Abstract
By enzymatically hydrolyzing the terminal phosphodiester bond at the 3′-ends of DNA breaks, tyrosyl-DNA phosphodiesterase (Tdp1) repairs topoisomerase-DNA covalent complexes and processes the DNA ends for DNA repair. To identify novel Tdp1 inhibitors, we developed a high-throughput assay that uses electrochemiluminescent (ECL) substrates. Subsequent to screening of 1981 compounds from the ‘diversity set’ of the NCI-Developmental Therapeutics Program, here we report that furamidine inhibits Tdp1 at low micromolar concentrations. Inhibition of Tdp1 by furamidine is effective both with single- and double-stranded substrates but is slightly stronger with the duplex DNA. Surface plasmon resonance studies show that furamidine binds both single- and double-stranded DNA, though more weakly with the single-stranded substrate DNA. Thus, the inhibition of Tdp1 activity could in part be due to the binding of furamidine to DNA. However, the inhibition of Tdp1 by furamidine is independent of the substrate DNA sequence. The kinetics of Tdp1 inhibition by furamidine was influenced by the drug to enzyme ratio and duration of the reaction. Comparison with related dications shows that furamidine inhibits Tdp1 more effectively than berenil, while pentamidine was inactive. Thus, furamidine represents the most potent Tdp1 inhibitor reported to date.
INTRODUCTION
Discovered by Nash and coworkers in 1996, tyrosyl DNA phosphodiesterase I (Tdp1) belongs to the phospholipase D superfamily of phospholipids hydrolyzing enzymes (1,2). Functionally, Tdp1 is part of the DNA repair complex that resolves the irreversible topoisomerase I (Top1)-DNA cleavage complexes by catalyzing the hydrolysis of 3′-phosphotyrosyl bonds (3). In addition to the removal of peptides bound via a 3′ phosphotyrosyl linkage, Tdp1 can catalyze the cleavage of other chemical bonds such as a phosphohistidine bond (4). Tdp1 can also remove a 3′-phosphoglycolate or biotin-linked substrate and act at 3′-abasic sites (4,5). Tdp1 thus participates in the repair of a variety of 3′ adducts/base damages from DNA. An interaction of Tdp1 with DNA ligase III (6) and XRCC1 (7), members of the base excision repair (BER) complex has also been demonstrated. More recently, Tdp1 has also been implicated in the repair of topoisomerase II (Top2)-mediated DNA damage as bacterially expressed yeast Tdp1p processed the 5′ phosphotyrosyl linkage of a peptide derived from yeast Top2 covalently to DNA (8). Therefore, Tdp1 may function in multiple DNA repair pathways.
Tdp1 is physiologically important since a point mutation in the TDP1 gene causes the neurological disorder called spinocerebellar ataxia with axonal neuropathy (SCAN1) (9). SCAN1 cells exhibit hypersensitivity to camptothecin (CPT), a potent Top1 inhibitor (6,10,11). Moreover, overexpression of a human or yeast Tdp1 fusion protein has been shown to alleviate some of the effects of CPT treatment (12,13). These observations suggest that inhibitors of Tdp1 could act synergistically with CPT in a combined anticancer therapeutic regimen. Additionally, hypersensitiveness to CPT in Tdp1-defective yeast was conditional to deficiencies in the checkpoint (Rad9) and 3′-endonucleases (Mus81/Eme1) pathways (14–16). Thus, in principle, therapeutic selectivity can be achieved by combining Top1 and Tdp1 inhibitors as a significant number of tumors have defective DNA repair and checkpoint pathways (17).
As Tdp1 inhibitors in association with Top1 inhibitors could confer a selective advantage for cancer chemotherapy, we began searching recently for Tdp1 inhibitors (18). Currently, the only reported inhibitors of Tdp1 are vanadate, tungstate, aminoglycoside antibiotics and ribosome inhibitors (18,19). Our initial aim was to develop a high-throughput assay that would provide a sensitive, reliable and a rapid method to screen chemical libraries for novel Tdp1 inhibitors.
Here, we report the development of a sensitive high-throughput electrochemiluminescent (ECL) assay to identify novel inhibitors of Tdp1. Identified by the ECL assay, the dication furamidine (DB75, NSC 305831) was further studied to determine its molecular interactions with recombinant Tdp1 and its DNA substrates.
MATERIALS AND METHODS
Drugs and reagents
The 1981 compounds of the diversity set were obtained from the Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI), NIH. Berenil and Pentamidine were purchased from Sigma-Aldrich (St. Louis, MO, USA). High-performance liquid chromatography-purified oligonucleotides and tyrosyl nucleotides were purchased from the Midland Certified Reagent Co. (Midland, TX, USA).
Preparation of human Tdp1
Human Tdp1 expressing plasmid pHN1910 (a gift from Dr Howard Nash, Laboratory of Molecular Biology, National Institute of Mental Health, National Institutes of Health) was constructed using vector pET-15b (Novagen, Madison, WI, USA) with full-length human Tdp1 and an additional His-tag sequence of MGSSHHHHHHSSGLVPRGSHMLEDP in its N terminus. The His-tagged human Tdp1 was purified from Novagen BL21 cells using chelating sepharose™ fast flow column (Amersham Biosciences, Piscataway, NJ, USA) according to the company's protocol. The collected fractions were assayed immediately for Tdp1 activity. Fractions that showed Tdp1 activity were pooled and dialyzed in 20% glycerol, 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM β-mercaptoethanol and 2 mM EDTA. Dialyzed samples were aliquoted and stored at −80°C. Tdp1 concentration was determined using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA, USA). Tdp1 purity was determined as a single ∼70 kDa band representing over 95% of the detectable proteins stained by Coomassie after SDS–polyacrylamide gel electrophoresis (SDS-PAGE).
