Abstract
Vertebrate and invertebrate eye development require the activity of several evolutionarily conserved genes. Among these the Pax-6 genes play a major role in the genetic control of eye development. Mutations in Pax-6 genes affect eye development in humans, mice, and Drosophila, and misexpression of Pax-6 genes in Drosophila can induce ectopic eyes. Here we report the identification of a paired-like homeobox gene, DRx, which is also conserved from flies to vertebrates. Highly conserved domains in the Drosophila protein are the octapeptide, the identical homeodomain, the carboxyl-terminal OAR domain, and a newly identified Rx domain. DRx is expressed in the embryo in the procephalic region and in the clypeolabrum from stage 8 on and later in the brain and the central nervous system. Compared with eyeless, the DRx expression in the embryo starts earlier, similar to the pattern in vertebrates, where Rx expression precedes Pax-6 expression. Because the vertebrate Rx genes have a function during brain and eye development, it was proposed that DRx has a similar function. The DRx expression pattern argues for a conserved function at least during brain development, but we could not detect any expression in the embryonic eye primordia or in the larval eye imaginal discs. Therefore DRx could be considered as a homolog of vertebrate Rx genes. The Rx genes might be involved in brain patterning processes and specify eye fields in different phyla.
Keywords: gene isolation, Rx domain, brain development
The development of the eye is a process requiring precise patterning and cell fate decisions. This is most obvious in the Drosophila compound eye with its hexagonal array of approximately 800 ommatidia (1). Already in the embryo the imaginal primordia for the eye are set aside and proliferate during larval stages to form the eye–antennal imaginal discs. These discs develop during metamorphosis into adult eye structures and parts of the head. During early stages of eye development a large number of different transcription factors contribute to the pattern formation processes (2). Many of them belong to the class of homeobox genes and Pax genes and are highly conserved during evolution.
The most striking example is the functional conservation of the Pax-6 genes during eye development. Pax-6 genes are characterized by a 128-amino acid paired domain and a second DNA-binding domain, a homeodomain. All Pax-6 genes identified so far seem to have a function during eye development. This is demonstrated by the Small eye mutation in mice (3–5), the Aniridia mutation in humans (6), and the eyeless mutation in Drosophila (7). The hypothesis that eyeless is a master control gene for eye development in Drosophila was put forward on the basis of experiments showing that targeted expression of the eyeless gene in different imaginal discs can induce the formation of ectopic eyes on legs, wings, and antennae (8). In addition, Pax-6 genes from mouse, ascidians, and squid can fulfil the same function and induce ectopic eyes in Drosophila. (8–10).
In addition to Pax-6 genes, other genes expressed during early eye development are conserved between flies and vertebrates, such as the homeobox gene sine oculis (11, 12) and the nuclear gene eyes absent (13) in Drosophila. These genes are downstream of eyeless, suggesting that a large part of the genetic cascade regulating eye development has been conserved (14). Recently another vertebrate homeobox gene, Rx (15, 16) or rax (17), was identified. It belongs to the class of paired-like homeobox genes and is expressed in the forebrain and in the developing retina. Misexpression experiments with the Xenopus Rx gene result in the production of ectopic retinal tissue in the frog. Even more informative are mouse embryos carrying a null allele of the Rx gene: these animals do not form optic cups and as a consequence do not develop eyes (16). This phenotype is similar to Small eye and argues for a important role of the Rx genes in the establishment and maintenance of the retinal fate.
Here we describe the isolation of an Rx gene from Drosophila, referred to as DRx. The homeodomains of the Drosophila and the Xenopus Rx genes are identical, suggesting that DRx is a homolog of the vertebrate Rx genes. The Drosophila gene is expressed during early embryonic development in the procephalic region and the clypeolabrum and later in the brain and the central nervous system. The sequence conservation and expression pattern of DRx suggest an important role of the gene during brain development in Drosophila. However, no expression has been detected in the eye primordia of the embryo or in larval eye imaginal discs.
