Abstract
Small cytoplasmic RNA (scRNA) of Bacillus subtilis is the RNA component of the signal recognition particle. scRNA is transcribed as a 354-nt precursor, which is processed to the mature 271-nt scRNA. Previous work demonstrated the involvement of the RNase III-like endoribonuclease, Bs-RNase III, in scRNA processing. Bs-RNase III was found to cleave precursor scRNA at two sites (the 5′ and 3′ cleavage sites) located on opposite sides of the stem of a large stem-loop structure, yielding a 275-nt RNA, which was then trimmed by a 3′ exoribonuclease to the mature scRNA. Here we show that Bs-RNase III cleaves primarily at the 5′ cleavage site and inefficiently at the 3′ site. RNase J1 is responsible for much of the cleavage that releases scRNA from downstream sequences. The subsequent exonucleolytic processing is carried out largely by RNase PH.
INTRODUCTION
Stable RNAs are synthesized in precursor forms that are then processed by endonucleases and exonucleases to mature forms (1). Maturation of stable RNA in Bacillus subtilis is known to involve the following endonucleases: RNase M5 catalyzes 5S rRNA maturation (2); RNase P cleaves tRNA precursors to generate the mature 5′ end (3); Bs-RNase III, the B. subtilis version of RNase III, is involved in rRNA processing (4); RNase Z is required for cleavage of CCA-less tRNA precursors (5); and RNase J1 has recently been shown to be involved in 16S rRNA processing (6).
Following endonuclease cleavage, trimming by 3′-to-5′ exoribonucleases is often required to produce the mature RNA species. Deutscher and colleagues have used mutant strains of E. coli that are missing one or more of the eight known 3′-to-5′ exonuclease activities to demonstrate a remarkable ability of many exonucleases to participate in maturation of tRNAs (7). We have constructed mutant strains of B. subtilis that are deficient in one or more of the four known 3′-to-5′ exoribonucleases in this organism: polynucleotide phosphorylase (PNPase), RNase R, RNase PH and YhaM (8). We have shown that RNase PH plays a significant role in CCA-containing tRNA maturation, although both RNase R and YhaM can also perform this function (9). PNPase did not appear to be involved in tRNA maturation.
Bacillus subtilis small cytoplasmic RNA (scRNA) is the functional homolog of the E. coli 4.5S RNA, which is a constituent of the SRP-like complex of E. coli that is involved in protein targeting (10). In E. coli, 4.5S RNA precursor is cleaved endonucleolytically by RNase P (11), and final maturation is accomplished by exonuclease RNase T and, to a lesser extent, RNase PH (12). Bacillus subtilis scRNA is transcribed as a 354-nt precursor RNA that, based on computer predictions, contains a short 5′-terminal stem-loop, a longer internal stem-loop structure that includes a large 255-nt loop, and a 3′-terminal stem-loop that functions as the transcription terminator (Figure 1). The mature scRNA is 271 nt, with a 5′ end mapping to the site of Bs-RNase III cleavage [site ‘A’ in Figure 1; (13,14)]. The 3′ end of mature scRNA (asterisk in Figure 1) maps 4 nt upstream of a second Bs-RNase III cleavage site (site ‘b’ in Figure 1). It has been proposed that scRNA maturation is the result of cleavage by Bs-RNase III at the A and b sites, followed by exonuclease trimming at the 3′ end to remove the last 4 nt (14). Our results suggest that 3′ exonucleases can act on precursor scRNA from a site other than the downstream Bs-RNase III cleavage site.
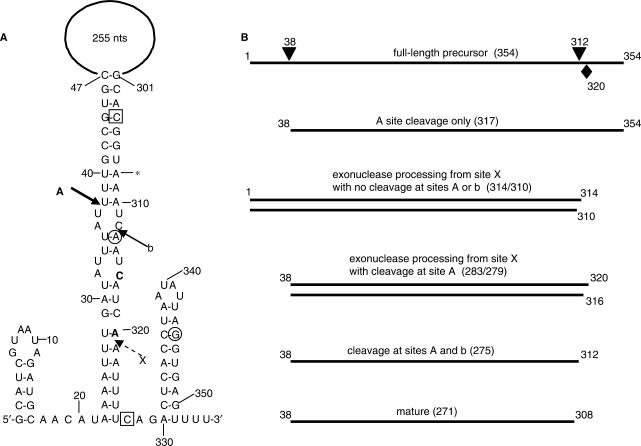
Figure 1.
(A) Schematic diagram of scRNA with nucleotide sequence of 5′ and 3′ ends. The large internal loop of 255 nt is not drawn to scale. Solid arrows show location of Bs-RNase III cleavage sites A and b. Proposed cleavage site X is indicated by dashed arrow. Asterisk denotes 3′ end of mature scRNA. The bold C and A nucleotides are at the 3′ end of the 279- and 283-nt products, respectively. Boxed nucleotides mark the ends of the sequence that is complementary to the 3′-proximal oligonucleotide probe (shown schematically in Figure 6B). Circled nucleotides mark the ends of the sequence that is complementary to the 3′-terminal oligonucleotide probe (shown schematically in Figure 6C and D). (B) Line diagrams of RNA products detected by various probes. Arrowheads above full-length line indicate sites of Bs-RNase III cleavage. Diamond below full-length line indicates site X. Numbers at each end of the line indicate 5′ and 3′ ends of RNAs. Above each line is the likely origin of the RNA species, with the size of the RNA in parentheses. (The sizes of the RNAs are those predicted from earlier data and calculated from the data shown in Figures 2, 3 and 6. The degree of precision from these data is ±2 nt, as mentioned in the text.) For clarity, the 314- and 310-nt RNAs, as well as the 283- and 279-nt RNAs, are grouped.
