Abstract
The tumor suppressor, p53, plays critical roles in the cell cycle progression, DNA repair and apoptosis. The PIAS proteins (protein inhibitor of activated STAT) were originally identified as inhibitors of the JAK-STAT pathway. Subsequently, crosstalk between the PIAS proteins and other signaling pathways has been shown to be involved in various cellular processes. Particularly, previous studies have demonstrated that PIAS proteins regulate p53-mediated transcription through sumoylation. hZimp10, also named zmiz1, is a novel PIAS-like protein and functions as a transcriptional co-activator. We recently identified p53 to be an hZimp10 interacting protein in the yeast two-hybrid screen. The interaction between p53 and hZimp10 was confirmed by GST pull-down and co-immunoprecipitation assays. Co-localization of p53 and hZimp10 proteins was also observed within cell nuclei by immunostaining. Moreover, we show that expression of exogenous hZimp10 enhances the transcriptional activity of p53 and knockdown of endogenous hZimp10 reduces the transcriptional activity of p53. Furthermore, using chromatin immunoprecipitation assays, we demonstrate that hZimp10 binds to p53 on the p21 promoter. Finally, p53-mediated transcription is significantly impaired in Zimp10 null embryonic fibroblasts. Taken together, these results provide the first line of evidence to demonstrate a role for Zimp10 in regulating p53 function.
INTRODUCTION
The tumor suppressor protein p53 is a sequence-specific DNA-binding transcription factor that regulates the cell cycle checkpoint pathway in response to DNA damage (1). The p53 gene is the most frequent target for genetic alterations in cancer, with mutations occurring in ∼50% of all human tumors (2,3). The tumor suppressor functions of p53 are directly linked to its ability to control the expression of gene products implicated in cell cycle arrest and apoptosis (4,5). p53 binds as a tetramer to specific response elements located in the transcriptional control regions of p53 target genes, which initiates the recruitment of other transcriptional co-regulators to assemble a transcriptional complex that initiates RNA synthesis (1,6). A broad spectrum of p53 downstream target genes have been identified to be controlled by p53 in a positive or negative manner (7). The fact that a variety of pathways are mediated by these p53 target genes demonstrates the role of p53 as an integrator of diverse cellular signals.
The p53 protein contains two N-terminal activation domains, a DNA-binding domain, and a C-terminal oligomerization domain (1). The critical step for p53-mediated transcriptional activation is facilitated by the ability of p53 to simultaneously bind to specific DNA sequences and recruit CBP/p300 and other transcriptional co-regulators to p53-responsive promoters. CBP/p300 recruitment appears to concomitantly bring the general transcription machinery, including TFIIB and TBP, and RNA polymerase II to the target promoters (8,9). Although a wealth of information exists concerning p53, it is unclear about the actual mechanism by which this critical tumor suppressor protein directly interacts with its target genes and co-regulators to mediate its transcriptional activity.
The PIAS proteins (protein inhibitor of activated STAT) were first identified as transcriptional co-regulators of the JAK-STAT pathway (10). PIAS1 and PIAS3 can inhibit the activity of STAT1 and STAT3, respectively (11–13). Recent studies imply that the PIAS proteins may play a role in chromatin modulation through sumoylation (14,15). Sequence analysis indicates that the SUMO E3 ligase RING domain shares significant homology with the Miz domain of PIAS proteins (16). Several PIAS proteins, such as PIASxα, xβ, 1 and 3, have been shown to interact with SUMO-1 and Ubc9 to sumoylate a variety of transcriptional factors and other regulatory proteins (17–23). Particularly, it has been shown that the transcriptional activity of p53 can be regulated by PIAS through sumoylation (24,25).
hZimp10 and hZimp7, also named zmiz1 and zmiz2, respectively, are novel PIAS-like proteins that share a ring finger domain, termed Miz (msx-interacting zinc finger), with other PIAS proteins (26,27). This domain has been shown to be important for PIAS-target protein interactions and post-translational modifications (15). A novel Drosophila gene, termed tonalli (tna), appears to be the ortholog of hZimp7 and 10 (28). The protein encoded by tna genetically interacts with the SWI2/SNF2 and Mediator complexes, implying a potential role for the hZimp proteins in transcription. To further explore their roles in transcription, we performed a yeast two-hybrid screen to seek out potential interacting proteins of hZimp7 and 10. Intriguingly, p53 was identified in the screen. Using different in vitro and in vivo approaches, we demonstrated that hZimp10 physically interacts with the p53 protein, and through the interaction hZimp10 augments p53-mediated transcription. These data elucidate a link between hZimp10 and p53 and demonstrate that hZimp10 is a transcriptional co-regulator of p53.