MATERIALS AND METHODS
General Methods.
Isolation of DNA from λ phages and plasmids, restriction endonuclease digestions, gel electrophoresis of DNA, labeling of DNA, and Southern blot analysis were performed as described by Sambrook et al. (18). The phage λ W60 was isolated from a genomic Drosophila Canton-S library prepared in the EMBL4 vector (kindly provided by V. Pirrotta, Univ. of Geneva, Switzerland), the other genomic phages were isolated from a genomic ey2 library in the λ Fix vector (7). Genomic DNA fragments isolated from phages were subcloned in Bluescript vectors (Stratagene).
Isolation of cDNAs.
Embryonic and larval oligo(dT)-primed cDNA was synthesized by using the Marathon cDNA amplification kit (CLONTECH). Starting materials was poly(A)+ RNA from the Drosophila strain Canton-S (CLONTECH). The DRx cDNAs were amplified by PCR using gene-specific primers. Additional 5′ and 3′ rapid amplification of cDNA ends (RACE) reactions were performed to obtain longer cDNA clones. All cDNA clones isolated by PCR were subcloned in the pCR2.1 vector (Invitrogen) by means of AT cloning.
DNA Sequencing and Sequence Analysis.
DNA was sequenced by the dideoxynucleotide procedure of Sanger et al. (19). Sequencing was done on both strands of the DNA with the Sequenase Version 2.0 DNA sequencing kit from United States Biochemical. Overlapping deletions were generated by using the exonuclease III–S1 method as described by the supplier (Pharmacia). In addition, gene-specific primers deduced from previously determined sequences were used. Sequences were analyzed by using the HUSAR/GCG sequence analysis software package from the University of Heidelberg.
In Situ Hybridization.
In situ hybridization to whole mount embryos was performed as described by Tautz and Pfeifle (20) with modifications (21). In the labeling reaction, a random primer concentration of 5 mg/ml was used and the reaction was incubated overnight at 14°C, then 2 units of the Klenow fragment of DNA polymerase were added, and the reaction was allowed to continue for 4 h at room temperature. The anti-digoxigenin antibody was preabsorbed with a large volume of fixed embryos in a 1:100 dilution overnight at 4°C.
RESULTS
Identification of the W60 Locus.
In an attempt to analyze the 5′ region of the Orthopedia gene (ref. 22; U.W., U. Kloter, and W.J.G., unpublished results) in section 57B of the second chromosome, we isolated additional genomic phages and performed in situ hybridization experiments to determine the presence of nearby transcription units. Using two central EcoRI fragments of 7.5 and 3.5 kb from phage λW60 (Fig. 1) located 11 kb upstream of the Orthopedia gene revealed identical expression patterns in Drosophila embryos. The expression patterns were distinct from those of Orthopedia, indicating that there was a neighboring gene, which we named W60. In contrast to the EcoRI fragments mentioned above, the two adjacent EcoRI fragments of λW60 were negative in these experiments. W60 signals were detected in the procephalic region from stage 8 on and later in the brain, roughly in a region from which the primordia of the eye–antennal discs originate. This opened the possibility that W60 might be involved in eye development, possibly as a regulator of eyeless.
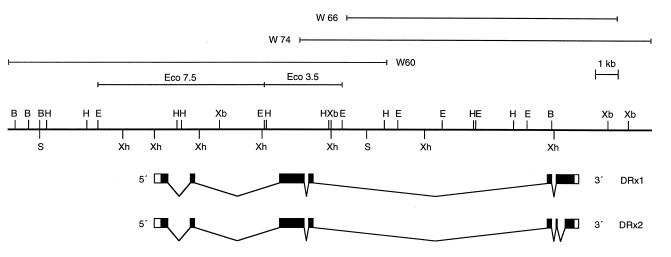
Figure 1.
