Abstract
The unusual DNA base β-d-glucosyl-hydroxymethyluracil, called “J,” replaces ≈0.5–1% of Thy in DNA of African trypanosomes but has not been found in other organisms thus far. In Trypanosoma brucei, J is located predominantly in repetitive DNA, and its presence correlates with the silencing of telomeric genes. Using antibodies specific for J, we have developed sensitive assays to screen for J in a range of organisms and have found that J is not limited to trypanosomes that undergo antigenic variation but is conserved among Kinetoplastida. In all kinetoplastids tested, including the human pathogens Leishmania donovani and Trypanosoma cruzi, J was found to be abundantly present in the (GGGTTA)n telomere repeats. Outside Kinetoplastida, J was found only in Diplonema, a small phagotrophic marine flagellate, in which we also identified 5-MeCyt. Fractionation of Diplonema DNA showed that the two modifications are present in a common genome compartment, which suggests that they may have a similar function. Dinoflagellates appear to contain small amounts of modified bases that may be analogs of J. The evolutionary conservation of J in kinetoplastid protozoans suggests that it has a general function, repression of transcription or recombination, or a combination of both. T. brucei may have recruited J for the control of genes involved in antigenic variation.
In the nuclear DNA of Trypanosoma brucei, ≈0.5–1% of Thy is replaced by the modified base β-d-glucosyl-hydroxymethyluracil (β-gluc-HOMeUra) (1). This base that we call “J” was detected initially by 32P-nucleotide postlabeling combined with two-dimensional TLC (2D-TLC) (2), and we used this technique to show that approximately one-half of the cellular J is present in both strands of the telomeric (GGGTTA)n repeats (3). To map the location of J more precisely, we have generated antisera that immunoprecipitate J-containing duplex DNA and that detect this DNA with high sensitivity and specificity on dot blots (4). We have used these antisera to demonstrate that J is present in other repetitive DNA sequences but not in housekeeping genes or transcribed repeats (4). Moreover, we have shown that J is responsible for the blocked restriction sites that are present in silent telomeric variant surface glycoprotein (VSG) genes but not in actively transcribed VSG genes (4–6). This result has linked J to the transcriptional control of VSG genes.
Thus far, J has been detected only in African trypanosome species that undergo antigenic variation (2). The availability of antibodies acting against J has prompted us to reinvestigate whether J is also present in other organisms. With anti-J–DNA immunoblots, approximately one J per 107 bases can be detected, which is ≈1,000-fold more sensitive than 32P-postlabeling (4). We have used this assay in combination with anti-J immunoprecipitation and 32P-postlabeling to analyze a range of eukaryotic DNAs. Special attention was paid to the kinetoplastid flagellates and to dinoflagellates. The order Kinetoplastida encompasses both free-living and parasitic flagellated protozoans and includes T. brucei. Kinetoplastids represent one of the earliest lineages of mitochondria-containing eukaryotic cells (7) and are distinguishable from other protozoa by the presence of kinetoplast DNA, an unusual type of mitochondrial DNA near the base of the flagellum (8). Dinoflagellates are of special interest because some of them have replaced a high fraction of Thy with HOMeUra in their DNA (9–11). Because we have obtained indirect evidence for T. brucei that HOMeUra is a precursor of J (ref. 12 and unpublished results), these organisms should be candidates for having glucosylated HOMeUra (J).
MATERIALS AND METHODS
Cells and DNA Analysis.
DNA was derived from: T. brucei brucei (427); Trypanosoma congolense (WG81 and TSW13 bloodstream forms and WG81 procyclics); Trypanosoma vivax (Y58); Crithidia fasciculata (= C. luciliae); Leishmania donovani (HU3); Leishmania tarentolae (tarVIa); Trypanosoma cruzi (CL-Brener and Sylvio X10/6); Trypanoplasma borreli (Tt-JH); Drosophila melanogaster (whole wild-type fly); Sf9 (Spodoptera frugiperda cell line); Caenorhabditis elegans (whole worm, Bristol N2); Saccharomyces cerevisiae [M398 (matα, ura3–52, trpΔ1, his3Δ200, leu2Δ1, trkΔ1), BJ1991 (matα, leu2, ura3–52, trp-1, prb1, and pep4–3)]; Prorocentrum micans (Ehr); Crypthecodinium cohnii (Whd); Toxoplasma gondii (Pg4II tachyzoites); Plasmodium falciparum (mixed population); Entamoeba histolytica (HM-1); Diplonema (= Isonema papillatum ATCC 50162); Trichomonas vaginalis (ATCC 30001); Giardia lamblia (WBC5); Petunia corollas (V26 and IRc2); and Escherichia coli and calf thymus DNA, which were purchased from Sigma. Mammalian DNA samples enriched for telomeric repeats were prepared as described (13, 14). T. brucei bloodstream form and procyclic trypanosomes were grown as described (4). T. cruzi epimastigotes and L. donovani promastigotes were cultured axenically without feeder cells. T. cruzi bloodstream trypomastigotes were grown in monolayers of African green monkey kidney (Vero) cells and were isolated after they lysed the infected host cells. L. donovani amastigotes were grown in hamsters and were isolated from spleens and livers. 32P-nucleotide postlabeling combined with 2D-TLC was done as described (2). Briefly, DNA was digested to 3′-monophosphates, which were 5′-labeled (32pdNp) and subsequently 3′-dephosphorylated (32pdN). Chromatograms were scanned and nucleotide spots were quantitated with a PhosphorImager (FUJIX Bas 2000, Tokyo). The quantitations shown are based on one experiment, and quantitation of J was corrected for incomplete recovery by postlabeling. Postlabeling of synthesized standards has shown that the labeling efficiency of J is 50% (F.v.L. and P.B., unpublished results). Chemical deamination and elution of nucleotides from TLC sheets was done essentially as described (1, 15). Blot and hybridization procedures are described in ref. 3.