High-throughput electrochemiluminescent assay
Our electrochemiluminescent (ECL) assay is based on the BioVeris (BV) ECL technology developed by BioVeris, Inc. (Gaithersburg, MD, USA). ECL is based on the use of ruthenium labels (BV-TAG™), designed to emit light when stimulated. These labels, together with a specific instrumentation (M-SERIES® Analyzer), provided our platform for biological measurements. This technology has been successfully applied to drug discovery, pharmaceutical industry, clinical and industrial detection (20,21). The present report is the first application of the ECL biotechnology to Tdp1.
Preparation of the ECL BV-14Y substrate
The 5′-biotinylated 14Y DNA substrate (sequence shown in Figure 3A) was obtained from Midland Certified Reagent and coupled to an NHS ester BV-Tag (BioVeris Inc.) to generate the ECL substrate BV-14Y. Coupling was achieved by incubating 175 µl of 5′-biotinylated 14Y DNA at 200 µM in phosphate buffered saline (PBS), pH 7.4 with 25 µl of NHS-ester BV-Tag (BioVeris Inc.) at 3 µg/µl in 100% dimethyl sulfoxide (DMSO). After 30 min at room temperature under agitation, the coupling reaction was loaded onto a mini Quick Spin Oligo column (Roche Diagnostics, Indianapolis, IN, USA) pre-equilibrated with 3 volumes of PBS, pH 7.4 containing 0.075% (w/v) sodium azide (Sigma-Aldrich, St. Louis, MO, USA). The recovered fraction was aliquoted and stored at −20°C at 10 µM in PBS.
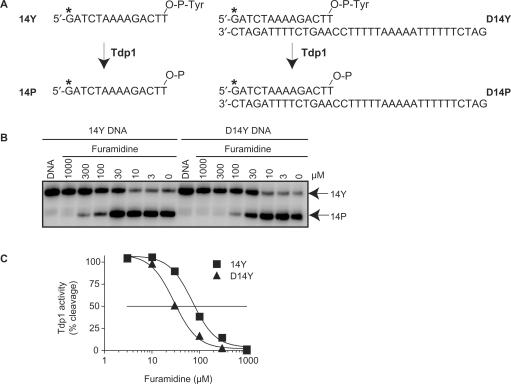
Figure 3.
Inhibition of Tdp1 activity by furamidine. (A) Schematic representation of the Tdp1 biochemical assays. The partially duplex oligopeptide D14Y or single-stranded 14Y were used as substrates. 32P-Radiolabeling (*) was at the 5′ terminus of the 14-mer strand. Tdp1 catalyzes the hydrolysis of the 3′-phosphotyrosine bond and converts 14Y and D14Y to an oligonucleotide with 3′-phosphate, 14P or D14P, respectively. (B) Representative gel showing Tdp1 inhibition by furamidine with single strand (14Y) and partially duplex (D14Y) substrates. Reactions were performed at 25°C for 20 min. Arrows indicate the 3′-phosphate oligonucleotide product (14P) that runs quicker than the corresponding tyrosyl oligonucleotide substrate (14Y) in a denaturing PAGE (18). The duplex D14Y substrate and D14P product are detected on the gel by their corresponding labeled single strands (14Y and 14P), as they are no longer annealed under the denatured conditions. (C) Densitometry analysis of the gel shown in panel B. Tdp1 activity was calculated as the percentage of 14Y converted to 14P as a function of the concentration of furamidine. The horizontal line corresponds to 50% inhibition of Tdp1 activity.
Linking of the ECL substrate to streptavidin magnetic beads
A substrate stock solution for 500 reactions was prepared by mixing 40 µl of 10 µM ECL BV-14Y substrate with 500 µl of streptavidin magnetic beads (Dynabeads M-280, BioVeris Inc.) and incubating for 30 min at 25°C under constant agitation. After magnetic separation and a two-volume wash with the Tdp1 assay buffer (50 mM Tris-HCl, pH 8.0, 80 mM KCl, 2 mM EDTA and 1 mM DTT), the linked ECL substrate was resuspended in 500 µl of assay buffer at a concentration of 800 nM. The linked ECL substrate can be stored at 4°C for up to a month without loss of activity.
ECL assay
The linked ECL BV-14Y substrate at a concentration of 0.8 nM was incubated with 1 nM Tdp1 in the absence or presence of the drugs (10 µM) to be tested. Reactions were carried out in 96-well plates at a final volume of 100 µl/well in assay buffer for 60 min at 37°C. The reactions were stopped by adding 1 volume of stop buffer (25 mM MES pH 6.0). Plates were read with an M-SERIES® M8 analyzer (BioVeris Inc.) and the ECL arbitrary units were plotted using the Prism software (GraphPad Software, San Diego, CA, USA).
Preparation of Tdp1 substrates for gel assays
High-performance liquid chromatography-purified oligonucleotides 14Y (see Figure 3A) (7) and 14Y-CC (see Figure 5A) were labeled at their 5′-end with [γ-32P]ATP (PerkinElmer Life and Analytical Sciences, Boston, MA, USA) by incubation with 3′-phosphatase-free T4 polynucleotide kinase (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's protocols. Unincorporated nucleotides were removed by Sephadex G-25 spin-column chromatography (mini Quick Spin Oligo columns; Roche Diagnostics). For the production of the oligonucleotide duplexes D14Y, the 32P-radiolabeled 14Y oligonucleotide was mixed with the complementary oligonucleotide (see Figure 3A) at equal molar ratios in annealing buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM MgCl2), heated to 96°C, and allowed to cool down slowly (over 2 h) to room temperature.