MATERIALS AND METHODS
Yeast two-hybrid system
Yeast two-hybrid experiments were performed as described previously (29). The DNA fragments containing truncated hZimp10, hZimp7 or hLZTS2 were fused in frame to the GAL4 DBD in the pGBKT7 vector (CLONTECH Laboratories, Inc., Palo Alto, CA, USA). Truncated p53 (amino acids: 251–383) was fused to the GAL4 TAD in the pVP16 vector (CLONTECH). The constructs were transformed into the modified yeast strain PJ69-4A (30). Transformants were selected on Sabouraud Dextrose medium lacking tryptophan, leucine and/or adenine. The specificity of the interaction with p53 was measured by a liquid β-galactosidase (β-gal) assay (29).
Recombinant DNAs
Full-length hZimp10 and hZimp7 cDNA was identified and sub-cloned into pcDNA3-FLAG vector as described previously (26,27). Subsequently, truncated mutants of hZimp10 or hZimp7 were generated and sub-cloned into the pGBKT7 vector containing a GAL4 DBD for the yeast two-hybrid assay or into pGEX4T3 for making GST fusion proteins. Truncated p53 (amino acids: 251–383) was cloned into the pVP16 vector containing the transcriptional activation domain of VP16. Double-stranded oligonucleotides corresponding to human hZimp7 (Z7-1: 5′-GGACTGCATTATAAGCCTAC-3′, Z7-2: 5′-GGACACCAGGACTACACACC-3′, and Z7-3: 5′-GGTGGAGCAGACAGCTATCA-3′) and hZimp10 (Z10-1: 5′-GGCCTCCATTACATCACAGT-3′, Z10-2: 5′-GGCAGCAGCAGCAGTTCTCA-3′ and Z10-3:5′-GGCACCAACTCCAACGACTA-3′) were cloned into the pBS/U6 vector to generate the short hairpin RNA (shRNA) (31). Subsequently, the U6 promoter and the hZimp7 or hZimp10 shRNA sequences were PCR amplified and transferred into the pLentiSuper vector (Invitrogen, Carlsbad, CA, USA). The viral vector was co-transfected with other packaging plasmids into human embryonic kidney 293T cells for the virus production (32). The pGL2hmdm-HX-Luc reporter was a kind gift from Dr Moshe Oren (The Weizmann Institute of Science, Israel). The p21-Luc reporter, pCMV-NEO-Bam p53 and the p53-induced promoter/reporter PG13-Luc were generously given by Dr Bert Vogelstein (Johns Hopkins, Baltimore, MD, USA). The hZimp10 adenoviral expression vector was cloned into the pAdTrack shuttle vector (33). The plasmids were then cleaved with PmeI, and transformed into BJ5183 cells that contain pAdEasy-1 vector. Adenoviral vectors were amplified in DH5α cells, and propagated in HEK293 cells. Viral titers were determined using plaque assays.
Cell cultures and transient transfections
The HCT116 p53+/+ and HCT116 p53−/− human colon carcinoma cell lines are gifts from Dr Bert Vogelstein, which were maintained in McCoy's medium with 10% fetal bovine serum (FBS, HyClone Laboratories). The SaOS-2 and U2OS cell lines are gifts of Dr Giannino Del Sal (Laboratorio Nazionale C.I.B. Italy) and were cultured in DMEM with 10% FBS. The human breast cancer cell line, MCF7, and embryonic kidney cell line, HEK293, were grown in 10% FBS-DMEM.
Transient transfections were carried out using LipofectAMINE2000 (Invitrogen, Carlsbad, CA, USA). Approximately 1.5 × 104 cells were seeded into a 48-well plate 16 h before transfection. Approximately 200 ng of total plasmid DNA per well were used in transfection. Total cell lysates were collected 8–12 h after transfection and then luciferase and β-gal activities were measured in a Monolight 3010 luminometer (Pharmingen, San Diego, CA, USA). Luciferase activity was normalized by β-gal in the same samples and reported as relative light units (RLU). Individual transfection experiments were done in triplicate and the results are reported as mean RLU (±SD) from representative experiments.