MATERIALS AND METHODS
Bacterial strains
Bacillus subtilis exoribonuclease mutant strains were derivatives of the parent strain BG1, which is trpC2 thr-5 and which is designated ‘wild type’ in this study. Construction of these strains has been described previously (8), as has isolation of the rncS deletion strain (4). Construction of various endonuclease mutant strains, using an rnr rph yhaM triple exonuclease mutant as host, is described in the text.
Northern blot analysis
RNA was isolated by hot phenol extraction from B. subtilis cultures grown to mid-logarithmic phase in minimal medium containing Spizizen salts with 0.5% glucose, 0.1% casamino acids, 0.001% yeast extract, 50 μg/ml tryptophan and threonine, and 1 mM MgSO4, as described (15). Northern blot analysis of RNA separated on 6% denaturing polyacrylamide gels or sequencing gels was done as previously described (16). For the triple mutant strains containing endonuclease mutations, a rich culture medium was used, containing 1% yeast extract, 2% tryptone, 1% NaCl, 1% glucose and with or without 1 mM IPTG. Cultures were grown until an OD600 of 0.6, and RNA was isolated by hot phenol extraction, as above. The scRNA riboprobe was synthesized by T7 RNA polymerase (Ambion) in the presence of [α-32P]UTP, using as template an isolated PCR fragment containing the scRNA large internal loop sequence. 5′-end-labeled oligonucleotide probes were prepared using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. To control for RNA loading in northern blot analyses of scRNA processing and of ΔermC mRNA half-life, membranes were stripped and probed for 5S rRNA, as described (17). Size markers on sequencing gels were sequencing reactions done on single-stranded M13mp18 DNA. For the northern blot shown in Figure 6D, the size marker was TaqI-digested plasmid pSE420, as described previously (8).
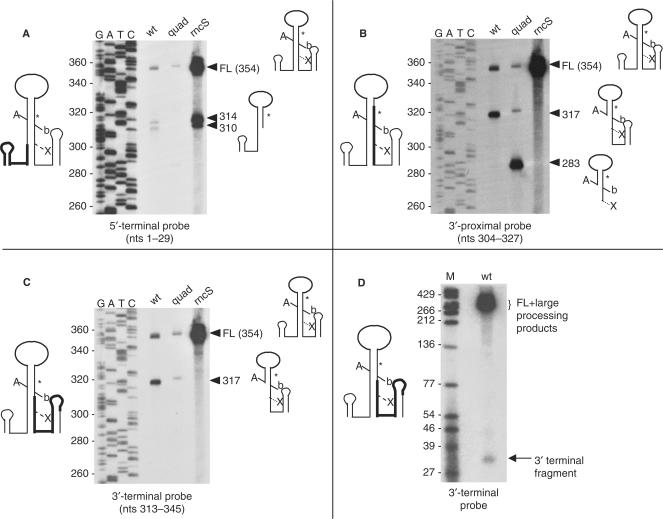
Figure 6.
Northern blot analyses of scRNA processing products using different oligonucleotide probes. In panels A–C, RNA from the following strains was analyzed: wt, wild type; quad, quadruple exonuclease mutant; rncS, rncS deletion strain. The probe used is written below each blot, together with a schematic diagram of scRNA on the left side of the blot showing with bold lines the extent of complementarity to each probe. Sequencing reaction that provided the nucleotide size marker is at left. Arrowheads with sizes of bands detected are on the right, together with schematic diagrams of the RNAs detected. In panel D, the 3′-terminal probe was used to detect a downstream fragment of endonuclease cleavage in the wild-type strain.
Quantitation of radioactivity in bands on northern blots was done with a Storm 860 PhosphorImager instrument (Molecular Dynamics). ΔermC mRNA half-life was determined by a linear regression analysis of percent RNA remaining versus time.
Bs-RNase III assay
Preparation of B. subtilis cell extracts was essentially as described previously (15), except that the dialysis buffer was 20 mM Tris-HCl (pH 8.0), 60 mM KCl, 5% glycerol, 0.1 mM EDTA, 1 mM DTT and 0.2 mM phenylmethylsulfonyl fluoride. Conditions for Bs-RNase III cleavage and analysis of results were as described (14).
RESULTS
Evidence for an alternative pathway of scRNA processing
scRNA processing was analyzed first in the single exoribonuclease mutant strains, each of which was deficient for one of the four known 3′-to-5′ exonucleases. We expected that we might observe a deficiency in the exonucleolytic processing that is thought to initiate after Bs-RNase III cleavage at site b (Figure 1), resulting in an accumulation of the 275-nt RNA. RNA was isolated from the wild-type and mutant strains, and the RNA was separated on a high-resolution denaturing polyacrylamide gel (‘sequencing gel’) alongside a sequencing ladder that served as a size marker. The sequencing gel was electroblotted and the membrane was probed with an scRNA riboprobe that was complementary to the internal 255-nt loop portion. Additional experiments with 5′- and 3′-specific, end-labeled oligonucleotide probes allowed unambiguous identification of the RNA bands detected by the riboprobe.
