Abstract
Most new genes arise by duplication of existing gene structures, after which relaxed selection on the new copy frequently leads to mutational inactivation of the duplicate; only rarely will a new gene with modified function emerge. Here we describe a unique mechanism of gene creation, whereby new combinations of functional domains are assembled at the RNA level from distinct genes, and the resulting chimera is then reverse transcribed and integrated into the genome by the L1 retrotransposon. We characterized a novel gene, which we termed PIP5K1A and PSMD4-like (PIPSL), created by this mechanism from an intergenic transcript between the phosphatidylinositol-4-phosphate 5-kinase (PIP5K1A) and the 26S proteasome subunit (PSMD4) genes in a hominoid ancestor. PIPSL is transcribed specifically in the testis both in humans and chimpanzees, and is post-transcriptionally repressed by independent mechanisms in these primate lineages. The PIPSL gene encodes a chimeric protein combining the lipid kinase domain of PIP5K1A and the ubiquitin-binding motifs of PSMD4. Strong positive selection on PIPSL led to its rapid divergence from the parental genes PIP5K1A and PSMD4, forming a chimeric protein with a distinct cellular localization and minimal lipid kinase activity, but significant affinity for cellular ubiquitinated proteins. PIPSL is a tightly regulated, testis-specific novel ubiquitin-binding protein formed by an unusual exon-shuffling mechanism in hominoid primates and represents a key example of rapid evolution of a testis-specific gene.
Sequence comparisons of completed genomes (Waterston et al. 2002; Gibbs et al. 2004; Mikkelsen et al. 2005) have revealed that de novo emergence of novel genes is rare. The majority of new genes arise by inadvertent plagiarism of existing genomic structures, most often by duplication and subsequent divergence of whole genes or gene modules (Ohno 1970; Long et al. 2003). These duplications often occur as part of larger segmental duplications (Samonte and Eichler 2002), but intronless copies of individual genes can also be disseminated throughout genomes by the activity of the LINE-1 (L1) retrotransposon (Ostertag and Kazazian 2001; Babushok and Kazazian 2007; Babushok et al. 2007). In the presence of the original prototype gene, faithfully duplicated whole genes are rapidly inactivated by mutations as selective pressures are relaxed; only a rare replica will survive to acquire a novel function (Kimura 1983; Samonte and Eichler 2002). In contrast, while acquisition of novel functional domains (typically as alternative exons or products of intergenic recombination) allows rapid and creative functional adaptations, it also carries a barrier to fixation, as modifications to the original gene may have a negative impact on its function and host fitness. Here, we describe another mechanism of gene creation, whereby new combinations of functional domains are assembled on the RNA level, and the resulting chimera is then introduced into the genome by L1 reverse transcriptase. We characterized a novel gene, which we called PIP5K1A and PSMD4-like (PIPSL), that was created by this mechanism in the hominoid lineage.
PIPSL was first reported as an intronless genomic copy of a chimeric RNA in a human genome survey of readthrough transcripts. A chimeric PIP5K1A-PSMD4 RNA was created by intergenic splicing between the neighboring PIP5K1A and PSMD4 (also known as S5a) genes. To be consistent with the nomenclature used in previous functional studies of PSMD4, we refer to PSMD4 as S5a. Unexpectedly, the transcript PIP5K1A-S5a mapped to two different genomic locations: the original chromosome (Chr) 1 location of the parental PIP5K1A and S5a genes, as well as another site, on Chr 10, where it was found as an intronless copy (referred to as PIP5K1A-PSMD4) (Akiva et al. 2006).
The two parental genes giving rise to the chimeric transcript, PIP5K1A and S5a, are both well conserved in eukaryotes. The human PIP5K1A gene (called PIP5K1b in the mouse) encodes the alpha isoform of phosphatidylinositol 4-phosphate 5-kinase type I (PIP5K). PIP5 kinases associate with the plasma membrane (Bazenet et al. 1990; Homma et al. 1998), where they phosphorylate the D-5 inositol position of the phosphatidylinositol 4-phosphate (PI4P) in the final step of phosphatidylinositol 4,5-bisphosphate (PIP2) synthesis, a phospholipid regulator of multiple cellular processes including cytoskeleton assembly, platelet activation, and vesicle trafficking (Doughman et al. 2003; Oude Weernink et al. 2004). While the kinase domain of all PIP kinases is well conserved, the distinct substrate specificities and cellular localizations are thought to be determined by the type-specific 20–25 amino acid (aa) activation loop (Kunz et al. 2000) and divergent C-terminal regions (Di Paolo et al. 2002; Ling et al. 2002).
The S5a gene encodes a non-ATPase subunit of the eukaryotic 26S proteasome, a highly conserved multiprotein complex responsible for the regulated degradation of proteins modified by polyubiquitin chains (Coux et al. 1996). S5a was originally identified for its binding of polyubiquitin chains that mimicked the 26S proteasome (Deveraux et al. 1994; Ferrell et al. 1996; Piotrowski et al. 1997; Beal et al. 1998). Deletion analyses of S5a identified two conserved 20–30 aa hydrophobic regions at the protein’s C terminus, sufficient for polyubiquitin binding (Young et al. 1998). These regions define the general e-e-e-Φ-X-X-A-X-X-X-S-X-X-e ubiquitin-interacting motif (UIM; Φ denotes a hydrophobic and e an acidic residue) found in a variety of Ub-interacting proteins including S5a itself, ubiquitin ligases, deubiquitinating enzymes, and proteins in receptor endocytosis pathways (Hofmann and Falquet 2001; Swanson et al. 2003). Both the exact sequence of UIMs, including substitutions at conserved residues, as well as the number and position of UIMs within the protein affect the affinity for polyubiquitinated proteins (Young et al. 1998; Miller et al. 2004).
Here we characterize the structure, evolution, transcription, translation, and function of the novel PIPSL gene. We show that the chimeric PIPSL gene formed by L1-mediated retrotransposition in a hominoid ancestor of a readthrough, intergenically spliced transcript between the PIP5K1A and S5a genes. PIPSL is transcribed specifically in the testis in both humans and chimpanzees and is post-transcriptionally repressed by different mechanisms in these primate lineages. It was subject to strong positive selection after its formation, diverging from both parental genes. PIPSL lost the lipid kinase activity of PIP5K1A, likely because of a mutation in the ATP-binding pocket. However, it maintains significant affinity for cellular ubiquitinated proteins. This tightly regulated novel testis-specific ubiquitin-binding protein formed by an exon-shuffling mechanism is an important example of rapid evolution of a testis-specific gene.
Results
The PIPSL gene arose by L1-mediated retrotransposition in hominoids
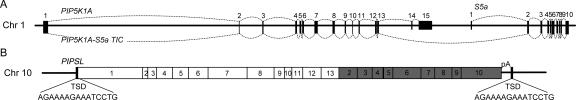
The human PIPSL gene is located on Chr 10 and is comprised of the first 13 exons of the 15-exon PIP5K1A gene joined to the last nine exons of the adjacent 10-exon S5a gene (Fig. 1). The two “parental” genes are located in tandem on Chr 1, separated by 5.2 kb, and normally form a low-abundance readthrough transcript spliced between PIP5K1A exon 13 and S5a exon 2 (Fig. 2B), both in the same phase, allowing for in-frame fusion of the greater part of both proteins (for intron analysis, see Supplemental Table S1). The event that produced this PIP5K1A-S5a Transcription-Induced Chimeric mRNA (PIP5K1A-S5a TIC) is typical of intergenic splicing, going from an existing splice donor in PIP5K1A to the first splice acceptor of S5a, with <8.5 kb separating the two genes (Akiva et al. 2006; Parra et al. 2006).
Figure 1.