Anti-J–DNA Immunoblot.
DNA was blotted onto nitrocellulose, and the filters were baked for 2 hr at 80°C and blocked for 2 hr in TBST (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.02% Tween-20) with 5% milk powder. After three washes with TBST, the blots were incubated for 2 hr with antiserum 539αJ (4), diluted 1:10,000-fold in TBST with 2% milk powder, and then washed three times with TBST. Immunodetection was performed by using a horseradish peroxidase-conjugated sheep–anti-rabbit antibody (Netherlands Red Cross Blood Transfusion Service, The Netherlands) diluted 1:10,000-fold in 2% milk powder in TBST in combination with enhanced chemiluminescence (Amersham). All DNA samples were analyzed on Southern blots (200 ng of DNA) and dot blots (1 μg of DNA).
Anti-J Immunoprecipitation.
Two micrograms of sonicated DNA (0.5–3 kb) was added to 5 μl of antiserum 538αJ (4) in a final volume of 500 μl of IP buffer [TBST with 2 mM EDTA (TBSTE)/0.1 mg tRNA/ml/1 mg BSA/ml] and incubated for 2 hr at room temperature. Twenty microliters of ProtA beads (Repligen) were washed twice with TBSTE, preblocked for 30 min in 100 μl of IP buffer, and incubated for 1 hr with the IP reaction. The bead–antibody–DNA complexes were washed four times with TBSTE and finally proteinase-K-treated at 56°C to release the bound DNA, which was phenol-extracted and ethanol-precipitated with 20 μg of glycogen. For immunoprecipitation of 32P-labeled nucleotides from the kinase reaction in the postlabeling assay, 10 μl of ProtA beads was washed twice in PBS, resuspended in 200 μl of PBS with 0.5 mg of BSA/ml and incubated with 3 μl of 539αJ serum. Unbound antibodies were removed by three washes with PBS. The antibody–bead complexes were resuspended in 10 μl of PBS and added to 40 μl of a 5-fold-diluted nucleotide kinase reaction. Nucleotides in the supernatant (10 μl) were 3′-dephosphorylated and analyzed by 2D-TLC (2).
RESULTS
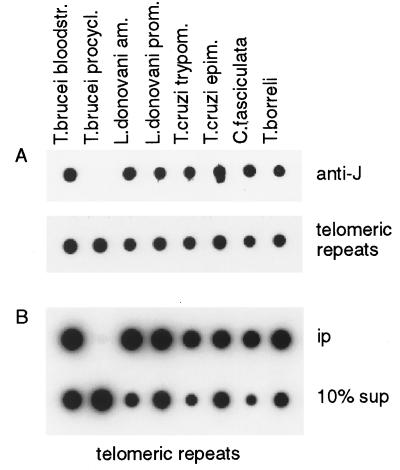
J-specific antisera can be used to detect low levels of J in DNA on dot or Southern blots (4). Using these antisera, we analyzed the two major subgroups within the Kinetoplastida: the trypanosomatids, which are obligate parasites, and the early diverging sister group bodonids/cryptobiids, which consists of both free-living and parasitic protists (8). Dot blots were made with DNA of the trypanosomatids T. brucei, T. cruzi, and Leishmania, which parasitize two hosts, C. fasciculata, which is parasitic only in insects and of the cryptobiid fish parasite T. borreli. Of the digenetic trypanosomatids, we analyzed both the mammalian and the insect stages because J had been found only in the bloodstream form of T. brucei (2). Genomic DNA of all of the kinetoplastid genera tested bound the J-specific antibody (Fig. 1A). Of interest, developmental regulation of the DNA modification was seen only for T. brucei (Fig. 1A) and for T. congolense, another African trypanosome analyzed (results not shown). The results of the immunoblots were confirmed by a 32P-postlabeling experiment (Table 1). The detection level of J with 32P-postlabeling is ≈0.02%. Unambiguous detection of J was facilitated by efficient enrichment for J by anti-J immunoprecipitation (αJ-IP) of sonicated DNA, followed by 32P-postlabeling (Table 1 and see below). Anti-J immunoprecipitation also resulted in enrichment for HOMeUra (Table 1). It is not known whether this HOMeUra is the unglucosylated precursor of J or a degradation product of it, but indirect evidence strongly suggests that HOMeUra may be an intermediate in the synthesis of J (F.v.L. and P.B., unpublished results and ref. 12).