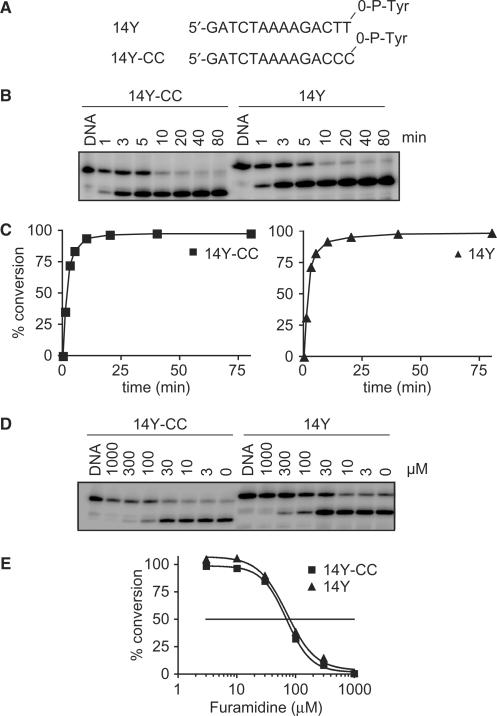
Figure 5.
Inhibition of Tdp1 by furamidine is independent of the presence of thymidines at the 3′-end of the substrate. (A) Sequences of the oligonucletotide substrates 14Y and 14Y-CC, which differ in their 3′-terminal bases (–TT or –CC) linked to the phosphotyrosine. (B) Reactions (100 µl) containing either 25 nM 14Y or 14Y-CC and 5 ng of Tdp1 were incubated at 25°C. Aliquots were taken at the indicated times (min). Reaction products were analyzed by denaturing PAGE. (C) Densitometry analysis of the gel shown in B. Tdp1 activity measured as the percentage of DNA substrates 14Y-CC (left panel) or 14Y (right panel) converted to their corresponding products as a function of reaction time. (D) Reactions (20 µl) containing 25 nM 14Y or 14Y-CC and 1 ng Tdp1 were carried out in the presence of indicated concentrations (µM) of furamidine at 25°C, pH 8, for 20 min. A representative gel is shown. (E) Densitometry analysis of the gel shown in D. Tdp1 activity was calculated as the percentage of DNA substrates 14Y or 14Y-CC converted to their product. The horizontal line corresponds to 50% inhibition of Tdp1 activity.
Tdp1 gel assays
Unless indicated otherwise, Tdp1 assays were performed in 20 µl mixtures in Tdp1 assay buffer (see above) plus 40 µg/ml bovine serum albumin. Twenty five nanomolar 5′-32P-labeled substrate (14Y or 14Y-CC or D14Y) was reacted with 1 ng of Tdp1 (0.7 nM) in the absence or presence of inhibitor for 20 min at 25°C. Reactions were stopped by the addition of 60 µl of gel loading buffer [96% (v/v) formamide, 10 mM EDTA, 1% (w/v) xylene cyanol and 1% (w/v) bromphenol blue]. Twelve-microliter aliquots were resolved in 20% denaturing polyacrylamide (AccuGel; National Diagnostics, Atlanta, GA, USA) (19:1) gel containing 7 M urea. After drying, gels were exposed overnight to PhosphorImager screens (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). Screens were scanned, and images were obtained with the Molecular Dynamics software. Densitometry analyses were performed using the ImageQuant 5.2 software (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). Tdp1 activity was determined by measuring the fraction of substrate converted into 3′-phosphate DNA product by densitometry analysis of the gel image (3). Figures show representative results that were consistently reproduced at least three times.
Surface plasmon resonance analysis
Binding experiments were performed on a Biacore 2000 instrument (Biacore Inc., Piscataway NJ, USA). 5′ biotinylated stem-loop (biotin-GATCTAAAAGACTTTCTCAAGTCTTTTAGATC) and single-stranded oligonucleotides (biotin-GATCTAAAAGACTT) were synthesized by IDT (Coralville, IA, USA). Stem-loop oligonucleotides were annealed by heating to 90°C for 5 min followed by snap cooling on ice for 15 min. Biotinylated oligonucleotides were immobilized to neutravidin-coated sensor chips as described previously (22). Approximately 5000 RU's of neutravidin was attached to all flow cells on the sensor chips. Oligonucleotides were reconstituted in buffer consisting of 10 mM Tris, pH 7.5, 300 mM NaCl and 1 mM EDTA. Single-stranded and stem-loop oligonucleotides were injected over flow cell 2 and 4, respectively until approximately 500 RU's of oligonucleotide were captured on the chip surface. Furamidine was diluted into running buffer [10 mM MES, 100 mM NaCl, 1 mM EDTA, 5% DMSO (v/v) pH 6.25] and injected over all flow cells at 20 µl/min at 25°C. Following compound injections, the surface was regenerated with a 10 s 1 M NaCl injection followed by a 10 s running buffer injection. A DMSO calibration curve was included to correct for refractive index mismatches between the running buffer and compound dilution series. Data was analyzed using the Scrubber software version 2 (David Myszka, University of Utah) and the equilibrium binding of furamidine was fit to either a single-site or two-site steady state binding model (Figure 4C and D). The equation used to fit the single-site steady state binding model was:
where B = bound complex, Rmax = total concentration of ligand, F = total concentration of analyte, NS = non-specific binding and KD = equilibrium dissociation constant. The equation used to fit the two-site steady state binding model was:
where B = sum of bound species for the two types of complex, Rmax1 and Rmax2 = total concentration to the two species of ligand, F = total concentration of analyte, KD1 and KD2 = equilibrium dissociation constants for the two types of bound complex and NS = non-specific binding.
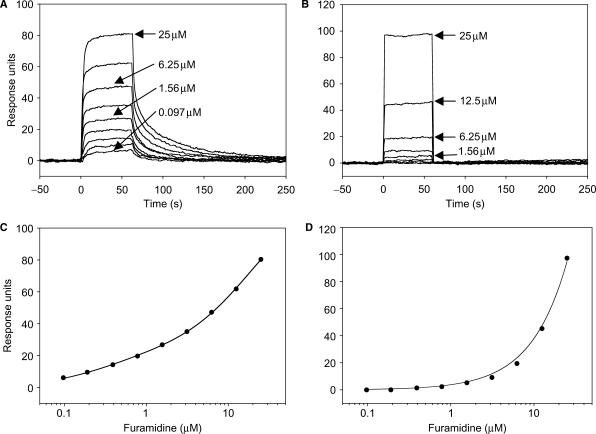
Figure 4.