(A) Neighboring 15-exon PIP5K1A and 10-exon S5a genes on Chr 1 are spliced to form PIP5K1A, S5a, and PIP5K1A-S5a TIC mRNAs. (Black rectangles) Exons; (curved lines) splicing. (B) PIP5K1A-S5a TIC was retrotransposed by L1 to create the PIPSL gene on Chr 10. (TSD) Target site duplication; (pA) A-rich repeat. (White rectangles) Regions corresponding to PIP5K1A exons; (gray rectangles) regions corresponding to S5a exons.
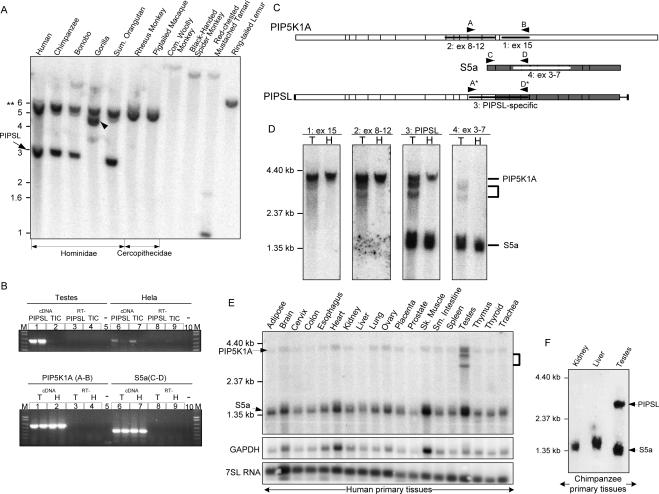
Figure 2.
(A) Southern hybridization was performed on HindIII-digested primate DNA using a 32P-labeled probe to the S5a gene. The 3-kb band corresponds to PIPSL; it migrates at 4.8 kb in Gorilla because of an RFLP (arrowhead). The 5.9-kb and 5.6-kb doublet (**) corresponds to S5a and its Chr 21 pseudogene. Supplemental Figure S7 depicts probe locations. (B) Shown are duplicate RT-PCRs on total RNA from human testis or HeLa cells using the primers depicted in C. In the top gel of B, A* and D* primers specific for PIPSL amplified PIPSL from testis (lane 1), but not HeLa cDNA (lane 6). A and D primers specific for the PIP5K1A-S5a Transcription-Induced Chimera (TIC) amplified only trace amounts of TIC from testis (lane 2) and HeLa (lane 7) cDNAs. In the bottom gel, parental PIP5K1A (lanes 1,2) and S5a (lanes 6,7) transcripts were amplified from the same testis (T) and HeLa (H) cDNAs, serving as internal controls. (Lanes 3,4,8,9) RT-PCRs lacking reverse transcriptase; (lanes 5,10) no template PCRs; (M) 1-kb Plus DNA ladder. (C) Primers used in RT-PCRs are shown by arrowheads (A, B, C, D, A*, and D*). Probes used in Northern blots are shown by black (ex 15, ex 8–12, and PIPSL-specific) and white (ex 3–7) lines. (D) Northern blot of PIPSL in human testis (T) showing two PIPSL transcripts of ∼3.0–3.6 kb (bracket). HeLa RNA (H) is used as a negative control. PIPSL is visible with probe 2 in PIP5K1A exons 8–12, PIPSL-specific probe 3 spanning both parental genes, and probe 4 in S5a exons 3–7, but not with negative-control probe 1 to PIP5K1A exon 15. The 3.7-kb PIP5K1A and 1.3-kb S5a mRNAs serve as internal controls. (E) Northern blot on RNAs from multiple human tissues with PIPSL-specific probe 3 shows PIPSL transcripts only in the testis. PIP5K1A and S5a are ubiquitously present. To confirm comparable loading, the blot was stripped and reprobed for GAPDH and 7SL RNAs. (F) A Northern blot on RNAs from several chimpanzee tissues with PIPSL-specific probe 3 shows ∼3.0-kb PIPSL transcripts only in the testis. Internal control parental S5a is ubiquitously present. The human PIPSL-specific probe 3 does not hybridize to the chimpanzee PIP5K1A transcript.
To determine whether L1-mediated retrotransposition of PIP5K1A-S5a TIC was responsible for creation of the PIPSL gene, we examined the genomic site of PIPSL for signs of L1-mediated integration. The intronless 3.3-kb PIPSL gene is integrated at a canonical L1 insertion site 5′-TTCT’GA-3′, terminates in an 80-bp A-rich repeat (GAAA)n, and is flanked by 15-bp target site duplications, all typical of L1 retrotransposition (Ostertag and Kazazian 2001). Concordant divergence of both parts of the PIPSL gene (2.31% and 2.26%) underscores their integration in a single event. Moreover, older processed pseudogenes of PIP5K1A and S5a in the human genome confirm that both genes are expressed at a time and tissue site compatible with L1-mediated retrotransposition.
We used three different methods to estimate the age of the PIPSL insertion: BLAT (Kent et al. 2002) searches of completed genomes, Southern analysis, and calculations using nucleotide divergence. From BLAT, PIPSL was found on the homologous chromosome in the chimpanzee, but not in the rhesus monkey, mouse, and rat genomes. Southern analysis revealed the PIPSL gene throughout the Hominidae lineage up to Orangutan (∼14 million years ago [Mya]); it was absent in the two Cercopithecidae tested, Rhesus monkey and Pigtailed macaque (∼25 Mya) (Fig. 2A) (Goodman et al. 1998). The hominoid origin of PIPSL was further corroborated by molecular age estimates of 15–19 Mya, assuming a neutral mutation rate of 1.2–1.5 × 10−9/nt/yr (Yi et al. 2002).
PIPSL gene has testis-specific transcription in humans and chimpanzees
In order to distinguish between the nearly identical PIPSL and PIP5K1A-S5a TIC transcripts, we carried out RT-PCRs using oligonucleotides specific for one or the other of the transcripts; the identities of RT-PCR products were confirmed by sequencing (Fig. 2B). We detected high levels of PIPSL RNA in the testis, but only trace levels in HeLa cells (Fig. 2B), 143B cells, and 293T cells (data not shown). In contrast, only trace levels of PIP5K1A-S5a TIC were present in testis and cultured cells. PIP5K1A and S5a genes were highly transcribed in all tissues tested.
Having determined that in the testis the PIPSL transcript is significantly more abundant than PIP5K1A-S5a TIC, allowing us to distinguish between them, we used Northern analysis to determine the size and abundance of PIPSL relative to the parental transcripts (Fig. 2D). In agreement with RT-PCR results, two distinct PIPSL transcripts of ∼3.0–3.6 kb were detected in testis, but not in HeLa RNA, while PIP5K1A and S5a RNAs were present in both. Using a chimeric PIPSL-specific probe to check PIPSL transcription in multiple human tissues (Fig. 2E), we found abundant PIPSL RNA in the testis, but undetectable levels elsewhere. PIP5K1A and S5a were ubiquitously transcribed. Consistent with the testis specificity of PIPSL, the sole PIPSL GenBank transcript (BC068549) also originated in testis (Strausberg et al. 2002). The chimpanzee PIPSL gene is also transcribed, and, similar to human PIPSL, its transcription appears to be limited to testis (Fig. 2F).
5′-RACE mapping of human PIPSL transcription start sites revealed three adjacent starts ∼40 bp upstream of the PIP5K1A translation initiation AUG (Supplemental Fig. S1); the first one coincided with the BC068549 GenBank mRNA. A new TATAA sequence, ∼47 bp upstream of transcription starts, arose by mutation from TGTAA prior to human–chimp divergence, and a GC-rich region immediately precedes TATAA. Several canonical AATAAA polyadenylation (pA) sites are downstream from the PIPSL coding sequence, predicting two RNA sizes: 2868 bp for the original pA signal from S5a, and 3593/3636 bp for the next two. These sizes are consistent with the two PIPSL bands seen on the Northern blot of human PIPSL (Fig. 2D,E). Another, nonconventional TATAAA pA signal was used by the 3348-bp PIPSL transcript BC068549. In chimpanzees, the first polyadenylation site has a stronger predicted recognition sequence; consistent with this, the predominant PIPSL transcript is ∼3 kb on the Northern blot (Fig. 2E).