Figure 1.
Conservation of J in telomeric repeats of Kinetoplastida. (A) Dot blot with 200 ng of DNA of each kinetoplastid sample indicated was incubated with anti-J antiserum (anti-J). Abbreviations of the organisms and the life cycle stages are explained in Table 1. Bound antibodies were detected with a second antibody conjugated to horseradish peroxidase and were visualized by enhanced chemiluminescence. After stripping the blot, DNA loading was checked by hybridization by using a (GGGTTA)5-telomeric repeat oligo as a common probe (telomeric repeats). J also was found in L. tarentolae, L. donovani chagasi, L. brasiliensis, L. mexicana, and T. vivax (data not shown). (B) An anti-J immunoprecipitation of sonicated DNA. Modified DNA bound by antibody (ip) and 10% of the supernatant (10% sup) were blotted and analyzed by hybridization with the telomeric repeat probe.
Table 1.
Identification of J in kinetoplastids by 32P-postlabeling combined with anti-J immunoprecipitation
| Species | Host/vector | Stage | % J
|
% HOMeUra
|
% IP*
|
||
|---|---|---|---|---|---|---|---|
| Total | αJ-IP | Total | αJ-IP | Tel. rep. | |||
| T. brucei | mammal | bloodstream form | 0.12 | 1.6 | 0.04 | 0.2 | 16 |
| insect | procyclic | 0.00 | 0.0 | 0.02 | 0.0 | 0.1 | |
| L. donovani | mammal | amastigote | 0.02 | 2.9 | 0.01 | 0.1 | 47 |
| insect | promastigote | 0.04 | 2.1 | 0.03 | 0.2 | 22 | |
| T. cruzi | mammal | trypomastigote | 0.04 | 2.7 | 0.05 | 0.3 | 53 |
| insect | epimastigote | 0.02 | 1.2 | 0.03 | 0.2 | 18 | |
| C. fasciculata | insect | — | 0.08 | 1.9 | 0.03 | 0.4 | 59 |
| T. borreli | fish | bloodstream | 0.04 | 2.1 | 0.02 | 0.2 | 32 |
IP, immunoprecipitation; Tel. rep., telomere repeat.
Percentage of DNA fragments immunoprecipitated from the input.
In T. brucei, J is found predominantly in sequence repeats, with approximately one-half in the telomeric repeats (3). By using anti-J immunoprecipitations, we tested whether J is abundant also in the conserved hexameric telomere repeats of the other organisms. DNA of all samples was sonicated to a similar size range (0.5–3 kb), and fragments bound by the J-specific antibodies were analyzed by dot blot hybridization. In all kinetoplastids, except for insect form (procyclic) T. brucei, telomeric repeats were bound efficiently by the antibodies (Fig. 1B; Table 1). Because the efficiency of immunoprecipitation is determined by the density of modification (4), these results show that, in all the kinetoplastids studied here, the telomeric repeats are modified densely. Our results (Table 1 and data not shown) suggest that, in the other organisms, the fraction of base J in telomeric DNA is higher than in T. brucei, and this fraction might be regulated developmentally in L. donovani and T. cruzi, but this result has not been verified by quantitative chemical analysis (3) of purified telomeres.
Having found that J is conserved in kinetoplastids, we set out to screen a wide range of eukaryotes for the presence of J. Total genomic DNA from various origins was analyzed on Southern blots (Fig. 2) and dot blots (not shown). DNA loading was checked by staining the gel with ethidium bromide (Fig. 2), and the identity of the DNA samples was confirmed by determining the nucleotide composition by using 32P-postlabeling (not shown). No J was detected in mammalian tissue, tumor, or cell line DNA. Some of the mammalian DNAs tested are shown in Fig. 2. The detection level of J with DNA immunoblots is approximately one J per 107 bases (with 1 μg of DNA) (4), which corresponds to eight J residues in a T. brucei genome of 8 ×107 bp and 600 J residues in a mammalian genome of 6 × 109 bp. Even with human DNA ≈100-fold–enriched for telomeric repeats (13, 14), no antibody binding was detected (Fig. 2). We did not expect to find J in mammals because cells of vertebrate and (most) invertebrate animals contain HOMeUra-glycosylase activity (16). This DNA repair enzyme cleaves the base HOMeUra from the ribose moiety after which the apyrimidinic site is repaired by other enzymes of the base excision repair pathway (17, 18). Because J (β-gluc-HOMeUra) probably is synthesized via HOMeUra, expression of HOMeUra glycosylase probably is incompatible with biosynthesis of J.