Binding of furamidine to a 495 response units surface of a stem-loop duplex oligonucleotide (A) and 504 response units surface of a single-stranded oligonucleotide (B). The equilibrium level of binding was determined for each furamidine concentration for the duplex oligonucleotide (C) or the single-stranded oligonucleotides (D). The graphs represent a fit using a two binding-site model for the stem-loop duplex oligonucleotide (C) or a single binding-site model for the single-stranded oligonucleotide (D). Twofold increments of furamidine concentration (0.097, 0.19, 0.39, 0.78, 1.56, 3.125, 6.25, 12.5 and 25 µM) were used.
RESULTS
Novel high-throughput electrochemiluminescent (ECL) assay to screen for Tdp1 inhibitors
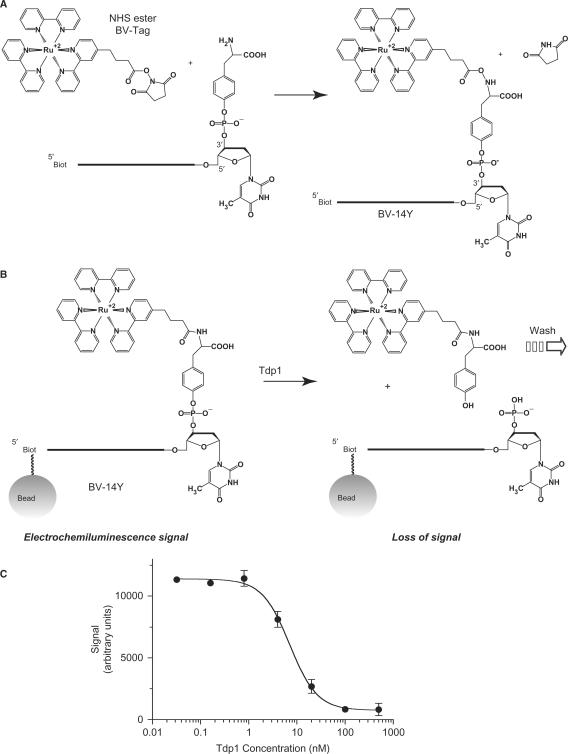
To discover inhibitors of Tdp1, we developed a novel ECL high-throughput assay (for details see Materials and Methods section and Figure 1). An ECL substrate for Tdp1 was generated after coupling a ruthenium-containing tag (BV-Tag) to the 3′-end of a 14-mer oligonucleotide with a 5′-biotin and a 3′-tyrosyl moiety (Figure 1A) (3,7). In the presence of Tdp1, the tyrosyl-bound BV-Tag is hydrolyzed from the ECL substrate (BV-14Y DNA) and washed away, leading to a loss of the electroluminescent signal (Figure 1B and C). In this assay, a potential Tdp1 inhibitor would be detected as preventing this loss of signal with a level of signal retention reflective of the potency of the putative inhibitor. Thus, in our high-throughput ECL assay, active compounds were identified as restoring to the control levels (without Tdp1) the signal lost in the presence of Tdp1.
Figure 1.
High-throughput electrochemiluminescene assay developed to identify novel Tdp1 inhibitors. (A) Generation of the electrochemiluminescent (ECL) substrate (BV-14Y). The ruthenium-containing tag (NHS ester BV-Tag; from BioVeris Corp.) is coupled to the 3′-end of the tyrosyl-containing DNA substrate [14Y (sequence as in 3A) linked to a biotin at its 5′ end]. After coupling, the BV Tag is attached to the phosphotyrosine of the 14Y DNA forming the BV-14Y DNA after the release of a succinimide group. (B) Processing of the ECL substrate by Tdp1. The ECL substrate BV-14Y bound to magnetic beads was generated as described in the Materials and Methods section. Upon addition of Tdp1, the tyrosine-BV-Tag group is hydrolyzed away leaving behind a 3′ phosphate on the DNA bound to the beads. After stopping the reactions, the samples are analyzed by the ECL analyzer. During the analysis, only the magnetic beads are retained in the analysis chamber by magnetic field while the rest of the sample is washed away. In the absence of Tdp1, the ECL signal is maximum as the BV-14Y DNA is retained on the magnetic beads. Upon addition of Tdp1, the tyrosine-BV-Tag group is cleaved away resulting in the loss of ECL signal (arrow). Potential Tdp1 inhibitors prevent this loss of signal. (C) Signal response curve in the presence of increasing concentrations of Tdp1. The ECL signal is lost when the Tdp1 concentration is increased.
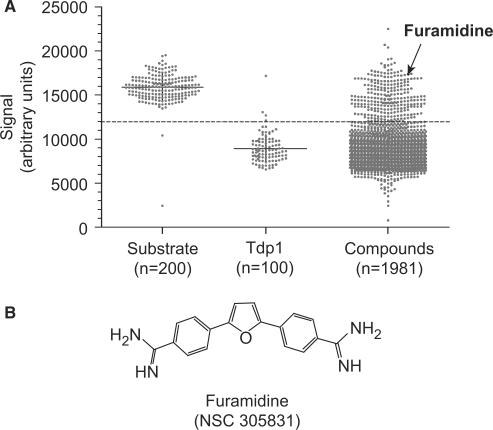
Using the high-throughput ECL assay, we screened the ‘Diversity Set’ from the Developmental Therapeutics Program (DTP) of the National Cancer Institute against recombinant human Tdp1. The diversity set consists of 1981 compounds representative of the chemical/structural diversity of the available repository of more than 140 000 chemicals (for details, see http://dtp.nci.nih.gov/branches/dscb/diversity_explanation.html). Of the 1981 compounds from the DTP diversity set, 169 positive hits were identified as inhibiting at least 70% of the Tdp1 activity at a fixed concentration of 10 µM (Figure 2A). Subsequent analyses as HPLC to evaluate chemical purity of the samples reduced the number of potential inhibitors of Tdp1 to 69 compounds. Further testing in gel-based assays led to the identification of 49 compounds as confirmed Tdp1 inhibitors. Thus, ∼8% of the NCI-DTP diversity set (1981 chemicals) were active against Tdp1 at 10 µM drug concentration in the ECL high-throughput assay. This assay is therefore a sensitive and efficient technique for the screening of novel Tdp1 inhibitors.