Molecular organization of the DRx locus. A restriction map of the cloned region from the DRx locus in section 57B of the second chromosome is shown. The extent of the isolated phages and the location of the two EcoRI fragments used to identify the gene are diagrammed above the restriction map. The seven exons of the DRx transcription unit and the two different cDNA forms are shown below the restriction map. Noncoding regions are indicated by white boxes, coding regions by black boxes. Restriction endonuclease sites: B, BamHI; E, EcoRI; H, HindIII; S, SalI; Xb, XbaI; Xh, XhoI.
Preliminary sequence analysis of the 3.5-kb genomic EcoRI fragment identified a longer ORF including an M or opa repeat (23, 24), characteristic for developmentally regulated genes and suggested that the sequence might derive from an exon of the W60 gene.
Isolation of W60 cDNAs by PCR.
Because the isolation of W60 cDNAs from a 3- to 12-h embryonic cDNA library with the 3.5-kb EcoRI fragment failed, a PCR approach was pursued. First, oligo(dT)-primed cDNA pools from embryonic and third instar larvae mRNA were generated with the Marathon cDNA amplification kit. Then primers were designed within the previously identified ORF and a 600-bp fragment from the embryonic cDNA pool was PCR amplified. With the information that the 600-bp region is indeed part of a W60 cDNA, we performed 5′ RACE and 3′ RACE reactions to isolate longer cDNAs. Several cDNA clones overlapping in the 600-bp region were identified and further characterized. Two cDNA clones from the 3′ RACE experiments differed in length by 0.4 kb, presumably representing alternatively spliced cDNA forms. The different types of cDNA clones were sequenced on both strands, and the resulting sequences were combined. To minimize errors in the cDNA sequence because of the multiple PCR amplifications, we designed gene-specific primers and determined the genomic sequence in parallel. This allowed us to analyze the exon–intron boundaries exactly (Fig. 1).
Sequence Conservations and Gene Structure of DRx.
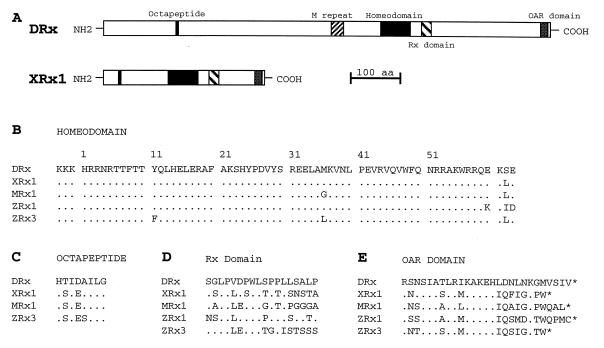
Two alternatively spliced transcripts of W60 were detected. The combined cDNA sequence of the longer splice form DRx 1 has a total length of 3.2 kb. The complete nucleotide sequence and the deduced amino acid sequence are shown in Fig. 2. The ORF starts at position 269, terminates at position 2974, and encodes a protein of 902 amino acids with a predicted molecular weight of 95,757 and an isoelectric point (pI) of 6.81. The sequence preceding the ATG fits only poorly to the consensus translation initiation sequence (25). The termination codon TAG found at position 903 is followed by a putative polyadenylation signal (AATAAA) and a 26-nucleotide poly(A) tract. In the carboxyl-terminal part of the deduced protein sequence, a paired-like homeodomain was identified which is 100% identical to the homeodomain of the Rx gene from Xenopus laevis, 98% identical to the Rx gene from mouse, and 97% identical to the Rx gene from zebrafish (Fig. 3B). Therefore, the gene we isolated is a Drosophila homolog of the vertebrate Rx genes, presumably the same gene as that mentioned by Mathers et al. (16). The same homeodomain sequence was also reported as bk50 in a screen for homeodomain proteins binding to a common Engrailed binding site (26). In contrast to paired-type homeodomains found in Pax genes that share a characteristic serine residue at position 50, paired-like homeodomains have a glutamine at this position. According to Mathers et al., we renamed W60 as DRx. Like the vertebrate genes, DRx has an octapeptide sequence in the amino-terminal part (Fig. 3C) and an OAR domain at the carboxyl terminus (Fig. 3E). Additional sequence conservations are found at the amino and carboxyl termini of the homeodomain and in a region between the homeodomain and the OAR domain. We designate this latter region as the Rx domain (Fig. 3D). However, the total DRx protein is more than twice as long as the corresponding proteins in vertebrates (Fig. 3A) and very rich in alanine (8.4%), glycine (9.9%), serine (10.4%), threonine (4.9%), and proline (10.0%) (% values are molar). Glutamine (7.8%) is mainly present in the form of an M or opa repeat.