Figure 2.
Screening for J in DNA with a zoo blot. Southern blot of a 1% agarose gel with ≈200 ng of total DNA of each sample was incubated with antiserum 539αJ, and bound antibodies were indirectly detected by enhanced chemiluminescence (anti-J). Black lines indicate the position of the slots. Lanes: 1, bloodstream form T. brucei; 2, procyclic (insect form) T. brucei; 3–6, human blood, sperm, breast tumor, and ovarian tumor; 7–8, mouse testis and lung; 9, calf thymus; 10–11, human cell lines HeLa and G401; 12–14, human DNAs enriched ≈100-fold for telomeric repeats; 12, telomeric tracts from HeLa cells;13, matrix-attached DNA from HeLa cells;14, telomeric tracts from G401 cells;15, D. melanogaster (also contained RNA); 16, Sf9 cells; 17, C. elegans; 18–19, S. cerevisiae strains M398 and BJ1991; 20, P. pastoris; 21, P. micans; 22, C. cohnii; 23, P. falciparum; 24, T. gondii; 25, E. histolytica; 26, Diplonema; 27, T. vaginalis; 28, G. lamblia; and 29, E. coli. Because no common probe was available, DNA loading was checked by the staining of the gel with ethidium bromide (EtBr).
J also was not detected in fly (D. melanogaster), nematode (C. elegans), plant (P. corollas), yeast (S. cerevisiae, Pichia pastoris, and Schizosaccharomyces pombe), E. coli DNA (Fig. 2; Table 2), or phage lambda DNA (not shown). Also, most of the nonkinetoplastid protozoa lacked detectable J. This outcome applied both to organisms without mitochondria that represent early branches of the eukaryotic tree, such as T. vaginalis, G. lamblia, and E. histolytica (Fig. 2), and to later branches, such as the apicomplexans P. falciparum, P. berghei (not shown), and T. gondii (Fig. 2). A phylogenetic tree with the organisms analyzed in this study is shown in Fig. 3. Nucleotide postlabeling suggested the presence of 6-MeAde in T. vaginalis DNA (not shown), but this result has not been verified by other methods.
Table 2.
Occurrence of J outside kinetoplastida
| DNA | J |
|---|---|
| Mammals | − |
| Human telomeres | − |
| Drosophila, Sf9 cells | − |
| C. elegans | − |
| P. corrolas | − |
| S. cerevisiae, P. pastoris | − |
| P. micans, C. cohnii | ? |
| Plasmodium, Toxoplasma | − |
| E. histolytica | − |
| Kinetoplastida | + |
| Diplonema (= Isonema) | + |
| T. vaginalis | − |
| G. lamblia | − |
?, P. mincas and C. cohnii reacted with anti-J antibodies on DNA blots, but other lines of evidence showed that positive signal probably is not caused by J.
Figure 3.
Phylogenetic tree of eukaryotes tested for the presence of J. This phylogenetic tree inferred from 16S-like rRNA sequence similarities is modified from Sogin (39). The position of Diplonema (dashed line) is still unclear (see text). J was found in the order Kinetoplastida and in Diplonema (underlined).