GSTpull-down assay
Expression and purification of GST fusion proteins were performed as described previously (34). Equal amounts of GST-fusion proteins coupled to glutathione sepharose beads were incubated with MCF7 cell lysate at 4°C for 2 h in binding buffer (20 mM Tris-HCl-pH 7.8, 180 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 0.5 mM ZnCl2, 10% glycerol, 0.1% NP-40, 0.05% dry non-fat milk, 1 mM DTT, 0.5 mM PMSF). Beads were carefully washed three times with binding buffer and then analyzed by SDS-PAGE followed by western blot analysis using a p53 antibody (DO-1: sc-126 Santa Cruz Biotechnology).
Cross-linking, immunoprecipitation and western blotting
The pcDNA3-HA-p53 with or without pcDNA3-FLAG-hZimp10 vector was transfected into HEK293 cells. After 48 h of transfection, cells were incubated with 0.5 mM dithio-bis (succinimidyl propionate) (DSP) in PBS to cross-link for 30 min, washed once with 10 mM Tris-HCl pH 7.6, and incubated with the same buffer for 5 min to neutralize DSP. The whole cell lysates were prepared as described previously (26), and incubated with different antibodies at 4°C with gentle rotation overnight. Then equilibrated Protein-A Sepharose beads were added for 1.5 h at 4°C, and then collected. The beads were washed, proteins eluted using 2× sample buffer (125 mM Tris-HCl pH 6.8, 4% SDS, 20% (v/v) glycerol, 0.004% bromphenol blue), and analyzed by western blot. Detection was performed with ECL reagents according to manufacturer's protocol using ECL Hyperfilm (Amersham).
RNA isolation, RT-PCR and quantitative PCR assays
MCF7 cells were infected with either the hZimp10 expression adenovirus or a control virus in 5% FBS-DMEM, then UV irradiated at 80 J/m2, and cultured in the medium for 9 h. Total RNA was then extracted using RNAWiz RNA isolation reagent (Ambion, TX; Cat#: 9736). The reverse transcription polymerase chain reaction (RT-PCR) was carried out as described previously (35). In a 50 µl PCR reaction, 1 μl of cDNA was amplified using 25 cycles of 45 s at 94°C, 30 s at 61°C (for p21) or 50°C (for GAPDH) and 30 s at 72°C. Primers for p21 (5′-ATGTCAGAACCGGCTGGGGAT-3′; 5′-GGAGTGGTAGAAATCTGTCATGC-3′), mdm2 (5′ –ATGTGCAATACCAACATGTCTGTACC-3′; 5′ –TTTGGTCTAACCAGGGTCTCTTGT-3′) and GAPDH (5′-CCATGGAGAAGGCTGGGG-3′; 5′-CAAAGTTGTCATGGATGACC-3′) were used in the PCR reaction. For quantitative PCR, cDNA samples were mixed with SYBR qPCR Super Mix Universal (Invitrogen) and specific primers in the MX 3005P thermocycler (Stratagene). Relative mRNA levels were calculated from the point where each curve crossed the threshold line as reported previously (36,37). Reactions were done in triplicate and the values were normalized by the GAPDH expression level.
Chromatin immunoprecipitation (ChIP) and re-ChIP assays
MCF7 or HEK293 cells were irradiated at 40 J/m2 using a UV Stratalinker 1800 (Stratagene) and then incubated at 37°C for 6 h. Subsequently, cells were treated with DSP to cross-link protein complexes and washed with Tris-wash buffer. After the last wash with PBS, cells were treated with formaldehyde and subjected to ChIP analysis as described previously (38). Briefly, cells were collected and washed sequentially with cold PBS, Wash Buffer I (0.25% Triton X100, 10 mM EDTA, 0.5 mM EGTA and 10 mM HEPES, pH 6.5), and Wash Buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and 10 mM HEPES, pH 6.5). Cells were then lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.1 and protease inhibitors). The chromatin was sheared to an average size of 800 bp by sonication and diluted 10-fold in ChIP dilution buffer (2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1, 1% Triton X-100), and then subjected to immunoprecipitation with either an anti-Zimp10 chicken antibody or anti-p53 antibody (Santa Cruz) for overnight at 4°C and recovered with goat α IgY-agarose (Aves Labs) or Protein A Sepharose (Amersham). The immunoprecipitates were serially washed with different TSE-based buffers and eluted for PCR analysis. For re-ChIP, the immunocomplexes were eluted with re-ChIP elution buffer (10 mM DTT), and the supernatant was diluted 1:40 in ChIP dilution buffer. Antibodies against the second proteins of interest were added, incubated at 4°C overnight, and collected by incubating with either goat anti-chicken IgYagarose or protein-A beads at 4°C for 2 h. In both ChIP and re-ChIP assays, the immuno-complexes were eluted from the beads through incubation with 10X bead volume of elution buffer (1% SDS, 0.1 M NaHCO3). Cross-links were reversed by incubating elution samples at 65°C for 6 h and chromatin fragments were purified with the PCR Purification Kit (Qiagen). ChIP and input DNA were analyzed by PCR using p21 promoter-specific primers, 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′, respectively (39,40). P21 PCR parameters were: 95°C for 5 min then 33 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 20 s. The samples were also amplified with GAPDH primers, 5′-CGGTGCGTGCCCAGTTG-3′ and 5′-GCGACGCAAAAGAAGATG-3′, as controls (41).