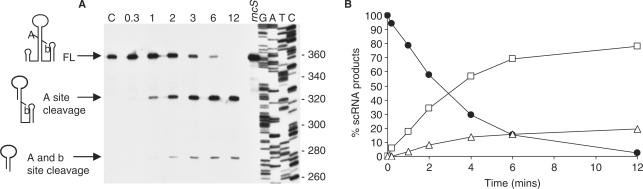
The results of this northern blot experiment are shown in Figure 2, lanes 1–5. (The relative sizes and extents of all bands detected in this blot are shown schematically in Figure 1B.) An intense band representing the mature scRNA ran at 271 nt, and a faint band representing the full-length scRNA precursor (354 nt) was detected. The 275-nt RNA was scRNA that had been cleaved at the 5′ and 3′ Bs-RNase III cleavage sites (A and b in Figure 1). The average amount of this 275-nt band, in most strains examined, was ∼20–25%, relative to total scRNA (Table 1). A 317-nt RNA band was an scRNA precursor that had been cleaved at the Bs-RNase III site A only, as determined by probing with 5′- and 3′-specific oligonucleotide probes (see Figure 6). Two very faint bands below the 317-nt band were barely visible on this exposure, but were clearly present on longer exposures. These were 314- and 310-nt RNAs that were not cleaved by Bs-RNase III and that retained the 5′ end of precursor scRNA (as demonstrated by a 5′-specific probe which was complementary to scRNA nucleotides 1–29; see Figure 6A). Importantly, we did not observe a band that corresponded to cleavage at the Bs-RNase III site b alone, which would be 312 nt. Although the absolute size of bands detected on these sequencing gel northern blots could not be determined to an accuracy of greater than ±2 nt, there was independent evidence that these 314- and 310-nt bands were not the result of Bs-RNase III cleavage, since they were also present in the strain that is deleted for the gene encoding Bs-RNase III (Figure 6).
Figure 2.
Sequencing gel northern blot analysis of scRNA processing pattern in single (lanes 2–5), double (lanes 6–8) and quadruple (lane 9) exoribonuclease mutants. The scRNA riboprobe, complementary to the large internal loop, was the probe. Below each lane is the genotype of the strain from which RNA was isolated. Sequencing reaction that serves as size marker is in the left four lanes, with nucleotide sizes indicated on the left. Identity of detected bands is indicated on the right. Schematic diagrams, from top to bottom, represent: full-length scRNA; scRNA cleaved at Bs-RNase III site A only; scRNA cleaved at Bs-RNase III sites A and b; and fully mature scRNA. Full-length and mature scRNA and products from Bs-RNase III cleavage are indicated by arrows; products that are thought to arise by processing after endonuclease cleavage at site X are indicated by lines without arrowheads.
Table 1.
Quantitation of scRNA forms (numbers are sizes of scRNA forms, in nucleotides) in wild-type and mutant strains
| Genotype | 283 | 279 | 275 | 271 |
|---|---|---|---|---|
| 1. wild type | 0 | 0 | 22 ± 1 | 74 ± 1 |
| 2. pnpA | 0 | 0 | 24 (25,23) | 73 (72,75) |
| 3. rnr | 0 | 0 | 20 (21,19) | 77 (76,78) |
| 4. rph | 2 (1,2) | 4 (3,5) | 25 (24,26) | 66 (65,66) |
| 5. yhaM | 0 | 0 | 21 (21,21) | 77 (77,77) |
| 6. pnpA rnr | 0 | 0 | 19 (22,16) | 77 (73,81) |
| 7. pnpA rph | 2 (1,2) | 3 (2,3) | 26 (27,25) | 67 (67,67) |
| 8. pnpA yhaM | 0 | 1 (1,0) | 28 (27,28) | 69 (68,70) |
| 9. rnr rph yhaM | 8 ± 2 | 37 ± 2 | 9 ± 2 | 46 ± 2 |
| 10. pnpA rph yhaM | 5 (7,2) | 4 (4,4) | 18 (22,15) | 71 (65,78) |
| 11. pnpA rnr yhaM | 0 | 0 | 16 (18,14) | 83 (81,84) |
| 12. pnpA rnr rph | 2 (2,1) | 3 (4,2) | 26 (18,33) | 69 (75,63) |
| 13.pnpA rnr rph yhaM | 26 ± 1 | 19 ± 1 | 25 ± 1 | 29 ± 3 |
Values shown in bold are percent of total scRNA detected. For strains where three or more experiments were performed, SDs are given. For other strains, the average of two experiments are shown, with the variation in the data in parentheses.
Surprisingly, in the strain deleted for RNase PH (lane 4), two additional bands were observed running above the 275-nt band. These were estimated to be 4 and 8 nt longer than the 275-nt precursor, and they did not contain the 5′-proximal part of precursor scRNA (i.e. they were generated following Bs-RNase III cleavage at site A). This result was not expected if scRNA processing occurred by cleavage at both Bs-RNase III cleavage sites, followed by exonucleolytic trimming. In that case, only two types of precursors would be observed: products of incomplete Bs-RNase III cleavage (e.g. the 317-nt band) and products that were between 275 and 271 nt in size. The observation of RNAs with sizes of 283 and 279 nt suggested RNase PH-dependent processing from a site downstream of the Bs-RNase III b site.
Analysis of scRNA processing in strains with multiple exoribonuclease deficiencies
scRNA processing was analyzed in double exonuclease mutant strains, which were deficient for PNPase and one of the other 3′-to-5′ exoribonucleases (Figure 2, lanes 6–8). The lack of RNase PH in the pnpA rph double mutant resulted in readily detectable 283- and 279-nt bands, with similar relative amounts to that of the rph mutant alone (compare Table 1, lines 4 and 7). A small amount of these additional bands was also detected in the pnpA yhaM double mutant (Table 1, line 8).