Figure 2.
Nucleotide and deduced amino acid sequence of the DRx homeobox gene. Nucleotides and amino acids of the cDNA DRx1 are numbered on the left side. The homeodomain, the octapeptide, and the OAR domain are boxed, and the Rx domain is underlined by a thick bar. The splice sites are indicated by arrowheads. The alternatively used splice site is indicated by an open arrowhead and the putative polyadenylation signal is underlined.
Figure 3.
Comparison of amino acid sequences of Drosophila and vertebrate Rx genes. (A) Schematic presentation of putative domains in the DRx and Xenopus Rx1 proteins. (B–E) Amino acid comparisons are shown between the homeodomain (B), octapeptide (C), Rx domain (D), and OAR domain (E) of DRx and vertebrate Rx genes from Xenopus laevis (XRx1), mouse (MRx1), and zebrafish (ZRx1 and ZRx3).
The genomic organization of DRx was analyzed by Southern blots with the cDNA as a probe to identify the various exon positions. The exon–intron boundaries were then determined by sequencing. For the 3′ part of the gene additional genomic phages were isolated, because this region was not represented in the initial phage λW60. The transcription unit of the DRx gene consists of seven exons spanning a genomic region of at least 18 kb (Fig. 1). The transcription initiation site remains to be determined. The homeodomain comprises two exons with an intron at position 44, a very common splice position for homeodomain proteins. The intron size between the two homeodomain exons is about 9 kb. Alternative splicing in the 3′ part of the gene results in a putative protein form that is 130 amino acids shorter (Fig. 2). Using gene-specific primers, we PCR amplified the regions between the different exons from embryonic as well as larval cDNA, but we could not find splice forms other than the ones already described.
DRx Expression.
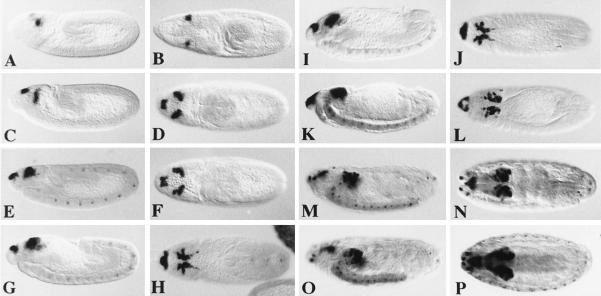
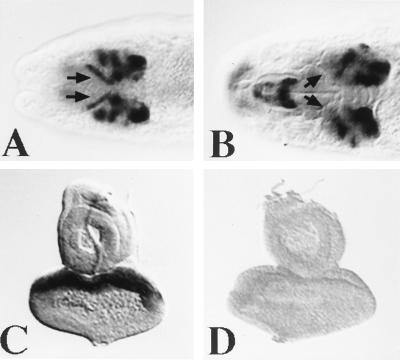
Drosophila embryos were examined for DRx expression by whole mount in situ hybridization. During the early stages of embryonic development, the syncitial and cellular blastoderm stage, no signals were detected. With the onset of gastrulation and germ-band extension at early stage 8 the first expression is seen in two dorsolateral spots in the procephalic region (Fig. 4B). At the end of stage 8 an additional signal is visible in a dorsal region (Fig. 4A) that later on will gives rise to the clypeolabrum. The DRx expression becomes more pronounced at stage 9, when the dorsolateral spots are increasing in size (Fig. 4 C and D). During extended germ-band stage, when the clypeolabrum becomes a distinct structure of the procephalon, cells expressing DRx are moving closer to the midline, and an additional expression in cells of the central nervous system is detected (Fig. 4 E and F). During stage 12, when the germ-band retracts and metamerization is clearly visible, the optic lobe starts to invaginate. The cells expressing DRx in the procephalon move even closer together, and the expression pattern splits at this stage and the clypeolabrum expression extends more laterally (Fig. 4 G–J). Because of the morphogenetic movements during head involution, DRx-positive cells in the clypeolabrum move inside the embryo (Fig. 4 K–N). At this stage expression is observed in the