Immunostaining
HEK293 cells were seeded overnight and synchronized with 0.5 mM mimosine (Sigma Chemical Co., St. Louis, MO, USA) as described previously (42). Cells were released from the mimosine block by washing three times with PBS and incubating in fresh 10% FBS-DMEM at 37°C for 12 h. Cells were fixed with 4% paraformaldehyde and immunostained with either an anti-Zimp10 or anti-p53 antibody followed by incubation with species-specific Alexafluor 488 and 594-conjugated secondary antibodies. Images were analyzed by confocal microscopy with 60× and 40× objectives.
Mouse embryonic fibroblasts
Mice heterozygous for a neomycin-disrupted allele of the Zimp10 gene were mated, and embryos were harvested from the females at embryonic day 9.5. Embryos were isolated in cold PBS and digested with 250 μl trypsin (0.05%) as described previously (34). Cells were directly plated into 48-well plates to adhere overnight, and used for transient transfection assays. To determine MEF genotype, embryo sacs isolated during the dissection were digested, genomic DNA was extracted, and the wild-type or mutant Zimp10 alleles were determined by PCR with specific primers.
RESULTS
Identification of p53 as a potential hZimp10 interacting protein
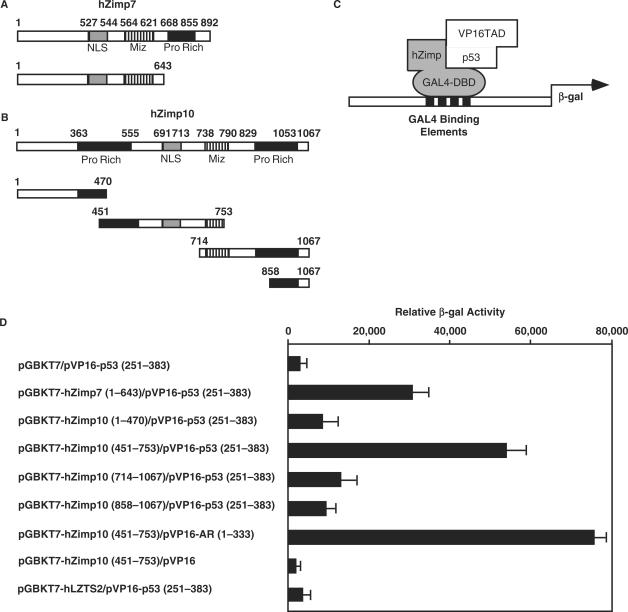
Using a bait construct containing the N-terminal region of hZimp7 (amino acids: 1–643), which is also highly conserved in hZimp10, we employed a modified yeast two-hybrid system to identify proteins that potentially interact with hZimp7 and/or hZimp10. Of 3.4 × 107 transformants, 123 grew under selective conditions and showed increased adenine and β-gal production in medium. Rescue of the plasmids and sequencing of the inserts revealed several different cDNAs. Most of them were transcriptional factors and PIAS proteins. Among these clones, a cDNA encoding the tumor suppressor p53, between amino acids 251 and 383, was identified. Since hZimp7 and hZimp10 share significant sequence similarity, we co-transformed the p53 clone with various constructs containing either GAL-DBD alone or the fusion proteins with the N-terminal fragment of hZimp7 (1–643 amino acids), and different truncations of hZimp10 (Figure 1A–C). The original bait construct, pGBKT7-hZimp7 (1–643aa), showed a specific interaction with pVP16-p53 (251–383aa) (Figure 1D). Interestingly, the construct containing the fragment of hZimp10 between amino acids 451–753 appeared to interact with the pVP16-p53 as well. The above results provide the first line of evidence to demonstrate an interaction between hZimp proteins and the tumor suppressor protein p53.
Figure 1.