Figure 2.
Identification of furamidine as a Tdp1 inhibitor by high-throughput electrochemiluminescene assay. (A) Graph representing the effect of the 1981 compounds from the NCI-DTP diversity set on Tdp1 activity. Each dot indicates a signal value for a tested sample at 10 µM drug concentration. The substrate electrochemiluminescence (arbitrary units) is lost in the presence of Tdp1 (‘n’ indicates the number of samples). The effect of 1981 compounds screened is represented. Positive Tdp1 inhibitors prevent the loss of ECL signal. Dashed line represents 50% inhibition of Tdp1 activity. Furamidine was among the most potent compounds that inhibited Tdp1 activity. (B) Chemical structure of furamidine.
The diamidine NSC 305831 (for structure, see Figure 2B), more commonly known as furamidine (DB75) (23), was among the most potent inhibitors of Tdp1 with complete restoration of the normal Tdp1 signal in the ECL assay (see Figure 2A). Furamidine was chosen among other positive hits because it belongs to a very interesting group of compounds. Furamidine and closely related heterocyclic amines are currently under clinical investigation as anti-parasitic agents (24). DB289, an orally available methamidoxime prodrug of furamidine is currently under phase III clinical trials as a trypanocidal agent against human African trypanosomiasis (25,26). These compounds have been well studied for their binding to double-stranded DNA. Previous studies have shown that furamidine binds duplex DNA in the DNA minor groove selectively at AT rich sites [(A/T)4] (23,27,28). Furamidine can also intercalate between GC base pairs of duplex DNA (23,29,30). Furamidine could therefore interfere with DNA processing enzymes such as Tdp1.
Because furamidine was able to inhibit Tdp1 activity with a single-stranded DNA substrate and because of its clinical relevance, we chose to study furamidine in more detail.
Furamidine inhibits Tdp1 activity both with duplex and single-stranded substrates but is more effective with the duplex substrate
Having identified furamidine as a novel Tdp1 inhibitor, we characterized its molecular effect on Tdp1 activity using gel-based assays (Figure 3) (18). Since both partially duplex and single-stranded DNA are substrates for Tdp1 (2,19,31), we compared the inhibition of Tdp1 by furamidine using the D14Y and 14Y substrates (sequences as shown in Figure 3A) (18). As demonstrated in Figure 3 (panels B and C) furamidine inhibits the processing of both the single and double-stranded substrates by Tdp1.
Since Tdp1 inhibition is slightly more effective in the duplex substrate (Figure 3B and C), DNA binding/intercalation (23,29,30) may contribute to furamidine's potency. However, inhibition of Tdp1 activity by furamidine is also evident in a single-stranded substrate. This is unlike aclarubicin, a known DNA intercalator that inhibits Tdp1 selectively only with double-stranded DNA (18). Thus, additional mechanism of Tdp1 inhibition by furamidine exists besides DNA intercalation or minor groove binding.
Binding of furamidine to a double- and single-stranded substrate
We next evaluated the ability of furamidine to directly interact with DNA in the absence of Tdp1. Surface plasmon resonance (SPR) analyses were carried out using single-stranded and double-stranded substrates (see Materials and Methods section). As expected, Figure 4A shows detectable binding of furamidine to duplex oligonucleotide at submicromolar concentrations (23,27–30,32). Furamidine rapidly reached a steady state binding level with duplex DNA but then disassociated more slowly. The equilibrium binding could only be fit using a two binding-site model with affinities of 0.33 and 19 µM (Figure 4C and equations described in the Materials and Methods section). This seems reasonable given that the sequence contains an AT rich site [(A/T)4] within the duplex oligonucleotide, which corresponds to a high affinity-binding site for a heterocyclic diamidine (23,27,28). The 14 base-pairs duplex probably also support additional compound binding at lower affinity sites.
The binding of furamidine to a single-stranded substrate was also examined by SPR. Figure 4B shows that furamidine both associates with and dissociates from a single-stranded substrate very rapidly in contrast with what was observed for the duplex. This observation most likely reflects the electrostatic interaction between the phosphate backbone and the charged compound. We estimate the Kd of furamidine to be ∼70 µM with single-stranded DNA (Figure 4D and equations described in the Materials and Methods section). This result is in agreement with the IC50 obtained for the same single-stranded substrate (Figure 3C). To elucidate whether the inhibition of Tdp1-mediated cleavage of the single-stranded DNA substrate could be due to a stabilization of a hairpin structure by furamidine, we plugged our sequence in the mfold web application (http://bioweb.pasteur.fr/). The most favorable folding option with three formed base pairs, gave a ΔG value of 0.32 kcal/mol at 20°C and a Tm value of 13.6°C, rendering this structure unstable under our reaction conditions (25°C). In addition, the fact that we were able to measure a dissociation rate with the duplex but not with the single-stranded oligonucleotide by SPR leads us to believe that furamidine does not stabilize any hairpin structure. We also evaluated the binding of furamidine to amine-coupled Tdp1 protein (data not shown) and found the interaction to be very weak with a Kd of >900 µM. Together, these experiments demonstrate that furamidine does bind DNA with a preference for a duplex substrate.