antennomaxillary complex. Staining in the medial edges of the two brain lobes, in the clypeolabrum, and in the antennomaxillary complex is then seen until the end of embryogenesis (Fig.4 O and P). DRx expression in the brain is similar to that of eyeless, but the expression patterns are not completely overlapping. However, in contrast to eyeless, no staining of the eye disc primordia per se is observed, when they become distinct structures during stage 16 (Fig. 5 A and B), nor is DRx expressed in imaginal discs of third-instar larvae (Fig. 5 C and D).
Figure 4.
Spatial distribution of DRx transcripts during Drosophila embryogenesis. Stages were determined according to Campos-Ortega and Hartenstein (42). In all views anterior is to the left (×60). (A and B) Lateral and dorsal views, respectively, of a stage 8 embryo. (C and D) Lateral and dorsal views of a stage 9 embryo. (E and F) Lateral and dorsal views of a stage 11 embryo. (G and H) Lateral and dorsal views of a stage 12 embryo. (I and J) Lateral and dorsal views of a stage 13 embryo. (K and L) Lateral and dorsal views of a stage 14 embryo. (M and N) Lateral and dorsal views of a stage 15 embryo. (O and P) Lateral and dorsal views of a stage 17 embryo.
Figure 5.
Comparison of DRx and eyeless expression in embryos and imaginal discs. (A) eyeless expression in a stage 16 embryo is visible in the brain and the embryonic eye primordia (arrows). (B) DRx expression is seen in the brain, but not in the embryonic eye primordia (arrows). (C) eyeless is expressed in the anterior part of the eye disc. (D) No DRx expression is visible in eye–antennal imaginal discs. [A and B, ×180; C and D, ×70.]
DISCUSSION
DRx Is a Drosophila Homolog of Vertebrate Rx Genes.
In this paper we describe the identification of a Drosophila homolog of vertebrate Rx genes. These genes encode highly conserved paired-like homeodomain proteins that are characterized by an octapeptide and the OAR domain, which might represent a transactivation domain (17). The Drosophila Rx gene encodes a protein that is more than twice as large as the vertebrate proteins, a feature that is also found in the Drosophila homologs of other vertebrate genes such as Pax-6/eyeless (7) or Pax-2/sparkling (27). Besides the already known highly conserved domains we identified another conserved region in the different proteins, the Rx domain. Rx genes isolated to date from mice and Drosophila seem to be single-copy genes, whereas Xenopus has two genes and the zebrafish, three (16).
The vertebrate Rx genes show an astonishing conservation of their expression patterns and probably of their function too. In mice the Rx gene is expressed at E7.5 (embryonic day 7.5) in the cephalic neural fold, at E8.5 in the forebrain region and the optic placodes, whereas from E10.5 on it is restricted to the developing eye and in later stages to the neuroretina. A similar expression pattern is seen in Xenopus in the forebrain, the optic cups, and later also in the neuroretina. In the zebrafish different functions seem to be fulfilled by the three different genes, because only two genes (ZRx1 and ZRx2) are expressed in the developing retina, and one gene (ZRx3) is expressed in the forebrain. By sequence comparison we cannot definitively say to which of the three genes DRx is most closely related.