PIPSL gene was subject to positive selection in a primate ancestor
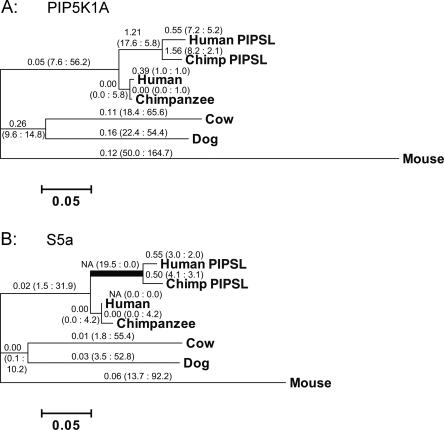
Both parts of the PIPSL gene accumulated mutations at a higher rate compared to their parental homologs (Fig. 3). There is a striking excess of nonsynonymous substitutions (n) over synonymous substitutions (s) in the C-terminal S5a region of PIPSL (Fig. 3B). To test the statistical significance of this difference, ancestral DNA sequences representing all internal nodes of the tree were reconstructed using the maximum-1ikelihood method. Reconstructed sequences of the parental S5a and PIP5K1A and the chimeric PIPSL genes in the primate ancestor were then used in pairwise nonsynonymous/synonymous calculations (Supplemental Methods).
Figure 3.
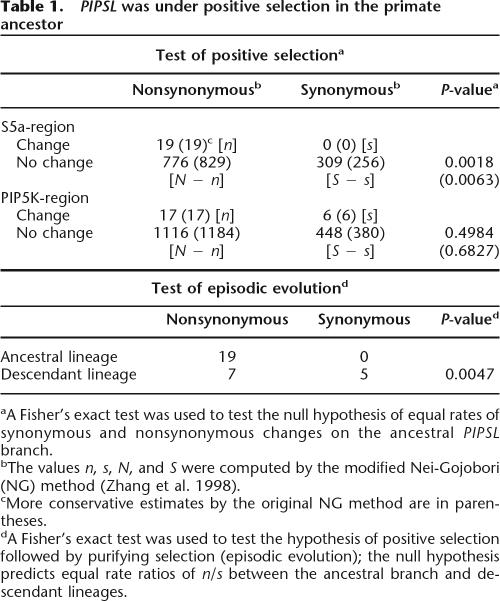
PIPSL experienced a burst of positive selection shortly after its creation (A). The phylogenetic tree of the PIP5K1A region (A) and the S5a region of the PIPSL gene (B). Branches are drawn in proportion to the total number of substitutions incurred, with nonsynonymous-to-synonymous substitution rate ratios (Ka/Ks), and the nonsynonymous (n) and synonymous (s) substitutions in parentheses (n : s) shown above each branch. Nineteen nonsynonymous and 0 synonymous changes occurred in the S5a region of PIPSL along the branch leading to the human/chimp split (B, indicated by bold line). The n/s ratio is significantly greater than its neutral expectation (P = 0.0018, Fisher’s exact test; Table 1), indicating that the PIPSL gene was under strong positive selection shortly after its formation. (NA) Not applicable (Ks = 0).
In the ancestral PIPSL gene lineage (from formation of PIPSL until the speciation of humans and chimpanzees), the N-terminal PIP5K1A region of PIPSL had substitution rates consistent with relaxed functional constraint or neutral evolution (Ka/Ks = 1.21, P = 0.4984; Fisher’s exact test; Table 1). However, there were 19 nonsynonymous and 0 synonymous differences between the inferred S5a and PIPSL ancestral sequences. The potential numbers of nonsynonymous and synonymous sites in the S5a region are 794.6 and 309.4, respectively. Thus, the n/s ratio (19/0) is significantly greater than its neutral expectation (N/S = 795/309, P = 0.0018; Fisher’s exact test; Table 1). The same conclusion was reached when more conservative estimates of N and S, as well as other transition/transversion ratios (R), were included in the calculation (Nei and Gojobori 1986; Zhang et al. 1998). These results strongly suggest that positive selection acted in the ancestral PIPSL evolution (see Discussion).
Table 1.
PIPSL was under positive selection in the primate ancestor
aA Fisher’s exact test was used to test the null hypothesis of equal rates of synonymous and nonsynonymous changes on the ancestral PIPSL branch.
bThe values n, s, N, and S were computed by the modified Nei-Gojobori (NG) method (Zhang et al. 1998).
cMore conservative estimates by the original NG method are in parentheses.
dA Fisher’s exact test was used to test the hypothesis of positive selection followed by purifying selection (episodic evolution); the null hypothesis predicts equal rate ratios of n/s between the ancestral branch and descendant lineages.
After speciation, both the human and chimpanzee PIPSL lineages show small Ka/Ks ratios (Fig. 3), with n/s of 19/0 for the ancestral branch and 7/5 for descendant lineages. The significant difference between the selective pressures on the ancestral and descendent lineages (P = 0.0047; Fisher’s exact test) (Fig. 3; Table 1) suggests that the positive selection was episodic: it occurred in the primate ancestor and was followed by nearly neutral evolution after speciation. While this is the first example of positive selection on a retrogene that arose from an intergenically spliced transcript, our data are consistent with the burst of adaptive evolution after gene duplication, previously observed in primates and Drosophila (Zhang et al. 1998; Jones and Begun 2005).
PIPSL translation is disrupted in humans and repressed in chimpanzees
Positive selection on the ancestral PIPSL gene indicates that the PIPSL gene has been translated and functional in the primate ancestor. To evaluate PIPSL translation in current-day humans and chimpanzees, we used three different approaches: sequence analysis of translation initiation contexts of human and chimpanzee PIPSL genes, in vitro transcription/translation assays, and Western blot analyses of human and chimpanzee tissues.
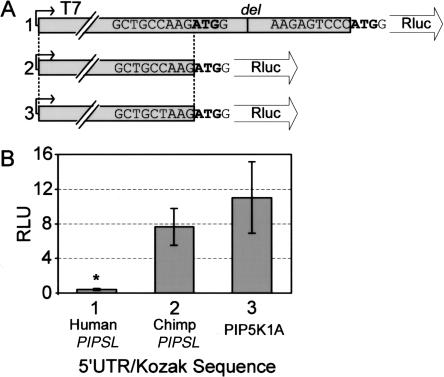
The complete 414-bp 5′-UTR region of the PIP5K1A was copied during retrotransposition and remains virtually identical in the ancestral PIPSL, positioning its open reading frame (ORF) within the original translation initiation context of PIP5K1A gene (Supplemental Fig. S1). A −4T → C substitution prior to the human/chimpanzee split brought the PIPSL Kozak region even closer to the consensus (Kozak 1986). This canonical Kozak is retained by chimp PIPSL, strongly suggesting that its translation efficiency is similar to the identically initiated PIP5K1A. In contrast, the human PIPSL gene suffered a single nucleotide deletion at +45 bp, causing an early frameshift. To check whether this deletion occurred recently and is polymorphic, we PCR-amplified the mutated region in 42 individuals (84 haploid genomes) from diverse ethnic groups (Supplemental Table S3). Sequencing of PCR products failed to detect any individuals without the deletion, indicating that it is fixed or has a very high frequency. The second in-frame AUG at +105 of the human PIPSL has a weak Kozak sequence (Kozak 1986), but, if used for translation initiation, would produce an almost complete PIPSL protein, short 35 amino acids.