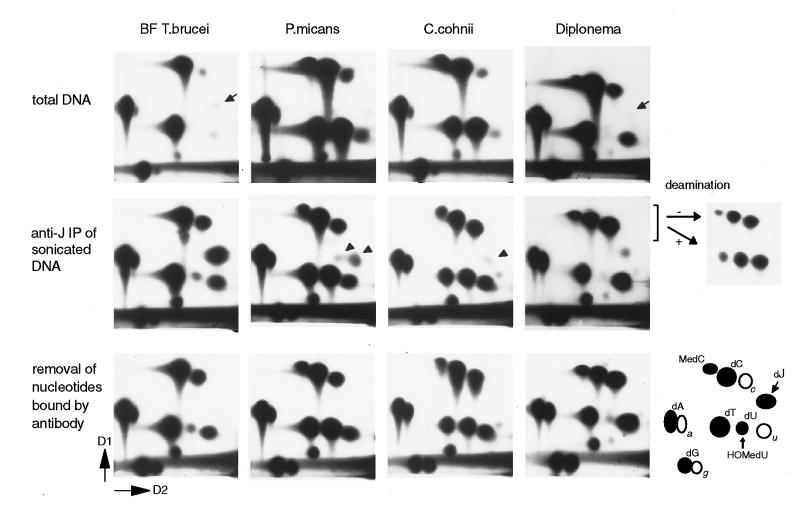
However, with immunoblots, a positive signal for J was found with DNA of the two free-living marine dinoflagellates C. cohnii and P. micans and with DNA of the phagotrophic marine flagellate Diplonema (= I. papillatum) (Figs. 2 and 3). To test whether the dinoflagellates and Diplonema indeed contain J, the DNAs were analyzed by 32P-postlabeling combined with 2D-TLC. Using total DNA, dJMP was detectable in Diplonema (≈0.02%) but not in P. micans or C. cohnii (Fig. 4). In agreement with previous studies, we found that these dinoflagellates contain 5-MeCyt and have replaced a substantial fraction of Thy with HOMeUra (10, 11, 19). Diplonema DNA also contained a spot that migrates at the position of 5-Me-dCMP (see below) whereas no DNA methylation was detected in T. brucei (Fig. 4) or other kinetoplastids (not shown).
Figure 4.
Detection of J in Diplonema and J-like modifications in dinoflagellates. Analysis of bloodstream form T. brucei, P. micans, C. cohnii, and Diplonema by 32P-nucleotide postlabeling combined with 2D-TLC (D1 and D2 are indicated). The position of the labeled 5′-deoxynucleotidemonophosphates (dN) is explained in the right bottom corner. a, c, u, and g indicate contaminating ribonucleotides. dU and HOMedU nucleotides comigrate under these conditions. (Top) Labeling of total DNA. J is indicated with small arrows. (Middle) Analysis of samples after anti-J immunoprecipitation of sonicated DNA fragments. Arrowheads indicate nucleotides in dinoflagellates that migrate close to, but differently from, J. To test whether the nucleotide close to dC in Diplonema is 5-Me-dCMP, it was isolated together with dCMP and CMP, chemically deaminated (+), and rerun on 2D-TLC, mixed with nondeaminated input (−). Deamination of 5-Me-dCMP, dCMP, and CMP results in dTMP, dUMP, and UMP, respectively. (Bottom) Labeled nucleotides from the Middle were incubated with anti-J antibodies coupled to ProtA beads to specifically remove nucleotides recognized by the antibodies. The supernatant was analyzed by 2D-TLC.
For a more sensitive J detection, we enriched for modified DNA by using anti-J immunoprecipitation of sonicated DNA (Fig. 4). Postlabeling of immunoprecipitated DNA showed an ≈13- and 58-fold–enrichment for dJMP in bloodstream form T. brucei and Diplonema, respectively. With immunoprecipitated DNA of P. micans and C. cohnii, we detected nucleotides that ran close to the J–nucleotide (Fig. 4, middle). However, the migration of these nucleotides relative to the other labeled ribo- and deoxynucleotides was slightly different from the migration of J–nucleotide (Fig. 4), as proven by mixing experiments (not shown). HOMedUMP was enriched ≈1.5-fold, and no significant enrichment was seen for 5-Me-dCMP. To test whether the J-like nucleotides are the ones recognized by the antibody, we incubated the labeled mononucleotides (from the postlabeling of immunoprecipitated DNAs) with antibody coupled to ProtA beads and analyzed the nonbound nucleotides from the supernatant. With T. brucei and Diplonema, this incubation resulted in specific removal of dJMP, and with the dinoflagellates, this incubation resulted in specific removal of the J-like nucleotides (Fig. 4, Bottom). HOMedUMP was not detectably reduced. This result showed that the antibody binding to C. cohnii and P. micans DNA is caused by the J-like nucleotides and not by others such as HOMedUMP.
Of interest, immunoprecipitation with Diplonema DNA not only enriched for dJMP but also for the 5-Me-dCMP spot (≈20-fold). This enrichment was mediated by J because the 5-Me-dCMP itself was not bound by antibody (Fig. 4, Bottom). To verify its identity, we isolated the nucleotide that migrated at the position of 5-Me-dCMP, together with dCMP and CMP, then chemically deaminated the nucleotides and reanalyzed them on 2D-TLC (Fig. 4). As expected, dCMP and CMP were converted into dUMP and UMP, respectively. The other nucleotide was converted into dTMP, which is expected for 5-Me-dCMP. We therefore conclude that Diplonema contains both J and 5-MeCyt, and these modifications reside close to each other.