Most informative for determining the function of Rx genes are loss of function mutants in mice and ectopic expression experiments in Xenopus. MRx null mutant mice have no visible eye structures, because the optic cups do not form and anterior brain structures are lacking. In contrast, ectopic expression of XRx in Xenopus embryos demonstrates the capacity of XRx to induce ectopic retinal pigment epithelium in the proximity of the anterior neural tube (16). The expression pattern of the DRx gene argues for similar function of this gene. As in vertebrates, DRx is expressed already in the early embryo in the procephalic region from which the eye primordia may originate.
Expression of DRx During Embryonic Brain Development.
Judging from the pattern of gene expression, a major function of DRx might be in brain development as found for the vertebrate homologs. In contrast to the ventral nerve cord, brain development in Drosophila is not so well understood. Because of the absence of morphological landmarks it is more difficult to assign individual brain neuromeres to specific segments and to determine which genes products control different processes of brain development. The establishment of a fate map for brain neuroblasts (28) and the analysis of phenotypes from genes expressed in the brain indicated that head gap genes such as buttonhead (29), empty spiracles (21), orthodenticle (30), and tailless (31) not only function in patterning processes of the epidermis but also play an important role in regionalizing the brain (32, 33). All these genes have overlapping expression domains in broad areas early during development, but their expression domains become much more distinct during germ-band extension. At this stage expression is seen in overlapping sets of brain neuroblasts, and mutations in head gap genes lead to specific defects in these groups of cells (32, 33). Because all of the head gap genes are expressed much earlier than DRx, they might be regulators of DRx during brain development. tailless is expressed in all protocerebral neuroblasts, and these neuroblasts are absent in tailless loss-of-function mutations (34). The originally uniform expression of tailless in the procephalic region is not uniform anymore during gastrulation and has centrally located high level (HL) and surrounding low level (LL) expression domains (33). This expression would argue for a subdivision of the brain or a specification of defined structures by different levels of tailless protein because the different expression domains are also regulated by different cis-regulatory elements in the tailless promoter (35). When one compares the expression of DRx at stage 9/10 with the expression of tailless, DRx is found only in a subset of the tailless-expressing cells. These cells seems to be in the HL tailless domain, an area of the brain where the first neuroblasts delaminate from the central protocerebral neuroectoderm. The head gap gene orthodenticle also is expressed in most protocerebral neuroblasts, and mutations in the orthodenticle gene also result in the absence of most structures of the protocerebrum (32, 33). Because DRx has a much more restricted expression domain compared with orthodenticle and tailless, it might act as a transcription factor to activate specific target genes in a defined subdomain of the brain.
Functional Conservation of Rx Genes During Evolution.
It is known that a variety of genes, most of them encoding transcription factors, are necessary for eye development in Drosophila and vertebrates (2). Among these Pax-6/eyeless seems to be extraordinary because of its capacity to induce ectopic eyes. In Drosophila it was shown that the homeodomain protein sine oculis and the nuclear protein eyes absent function downstream of eyeless (14, 36) during eye development, whereas in the embryo sine oculis acts in parallel to eyeless (14). Both genes are necessary for ectopic and normal eye development and are independent targets of eyeless (14). In vertebrates there are three eyes absent homologs (37) and three sine oculis homologs (38). All three vertebrate eyes absent genes are expressed in the developing eye, and their expression patterns partially overlap with Pax-6 and depend on Pax-6 expression in the lens and the nasal placode. From the three sine oculis homologs only Six3 is expressed during eye development (39), but an epistatic relationship between Pax-6 and Six3 cannot be determined because the optic vesicle, where Six3 is expressed in normal embryos, degenerates in Small eye mutant mice. Nevertheless, all the available comparative data argue for a conserved morphogenetic pathway during eye development involving Pax-6 and its target genes Eya and Six3.