Specific interaction between hZimp10 and p53. (A) and (B): The cDNA fragments containing different portions of human Zimp7, Zimp10 or human LZTS2 were fused to the GAL4-DBD in the pGBKT7 vector. Numbers correspond to amino acid residues. (C) A schematic representation of the yeast two-hybrid assay for mapping the interaction between p53, hZimp7 and hZimp10 proteins. (D) The pVP16-p53 containing the fusion protein of VP16-TAD and p53 (251–383) were co-transformed with pGBKT7 vector alone or different pGBKT7 fusion constructs. In addition, pVP16-AR (1–333) was included as a positive control, for the pGBKT7-hZimp10 (451–753). Transformed cells were plated on SD-Ade-Leu-Trp plates, and SD-Leu-Trp plates to monitor transformation efficiency. Three independent colonies were inoculated from each transformation experiment for subsequent liquid β-gal assays. The data for the liquid β-gal assays are shown as the mean ± S.D.
hZimp10 interacts with p53 in vitro and in vivo
The interactions between hZimp7 and hZimp10 with p53 were further assessed by GST pull-down experiments. Truncated hZimp7, hZimp10 and the PIAS1 SAP (Scaffold attachment factor, Acinus and PIAS) and MIZ domains were cloned in frame to generate GST fusion proteins. They were expressed upon IPTG induction and equalized on a SDS–PAGE gel with comassie blue staining (Figure 2A). MCF7 cell lysates were applied to each sample, which was immobilized onto a glutathione-sepharose matrix for the binding assay. The elutions from the above samples were analyzed by SDS-PAGE and western blot with a p53 antibody. PIAS1 and Zimp proteins contain a conserved Miz domain that shares similar sequences with the ring finger domain of MDM2 (24). Therefore, we tested whether the Miz domains of PIAS1 and Zimp10 interact with p53. Intriguingly, only a weak interaction was observed between the Miz domains and endogenous p53 proteins (Figure 2A). In addition to the Miz domain, the SAP (Scaffold attachment factor A/B, Acinus and PIAS) domain has also been shown to be involved in protein–protein interactions (43). Thus, we included it in our binding assays, although only slight binding activity above background was observed with this construct. Notably, the GST-fusion construct containing amino acids 451–753 of hZimp10 showed the strongest binding activity with p53. Since this region only covers a small portion of the Miz domain, this result suggests that the Miz domain of hZimp10 may not be required for the interaction with p53. In addition, the GST-fusion protein containing the N-terminal fragment of hZimp7 also showed an interaction with p53, which is consistent with the yeast two-hybrid results. Taken together, the above results show that the regions spanning hZimp10 amino acids 451 to 753 and hZimp7 amino acids 1–643 are mainly responsible for binding to p53.