To check whether the alternative +105 AUG could serve for translation initiation of human PIPSL, we performed in vitro transcription/translation reactions comparing translation efficiencies of a Renilla luciferase reporter in the 5′-UTR/Kozak context of PIP5K1A, or the human or chimpanzee PIPSL genes. As expected from their nearly identical translation contexts, the chimp PIPSL construct expressed similarly to PIP5K1A (69.5%, P = 0.07) (Fig. 4). In contrast, the translation of human PIPSL was exceedingly low, measuring only 3.6% of PIP5K1A (P < 0.001).
Figure 4.
(A) Diagram showing Renilla luciferase reporter constructs, in which the Renilla Kozak sequence is replaced by the 5′-UTR/Kozak regions from human PIPSL (1), chimp PIPSL (2), and PIP5K1A (3). (B) Average amounts of Renilla produced by in vitro transcription/translation reactions using the three constructs ±SD. For each construct, eight replicates were performed using six to eight independent DNA preparations. Human PIPSL translation was significantly reduced (P < 0.001).
Consistent with disrupted translation of the human PIPSL gene caused by a deletion at +45 bp, we were unable to detect PIPSL protein by Western blot in human testis lysates and teratocarcinoma cell lines (Supplemental Fig. S2A,B). Unexpectedly, we were also unable to detect the chimpanzee PIPSL protein in chimpanzee testicular tissue (Supplemental Fig. S2E). The lack of detectable chimpanzee PIPSL protein, despite (1) the abundant PIPSL RNA detected by Northern blot (Fig. 2E), (2) the efficient Kozak translation initiation context as seen in in vitro translation assays (Fig. 4), and (3) apparent protein stability when overexpressed in cultured cells from transfected, coding region-only chimpanzee PIPSL constructs (Fig. 6B,D, below; Supplemental Fig. S2E), suggests that native chimpanzee PIPSL is subject to post-transcriptional repression in vivo that is not recapitulated in our in vitro assay.
Figure 6.
PIPSL proteins lack PIP5 kinase activity, and bind cellular ubiquitinated proteins. (A) C-terminally HA-tagged human PIP5K1A (1), S5a (2) and PIPSL (3), and chimpanzee PIPSL (4) proteins were overexpressed in 293T cells, IP’d against HA epitope, and tested for phosphorylation of PI4P to produce PIP2. (B) The amounts of IP’d proteins were analyzed by an anti-HA Western blot. (C) Coimmunoprecipitation of HA-tagged human PIP5KA (1), S5a (2), S5AL (3), chimpanzee PIPSL (4), and untransfected 293T cells (5) with cellular ubiquitinated proteins (bracket). After anti-HA IP, proteins were immunoblotted for ubiquitin (C) or for HA-tag (D). (E) Anti-Ub Western blot of cellular lysates to demonstrate equal levels of endogenous ubiquitinated proteins. (F) Equal loading of lysates confirmed by Western against S10a. (G) Average band intensities of ubiquitinated protein from three independent co-IP experiments were normalized by HA-tagged protein amounts and scaled to the same overall blot intensity. Error bars denote ±1 SD. (*) Immunoglobulin chains; (**) a nonspecific band.
Both the strong positive selection on the ancestral PIPSL gene and the translational disruption/repression seen in the descendant human and chimpanzee lineages are consistent with rapid evolution of reproductive genes across evolutionary taxa, including higher primates (Wyckoff et al. 2000; Swanson and Vacquier 2002; Clark and Swanson 2005) (see Discussion). In order to gain better understanding of the potential effects of the PIPSL protein, we used four different approaches to study the PIPSL protein function: (1) sequence analyses of functional domains, (2) studies of cellular localization and assays of parental gene function, (3) PIP5 kinase activity of PIP5K1A, and (4) ubiquitinated protein binding of S5a.
Domain analysis of PIPSL proteins
The N-terminal region of the PIPSL proteins is 94.6%–94.8% identical to PIP5K1A (Supplemental Fig. S3) and includes the kinase catalytic “core” region (amino acids 65–436) (Rao et al. 1998; Galiano et al. 2002) and two small deletions: a 15-aa deletion of the nonconserved C terminus due to skipped exons 14–15 (Δ535–549), and a 5-aa internal deletion (Δ61–65) partially overlapping the kinase core. Human PIPSL has an additional Δ1–35 deletion at the nonconserved N terminus. A few mutations alter residues conserved or conservatively substituted among PIP kinases: Δ65K, K66G, R138W, D146S(chimp), I180V(hu), and R427W. Both of the KK regions previously shown to mediate membrane localization (KK400–401, KK438–439) (Kunz et al. 2000; Arioka et al. 2004) remain intact; however, several basic residues in the N-terminal region, postulated to interact with the cellular membrane (Rao et al. 1998), have been mutated (ΔK65, K66G, R93S, and R138W). The three known catalytic residues (K181, D309, and D391 of PIP5K1A) (Ishihara et al. 1998; Rao et al. 1998) are preserved, and the activation loop (amino acids 396–417), important for substrate specificity (Kunz et al. 2000), remains relatively unscathed with one mutation each in nonconserved residues [V410I(hu) and G413R(chimp)]. Importantly, both PIPSLs share a nonconservative Q184R substitution in the close vicinity of K181; K181 corresponds to the ATP-binding Lys in protein kinases (Knighton et al. 1991), and its mutation in PIP5K1A abolishes catalytic activity (Rao et al. 1998). Further structural data are necessary to determine the significance of the Q184R mutation.
PIPSL proteins are 93.2%–94.0% identical to S5a and lack only its nine N-terminal amino acids because of the skipped exon 1 (Supplemental Fig. S4). Consistent with accelerated evolution of early primate and chimpanzee PIPSL genes, a strikingly high 19 mutations are shared between human and chimp, with an additional three human, and six chimp-specific changes. A high density of substitutions [R542G(hu), I560T, A580D, R595C(chimp), T613M, R616H, K628N, M632I, S640N, D648N(hu), R655C] falls into the poorly characterized N terminus of S5a, proposed to be involved in the association with 19S proteasome and other proteins (Anand et al. 1997; Young et al. 1998). Importantly, a cluster of mutations is within the two UIM regions, which mediate S5a binding to ubiquitinated proteins (Young et al. 1998): the two invariant residues of UIM1 are conservatively substituted (A744V and S748F), with four additional changes [Q811K, A813V, Y814C, and Q821R(chimp)] altering nonconserved residues in UIM2. These substitutions suggest reduced ubiquitin-binding compared to the parental S5a UIMs; however, PIPSL UIMs are still within the broad spectrum of UIM sequences of known ubiquitin receptors (Swanson et al. 2003; Miller et al. 2004) and are expected to bind ubiquitinated proteins.
Unlike PIP5K1A and S5a, PIPSL proteins are cytoplasmic
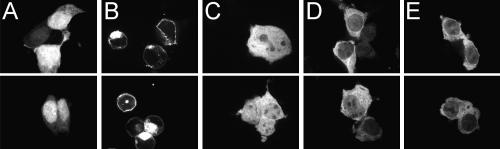
To determine whether PIPSL proteins mimic cellular localization of either of their parental homologs, we transiently transfected 293T cells with N-terminal GFP-fusion constructs of human PIP5K1A, S5a and PIPSL, and chimpanzee PIPSL (Fig. 5). In agreement with prior reports (Bazenet et al. 1990; Homma et al. 1998), PIP5K1A was located at the plasma membrane (Fig. 5B). S5a protein was found throughout the cell, with possible nucleolar exclusion. Notably, both parental localizations were lost by the cytoplasmic human and chimpanzee PIPSL proteins (Fig. 5D,E). To assess the relative contributions to PIPSL localization of its N-terminal PIP5K1A and C-terminal S5a regions, the two parts were analyzed separately (Supplemental Fig. S5). Strikingly, the N-terminal PIP5K1A region localized as the full human PIPSL protein, while the C-terminal S5a region reverted to the localization pattern of its parental S5a protein.