How does the expression of Rx genes correlate with their putative function in the genetic cascade mentioned above? Compared with Pax-6, the expression of the mouse Rx gene in the forebrain is much more restricted to cells that contribute to the optic vesicles. Other vertebrate genes known to be expressed in the anterior neural plate and neural fold, such as otx2 (40), the vertebrate homolog of orthodenticle, and Six3, have larger expression domains as compared with Rx and are involved in additional patterning processes. Because Rx is expressed earlier than Pax-6, it was proposed that Rx might directly or indirectly regulate Pax-6 during optic vesicle formation (17).
DRx expression starts also earlier than eyeless expression in Drosophila embryos, and the expression domains of both genes partially overlap in the head region. Because of the early expression pattern of DRx at stage 10, Mathers et al. (16) proposed that this expression would be in the eye disc primordia and implied that the function of Rx genes during eye development might be conserved from vertebrates to invertebrates. However, we have recently identified a second Pax-6 gene in Drosophila called twin-of-eyeless (T. Czerny, G. Halder, P. Callaerts, U. Kloter, W.J.G., and M. Busslinger, unpublished results) that is expressed at the blastoderm stage, earlier than DRx, which makes it unlikely that DRx acts upstream of Pax-6. Furthermore, it is not clear from which cells the eye disc primordia originate, because of their composite origin and the absence of specific markers that would allow one to trace them back to their origin. The earliest known markers are expressed at stage 14, and even then the assignment is tentative (41). At stage 16, when the embryonic eye disc primordia are morphologically visible and show strong eyeless expression, DRx expression is not detectable in these structures (Fig. 5), but the gene is still expressed in the brain. We cannot exclude the possibility of an earlier expression in the eye precursor cells.
Alternatively, only the DRx function in the brain might be evolutionarily conserved. Rx mutant mouse embryos lack the forebrain in severe cases, and the midbrain is also affected. DRx, on the other hand, is expressed in the protocerebrum, the most anterior portion of the Drosophila brain. This expression is in a comparable region of the brain, but the determination of DRx function awaits the analysis of DRx mutants. Of course we cannot rule out the possibility of a second Rx homolog in Drosophila that, as in zebrafish, might take over the eye specific function.
The different modes of eye development in vertebrates and Drosophila also could account for differences between Rx genes and DRx expression. However, the fact that DRx expression is confined to the brain suggests that its main function is in brain rather than eye development.
Acknowledgments
We thank A. Preiss for constant support and critical reading of the manuscript, B. Johannes and C. Alexief-Damianof for help in preparing the figures, U. Kloter for confirming the in situ hybridization experiments to imaginal discs, and W. Staiber for the excellent photographic work. We acknowledge F. Hirth, R. Leemans, and H. Reichert for helpful discussions. This work was initiated at the Biozentrum in Basel and was supported by the Kantons Basel, the Swiss National Science Foundation, and a grant from the Deutsche Forschungsgemeinschaft to U.W. (WA 556/4-1).
ABBREVIATION
- RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ223300).