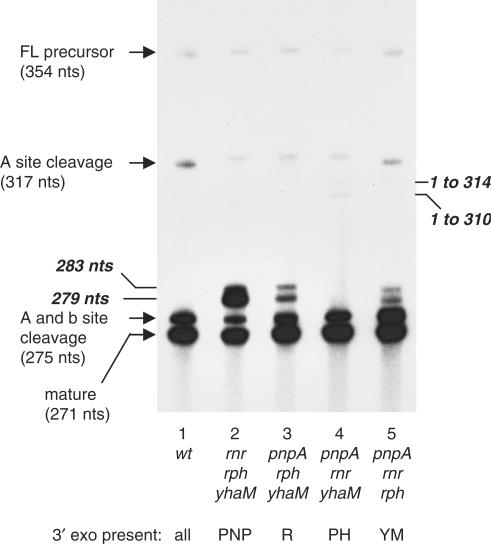
Next, processing of scRNA was analyzed in strains deleted for three of the four exoribonucleases, i.e. containing only one of the known 3′-to-5′ exonucleases (Figure 3). In the strain that had only RNase PH, the processing pattern was identical to wild type (compare lanes 1 and 4, Figure 3). This result firmly established RNase PH as the major 3′-to-5′ exonuclease involved in scRNA processing.
Figure 3.
Sequencing gel northern blot analysis of scRNA processing pattern in triple exoribonuclease mutants. Below each lane is the genotype of the strain from which RNA was isolated. For clarity, the single 3′ exoribonuclease known to be present in each mutant strain is indicated below the genotype (PNP, PNPase; R, RNase R; PH, RNase PH; YM, YhaM). Full-length and mature scRNA and products from Bs-RNase III cleavage are indicated by arrows; products that are thought to arise by processing after endonuclease cleavage at site X are indicated by lines without arrowheads.
Strikingly, in the strain that had only PNPase (Figure 3, lane 2), the 279-nt RNA now became the dominant precursor product, and there was almost as much of this precursor RNA as there was of the fully processed scRNA (Table 1, line 9). This result indicated clearly that cleavage at the Bs-RNase III b site was not a primary step in processing of the downstream side of the scRNA stem structure. Rather, 3′ exonuclease activity starting from either the native 3′ end or from a site generated by endonuclease cleavage downstream of the Bs-RNase III b site was required for efficient scRNA processing. PNPase processivity, initiating at a downstream site, was severely inhibited at the 279-nt position. Whatever the reason for the block to PNPase processivity, the other 3′ exonucleases were more efficient than PNPase at digesting through this point (see Discussion section).
The strain that contained only YhaM (Figure 3, lane 5) showed the same level of 283- and 279-nt RNAs as the single and double mutant strains that had significant amounts of these RNAs (Table 1, lines 4, 7 and 12). This result demonstrated that, in the absence of RNase PH, YhaM was capable of relatively efficient scRNA processing. (Actually, the two bands that were detected above the 275-nt precursor in the YhaM-only strain ran slightly faster than the 283- and 279-nt RNAs. We have not investigated this further.)
The strain that contained only RNase R (Figure 3, lane 3) had somewhat higher levels of the 283- and 279-nt RNAs (Table 1, line 10), suggesting that RNase R was not as efficient as RNase PH or YhaM in processing scRNA precursor.
Finally, results from the quadruple mutant strain (Figure 2, lane 9) showed that this strain had the least amount of mature scRNA and the highest amount of the 283-nt precursor RNA (Table 1, line 13). The presence of mature scRNA even in this mutant strain implies the existence of one or more 3′ exonucleases in B. subtilis in addition to the ones tested here.
scRNA transcription terminator structure functions as a block to 3′ exonuclease decay
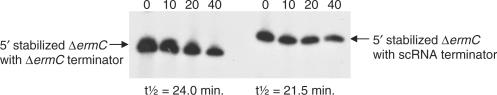
We tested whether the native 3′ end of scRNA was a possible initiation site for 3′ exonuclease processing. The scRNA 3′ end sequence (nts 327–354) is predicted to form a stable stem-loop structure (Figure 1) with a free energy of −14.2 kcal mol−1, which is typical of a transcription terminator sequence. We wished to determine whether the scRNA terminator structure could function as a barrier to 3′ exonuclease processivity in vivo, thus making it unlikely that 3′ exonuclease processing commences at the native 3′ end. For this experiment, the scRNA terminator structure was used to replace the 3′ terminator structure of a known stable mRNA. We have constructed a derivative of ΔermC mRNA that is quite stable, due to the presence of a strong 3′ transcription terminator structure, which has a predicted free energy of −22 kcal mol−1, and an inserted 5′-terminal secondary structure [‘ΔermC + 14/7A’ in (18)]. Because this mRNA is protected at both the 5′ and 3′ ends, it has a long half-life of more than 20 min. We reasoned that, if the scRNA 3′ end structure was a poor barrier to 3′ exonuclease processivity, replacing the 3′ terminator structure of the stable ΔermC mRNA with the 3′ terminator structure of scRNA would result in an unstable RNA, since protection at the 5′ end would be rendered irrelevant by rapid degradation from the 3′ end. Such a chimeric mRNA was constructed, and northern blot analysis was used to assess mRNA half-life (Figure 4). A small but not significant difference in the half-lives was observed, indicating that the scRNA 3′ end provides a strong barrier to 3′ exonuclease activity as does the ΔermC 3′ end. Thus, we hypothesized that 3′ exonucleolytic processing of scRNA begins at an endonuclease cleavage site, located in the 3′-proximal portion of scRNA, downstream of the Bs-RNase III b site. The approximate location of this site is labeled site X in Figure 1.
Figure 4.
Northern blot analysis of decay of stable ΔermC derivative (left) and stable ΔermC derivative with scRNA terminator sequence (right). Times (minutes) after rifampicin addition are indicated above each lane. The replacement of the ΔermC terminator with the scRNA terminator results in an increase in size of ∼15 nt. The half-lives for the two RNAs are shown below the blot (average of three experiments).