Inhibition of Tdp1 by furamidine is independent of the presence of thymidines at the 3′-end of the substrate sequence
Furamidine has previously been shown to exhibit a strong preference for A/T sequences (29,30). Since the co-crystal studies showed Tdp1 interaction with the last 4 nt at the 3′-end of the oligonucleotide substrate (33,34) and those 4 bases were ACTT in our 14Y oligonucleotide, we evaluated the effect of altering the sequence of the oligonucleotide substrate on the inhibition of Tdp1 by furamidine. The terminal thymine dinucleotide (–TT) of the 14Y oligonucleotide was replaced with a cytosine dinucleotide (–CC) to generate the 14Y-CC oligonucleotide (see Figure 5A). The ability of Tdp1 to process either the 14Y or 14Y-CC substrates was similar (Figure 5B and C). Kinetic plot analysis shows the processing of either substrate almost completely within 10 min of the reaction time and at the same rate by 1 ng of Tdp1 (Figure 5C). Upon addition of varying concentrations of furamidine, the processing of both the substrates 14Y and 14Y-CC by Tdp1 was inhibited to the same degree (Figure 5D and E). Thus, presence of a TT dinucleotide at the 3′ terminus of the DNA is not critical for Tdp1 inhibition by furamidine.
Furamidine inhibits Tdp1 more effectively than berenil and pentamidine
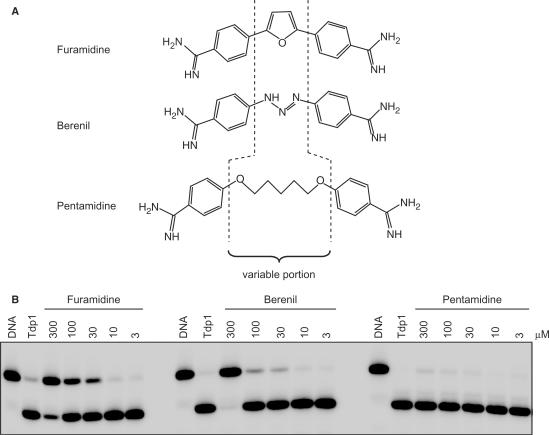
Structurally, furamidine can be considered as a bisbenzamidine derivative (Figure 6A) (35) and belongs to a family of diamidines (23). Of the several diamidines, berenil and pentamidine are clinically active and used against parasitic diseases (25,36–38). Though furamidine, pentamidine and berenil share similarities in structure (Figure 6A), they differ in their central moiety being a furan unit for furamidine, a triazene in berenil and a pentyldioxy chain in pentamidine (represented by the ‘variable portion’ in Figure 6A) (23).
Figure 6.
Differential activities of furamidine, berenil and pentamidine against Tdp1. (A) Chemical structures of furamidine, berenil and pentamidine. Dashed lines indicate the variable chemical moiety. (B) Reactions were performed with the indicated concentrations (µM) of furamidine, berenil and pentamidine. Reactions were for 20 min at pH 8.0 and 25°C in the presence of 25 nM 14Y substrate and 1 ng of Tdp1. Samples were separated on a 20% urea–PAGE gel and visualized.
To evaluate the contribution of the central furan portion of furamidine for Tdp1 inhibition, we tested the three analogs for their ability to inhibit Tdp1 activity. Figure 6B shows that pentamidine did not inhibit Tdp1 activity under our assay conditions, and berenil showed some activity, albeit at a high concentration (300 µM). Under the same conditions, furamidine exhibited an inhibition of Tdp1 activity at 30 µM (Figure 6B) and therefore is the most potent of the three bisbenzamidines examined.
Inhibition of Tdp1 by furamidine is dependent on both reaction duration and Tdp1 concentration
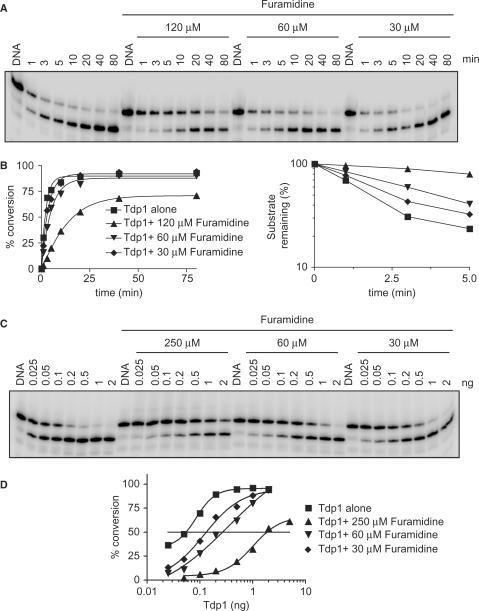
The hallmark of reversible inhibitors is that when the inhibitor concentration drops, enzyme activity is restored. Our initial gel assays (Figure 3) were performed at a fixed time (20 min) under conditions where Tdp1 almost fully converted the substrate in the absence of inhibitor (1 ng, pH 8.0). We next evaluated how reaction time and Tdp1 concentration affected furamidine's ability to inhibit Tdp1.
As shown in Figure 7A and B (A, left; and squares in B), 1 ng of Tdp1 converted ∼50% (t1/2) of the 14Y substrate within ∼1.9 min. Thus, we wished to determine how furamidine affected the kinetics of Tdp1 activity. Tdp1 activity was slowed down as the concentration of furamidine increased (Figure 7A). Kinetic plots (Figure 7B) demonstrated that furamidine increased the conversion half-time (t1/2) of the 14Y substrate from 1.9 min in the absence of drug to 2.7 min in the presence of 30 µM furamidine (diamond in Figure 7B) and 4.4 min in the presence of 60 µM furamidine (inverted triangle in Figure 7B). Additionally, increasing Tdp1 concentration was able to overcome Tdp1 inhibition by furamidine (Figure 7C and D). The 50% inhibition of Tdp1 activity observed by 30 µM furamidine with 0.1 ng of Tdp1 was almost completely reversed by increasing the concentration of Tdp1 to 1 ng (Figure 7C and diamond in Figure 7D). Similar effects were seen with 60 µM and 250 µM furamidine (Figure 7C and D). Thus, free Tdp1 competes with furamidine. Together, these results suggest that furamidine produces reversible and competitive inhibition of Tdp1.
Figure 7.