References
- 1.Wolff T, Ready D F. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 2.Freund C, Horsford D J, McInnis R R. Hum Mol Genet. 1996;5:1471–1488. doi: 10.1093/hmg/5.supplement_1.1471. [DOI] [PubMed] [Google Scholar]
- 3.Hogan, B., Hirst, E. M. A., Horsburgh, G. & Hetherington, C. M. (1988) Development (Cambridge, U.K.) 103, Suppl., 115–119. [DOI] [PubMed]
- 4.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 5.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 6.Ton C C, Hirvonen H, Miwa H, Wei M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, Royer-Pokora B, Collins F, Swaroop A, Strong L C, Saunders G F. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 7.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 8.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 9.Tomarev S I, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Proc Natl Acad Sci. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glardon S, Callaerts P, Halder G, Gehring W J. Development (Cambridge, UK) 1997;124:817–825. doi: 10.1242/dev.124.4.817. [DOI] [PubMed] [Google Scholar]
- 11.Cheyette B N R, Green P J, Martin K, Garren H, Hartenstein V, Zipursky S L. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 12.Serikaku M A, O’Tousa J E. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonini N M, Leiserson W M, Benzer S. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 14.Halder, G., Callaerts, P., Flister, S., Walldorf, U., Kloter, U. & Gehring, W. J. (1998), Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 15.Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. Mech Dev. 1997;61:187–198. doi: 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- 16.Mathers P H, Grinberg A, Mahon K A, Jamrich M. Nature (London) 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa T, Kozak C A, Cepko C L. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 21.Walldorf U, Gehring W J. EMBO J. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simeone A, D’Apice M R, Nigro V, Casanova J, Graziani F, Acampora D, Avantaggiato V. Neuron. 1994;13:83–101. doi: 10.1016/0896-6273(94)90461-8. [DOI] [PubMed] [Google Scholar]
- 23.McGinnis W, Levine M, Hafen E, Kuroiwa A, Gehring W J. Nature (London) 1984;308:403–408. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- 24.Wharton K A, Yedvobnick B, Finnerty V G, Artavanis-Tsakonas S. Cell. 1985;40:55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 25.Cavener D R. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalionis B, O’Farrell P H. Mech Dev. 1993;43:57–70. doi: 10.1016/0925-4773(93)90023-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu W, Noll M. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younossi-Hartenstein A, Nassif C, Hartenstein V. J Comp Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Wimmer E, Jäckle H, Pfeifle C, Cohen S. Nature (London) 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 30.Finkelstein R, Smouse D, Capaci T M, Spradling A C, Perrimon N. Genes Dev. 1990;4:1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- 31.Pignoni F, Baldarelli R M, Steingrimsson E, Diaz R J, Patapoutian A, Merriam J R, Lengyel J A. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- 32.Hirth F, Therianos S, Loop T, Gehring W J, Reichert H, Furukubo-Tokunaga K. Neuron. 1995;15:769–778. doi: 10.1016/0896-6273(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 33.Younossi-Hartenstein A, Green P, Liaw G-J, Rudolph K, Lengyel J A, Hartenstein V. Dev Biol. 1997;182:270–283. doi: 10.1006/dbio.1996.8475. [DOI] [PubMed] [Google Scholar]
- 34.Strecker T R, Merriam J R, Lengyel J A. Development (Cambridge, UK) 1988;102:721–734. doi: 10.1242/dev.102.4.721. [DOI] [PubMed] [Google Scholar]
- 35.Rudolph K M, Liaw G-J, Daniel A, Green P, Courey A J, Hartenstein V, Lengyel J A. Development (Cambridge, UK) 1997;124:4297–4308. doi: 10.1242/dev.124.21.4297. [DOI] [PubMed] [Google Scholar]
- 36.Bonini N M, Bui Q T, Gray-Board G L, Warrick J M. Development (Cambridge, UK) 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 37.Xu P-X, Woo I, Her H, Beier D R, Maas R L. Development (Cambridge, UK) 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- 38.Oliver G, Wehr R, Jenkins N A, Copeland N G, Cheyette B N R, Hartenstein V, Zipursky S L, Gruss P. Development (Cambridge, UK) 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 39.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development (Cambridge, UK) 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 40.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice M R, Nigro V, Boncinelli E. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartenstein V, Jan Y N. Roux’s Arch Dev Biol. 1992;201:194–220. doi: 10.1007/BF00188752. [DOI] [PubMed] [Google Scholar]
- 42.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. 2nd Ed. Berlin: Springer; 1997. [Google Scholar]