Quantitative analysis of Bs-RNase III activity at the A and b cleavage sites
If 3′ processing that initiates at site X was an important step in formation of mature scRNA, then Bs-RNase III cleavage at site b would likely contribute only in a minor way to scRNA processing. To assess quantitatively the relative efficiency of cleavage at the A and b sites, uniformly labeled precursor scRNA was prepared and was incubated with protein extracts prepared from wild-type or rncS mutant strains. A time course of the Bs-RNase III cleavage reaction is shown in Figure 5. Cleavage at the A site predominated: at later time points when 100% of the molecules were cleaved at the A site, <20% of the molecules were also cleaved at the b site. We therefore hypothesize that, in vivo, cleavage at the b site is inefficient, so that rapid accumulation of fully mature scRNA requires processing by 3′ exonucleases of an scRNA precursor that retains sequences downstream of the b site.
Figure 5.
(A) Products of Bs-RNase III activity on uniformly labeled scRNA, in B. subtilis protein extracts. Control lane, C, no extract added. Time (in minutes) of incubation with rncS+ extract is indicated at the top of each lane. Lane marked ‘rncS’ indicates incubation for 12 min with extract prepared from strain that is deleted for the rncS gene. Schematics of scRNA products are on the left. Size markers (nucleotides) are on the right. (B) Quantitation of percent of full-length scRNA (solid circles), scRNA cleaved at A site (squares) and scRNA cleaved at A and b sites (triangles).
Processing intermediates in a Bs-RNase III-deficient strain
Although Bs-RNase III is considered an essential enzyme in B. subtilis, we have isolated a rare mutant strain (presumably containing a second-site suppressor) that survived deletion of rncS, the gene encoding Bs-RNase III (4). Figure 6A shows the pattern of scRNA processing in the rncS strain, as detected by a 5′-end specific probe (complementary to nts 1–29). As expected, there was a massive accumulation of full-length pre-scRNA in this strain, as well as readily detectable 314- and 310-nt products. In experiments not shown here, the riboprobe that was used in the experiments shown in Figures 2 and 3 was used to demonstrate that no mature scRNA could be detected in the rncS strain, as we have observed previously (4). The 314- and 310-nt RNAs were only faintly visible in the wild-type strain, and they contained the 5′ end of scRNA, since the probe used in this experiment was a 5′-end specific probe. If we assume that the 5′ end of the 314- and 310-nt RNAs is at +1, then their 3′ ends map within a few nucleotides of the 3′ ends of the 283- and 279-nt RNAs seen in the 3′ exonuclease mutants. (The Bs-RNase III A site is at +37.) This is consistent with the suggestion above that 3′ exonuclease processing of scRNA starts downstream of the Bs-RNase III b site, at an endonuclease cleavage site.
The blot shown in Figure 6A was stripped and then probed with a labeled oligonucleotide that was complementary to nts 304–327 (boxed nucleotides in Figure 1). The 314- and 310-nt RNAS were not detected by this probe (Figure 6B), because there was only partial overlap between the 3′ ends of these RNAs and the probe. The 317-nt RNA, which represents scRNA cleaved at the Bs-RNase III site A only, was detected by this probe in the wild-type strain but not in the rncS mutant, as expected. The results in the quadruple mutant strain are discussed below. The blot was stripped again and then probed with a labeled oligonucleotide that was complementary to nts 313–345 (circled nucleotides in Figure 1). This probe detected full-length RNA, as well as the 317-nt RNA arising from cleavage at site A, but no other RNAs were detected (Figure 6C).
Direct evidence for 3′-proximal endonuclease cleavage
If endonuclease cleavage occurred at site X, then the downstream fragment generated by such cleavage should be stable enough to detect, as it would have the exonuclease-resistant transcription terminator at its 3′ end. The 3′-terminal probe, complementary to scRNA nts 313–345, was used in a northern blot analysis of RNA isolated from the wild-type strain, with conditions designed to detect very small RNA fragments. The result in Figure 6D showed that, indeed, a fragment of ∼35 nt could be detected. This indicated clearly the existence of an endonuclease cleavage site at 320 nt, i.e. site X (Figure 1). Cleavage at site X, together with cleavage at the Bs-RNase III A site, and without cleavage at the Bs-RNase III b site, would give a 283-nt RNA. Such a product was not detectable in the wild-type strain, but was the largest precursor product detected in the exoribonuclease mutant strains that lacked RNase PH, and was also the most abundant precursor detected in the quadruple mutant strain, when the internal riboprobe was used (Figures 2 and 3; Table 1). We reasoned that the upstream product of cleavage at 320 nt would be labile in the wild type, as it would have an unprotected 3′ end. In the exoribonuclease mutants, however, this upstream cleavage product would be more long-lived (and detectable) since there would be limited trimming from its 3′ end. The results with the 3′-proximal oligonucleotide probe (Figure 6B) confirmed this expectation. This oligonucleotide was expected to hybridize well with the 283-nt RNA but not with shorter RNAs (e.g. 279-, 275- and 271-nt). When total RNA from the quadruple mutant strain was probed with this oligonucleotide, a prominent 283-nt band was observed (Figure 6B). In fact, upon long exposures, the same band was detectable even in the wild-type strain (not shown). The sum of the two fragments detected in Figure 6B and D is 35 + 283 = 318, which is approximately the same size as the scRNA that is cleaved at the Bs-RNase III A site only (measured to be 317 nt). Detection of these two products is consistent with endonuclease cleavage at site X.