DISCUSSION
Using a sensitive anti-J immunoblot assay, in combination with anti-J immunoprecipitation and 32P-nucleotide postlabeling, we have found that the modified base J is present not only in African trypanosomes but in all kinetoplastid genera tested, including mammalian, fish, and insect parasites and mono- and digenetic species (8). These results suggest that J already was present in the ancestral kinetoplastid, which has been estimated to have existed ≈500 million years ago (7, 20). We also find J in the free-living, phagotrophic flagellate Diplonema (21, 22). The phylogenetic position of Diplonema is under discussion (21, 22). Its morphological characteristics and euglenoid movements suggest that it is a member of the flagellate group Euglenozoa, which also contains the Kinetoplastida. Comparison of small subunit rRNA sequences suggests, however, that Diplonema may be monophyletic with kinetoplastids but is not a member of the kinetoplastid clade (Fig. 3 and D. Maslov, S. Yasuhira, and L. Simpson, personal communication). This comparison is further supported by other characteristics of the DNA. First, the (GGGTTA)5-telomeric repeat probe only weakly hybridized to Diplonema DNA (not shown) whereas it hybridized well to the telomeric repeats of all kinetoplastids (Fig. 1). Second, the DNA of Diplonema is methylated, whereas no 5-MeCyt is detectable in kinetoplastids (Fig. 4).
Whereas no J was detected in eukaryotes that represent early branches of the eukaryotic tree, in dinoflagellates, we found three modified nucleotides that are recognized by the anti-J antiserum but that run differently from J in our 2D-TLC system. Whether these spots represent analogues of J and whether dinoflagellates contain, in addition, low levels of J not detected in our experiments remain to be verified. Dinoflagellates, ciliates, and apicomplexans (Toxoplasma and Plasmodium) belong to the same late evolutionary protist grouping, the Alveolates (23–26), but no J was found in the apicomplexans. For the moment, J-like bases seem therefore to be restricted to a limited set of protozoa.
Teebor and coworkers (27) have proposed a role for HOMeUra-glycosylase in the maintenance of 5-MeCyt in DNA, but our results with dinoflagellates and Diplonema show that HOMeUra and 5-MeCyt can coexist in the same DNA. In fact, 5-MeCyt is even enriched in DNA fragments of Diplonema containing J, suggesting that these two minor bases mark similar DNA stretches and may (in part) play a similar role.
In T. brucei, J is clustered in and around nontranscribed repetitive sequences. These include telomeric repeats (3), sequence repeats in and upstream of telomeric VSG gene expression sites, and the 177-bp repeats that make up the central part of minichromosomes (4). Clustering of J also is found in the DNA of the other organisms studied here, as immunoprecipitation of sonicated DNA with anti-J antibodies substantially enriched for J (Table 1; Fig. 4). In Kinetoplastida other than T. brucei, at least part of this clustered J is present in telomeric repeats (Fig. 1; Table 1). Whether other repetitive sequences in these organisms also contain J remains to be determined.
The function of J is not known, but three possibilities can be considered: repression of transcription, repression of recombination, or a combination of both. J previously has been suggested to be involved in transcriptional control VSG gene expression sites in bloodstream form T. brucei. In these parasites, inactive telomeric VSG genes contain blocked recognition sites for the restriction endonucleases PstI and PvuII, and these restriction sites become cleavable when the genes are activated (5, 6). It is now clear that the restriction sites are blocked because of the presence of J (4). DNA modification has not been found in inactive VSG genes with a chromosome-internal location (4, 5). The strict correlation between J and silenced VSG gene expression sites has led to a model in which the reversible activation and inactivation of expression sites would be determined by a competition between transcription and modification (4, 5). Alternatively, J could interfere with transcription elongation in silent expression sites. Cytosine methylation inhibits elongation of transcription in Neurospora crassa (28) and has been suggested to do so in Ascobolus (29, 30). A central role for J in the control of silencing of expression sites also is supported by the absence of modification in insect form T. brucei, in which all VSG gene expression sites are switched off by a stage-specific mechanism.
Clearly, the results presented here show that J is not limited to trypanosomes that undergo antigenic variation and therefore demonstrate that J has not evolved for the control of VSG genes. Furthermore, the other kinetoplastids are not known to undergo transcriptional silencing. It is therefore possible that the primary function of J is not in regulation of transcription of specific genes but in control of repetitive DNA elements. In T. brucei, J is found predominantly in and around highly repetitive DNA such as the telomeric 50-bp, 70-bp, and 177-bp repeats (3, 4) and is also present in chromosome-internal middle repetitive DNA such as the spliced-leader gene repeats (unpublished results). The presence of J might suppress recombination between repetitive sequences in nonhomologous positions in different chromosomes. However, J also could be involved in transcriptional repression of repetitive sequences. It recently has been suggested that (reversible) cytosine methylation is not primarily involved in developmental gene control but in suppression of intragenomic parasitic sequences such as retroviruses, Alu elements, and transposons. Methylation and suppression of these repetitive elements is suggested to be necessary for maintaining the integrity of the genome (31, 32), and J might have a similar function in Kinetoplastida.