Figure 2.
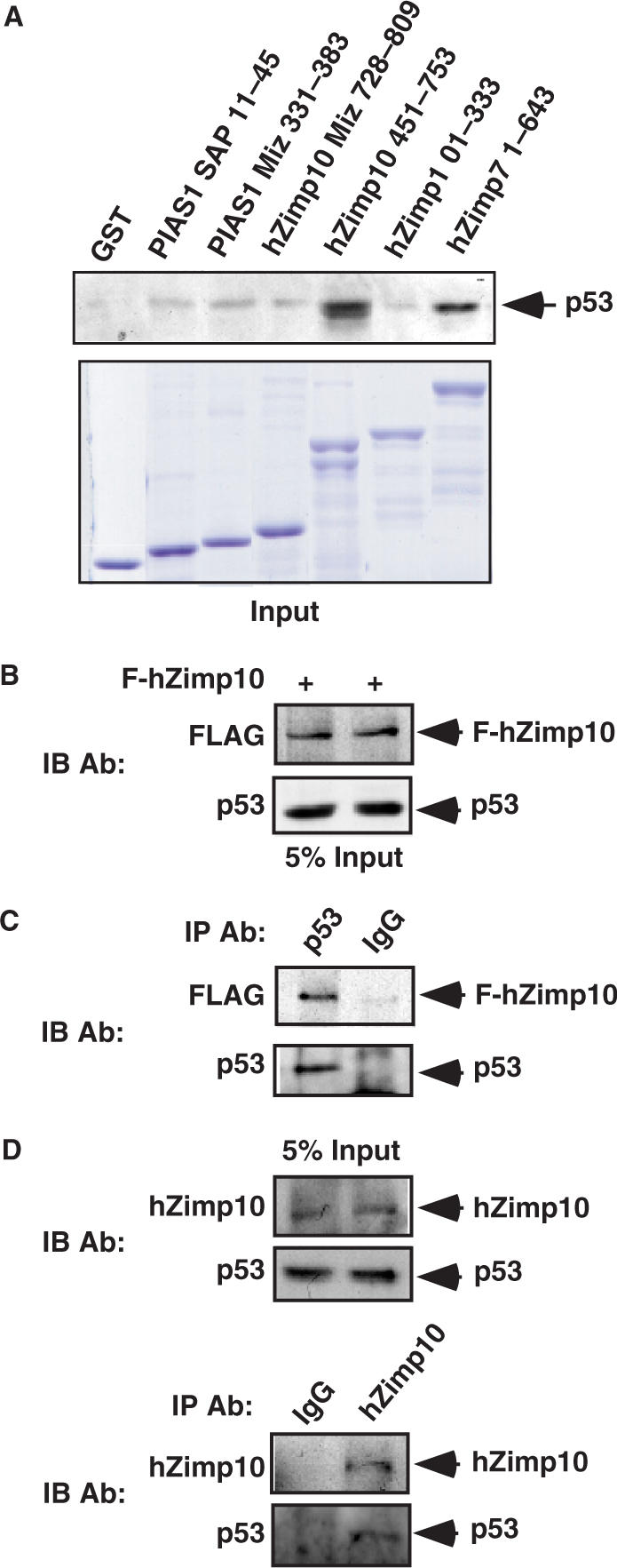
Physical interaction between hZimp10 and p53 in vitro and in intact cells. (A) Equal amounts of GST-PIAS1 MIZ domain, GST-PIAS1 SAP domain, GST-hZimp10 MIZ (728–809 amino acids), GST-hZimp10 Central (451–753 amino acids), GST-hZimp10 N-terminal (1–333 amino acids) and hZimp7 N-terminal (1–643 amino acids) fusion proteins were used to pull down endogenous p53 in MCF7 cells. GST protein alone was used as a negative control. Equal amounts of the above GST proteins were analyzed on SDS-PAGE. Material bound to GST columns was subjected to SDS-PAGE and western blot with p53-specific antibody. (B) HEK293 cells were transfected with pcDNA3-HA-p53 (0.1 μg) and pcDNA3-FLAG-tagged hZimp10 (1.8 μg). Here, 7.5% of the total lysate volume (input) was probed with anti-FLAG antibody or anti-p53 antibody. (C) Cell lysates were then immunoprecipitated with the anti-p53 antibody or normal IgG, and analyzed on SDS-PAGE by Flag antibody or p53 antibody. (D) Five percent of the initial lysate volume (input) or Zimp10 and IgG immunoprecipitates were analyzed by western blot using either hZimp10 or p53 antibody.
To confirm that hZimp10 interacts with p53 in intact cells, co-immunoprecipitation assays were carried out to detect potential protein complexes. Initially, we co-transfected FLAG-tagged hZimp10 with a p53 expression vector in HEK293 cells (Figure 2B). Whole cell lysates containing FLAG-hZimp10 and p53 proteins were immunoprecipitated with normal mouse IgG or a anti-p53 antibody. As shown in Figure 2C, Flag-hZimp10 proteins were only detected in the p53 immunoprecipitate but not in normal IgG immunoprecipitate. These data indicate that p53 can form a protein complex with hZimp10 in intact cells.
Next, we further evaluated the interaction between endogenous hZimp10 and p53 proteins in HEK293 cells. With specific antibodies against p53 and hZimp10, we detected the expression of both proteins in HEK293 cells (Figure 2D). Immunoprecipitation of the whole cell lysates with a homemade hZimp10 antibody (26) revealed that p53 forms a protein complex with hZimp10, which provides evidence to demonstrate that hZimp10 and p53 interact endogenously.