Figure 5.
Intracellular localization of GFP-fused PIP5K1A, S5a, and PIPSL proteins in 293T cells was observed by GFP fluorescence with confocal microscopy. (A) GFP control is seen throughout the cell, with nuclear predominance; (B) PIP5K1A is on the plasma membrane. (C) S5a is throughout the cell with cytoplasmic predominance and possibly nucleolar exclusion; (D) Human and (E) chimpanzee PIPSL proteins are cytoplasmic. A Western of IP’d GFP fusion proteins is shown in Supplemental Figure S6B.
PIPSL has negligible PIP5 kinase activity
To assess PIP5 kinase activity of PIPSL, human PIP5K1A, S5a and PIPSL, and chimpanzee PIPSL proteins tagged at their C termini with an HA epitope were expressed in 293T cells, isolated by immunoprecipitation (IP) against HA, and assayed for ability to phosphorylate PI4P, the preferred substrate of PIP5K1A (Zhang et al. 1997b). While PIP5K1A protein demonstrated clear PIP5 kinase activity, negligible phosphorylation was detected with similar amounts of PIPSL proteins (Fig. 6A,B). The results were confirmed using N-terminal GFP-fusion constructs (Supplemental Fig. S6A,B), ruling out artifactual loss of activity caused by tagging. PIPSL kinase activity was not restored when its C-terminal S5a region was removed, indicating that loss of activity was caused by mutations internal to the kinase “core,” and not by the C-terminal S5a fusion (Supplemental Fig. S6C).
PIPSL binds ubiquitinated proteins
To determine if PIPSL can bind ubiquitinated proteins, we performed co-IP experiments with C-terminally HA-tagged PIPSL constructs in 293T cells. Both human and chimpanzee PIPSL bound numerous ubiquitinated proteins in cellular lysates (Fig. 6C), but, in agreement with a predicted decrease in Ub affinity, binding by PIPSL proteins was decreased to 14%–16% of S5a (P ≤ 0.05) when normalized for protein amount. The PIPSL affinity for ubiquitinated proteins relative to S5a is roughly similar to that of known cellular ubiquitin interactors, such as epsin, eps15, eps15R, HSJ1, Hrs, USP25, and KIAA1386 (Polo et al. 2002; Miller et al. 2004). Ubiquitin binding by PIPSL was significantly higher than background binding by the negative control PIP5K1A (P < 0.01). As expected from functional domain analysis, separate co-IPs using the PIP5K1A and S5a parts of the human PIPSL gene confirmed that Ub binding is mediated only by the S5a region (data not shown).
Discussion
Here we demonstrate that a readthrough transcript joining the PIP5K1A and S5a genes was retrotransposed by L1 in a hominoid to create a testis ubiquitin-binding protein PIPSL. The PIPSL gene is actively transcribed in testis in humans and chimpanzees and is translationally repressed in both primate lineages by independent mechanisms. The creation of PIPSL required a concerted action of multiple rare processes and is highly unusual. Indeed, PIPSL is the only known example of a new gene produced by L1-mediated retrotransposition of an exon-shuffled transcript.
First, the PIP5K1A-S5a TIC RNA was formed by cotranscription and intergenic splicing, a rare occurrence in 4%–5% of tandem gene pairs (Akiva et al. 2006; Parra et al. 2006). We and others (Akiva et al. 2006) detected trace levels of PIP5K1A-S5a TIC by RT-PCR, too low to detect by Northern blot. The splicing in PIP5K1A-S5a TIC occurred between same-phase introns allowing for parental gene fusion; in contrast, 56%–64% of all TICs result in frameshifts and premature stop codons, and are expected to be degraded by Nonsense Mediated Decay (Akiva et al. 2006; Parra et al. 2006).
Second, the entire PIP5K1A-S5a TIC was retrotransposed by L1 in a process that truncates and rearranges the majority of integrants (Szak et al. 2002), and greatly disfavors retrotransposition of non-L1 RNAs (Esnault et al. 2000; Wei et al. 2001). To be inherited, the PIPSL insertion had to be represented in the germline. Furthermore, it was most likely beneficial or neutral to the host to become fixed in the ancestral primate population. Notably, PIPSL integration occurred at the tail end of the burst of L1 activity in ancestral primates, when hundreds of other mRNAs were mobilized by L1 to form fixed retrogenes and pseudogenes (Ohshima et al. 2003; Marques et al. 2005; Vinckenbosch et al. 2006).
Third, PIPSL had to be transcribed, an ability traditionally accorded only to pseudogenes that aberrantly include their original promoter or that fortuitously land downstream from another promoter (Mighell et al. 2000). It is now clear that, in fact, up to a third of all pseudogenes are transcribed, most of these in testis or spermatogenic cells (Kleene et al. 1998; Harrison et al. 2005; Marques et al. 2005; Sakai et al. 2006; Vinckenbosch et al. 2006). Testis specificity is likely caused by the transcriptionally permissive environment in the testis, where the components of the Pol II holoenzyme complex are known to increase 30–1000-fold (Schmidt and Schibler 1995, 1997), allowing for high transcription levels from non-promoter and weak promoter sequences (Ossipow et al. 1995; Schmidt 1996). This, in turn, enables efficient testicular transcription of retrogenes like PIPSL, which may lack a bone fide promoter to drive transcription in other tissues (Kleene et al. 1998; Marques et al. 2005). The difference between the testis-specific transcription of PIPSL reported here and the ubiquitous expression of PIPSL reported earlier (Akiva et al. 2006) is perhaps due to different methods used in the two studies. Akiva and colleagues detected human PIPSL transcripts by RT-PCR, which can detect very small amounts of template. We also found trace amounts of PIPSL by RT-PCR in several cell types. However, our Northern blot analysis showed appreciable PIPSL RNA only in testis. Testis specificity of PIPSL transcription was observed in both humans and chimpanzees.
Importantly, several reports of transcribed, but not translated pseudogenes point to their role in RNA-mediated control of other genes, joining a larger category of non-coding RNAs (ncRNAs) (Korneev et al. 1999; Eddy 2001; Hirotsune et al. 2003; Lee 2003; Harrison et al. 2005; Pang et al. 2005; Sakai et al. 2006). For example, the transcript of the nitric oxide synthase (NOS) pseudogene represses translation of the homologous neuronal NOS gene (Korneev et al. 1999). Another well-characterized, but controversial (Gray et al. 2006), example is the transcribed pseudogene of the makorin 1 gene (MKRNP1), which may be necessary for the efficient expression of the makorin 1 gene (Hirotsune et al. 2003). It is conceivable that PIPSL may participate in RNA-mediated regulation of its homologs (e.g., parental PIP5K1A and S5a genes).
Finally, upon retrotransposition, the PIPSL gene retained an intact ORF within an efficient translation initiation context, allowing for the production of the PIPSL protein in a primate ancestor. Under positive selection, the PIPSL gene has undergone rapid evolution that caused functional divergence of PIPSL from the PIP5K1A and S5a parental genes, and drastically reduced the PIPSL translational ability in the two primate lineages to levels undetectable by Western blot. A deletion early in the human PIPSL gene reduced its translation to 3.6% of PIP5K1A as judged by in vitro translation assays; this decrease is larger than previously reported for similar immediate Kozak sequences (Kozak 1986). In all likelihood, an additional drop in translation efficiency is due to interference from the original Kozak sequence just upstream. In contrast, the chimpanzee PIPSL gene maintains an optimal Kozak translation initiation context in vitro. However, endogenous PIPSL protein cannot be detected in chimpanzee testicular tissue despite high levels of mRNA. It is highly unlikely that PIPSL protein was degraded during lysate preparation, since our internal controls (the parental S5a as well as other housekeeping proteins) are intact in the same sample. While the PIPSL protein is stable when expressed from transfected constructs containing only the coding region of the chimpanzee PIPSL, further evidence, obtained from experimental systems able to fully recapitulate protein degradation processes in chimpanzee testis, will be required to formally exclude the possibility that the chimpanzee PIPSL protein is expressed but rapidly degraded in vivo.