Finally, the effect of J in Kinetoplastida and Diplonema might be mediated by the assembly of a specialized chromatin structure on modified DNA that might affect both repression of transcription and recombination. This effect also has been suggested for the chromatin of the silent mating-type cassette region in S. pombe (33). In S. cerevisiae, it has been shown that silencing proteins are involved in the maintenance of genome integrity through their role in repair of double-strand DNA breaks by nonhomologous end-joining (34). Methylation-dependent transcriptional repression in mammals also has been suggested to require chromatin proteins (35–37). In that case, African trypanosomes might have recruited the global repression mechanism associated with J for maintaining VSG gene expression sites in a silenced state or even for inducing that state. This model is supported by the remarkable life cycle stage-specific regulation of J biosynthesis, which only has been found for T. brucei and not for the other kinetoplastids. We have found recently that J synthesis stops when T. brucei differentiates from the bloodstream form to the insect form and that J is diluted out rather than actively removed (38). Why this process occurs only in African trypanosomes and not in the others is unclear. The conservation of the complex modification β-d-glucosyl-HOMeUra suggests that J is important for T. brucei, T. cruzi, and Leishmania, causing severe diseases—sleeping sickness, Chagas’ disease, and leishmaniasis, respectively. Because J is absent from their mammalian hosts, the biosynthesis of J might be a potential target for drug design.
Acknowledgments
We thank the following people for DNA samples: Dr. Rodney Adam (G. lamblia, University of Arizona, Tucson); Moniek van Beest and Dr. Hans Clevers (D. melanogaster, University of Utrecht, The Netherlands); Dr. Albert Cornelissen (P. falciparum, University of Utrecht); Francesca Fase-Fowler (C. fasciculata, L. tarentolae, Netherlands Cancer Institute, Amsterdam); Arno Flooren and Dr. Laura van ’t Veer (S. cerevisiae, Netherlands Cancer Institute); Lisa Garside (T. vivax, T. congolense, University of Bristol); Dr. Patricia Johnson (T. vaginalis, University of California, Los Angeles); Dr. John Kelly (L. donovani, L. mexicana, L. brasiliensis, London School of Tropical Medicine); Dr. Marck Koolen (T. gondii, Akzo Nobel, Boxtel, The Netherlands); Dr. Titia de Lange (mammalian DNA samples, Rockefeller University, New York); Drs. Henri van Luenen, Ivo de Baere, and Chris Vos (C. elegans, P. pastoris, Sf9 cells, Netherlands Cancer Institute); Dr. Paul Michels (T. borreli, Diplonema, International Institute of Cellular and Molecular Pathology, Brussels); Joshua Rogers and Dr. William Petri (E. histolytica, University of Virginia, Charolottesville, VA); Adriënne Vissers and Jan Kooter (P. corrolas, University of Amsterdam, The Netherlands); and Dr. Andrew Waters (P. berghei, Leiden University, The Netherlands). We thank Dr. Dmitri Maslov (University of California, Riverside); Dr. Larry Simpson (Howard Hughes Medical Institute, University of California, Los Angeles), and Dr. Shinji Yasuhira (Tohoku University, Sendai, Japan) for sharing unpublished results and Dr. Magali Berberof, Inês Chaves, Dr. Mike Cross, Anita Dirks, Dr. Dennis Dooijes, Herlinde Gerrits, Dr. Gloria Rudenko, Dr. Ronald Plasterk, Dr. Marie-Odile Soyer-Gobillard, and Dr. Titia de Lange for helpful discussions and critical reading of the manuscript. This work was supported by grants from the Netherlands Foundation for Chemical Research, with financial support of the Netherlands Organization for Scientific Research.
ABBREVIATIONS
- HOMeUra
hydroxymethyluracil
- HOMedU
hydroxymethyldeoxyuridine
- J
β-d-glucosyl-HOMeUra
- dJ
β-d-glucosyl-HOMedU
- VSG
variant surface glycoprotein
- 2D-TLC
two-dimensional-TLC
Footnotes
A commentary on this article begins on page 2037.