Identity of the endonuclease that cleaves at site X
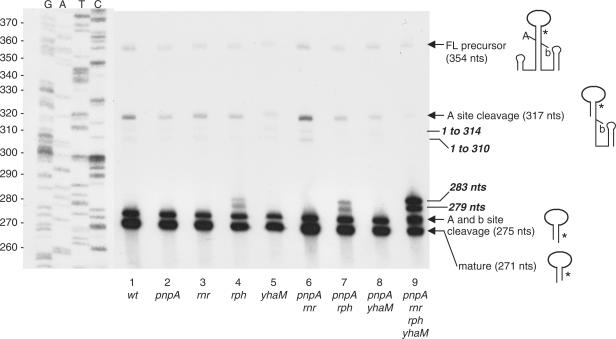
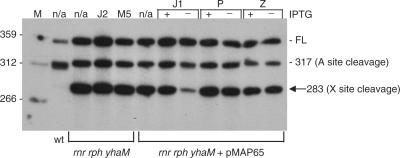
Candidate endonucleases that could be responsible for cleavage at site X included: RNase J1, RNase J2, RNase M5, RNase P and RNase Z. Strains were constructed that could test the involvement of these endonucleases in scRNA processing. The host for these constructions was the triple exonuclease mutant that contained only PNPase (i.e. the rnr rph yhaM strain, see Figure 3, lane 2). In this strain, the 283-nt band was easily detectable, and, from our analysis so far, we hypothesized that the presence of this band was associated with cleavage at site X. The continued presence of PNPase in this strain was beneficial, since strains lacking PNPase are deficient in competence (19), which would make strain construction difficult. The PNPase-containing triple mutant strain was transformed with chromosomal DNA from strains containing gene disruptions of RNase J2 (20) or RNase M5 (21), and from strains with conditionally expressed RNase J1 (20), RNA component of RNase P (22) or RNase Z (5). In the three latter cases, the endonuclease is essential, so expression was under control of the IPTG-inducible pspac promoter. The triple mutant strain transformants with a conditionally expressed endoribonuclease also contained plasmid pMAP65 (23), which provided additional copies of the lac repressor to reduce as much as possible expression in the absence of IPTG.
RNA was isolated from the five triple mutant strains carrying endonuclease mutations. Northern blot analysis was performed using a low-resolution polyacrylamide gel to separate the larger scRNA species. The blot was probed with the 3′-proximal oligonucleotide probe, which detects the 283-nt band (see Figure 6B). The results in Figure 7 showed that an effect on the 283-nt band was observed only in the strain with the RNase J1 mutation, grown in the absence of IPTG.
Figure 7.
scRNA processing in endonuclease mutant strains. Northern blot analysis on scRNA products, probed with the 3′-proximal probe. M, marker lane. Host strains are indicated underneath the blot: rnr rph yhaM is the triple mutant strain containing PNPase only; pMAP65 carries a copy of the lacI gene. Mutant endonucleases are indicated above the blot: J2, RNase J2; M5, RNase M5; J1, RNase J1; P, RNA component of RNase P; Z, RNase Z. For the latter three strains, the presence or absence of inducer IPTG is indicated. Identity of detected bands is shown on the right.
DISCUSSION
The most straightforward explanation of our results is that cleavage on the downstream side of the scRNA stem structure is accomplished not only by Bs-RNase III but also by RNase J1. In other words, release of scRNA from upstream and downstream precursor sequences can be accomplished either by two Bs-RNase III cleavages or by an upstream Bs-RNase III cleavage and a downstream RNase J1 cleavage. The data suggest that the latter pathway may predominate. Exonucleolytic trimming to the mature form is achieved primarily by RNase PH. The other three known exoribonucleases can perform this processing, albeit less efficiently. Such redundancy is similar to what we have observed in previous work on the processing of tRNA in B. subtilis (9), where RNase PH played the major role in tRNA 3′ end maturation, with other exonucleases also able to fill this role in its absence.
The data suggest that a significant portion of precursor scRNA is cleaved at site X: In the triple mutant strain that is missing RNase PH and in the quadruple mutant strain, a full 45% of the total scRNA is in the 283- or 279-nt form (Table 1, lines 9 and 13). These forms represent scRNAs that have not been cut by Bs-RNase III at site b. The 279-nt RNA is presumably a product of limited digestion from the 3′ end of the 283-nt RNA. The structural representation of scRNA in Figure 1 does not suggest a reason why digestion should be impeded at this point (see below). The 275-nt RNA is presumably the result of Bs-RNase III cleavage at sites A and b. Interestingly, the relative amount of the 275-nt RNA in almost all strains examined (Table 1) is similar to what was observed in the in vitro experiment (Figure 5).
The ability of E. coli RNase III to cleave on both sides of a target stem depends on the structure of the stem. In experiments with T7 phage RNA substrates, stems with an internal ‘bubble’ tend to be cut on the downstream side only, or primarily, while substrates with extensively base-paired stems tend to be cut on both sides of the stem (24,25). Using SP82 phage target RNAs, which have internal loop sequences, we have found that Bs-RNase III cleaves such RNAs at only one site in the downstream sequence of the predicted stem structure (26). We are unaware of a report describing a case where a natural substrate of E. coli RNase III is cleaved preferentially at the upstream site. A variant of T7 R1.1 RNA, in which the asymmetric internal loop sequence was rotated, did change the cleavage specificity to recognize primarily the upstream site (25). This latter result demonstrated that RNase III, in principle, can cleave efficiently at the upstream site of a substrate with a particular configuration. In the case of B. subtilis scRNA, early evidence from S1 mapping studies indicated that the upstream site was cleaved first (13), and cleavage of scRNA in vitro with purified Bs-RNase III showed a preference for cleavage at the upstream site (14). Our in vitro results (Figure 5), and the lack of a 312-nt fragment that would represent cleavage at site b, support the notion that Bs-RNase III cleaves efficiently at site A and inefficiently at site b.