Competitive inhibition of Tdp1 by furamidine. (A) A 100-µl reaction mixture containing 25 nM 14Y and 5 ng of Tdp1 was incubated at pH 8.0 at 25°C in the absence of drug, or in the presence of the indicated concentrations of furamidine. Aliquots were taken at the indicated times (min). Reaction products were analyzed by denaturing PAGE. (B) Densitometry analysis of the gel shown in A. Tdp1 activity measured as the percentage of DNA substrate 14Y converted to 14P (left panel) or substrate 14Y remaining (right panel) as a function of reaction time. (C) Reactions (20 µl) containing 25 nM 14Y and the indicated amounts (ng) of Tdp1 were carried out in the absence or presence of increasing concentrations of furamidine for 20 min. A representative gel is shown. (D) Densitometry analysis of the gel shown in C. Tdp1 activity was calculated as the percentage of DNA substrate 14Y converted to 14P. The horizontal line corresponds to 50% inhibition of Tdp1 activity.
DISCUSSION
Until now, Tdp1 activity has been measured by coupling a 3′-phosphotyrosine to DNA or by isolating small peptide fragments covalently linked to DNA, and resolving the reaction product (3′-phosphate DNA) from substrate in polyacrylamide gel (1–3,8,34,39). In 2002, Cheng et al. (40) used chromagenic substrates like the 3′-(4-nitro)phenyl phosphate DNA (40). However, spectrophotometric detection of 4-nitrophenol required high concentrations of Tdp1 because of its relatively poor extinction coefficient. More recently, fluorescence-based assays have been designed using oligonucleotide and nucleotide substrates containing 3′-(4-methylumbelliferone)-phosphate (DNA-MUP) (41). Tdp1-mediated cleavage releases the 4-methylumbelliferone from the DNA, and the resulting fluorescence can be quantitated (41). Our ‘mix and read’ high-throughput electrochemiluminescent (ECL) assay offers two main advantages. First, the ECL tag (BV-TAG) on the DNA substrate undergoes many excitation/emission cycles, which amplify the output signal, and increases sensitivity (Figure 1C). Since the process is selective for the BV-Tag, the background signal typical of fluorescent methods is eliminated. Second, positive hits are inferred by a gain/restoration of signal as opposed to previous techniques that rely on loss of signal, thereby providing high sensitivity and reliability. More than 70% of the compounds identified by our ECL screen at a single concentration were confirmed as inhibitors of Tdp1 by gel assays. Additionally, the ECL assay is quick, as the high-throughput configuration can process a 96-well plate in ∼10 min. Subsequent to screening the DTP diversity set by the ECL assay (Figure 2), the antiparasitic furamidine (23) was identified as a positive hit for Tdp1 inhibition and further characterized by gel-based assays.
Our results demonstrate that furamidine inhibits the activity of recombinant human Tdp1 at low micromolar concentrations both with single- and double-stranded DNA substrates (see Figures 2A and 3B). Moreover, we provide evidence that furamidine binds single-stranded DNA (see Figure 4B and D). Our results also show that furamidine exhibits no preference for a TT sequence over a CC at the terminus of a single-stranded oligonucleotide for Tdp1 inhibition (see Figure 5). Interestingly, another diamidine, pentamidine was inactive against Tdp1 (see Figure 6) despite the fact that furamidine and pentamidine have an overall similar structural curvature, that closely matches the curvature of the DNA minor groove (23). We also found that berenil was approximately one order of magnitude less effective than furamidine at inhibiting Tdp1. This suggests that the furan linker in furamidine contributes to furamidine's inhibition of Tdp1 activity. We believe that DNA binding is part of the furamidine's mechanism of action. Further studies such as crystallization will be required to fully elucidate its complete mechanism of action. It is plausible that furamidine could form a ternary complex by binding at the interface created by the DNA substrate and the enzyme (42,43).
To date, vanadate, tungstate, aminoglycoside antibiotics and ribosome inhibitors are the only known inhibitors of Tdp1 (18,19). Though vanadate and tungstate bound to Tdp1 provided insights into substrate binding and catalytic mechanism of Tdp1 (19,44), they are general inhibitors of a variety of enzymes involved in phosphoryl transfer. Moreover, vanadate, tungstate, aminoglycoside antibiotics and ribosome inhibitors inhibit Tdp1 activity in biochemical assays only at high (millimolar) concentrations (18). The present study demonstrates that furamidine is by far the most potent inhibitor of Tdp1 reported to date. However, determining the effect of furamidine on Tdp1 in cellular systems would be very hard to interpret as furamidine may have additional target(s) because of its DNA-binding activities. Nevertheless, furamidine presents a prototype, to explore the possibility of generating new chemotypes that would specifically target Tdp1. Evaluation and characterization of other novel Tdp1 inhibitors identified by our ECL screen are ongoing. Our present study with furamidine demonstrates that the ECL assay is a useful tool for the high-throughput screening of Tdp1 inhibitors.