The absence of chimpanzee PIPSL translation product is most consistent with post-transcriptional repression, possibly through its 5′- or 3′-UTR sequences. Potential mechanisms of post-transcriptional repression include microRNA-mediated translational repression (for review, see Bartel 2004), repression by interactions with complementary mRNAs (e.g., parental PIP5K1A or S5a) (Korneev et al. 1999), and competitive inhibition of the 5′ m7G-cap recognition by the cellular translational machinery (Lasko et al. 2005). Post-transcriptional repression is frequently involved in tight temporal regulation of developmentally important genes, such as the heterochronic gene Lin-14 in Caenorhabditis elegans (Wightman et al. 1993) and HOX genes in mammals (Naguibneva et al. 2006). Future work on post-transcriptional repression of the chimpanzee PIPSL gene will be important to better understand its role in primate reproductive biology.
Positive selection leading to functional divergence and translation repression of PIPSL is reminiscent of positive selection and rapid evolution of reproductive genes (Wyckoff et al. 2000; Swanson and Vacquier 2002; Clark and Swanson 2005), thought to be driven by a combination of sperm competition, sexual selection, and sexual conflict (Swanson and Vacquier 2002). As was observed in the case of the human PIPSL, positive selection is known occasionally to increase the frequency of frameshift mutations in reproductive genes (Gasper and Swanson 2006). This paradoxical effect could be due either to the creation of a novel beneficial function by a frameshifted gene, such as the protection from severe sepsis resistance by the frameshifted caspase 12 (Wang et al. 2006; Xue et al. 2006), or to the elimination of a new deleterious function that may have arisen in the process of ongoing adaptation to the changing reproductive environment. Conversely, with altered environmental pressures, repair, expression, and a new physiologic role of a previously non-expressed, inactivated pseudogene has also been reported (Trabesinger-Ruef et al. 1996).
While, as evidenced by its fixation in the population, the PIPSL gene was likely beneficial or neutral after its formation, it may have subsequently become detrimental because of mutational changes and/or altered environment. This could have been caused by the overexpression toxicity (Galiano et al. 2002) or dominant-negative effects of the PIPSL protein containing both parental gene functions. In this model, any mutations decreasing functionality of either part of the PIPSL gene or its translation would be predicted to decrease the potential detrimental effect and would be selected for and fixed in the population. After the elimination of kinase activity in early hominoids, selection on the PIP5K1A region was likely relaxed. In contrast, the C-terminal S5a region remained under strong positive selection, accumulating a high density of mutations in the conserved N terminus, UIM1 and UIM2. The parental S5a is thought to mediate the recognition of ubiquitinated proteins by the proteasome (Coux et al. 1996) and is one of the most avid binders of polyubiquitinated proteins (Lam et al. 2002; Miller et al. 2004). Thus, it would not be surprising if a functional S5a-like gene could have profound effects on intracellular protein trafficking, endocytosis, and regulated protein degradation and could serve as a potent substrate for positive selection. While PIP5K moiety of PIPSL is kinase inactive, its residual activity (e.g., protein binding) may confer additional functional nuances to PIPSL, perhaps, affecting a unique cellular process, not normally involving either of its parental proteins.
Methods
Detailed descriptions of antibodies, primers, cloning, informatics, evolutionary analysis, Southern, RT-PCR, 5′-RACE, Northern blot, deletion genotyping, transcription/translation, cell culture, immunofluorescence, kinase assay, and IP/Western are in the Supplemental Methods.
RT-PCR and Northern blot
For RT-PCR, Human Testicle Total RNA (Ambion) or total HeLa RNA were reverse transcribed with M-MLV RT (Promega) and used in PCR with Taq polymerase (Promega). For Northern blots, 8 μg of human Testicle Total RNA, FirstChoice Human Total RNA Survey Panel (Ambion), total HeLa RNA, or total RNA from chimpanzee kidney, liver, or testicle tissues (Department of Veterinary Medicine & Surgery, UT M.D. Anderson Cancer Center, Houston, TX) was used for hybridization in a NorthernMax-Gly system (Ambion), with [32P]dATP random-labeled DNA probes made with Strip-EZ DNA kit (Ambion).
Evolutionary analysis
The ancestral sequences at all interior nodes of the PIPSL tree were inferred by the likelihood-based Bayesian method (Yang et al. 1995). The numbers of synonymous (s) and nonsynonymous (n) substitutions for each tree branch were then estimated (Nei and Gojobori 1986). The numbers of potential synonymous sites (S) and nonsynonymous sites (N) were computed by the modified Nei-Gojobori (NG) method and the original NG method (Nei and Gojobori 1986; Zhang et al. 1998). The statistical significance of the difference between n/s and N/S was evaluated using the Fisher’s exact test (Zhang et al. 1997a).
Acknowledgments
We thank Donna George, Julie Liu, and Jeongsik Yong for helpful suggestions on co-IP and immunoblotting conditions, and members of the Kazazian lab for critical discussions. This study was supported by the grants from the U.S. National Institutes of Health to H.H.K. and C.S.A, and grants from the Ministry of Japan to K.O.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6252107
References
- Akiva P., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R., Diber A., Shemesh R., Novik A., Sorek R., Shemesh R., Novik A., Sorek R., Novik A., Sorek R., Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand G., Yin X., Shahidi A.K., Grove L., Prochownik E.V., Yin X., Shahidi A.K., Grove L., Prochownik E.V., Shahidi A.K., Grove L., Prochownik E.V., Grove L., Prochownik E.V., Prochownik E.V. Novel regulation of the helix–loop–helix protein Id1 by S5a, a subunit of the 26 S proteasome. J. Biol. Chem. 1997;272:19140–19151. doi: 10.1074/jbc.272.31.19140. [DOI] [PubMed] [Google Scholar]
- Arioka M., Nakashima S., Shibasaki Y., Kitamoto K., Nakashima S., Shibasaki Y., Kitamoto K., Shibasaki Y., Kitamoto K., Kitamoto K. Dibasic amino acid residues at the carboxy-terminal end of kinase homology domain participate in the plasma membrane localization and function of phosphatidylinositol 5-kinase gamma. Biochem. Biophys. Res. Commun. 2004;319:456–463. doi: 10.1016/j.bbrc.2004.04.187. [DOI] [PubMed] [Google Scholar]
- Babushok D.V., Kazazian H.H., Kazazian H.H.2007Progress in understanding the biology of the human mutagen LINE-1 Hum. Mutat. 28 :527–539. [DOI] [PubMed] [Google Scholar]
- Babushok D.V., Ostertag E.M., Kazazian H.H., Ostertag E.M., Kazazian H.H., Kazazian H.H. Current topics in genome evolution: Molecular mechanisms of new gene formation. Cell. Mol. Life Sci. 2007;64:542–554. doi: 10.1007/s00018-006-6453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bazenet C.E., Ruano A.R., Brockman J.L., Anderson R.A., Ruano A.R., Brockman J.L., Anderson R.A., Brockman J.L., Anderson R.A., Anderson R.A. The human erythrocyte contains two forms of phosphatidylinositol-4-phosphate 5-kinase which are differentially active toward membranes. J. Biol. Chem. 1990;265:18012–18022. [PubMed] [Google Scholar]