hZimp10 augments p53-mediated transcription
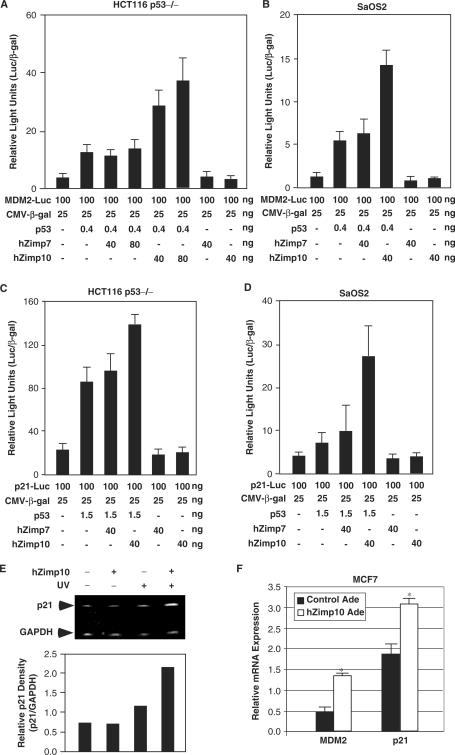
Since hZimp7 and 10 have been shown to act as transcriptional co-activators (26,27,34,44), we tested whether these Zimp proteins regulate p53-mediated transcription. In order to avoid potential confounding effects of endogenous p53, we chose p53-null cells for these experiments. A luciferase reporter driven by the MDM2 promoter (MDM2-Luc) was co-transfected with plasmids expressing p53, hZimp7 and hZimp10 into HCT116 p53−/− colon cancer cells in various combinations (Figure 3A). An approximate 2-fold increase of p53-mediated transcriptional activity above the baseline was observed when cells were transfected with 0.4 ng of p53 expression vector. The p53 activity was further increased ∼7- or 9-fold in the presence of 40 or 80 ng of hZimp10, respectively, above the baseline. In contrast, co-transfection of hZimp7 showed no significant effect (Figure 3A), which is consistent with the yeast two-hybrid and GST pull-down assays that showed only a weak interaction between p53 and hZimp7. There was no effect when hZimp7 or hZimp10 were transfected alone with the reporter plasmid, indicating that the Zimp10-mediated induction in reporter activity was indeed through p53 (Figure 3A). To confirm our findings, we repeated the transient transfection assay in SaOS2 osteosarcoma cells, another p53 negative cell line. A similar augmentation of hZimp10 on p53-mediated transcription was observed on the MDM2 promoter/reporter (Figure 3B). Moreover, we evaluated the effect of hZimp10 on p21 promoter, another downstream target of p53. A similar enhancement by hZimp10 was observed on p21 promoter in HCT116 p53−/− and SaOS2 cells (Figure 3C and D). These results provide evidence to demonstrate that hZimp10 augments p53-mediated transcription.
Figure 3.
hZimp10 augments p53-mediated transcription. (A) HCT116 p53−/− cells were transiently transfected in 48-well plates with 100 ng of MDM2-Luc, 25 ng of pSV40-β-gal, 0.4 ng of pCMV-p53, and where indicated, 40 or 80 ng of pcDNA3-FLAG-hZimp7 or pcDNA3-FLAG-hZimp10. The total amount of plasmid per well was normalized in all transfections by addition of pcDNA3 empty vector. Luciferase activity is reported as relative light units and represented as mean ± SD. (B) SaOS2 cells were transiently transfected in 48 wells as described above. (C) and (D) Transient transfection assays were performed in HCT116 p53−/− and SaOS2 cells with the p21-Luc reporter cells, respectively. (E) MCF7 cells were infected with either the hZimp10 expression adenovirus or a control virus, and then UV irradiated at 80 J/m2 as indicated. Semi-quantitative RT-PCR with p21 or GAPDH primers was performed on the total RNA isolated from each sample. PCR products were subjected to electrophoresis on a 5% acrylamide gel. The bar graph is a representation of the band intensity measured by densitometry. (F) Quantitative RT-PCR assays were performed to detect the levels of p21 and MDM2 transcripts in MCF cells that were either the hZimp10 expression adenovirus or a control virus. The detailed experimental procedures were described in the Materials and Methods section.
To investigate the effect of hZimp10 in regulating the transcriptional activity of endogenous p53, MCF7 cells, which possess wild-type p53, were infected with either hZimp10 expression adenoviruses or control GFP viruses, and treated with or without UV irradiation. The endogenous transcripts of the p53 target gene, p21, were then measured by semi-quantitative RT-PCR. The level of p21 transcript showed no significant change in cells in the absence of UV treatment (Figure 3E). However, in cells treated with UV irradiation, the level of p21 transcript was increased ∼1-fold in cells with ectopically expressed hZimp10 in comparison with the cells infected with GFP control viruses. The enhancement of hZimp10 on p53-meidated transcription in MCF7 cells was further evaluated by quantitative PCR assays. As shown in Figure 3F, the levels of MDM2 and p21 transcripts were higher in cells expressed exogenous hZimp10 proteins than control cells (P < 0.05). These results support our initial observation in the transient transfection experiments, and further demonstrate a functional role for hZimp10 in augmenting p53-mediated transcription.