In a previous report on turnover of seven small monocistronic mRNAs (8), no decay intermediates could be detected in a strain that contained only PNPase, whereas there was an accumulation of decay intermediates in any strain that was missing PNPase. A detailed analysis of rpsO mRNA showed that these intermediates had predicted secondary structure at their 3′ ends, suggesting that PNPase could degrade rapidly past such structures while other 3′ exonucleases could not. Thus it was surprising to find here that, with the exception of the quadruple mutant, the strain containing only PNPase accumulated by far the highest amount of immature scRNA molecules (Table 1, line 9). Interestingly, we have found previously an RNA substrate with a particular secondary structure that provides a strong block to PNPase processivity in vivo, but the same structure does not block other exonucleases in a strain that lacks PNPase (16). It is likely that full-length scRNA forms complex secondary and tertiary structures, with unpredictable consequences on processing. Furthermore, scRNA associates with at least three B. subtilis proteins—Ffh, FtsY and Hbsu (10). We note that the strain deleted for Bs-RNase III, in which almost 90% of the scRNA is in an unprocessed form and no mature scRNA is detectable, can still grow well, although it does shows a measurable growth defect (4). Thus, it is possible that even unprocessed scRNA interacts with its protein partners to function in protein trafficking, and such interactions could also affect RNA processing. In fact, the presence of bound proteins could explain the striking strong stop to processing at the 3′ end of scRNA (indicated by the asterisk in Figure 1). No products smaller than the mature 271-nt scRNA are detectable. Based solely on the 2D predicted structure, it is hard to understand why 3′ exonuclease processivity stops just at this site. Complex RNA structure and/or the presence of bound protein are likely to be factors in determining the ultimate 3′ end of the mature scRNA.
A closer inspection of the data in Table 1 reveals some noteworthy results. The triple mutant strain that contained only RNase PH gave no detectable 283- and 279-nt products, had a lower than normal amount of the 275-nt product, and had the highest amount of fully processed scRNA (Table 1, line 11). Compare these results with the data from the double mutant strain that contained RNase PH and RNase R (Table 1, line 8), where there was less of the fully processed scRNA, more of the 275-nt RNA, and detectable 283- and 279-nt RNAs. Paradoxically, the presence of an additional 3′ exonuclease resulted in a decreased amount of processing from 3′ ends. We speculate that RNase PH trimming is more efficient in a strain where other 3′ exonucleases are not present, since there is less competition for binding to the 3′ ends generated by endonuclease cleavage. Similar observations have been made in E. coli, where certain mRNAs and a regulatory RNA decay faster in an RNase II-deficient strain than in the wild type (27–29).
Another notable result seen in Table 1 was the small amount of 275-nt RNA in the triple mutant that contained only PNPase (Table 1, line 9; Figure 3, lane 2). This was significantly less than in all the other strains. We do not have a good explanation for this result. If the 275-nt RNA is generated solely by Bs-RNase III cleavage at sites A and b, one might not expect the amount of this RNA to be greatly affected by reducing the level of exonuclease activity present. However, if the 275-nt RNA could also be generated by cleavage at site X followed by exonuclease processing, one could understand why there is less of this RNA in a triple exonuclease mutant that contained only PNPase. However, if this were the case, then one would expect little 275-nt RNA in the quadruple mutant, and the data showed an amount of scRNA in this mutant that was similar to wild type.
The pattern of scRNA in the Bs-RNase III deficient strain was of interest. Besides the full-length scRNA, products of 314 and 310 nt were observed (Figure 6A), which we propose are products of cleavage at site X, followed by exonuclease trimming that is blocked at nts 314 or 310. (An RNA species that had its 3′ end at the same site [nt 308] as in mature scRNA was not observed in the Bs-RNase III-deficient strain. We conjecture that, in the wild type, exonucleolytic trimming to the mature 3′ end is aided by prior Bs-RNase III cleavage at the A site, which perturbs the double-stranded nature of this part of the molecule.) The 314- and 310-nt products were barely detectable in rncS+ strains (Figures 2 and 3; Figure 6A, wild-type and quadruple mutant lanes) but constituted ∼11% of the total scRNA in the rncS strain. If, as we have argued above, cleavage at site X is a significant component in scRNA processing, one might have expected to find a higher abundance of these products in a strain that was cleaved endonucleolytically only at site X. However, one needs to consider the effect of Bs-RNase III cleavage on the susceptibility of site X. In the full-length precursor form, the site X cleavage sequence is in a double-stranded state. If we assume that the endonuclease responsible for site X cleavage is single-strand specific, then little cleavage would occur here. After cleavage by Bs-RNase III at site A, an upstream product of 37 nt with an unprotected 3′ end is formed. It is likely that this first step in scRNA processing is followed by rapid degradation of the 37-nt 5′ fragment, which then leaves site X in a single-stranded form, rendering it susceptible to endonuclease cleavage. [In fact, using the 5′-terminal oligonucleotide probe, we could not detect the 37-nt 5′ fragment of Bs-RNase III cleavage at site A in the wild-type strain. This fragment was detectable, however, in the quadruple mutant strain (data not shown).] In the absence of Bs-RNase III cleavage at site A, site X is likely to be only partially in a single-stranded state and is therefore less susceptible to endonuclease cleavage. [This could also explain why cleavage at site X was not observed in vitro in the extract obtained from the rncS strain (Figure 5). In addition the in vitro conditions were optimized for Bs-RNase III cleavage, which are different from the conditions used by others for in vitro RNase J1 cleavage (6)].
