenable to both in vitro and in vivo use. In addition to the data presented, comparison of PTEN expression in IPTG treated UM42wt cells to that in five normal human urothelium samples by total protein concentration normalized western blotting, indicated that PTEN expression in UM42wt was within the same range as in the human samples (Herlevsen and Theodorescu, unpublished data). By inducing PTEN in UMUC-3 cells we show a reduced basal level of AKT phosphorylation at Threonine308. This reduction in AKT phosphorylation is presumed to translate to a significant reduction in AKT activity that is reflected by decreased tumorigenicity of the UM42wt clone.
Studies have previously revealed several PTEN protein interaction partners. For instance, the MAGI2 and MAGI3 multi PDZ-containing membrane localized proteins, with suggested scaffold properties [6; 7]. PTEN contains a PDZ-binding domain at its Carboxy-terminal end, and the regulation of PTEN is thought to be dependent on phosphorylation near these sites [8; 9]. Therefore the study of interaction partners has the potential to shed light on the regulation of PTEN activity. Through immunoprecipitations and subsequent MS analysis of the eluates from PTEN precipitations a panel of putative PTEN interaction partners was identified. One of the proteins identified in our screen was MVP, the major constituent of the vault complex which is the largest known ribonuclear complex. MVP constitutes about 70% of the molecular mass of the vault structure [10] and the structural integrity of vaults is dependent on MVP expression. Clinical observations suggest the importance of MVP in the intrinsic drug resistance phenotype [11]. Most importantly, MVP expression is in many cases a significant and independent predictor of drug response [11]. While no direct function for MVP has been described, reports suggest nuclear translocation of target proteins such as PTEN [12], nuclear exclusion of chemotherapeutic drugs [13] and as a scaffold protein downstream of the EGFR [14]. The PTEN / MVP interaction has been previously observed in a yeast two hybrid system, and co-immunoprecipitations have been performed in human cancer cells [12].
Paxillin is a phosphoprotein with multiple protein interaction domains and a well documented role in integrin based cellular adhesion and migration [15]. Paxillin has previously been detected in PTEN pulldowns [16]. However, it is not clear whether this interaction is due to a simultaneous interaction of PTEN and Paxillin with the Focal Adhesion Kinase (FAK), or whether the molecules interact directly with each other. Interestingly, FAK was not detected in our studies (Table 1, Supplementary Data Section). Phosphoinositol lipids have a central and well documented role in cell migration, and PTEN as a regulator of these intermediate signaling molecules has a central role in this process [17]. However the proposed role of PTEN as a protein phosphatase to FAK also has profound implications and relevance to cell migration and adhesion [18]. Taken together, the possibility that Paxillin and PTEN are part of the same molecular complex seems reasonable. Further studies would be needed to characterize the nature of such complexes.
No less than four proteins in the final gene set are proteins that are known to recombine in human tumors and form oncogenic chimeras. The EWS, FUS NPM and TFG genes share the common characteristic of having very low complexity regions. Such proteins could therefore be prone to give false positive results in precipitation assays such as those presented here. It is therefore interesting and intriguing that one of these proteins, TRK-fused gene (TFG), did not show any co-precipitation with the HA antibody in cells that did not express PTEN-HA, and was one of the three molecules that were confirmed as PTEN protein interaction partners in our study. To our knowledge this interaction is novel. As a consequence of its role in carcinogenesis as a chimera with predominantly TRK or ALK kinases [19] the gene has been predominantly studied as a chimera with these proteins. Initially TFG has been shown to interact with SH3 domains of Src, PLCgamma, and the p85 phosphatidylinositol 3-kinase subunit in Xenopus laevis [20]. TFG has recently been reported to interact with several proteins in human cell lines, such as SHP-1, TANK and NEMO and is suggested to function in the NFkappaB pathway [21]. It will be interesting to explore the presence and potential role for PTEN in such complexes.