References
- 1.Gommers-Ampt J H, van Leeuwen F, de Beer A L, Vliegenthart J F, Dizdaroglu M, Kowalak J A, Crain P F, Borst P. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 2.Gommers-Ampt J, Lutgerink J, Borst P. Nucleic Acids Res. 1991;19:1745–1751. doi: 10.1093/nar/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Leeuwen F, Wijsman E R, Kuyl-Yeheskiely E, van der Marel G, van Boom J H, Borst P. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Leeuwen F, Wijsman E R, Kieft R, van der Marel G A, van Boom J H, Borst P. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernards A, van Harten-Loosbroek N, Borst P. Nucleic Acids Res. 1984;12:4153–4170. doi: 10.1093/nar/12.10.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pays E, Delauw M F, Laurent M, Steinert M. Nucleic Acids Res. 1984;12:5235–5247. doi: 10.1093/nar/12.13.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes A P, Nelson K, Beverley S M. Proc Natl Acad Sci USA. 1993;90:11608–11612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vickerman K. In: The Diversity of the Kinetoplastid Flagellates. Lumsden W H R, Evans D A, editors. Vol. 1. London: Academic; 1976. pp. 1–34. [Google Scholar]
- 9.Rae P M. Proc Natl Acad Sci USA. 1973;70:1141–1145. doi: 10.1073/pnas.70.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rae P M. Science. 1976;194:1062–1064. doi: 10.1126/science.988637. [DOI] [PubMed] [Google Scholar]
- 11.Herzog M, Soyer M O, Daney de Marcillac G. Eur J Cell Biol. 1982;27:151–155. [PubMed] [Google Scholar]
- 12.Borst P, Gommers-Ampt J H, Ligtenberg M J, Rudenko G, Kieft R, Taylor M C, Blundell P A, van Leeuwen F. Cold Spring Harbor Symp Quant Biol. 1993;58:105–114. doi: 10.1101/sqb.1993.058.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Luderus M E, van Steensel B, Chong L, Sibon O C, Cremers F F, de Lange T. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lange T. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt G R, Cohen S S. Biochem J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boorstein R J, Levy D D, Teebor G W. Mutat Res. 1987;183:257–263. doi: 10.1016/0167-8817(87)90008-3. [DOI] [PubMed] [Google Scholar]
- 17.Barnes D E, Lindahl T, Sedgwick B. Curr Opin Cell Biol. 1993;5:424–433. doi: 10.1016/0955-0674(93)90007-d. [DOI] [PubMed] [Google Scholar]
- 18.Seeberg E, Eide L, Bjoras M. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 19.Steele R E, Rae P M. Nucleic Acids Res. 1980;8:4709–4725. doi: 10.1093/nar/8.20.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson L, Maslov D A. Curr Opin Genet Dev. 1994;4:887–894. doi: 10.1016/0959-437x(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 21.Porter D. J Protozool. 1973;20:351–356. [Google Scholar]
- 22.Triemer R E, Ott D W. Eur J Protistol. 1990;25:316–320. doi: 10.1016/S0932-4739(11)80123-9. [DOI] [PubMed] [Google Scholar]
- 23.Qu L H, Perasso R, Baroin A, Brugerolle G, Bachellerie J P, Adoutte A. BioSystems. 1988;21:203–208. doi: 10.1016/0303-2647(88)90014-7. [DOI] [PubMed] [Google Scholar]
- 24.Gajadhar A A, Marquardt W C, Hall R, Gunderson J, Ariztia-Carmona E V, Sogin M L. Mol Biochem Parasitol. 1991;45:147–154. doi: 10.1016/0166-6851(91)90036-6. [DOI] [PubMed] [Google Scholar]
- 25.Sogin M L, Morrison H G, Hinkle G, Silberman J D. Microbiologia. 1996;12:17–28. [PubMed] [Google Scholar]
- 26.Wolters J. BioSystems. 1991;25:75–83. doi: 10.1016/0303-2647(91)90014-c. [DOI] [PubMed] [Google Scholar]
- 27.Boorstein R J, Chiu L N, Teebor G W. Nucleic Acids Res. 1989;17:7653–7661. doi: 10.1093/nar/17.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roundtree M R, Selker E U. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry C, Faugeron G, Rossignol J L. Proc Natl Acad Sci USA. 1993;90:4557–4561. doi: 10.1073/pnas.90.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selker E U. Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- 31.Lengauer C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 33.Grewal S I, Klar A J. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 34.Tsukamoto Y, Kato J, Ikeda H. Nature (London) 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 35.Meehan R, Lewis J, Cross S, Nan X, Jeppesen P, Bird A. J Cell Sci (Suppl) 1992;16:9–14. doi: 10.1242/jcs.1992.supplement_16.2. [DOI] [PubMed] [Google Scholar]
- 36.Nan X, Campoy F J, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 37.Kass S U, Pruss D, Wolffe A P. Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 38.Blundell, P. A., van Leeuwen, F., Brun, R. & Borst, P. (1998) Mol. Biochem. Parasitol., in press. [DOI] [PubMed]
- 39.Sogin M L. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]