- Beal R.E., Toscano-Cantaffa D., Young P., Rechsteiner M., Pickart C.M., Toscano-Cantaffa D., Young P., Rechsteiner M., Pickart C.M., Young P., Rechsteiner M., Pickart C.M., Rechsteiner M., Pickart C.M., Pickart C.M. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- Clark N.L., Swanson W.J., Swanson W.J. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A.L., Tanaka K., Goldberg A.L., Goldberg A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Deveraux Q., Ustrell V., Pickart C., Rechsteiner M., Ustrell V., Pickart C., Rechsteiner M., Pickart C., Rechsteiner M., Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Di Paolo G., Pellegrini L., Letinic K., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P., Pellegrini L., Letinic K., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P., Letinic K., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P., Zoncu R., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P., Voronov S., Chang S., Guo J., Wenk M.R., De Camilli P., Chang S., Guo J., Wenk M.R., De Camilli P., Guo J., Wenk M.R., De Camilli P., Wenk M.R., De Camilli P., De Camilli P. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- Doughman R.L., Firestone A.J., Anderson R.A., Firestone A.J., Anderson R.A., Anderson R.A. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T., Maestre J., Heidmann T., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Ferrell K., Deveraux Q., van Nocker S., Rechsteiner M., Deveraux Q., van Nocker S., Rechsteiner M., van Nocker S., Rechsteiner M., Rechsteiner M. Molecular cloning and expression of a multiubiquitin chain binding subunit of the human 26S protease. FEBS Lett. 1996;381:143–148. doi: 10.1016/0014-5793(96)00101-9. [DOI] [PubMed] [Google Scholar]
- Galiano F.J., Ulug E.T., Davis J.N., Ulug E.T., Davis J.N., Davis J.N. Overexpression of murine phosphatidylinositol 4-phosphate 5-kinase type Ibeta disrupts a phosphatidylinositol 4,5 bisphosphate regulated endosomal pathway. J. Cell. Biochem. 2002;85:131–145. [PubMed] [Google Scholar]
- Gasper J., Swanson W.J., Swanson W.J. Molecular population genetics of the gene encoding the human fertilization protein zonadhesin reveals rapid adaptive evolution. Am. J. Hum. Genet. 2006;79:820–830. doi: 10.1086/508473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scott G., Steffen D., Worley K.C., Burch P.E., Steffen D., Worley K.C., Burch P.E., Worley K.C., Burch P.E., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Goodman M., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Schneider H., Shoshani J., Gunnell G., Groves C.P., Shoshani J., Gunnell G., Groves C.P., Gunnell G., Groves C.P., Groves C.P. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Gray T.A., Wilson A., Fortin P.J., Nicholls R.D., Wilson A., Fortin P.J., Nicholls R.D., Fortin P.J., Nicholls R.D., Nicholls R.D. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proc. Natl. Acad. Sci. 2006;103:12039–12044. doi: 10.1073/pnas.0602216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P.M., Zheng D., Zhang Z., Carriero N., Gerstein M., Zheng D., Zhang Z., Carriero N., Gerstein M., Zhang Z., Carriero N., Gerstein M., Carriero N., Gerstein M., Gerstein M. Transcribed processed pseudogenes in the human genome: An intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374–2383. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A., Yagami K., Wynshaw-Boris A., Yoshiki A., Wynshaw-Boris A., Yoshiki A., Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Falquet L., Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Homma K., Terui S., Minemura M., Qadota H., Anraku Y., Kanaho Y., Ohya Y., Terui S., Minemura M., Qadota H., Anraku Y., Kanaho Y., Ohya Y., Minemura M., Qadota H., Anraku Y., Kanaho Y., Ohya Y., Qadota H., Anraku Y., Kanaho Y., Ohya Y., Anraku Y., Kanaho Y., Ohya Y., Kanaho Y., Ohya Y., Ohya Y. Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J. Biol. Chem. 1998;273:15779–15786. doi: 10.1074/jbc.273.25.15779. [DOI] [PubMed] [Google Scholar]
- Ishihara H., Shibasaki Y., Kizuki N., Wada T., Yazaki Y., Asano T., Oka Y., Shibasaki Y., Kizuki N., Wada T., Yazaki Y., Asano T., Oka Y., Kizuki N., Wada T., Yazaki Y., Asano T., Oka Y., Wada T., Yazaki Y., Asano T., Oka Y., Yazaki Y., Asano T., Oka Y., Asano T., Oka Y., Oka Y. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- Jones C.D., Begun D.J., Begun D.J. Parallel evolution of chimeric fusion genes. Proc. Natl. Acad. Sci. 2005;102:11373–11378. doi: 10.1073/pnas.0503528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Kleene K.C., Mulligan E., Steiger D., Donohue K., Mastrangelo M.A., Mulligan E., Steiger D., Donohue K., Mastrangelo M.A., Steiger D., Donohue K., Mastrangelo M.A., Donohue K., Mastrangelo M.A., Mastrangelo M.A. The mouse gene encoding the testis-specific isoform of Poly(A) binding protein (Pabp2) is an expressed retroposon: Intimations that gene expression in spermatogenic cells facilitates the creation of new genes. J. Mol. Evol. 1998;47:275–281. doi: 10.1007/pl00006385. [DOI] [PubMed] [Google Scholar]
- Knighton D.R., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M., Xuong N.H., Taylor S.S., Sowadski J.M., Taylor S.S., Sowadski J.M., Sowadski J.M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Korneev S.A., Park J.H., O’Shea M., Park J.H., O’Shea M., O’Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kunz J., Wilson M.P., Kisseleva M., Hurley J.H., Majerus P.W., Anderson R.A., Wilson M.P., Kisseleva M., Hurley J.H., Majerus P.W., Anderson R.A., Kisseleva M., Hurley J.H., Majerus P.W., Anderson R.A., Hurley J.H., Majerus P.W., Anderson R.A., Majerus P.W., Anderson R.A., Anderson R.A. The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol. Cell. 2000;5:1–11. doi: 10.1016/s1097-2765(00)80398-6. [DOI] [PubMed] [Google Scholar]
- Lam Y.A., Lawson T.G., Velayutham M., Zweier J.L., Pickart C.M., Lawson T.G., Velayutham M., Zweier J.L., Pickart C.M., Velayutham M., Zweier J.L., Pickart C.M., Zweier J.L., Pickart C.M., Pickart C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- Lasko P., Cho P., Poulin F., Sonenberg N., Cho P., Poulin F., Sonenberg N., Poulin F., Sonenberg N., Sonenberg N. Contrasting mechanisms of regulating translation of specific Drosophila germline mRNAs at the level of 5′-cap structure binding. Biochem. Soc. Trans. 2005;33:1544–1546. doi: 10.1042/BST0331544. [DOI] [PubMed] [Google Scholar]
- Lee J.T. Molecular biology: Complicity of gene and pseudogene. Nature. 2003;423:26–28. doi: 10.1038/423026a. [DOI] [PubMed] [Google Scholar]
- Ling K., Doughman R.L., Firestone A.J., Bunce M.W., Anderson R.A., Doughman R.L., Firestone A.J., Bunce M.W., Anderson R.A., Firestone A.J., Bunce M.W., Anderson R.A., Bunce M.W., Anderson R.A., Anderson R.A. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- Long M., Betran E., Thornton K., Wang W., Betran E., Thornton K., Wang W., Thornton K., Wang W., Wang W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Marques A.C., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H., Vinckenbosch N., Reymond A., Kaessmann H., Reymond A., Kaessmann H., Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighell A.J., Smith N.R., Robinson P.A., Markham A.F., Smith N.R., Robinson P.A., Markham A.F., Robinson P.A., Markham A.F., Markham A.F. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hillier L.W., Eichler E.E., Zody M.C., Jaffe D.B., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Hillier L.W., Eichler E.E., Zody M.C., Jaffe D.B., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Eichler E.E., Zody M.C., Jaffe D.B., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Zody M.C., Jaffe D.B., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Jaffe D.B., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., Hellmann I., Lindblad-Toh K., Altheide T.K., Lindblad-Toh K., Altheide T.K., Altheide T.K., et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Miller S.L., Malotky E., O’Bryan J.P., Malotky E., O’Bryan J.P., O’Bryan J.P. Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J. Biol. Chem. 2004;279:33528–33537. doi: 10.1074/jbc.M313097200. [DOI] [PubMed] [Google Scholar]
- Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A., Souidi M., Cuvellier S., Harel-Bellan A., Cuvellier S., Harel-Bellan A., Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer; Berlin: 1970. [Google Scholar]
- Ohshima K., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N., Yada T., Gojobori T., Sakaki Y., Okada N., Gojobori T., Sakaki Y., Okada N., Sakaki Y., Okada N., Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipow V., Tassan J.P., Nigg E.A., Schibler U., Tassan J.P., Nigg E.A., Schibler U., Nigg E.A., Schibler U., Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Kazazian H.H., Kazazian H.H. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Oude Weernink P.A., Schmidt M., Jakobs K.H., Schmidt M., Jakobs K.H., Jakobs K.H. Regulation and cellular roles of phosphoinositide 5-kinases. Eur. J. Pharmacol. 2004;500:87–99. doi: 10.1016/j.ejphar.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Stephen S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Stephen S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Lenhard B., Hayashizaki Y., Mattick J.S., Hayashizaki Y., Mattick J.S., Mattick J.S. RNAdb—A comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Castelo R., Thomson T.M., Antonarakis S.E., Guigo R., Thomson T.M., Antonarakis S.E., Guigo R., Antonarakis S.E., Guigo R., Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski J., Beal R., Hoffman L., Wilkinson K.D., Cohen R.E., Pickart C.M., Beal R., Hoffman L., Wilkinson K.D., Cohen R.E., Pickart C.M., Hoffman L., Wilkinson K.D., Cohen R.E., Pickart C.M., Wilkinson K.D., Cohen R.E., Pickart C.M., Cohen R.E., Pickart C.M., Pickart C.M. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- Polo S., Sigismund S., Faretta M., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P., Sigismund S., Faretta M., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P., Faretta M., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P., Bossi G., Chen H., De Camilli P., Di Fiore P.P., Chen H., De Camilli P., Di Fiore P.P., De Camilli P., Di Fiore P.P., Di Fiore P.P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Rao V.D., Misra S., Boronenkov I.V., Anderson R.A., Hurley J.H., Misra S., Boronenkov I.V., Anderson R.A., Hurley J.H., Boronenkov I.V., Anderson R.A., Hurley J.H., Anderson R.A., Hurley J.H., Hurley J.H. Structure of type IIbeta phosphatidylinositol phosphate kinase: A protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- Sakai H., Koyanagi K.O., Imanishi T., Itoh T., Gojobori T., Koyanagi K.O., Imanishi T., Itoh T., Gojobori T., Imanishi T., Itoh T., Gojobori T., Itoh T., Gojobori T., Gojobori T. Frequent emergence and functional resurrection of processed pseudogenes in the human and mouse genomes. Gene. 2006;389:196–203. doi: 10.1016/j.gene.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Samonte R.V., Eichler E.E., Eichler E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Schmidt E.E. Transcriptional promiscuity in testes. Curr. Biol. 1996;6:768–769. doi: 10.1016/s0960-9822(02)00589-4. [DOI] [PubMed] [Google Scholar]
- Schmidt E.E., Schibler U., Schibler U. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development. 1995;121:2373–2383. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- Schmidt E.E., Schibler U., Schibler U. Developmental testis-specific regulation of mRNA levels and mRNA translational efficiencies for TATA-binding protein mRNA isoforms. Dev. Biol. 1997;184:138–149. doi: 10.1006/dbio.1997.8514. [DOI] [PubMed] [Google Scholar]
- Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Shenmen C.M., Schuler G.D., Altschul S.F., Schuler G.D., Altschul S.F., Altschul S.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J., Vacquier V.D., Vacquier V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson K.A., Kang R.S., Stamenova S.D., Hicke L., Radhakrishnan I., Kang R.S., Stamenova S.D., Hicke L., Radhakrishnan I., Stamenova S.D., Hicke L., Radhakrishnan I., Hicke L., Radhakrishnan I., Radhakrishnan I. Solution structure of Vps27 UIM–ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak S.T., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Boguski M.S., Landsman D., Boeke J.D., Landsman D., Boeke J.D., Boeke J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052.1–0052.18. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabesinger-Ruef N., Jermann T., Zankel T., Durrant B., Frank G., Benner S.A., Jermann T., Zankel T., Durrant B., Frank G., Benner S.A., Zankel T., Durrant B., Frank G., Benner S.A., Durrant B., Frank G., Benner S.A., Frank G., Benner S.A., Benner S.A. Pseudogenes in ribonuclease evolution: A source of new biomacromolecular function? FEBS Lett. 1996;382:319–322. doi: 10.1016/0014-5793(96)00191-3. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N., Dupanloup I., Kaessmann H., Dupanloup I., Kaessmann H., Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Grus W.E., Zhang J., Grus W.E., Zhang J., Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Kazazian H.H., Boeke J.D., Moran J.V., Boeke J.D., Moran J.V., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G., Ha I., Ruvkun G., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wyckoff G.J., Wang W., Wu C.-I., Wang W., Wu C.-I., Wu C.-I. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Xue Y., Daly A., Yngvadottir B., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Daly A., Yngvadottir B., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Yngvadottir B., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Sabeti P., Chen Y., Stalker J., Huckle E., Chen Y., Stalker J., Huckle E., Stalker J., Huckle E., Huckle E., et al. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am. J. Hum. Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Kumar S., Nei M., Kumar S., Nei M., Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S., Ellsworth D.L., Li W.H., Ellsworth D.L., Li W.H., Li W.H. Slow molecular clocks in Old World monkeys, apes, and humans. Mol. Biol. Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
- Young P., Deveraux Q., Beal R.E., Pickart C.M., Rechsteiner M., Ustrell V., Pickart C., Deveraux Q., Beal R.E., Pickart C.M., Rechsteiner M., Ustrell V., Pickart C., Beal R.E., Pickart C.M., Rechsteiner M., Ustrell V., Pickart C., Pickart C.M., Rechsteiner M., Ustrell V., Pickart C., Rechsteiner M., Ustrell V., Pickart C., Ustrell V., Pickart C., Pickart C. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a: A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kumar S., Nei M., Kumar S., Nei M., Nei M. Small-sample tests of episodic adaptive evolution: A case study of primate lysozymes. Mol. Biol. Evol. 1997a;14:1335–1338. doi: 10.1093/oxfordjournals.molbev.a025743. [DOI] [PubMed] [Google Scholar]
- Zhang X., Loijens J.C., Boronenkov I.V., Parker G.J., Norris F.A., Chen J., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Loijens J.C., Boronenkov I.V., Parker G.J., Norris F.A., Chen J., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Boronenkov I.V., Parker G.J., Norris F.A., Chen J., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Parker G.J., Norris F.A., Chen J., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Norris F.A., Chen J., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Chen J., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Thum O., Prestwich G.D., Majerus P.W., Anderson R.A., Prestwich G.D., Majerus P.W., Anderson R.A., Majerus P.W., Anderson R.A., Anderson R.A. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J. Biol. Chem. 1997b;272:17756–17761. doi: 10.1074/jbc.272.28.17756. [DOI] [PubMed] [Google Scholar]
- Zhang J., Rosenberg H.F., Nei M., Rosenberg H.F., Nei M., Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]