Initial results showed that RNase J1 was responsible for cleavage at site X (Figure 7). The difference in the level of the 283-nt band in RNA isolated from the RNase J1 mutant strain grown in the presence or absence of IPTG, in independent experiments, was 3–5-fold. It is likely that the pspac promoter is somewhat leaky, providing the cells with a low level of RNase J1 even in the absence of IPTG. To eliminate the possibility that the effect on scRNA processing that we see in the RNase J1 mutant is indirect, in vitro experiments will be needed to determine whether RNase J1 cleaves scRNA directly. However, such experiments may require reconstitution of scRNA with its bound proteins, as this likely affects the presentation of scRNA to RNase J1 in vivo. RNase J1 has now been implicated in the processing of two stable RNAs, 16S rRNA (6) and scRNA, as well as thrS and thrZ leader RNAs (20). Much work remains to determine whether RNase J1 is involved more generally in RNA processing and mRNA decay in B. subtilis.
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grant GM-48804 from the National Institutes of Health to D.H.B. We thank A. Nicholson for helpful discussions on RNase III processing and C. Condon for providing endonuclease mutant strains. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM-48804.
Conflict of interest statement. None declared.
REFERENCES
- 1.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sogin ML, Pace NR. In vitro maturation of precursors of 5S ribosomal RNA from Bacillus subtilis. Nature. 1974;252:598–600. doi: 10.1038/252598a0. [DOI] [PubMed] [Google Scholar]
- 3.Gardiner K, Pace NR. RNase P of Bacillus subtilis has a RNA component. J. Biol. Chem. 1980;255:7507–7509. [PubMed] [Google Scholar]
- 4.Herskovitz MA, Bechhofer DH. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol. 2000;38:1027–1033. doi: 10.1046/j.1365-2958.2000.02185.x. [DOI] [PubMed] [Google Scholar]
- 5.Pellegrini O, Nezzar J, Marchfelder A, Putzer H, Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britton R, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Deutscher MP. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 8.Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen T, Oussenko IA, Pellegrini O, Bechhofer DH, Condon C. Ribonuclease PH plays a major role in the exonucleolytic maturation of CCA-containing tRNA precursors in Bacillus subtilis. Nucleic Acids Res. 2005;33:3636–3643. doi: 10.1093/nar/gki675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herskovits AA, Bochkareva ES, Bibi E. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 2000;38:927–939. doi: 10.1046/j.1365-2958.2000.02198.x. [DOI] [PubMed] [Google Scholar]
- 11.Bothwell ALM, Garber RL, Altman S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5S RNA. J. Biol. Chem. 1976;251:7709–7716. [PubMed] [Google Scholar]
- 12.Li Z, Pandit S, Deutscher MP. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struck JCR, Hartmann RK, Toschka HY, Erdmann VA. Transcription and processing of Bacillus subtilis small cytoplasmic RNA. Mol. Gen. Genet. 1989;215:478–482. doi: 10.1007/BF00427046. [DOI] [PubMed] [Google Scholar]
- 14.Oguro A, Kakeshita H, Nakamura K, Yamane K, Wang W, Bechhofer DH. Bacillus subtilis RNase III cleaves both 5′- and 3′ sites of the small cytoplasmic RNA precursor. J. Biol. Chem. 1998;273:19542–19547. doi: 10.1074/jbc.273.31.19542. [DOI] [PubMed] [Google Scholar]
- 15.Deikus G, Babitzke P, Bechhofer DH. Recycling of a regulatory protein by degradation of the RNA to which it binds. Proc. Natl Acad. Sci. USA. 2004;101:2747–2751. doi: 10.1073/pnas.0307343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farr GA, Oussenko IA, Bechhofer DH. Protection against 3′-to-5′ RNA decay in Bacillus subtilis. J. Bacteriol. 1999;181:7323–7330. doi: 10.1128/jb.181.23.7323-7330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp JS, Bechhofer DH. Effect of translational signals on mRNA decay in Bacillus subtilis. J. Bacteriol. 2003;185:5372–5379. doi: 10.1128/JB.185.18.5372-5379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp JS, Bechhofer DH. Effect of 5′-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol. Microbiol. 2005;57:484–495. doi: 10.1111/j.1365-2958.2005.04683.x. [DOI] [PubMed] [Google Scholar]
- 19.Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 20.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Condon C, Brechemier-Baey D, Beltchev B, Grunberg-Manago M, Putzer H. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function. RNA. 2001;7:242–253. doi: 10.1017/s1355838201002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wegscheid B, Condon C, Hartmann RK. Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep. 2006;7:411–417. doi: 10.1038/sj.embor.7400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit M.-A, Dervyn E, Rose M, Entian K.-D, McGovern S, Dusko Ehrlich S, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 24.Robertson HD. Escherichia coli ribonuclease III cleavage sites. Cell. 1982;30:669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- 25.Calin-Jagerman I, Nicholson AW. Mutational analysis of an RNA internal loop as a reactivity epitope for Escherichia coli ribonuclease III substrates. Biochemistry. 2003;42:5025–5034. doi: 10.1021/bi030004r. [DOI] [PubMed] [Google Scholar]
- 26.Mitra S, Bechhofer DH. Substrate specificity of an RNase III-like activity from Bacillus subtilis. J. Biol. Chem. 1994;269:31450–31456. [PubMed] [Google Scholar]
- 27.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. Roles of RNase E, RNase II and PNPase in the degradation of rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–3377. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pepe CM, Maslesa-Galic S, Simons RW. Decay of the IS10 antisense RNA by 3′ exoribonucleases: evidence that RNase II stabilizes RNA-OUT against PNPase attack. Mol. Microbiol. 1994;13:1133–1142. doi: 10.1111/j.1365-2958.1994.tb00504.x. [DOI] [PubMed] [Google Scholar]
- 29.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]