Supplementary Material
ACKNOWLEDGEMENTS
See Supplementary Data Section.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- [2].Tsuruta H, Kishimoto H, Sasaki T, Horie Y, Natsui M, Shibata Y, Hamada K, Yajima N, Kawahara K, Sasaki M, Tsuchiya N, Enomoto K, Mak TW, Nakano T, Habuchi T, Suzuki A. Hyperplasia and Carcinomas in Pten-Deficient Mice and Reduced PTEN Protein in Human Bladder Cancer Patients. Cancer Res. 2006;66:8389–96. doi: 10.1158/0008-5472.CAN-05-4627. [DOI] [PubMed] [Google Scholar]
- [3].Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes Dev. 2001;15:1506–17. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Furnari FB, Huang HJ, Cavenee WK. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 1998;58:5002–8. [PubMed] [Google Scholar]
- [5].Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2000;72:4266–74. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- [6].Wu Y, Dowbenko D, Spencer S, Laura R, Lee J, Gu Q, Lasky LA. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J Biol Chem. 2000;275:21477–85. doi: 10.1074/jbc.M909741199. [DOI] [PubMed] [Google Scholar]
- [7].Wu X, Hepner K, Castelino-Prabhu S, Do D, Kaye MB, Yuan XJ, Wood J, Ross C, Sawyers CL, Whang YE. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci U S A. 2000;97:4233–8. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–8. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- [9].Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci U S A. 1999;96:10182–7. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–28. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mossink MH, van Zon A, Scheper RJ, Sonneveld P, Wiemer EA. Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene. 2003;22:7458–67. doi: 10.1038/sj.onc.1206947. [DOI] [PubMed] [Google Scholar]
- [12].Chung JH, Ginn-Pease ME, Eng C. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res. 2005;65:4108–16. doi: 10.1158/0008-5472.CAN-05-0124. [DOI] [PubMed] [Google Scholar]
- [13].Kitazono M, Sumizawa T, Takebayashi Y, Chen ZS, Furukawa T, Nagayama S, Tani A, Takao S, Aikou T, Akiyama S. Multidrug resistance and the lung resistance-related protein in human colon carcinoma SW-620 cells. J Natl Cancer Inst. 1999;91:1647–53. doi: 10.1093/jnci/91.19.1647. [DOI] [PubMed] [Google Scholar]
- [14].Kolli S, Zito CI, Mossink MH, Wiemer EA, Bennett AM. The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J Biol Chem. 2004;279:29374–85. doi: 10.1074/jbc.M313955200. [DOI] [PubMed] [Google Scholar]
- [15].Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol. 2000;2:E231–6. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
- [16].Haier J, Nicolson GL. PTEN regulates tumor cell adhesion of colon carcinoma cells under dynamic conditions of fluid flow. Oncogene. 2002;21:1450–60. doi: 10.1038/sj.onc.1205213. [DOI] [PubMed] [Google Scholar]
- [17].Sotsios Y, Ward SG. Phosphoinositide 3-kinase: a key biochemical signal for cell migration in response to chemokines. Immunol Rev. 2000;177:217–35. doi: 10.1034/j.1600-065x.2000.17712.x. [DOI] [PubMed] [Google Scholar]
- [18].Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–7. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- [19].Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, Lamant L, Falini B, Ott G, Mason DY, Delsol G, Campo E. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94:3265–8. [PubMed] [Google Scholar]
- [20].Ohan N, Sabourin D, Booth RA, Liu XJ. Xenopus laevis TRK-fused gene (TFG) is an SH3 domain binding protein highly expressed in the cement gland. Mol Reprod Dev. 2000;56:336–44. doi: 10.1002/1098-2795(200007)56:3<336::AID-MRD2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [21].Roccato E, Miranda C, Raho G, Pagliardini S, Pierotti MA, Greco A. Analysis of SHP-1-mediated down-regulation of the TRK-T3 oncoprotein identifies Trk-fused gene (TFG) as a novel SHP-1-interacting protein. J Biol Chem. 2005;280:3382–9. doi: 10.1074/jbc.M407522200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