ACKNOWLEDGEMENTS
This project has been supported in part by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research and by federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl Acad. Sci. USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SW, Burgin A.B., Jr, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debethune L, Kohlhagen G, Grandas A, Pommier Y. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3'-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 7.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst.) 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 8.Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc. Natl Acad. Sci. USA. 2006;103:8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 10.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme – DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst.) 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, Pouliot JJ, Boege F, Marko D. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 13.Nivens MC, Felder T, Galloway AH, Pena MM, Pouliot JJ, Spencer HT. Engineered resistance to camptothecin and antifolates by retroviral coexpression of tyrosyl DNA phosphodiesterase-I and thymidylate synthase. Cancer Chemother. Pharmacol. 2004;53:107–115. doi: 10.1007/s00280-003-0717-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl Acad. Sci. USA. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 16.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dexheimer ST, Antony S, Marchand C, Pommier Y. Anti-Cancer Agents in Medicinal Chemistry. 2007. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Z, Thibaut L, Jobson A, Pommier Y. Inhibition of human tyrosyl-DNA phosphodiesterase by aminoglycoside antibiotics and ribosome inhibitors. Mol. Pharmacol. 2006;70:366–372. doi: 10.1124/mol.105.021865. [DOI] [PubMed] [Google Scholar]
- 19.Davies DR, Interthal H, Champoux JJ, Hol WG. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J. Mol. Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 20.Bruno JG. Broad applications of electrochemiluminescence technoloy to the detection and quantitation of microbiological, biochemical and chemical analytes. Rec. Res. Devel. In Micro. 1997;1:25–46. [Google Scholar]
- 21.Deaver DR. A new non-isotopic detection system for immunoassays. Nature. 1995;377:758–760. doi: 10.1038/377758a0. [DOI] [PubMed] [Google Scholar]
- 22.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, et al. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson WD, Nguyen B, Tanious FA, Mathis A, Hall JE, Stephens CE, Boykin DW. Dications that target the DNA minor groove: compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr. Med. Chem. 2005;5:389–408. doi: 10.2174/1568011054222319. [DOI] [PubMed] [Google Scholar]
- 24.Soeiro MN, De Souza EM, Stephens CE, Boykin DW. Aromatic diamidines as antiparasitic agents. Expert Opin. Investig. Drugs. 2005;14:957–972. doi: 10.1517/13543784.14.8.957. [DOI] [PubMed] [Google Scholar]
- 25.Bouteille B, Oukem O, Bisser S, Dumas M. Treatment perspectives for human African trypanosomiasis. Fundam. Clin. Pharmacol. 2003;17:171–181. doi: 10.1046/j.1472-8206.2003.00167.x. [DOI] [PubMed] [Google Scholar]
- 26.Croft SL, Barrett MP, Urbina JA. Chemotherapy of trypanosomiases and leishmaniasis. Trends Parasitol. 2005;21:508–512. doi: 10.1016/j.pt.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Laughton CA, Tanious F, Nunn CM, Boykin DW, Wilson WD, Neidle S. A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity. Biochemistry. 1996;35:5655–5661. doi: 10.1021/bi952162r. [DOI] [PubMed] [Google Scholar]
- 28.Mallena S, Lee MP, Bailly C, Neidle S, Kumar A, Boykin DW, Wilson WD. Thiophene-based diamidine forms a "super" at binding minor groove agent. J. Am. Chem. Soc. 2004;126:13659–13669. doi: 10.1021/ja048175m. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Kumar A, Boykin DW, Bailly C, Wilson WD. Comparative thermodynamics for monomer and dimer sequence-dependent binding of a heterocyclic dication in the DNA minor groove. J. Mol. Biol. 2002;317:361–374. doi: 10.1006/jmbi.2002.5433. [DOI] [PubMed] [Google Scholar]
- 30.Bailly C, Dassonneville L, Carrasco C, Lucas D, Kumar A, Boykin DW, Wilson WD. Relationships between topoisomerase II inhibition, sequence-specificity and DNA binding mode of dicationic diphenylfuran derivatives. Anticancer Drug Des. 1999;14:47–60. [PubMed] [Google Scholar]
- 31.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 32.Mazur S, Tanious FA, Ding D, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. A thermodynamic and structural analysis of DNA minor-groove complex formation. J. Mol. Biol. 2000;300:321–337. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
- 33.Davies DR, Interthal H, Champoux JJ, Hol WG. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Chem. Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 34.Raymond AC, Rideout MC, Staker B, Hjerrild K, Burgin A.B., Jr. Analysis of human tyrosyl-DNA phosphodiesterase I catalytic residues. J. Mol. Biol. 2004;338:895–906. doi: 10.1016/j.jmb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Das BP, Boykin DW. Synthesis and antiprotozoal activity of 2,5-bis(4-guanylphenyl)furans. J. Med. Chem. 1977;20:531–536. doi: 10.1021/jm00214a014. [DOI] [PubMed] [Google Scholar]
- 36.Bray PG, Barrett MP, Ward SA, de Koning HP. Pentamidine uptake and resistance in pathogenic protozoa: past, present and future. Trends Parasitol. 2003;19:232–239. doi: 10.1016/s1471-4922(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 37.Whitelaw DD, Urquhart GM. Maternally derived immunity in young mice to infection with Trypanosoma brucei and its potentiation by Berenil chemotherapy. Parasite Immunol. 1985;7:289–300. doi: 10.1111/j.1365-3024.1985.tb00077.x. [DOI] [PubMed] [Google Scholar]
- 38.Werbovetz K. Diamidines as antitrypanosomal, antileishmanial and antimalarial agents. Curr. Opin. Investig. Drugs. 2006;7:147–157. [PubMed] [Google Scholar]
- 39.Woodfield G, Cheng C, Shuman S, Burgin AB. Vaccinia topoisomerase and Cre recombinase catalyze direct ligation of activated DNA substrates containing a 3′-para-nitrophenyl phosphate ester. Nucleic Acids Res. 2000;28:3323–3331. doi: 10.1093/nar/28.17.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng TJ, Rey PG, Poon T, Kan CC. Kinetic studies of human tyrosyl-DNA phosphodiesterase, an enzyme in the topoisomerase I DNA repair pathway. Eur. J. Biochem. 2002;269:3697–3704. doi: 10.1046/j.1432-1033.2002.03059.x. [DOI] [PubMed] [Google Scholar]
- 41.Rideout MC, Raymond AC, Burgin A.B., Jr. Design and synthesis of fluorescent substrates for human tyrosyl-DNA phosphodiesterase I. Nucleic Acids Res. 2004;32:4657–4664. doi: 10.1093/nar/gkh796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pommier Y, Cherfils J. Interfacial inhibition of macromolecular interactions: nature's paradigm for drug discovery. Trends Pharmacol. Sci. 2005;26:138–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr. Med. Chem. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 44.Davies DR, Interthal H, Champoux JJ, Hol WG. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure. 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]