Endogenous Zimp10 is required for p53-mediated transcription in vivo
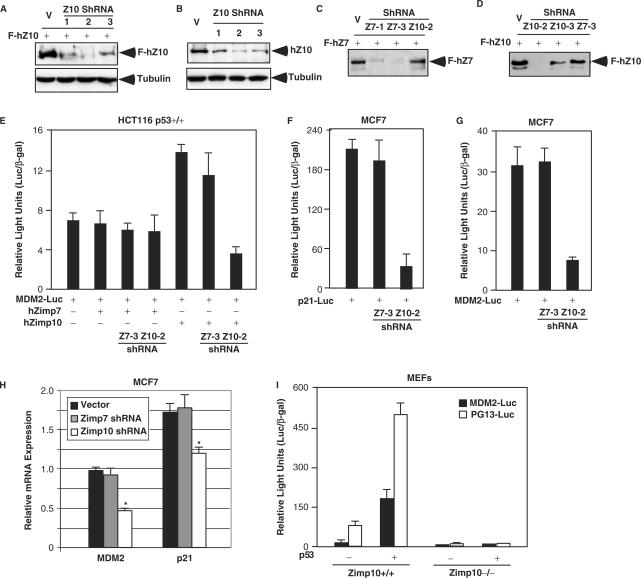
Next, we extended our study to investigate the involvement of endogenous hZimp10 in regulating the transcriptional activity of p53. We first generated three short hairpin RNA (shRNA) constructs for hZimp10 (31) and tested their knockdown effects on ectopically expressed hZimp10 in CV1 cells. All three hZimp10 shRNA constructs reduced the expression of FLAG-tagged hZimp10 protein (Figure 4A). However, there was no change in tubulin expression in the same samples. The hZimp10 shRNA construct 2 appeared most effective in the knockdown experiment using overexpressed hZimp10 and also significantly reduced the expression of endogenous hZimp10 protein in HEK293 cells (Figure 4B). In addition, we evaluated the specificity of shRNA vectors for hZimp7 and hZimp10 in MCF7 cells. As shown in Figure 4C and D, the shRNA vectors specific for hZimp7 or hZimp10 showed selective knockdown for their respective targets. In HCT116 p53+/+ cells, the hZimp10 shRNA construct selectively reduced hZimp10 enhancement of p53-mediated transcription (Figure 4E), which is consistent with the western blot results. Moreover, knockdown of endogenous hZimp10 expression by the hZimp10 shRNA vector in MCF7 cells resulted in a 5-fold reduction in p53-mediated transcription on both the p21-Luc and MDM2-Luc reporters (Figure 4F and G). In contrast, there was no change in p53 transcriptional activity in samples transfected with hZimp7 shRNA constructs. Using quantitative PCR assays, we further assessed the suppressive effect of the hZimp10 shRNA on the expression of endogenous p21 and MDM2 transcripts. The levels of p21 and MDM2 mRNAs were reduced in cells infected with hZimp10 shRNA-2 lentiviruses whereas cells either infected with the control viruses or hZimp7 shRNA viruses showed no effect (Figure 4H). Taken together, the above data indicate that endogenous hZimp10, but not hZimp7, plays an important role in the augmentation of p53-mediated transcription.
Figure 4.
Knockdown of hZimp10 expression reduces p53 transcription. (A) CV1 cells were transfected with 2 μg of pcDNA3-FLAG-hZimp10 (F-hZ10) and 200 ng of different pBS/U6-hZimp10-shRNA constructs (Z1, Z2 and Z3), or PBS/U6 vector only in 6-well plates. Whole cell lysates were prepared after 48 h of transfection, and analyzed by western blotting with either FLAG or Tubulin antibody. (B) Different hZimp10 shRNA constructs or PBS/U6 vector were transfected into HEK293 cells. Cells were harvested and cell lysates were analyzed as described in (A). (C) and (D) CV1 cells were transfected with 2 μg of pcDNA3-FLAG-hZimp10 or pcDNA3-FLAG-hZimp7 (F-hZ7) in the presence of 200 ng either hZimp10 or hZimp7 shRNA constructs as indicated in the figure to check for knockdown specificity. (E) Luciferase and β-gal activities were measured in whole cell lysates as described above. HCT116 p53+/+ or MCF7 cells were stably infected with either control lentivirus, hZimp7 shRNA lentivirus or hZimp10 shRNA lentivirus and transfected in 48-well plates with 100 ng of MDM2-Luc or 100 ng p21-Luc, 25 ng of pSV40-β-gal, 40 ng of pcDNA3-FLAG-hZimp10 or pcDNA3-FLAG-hZimp7. (H) The levels of p21 and MDM2 transcripts were assessed by quantitative RT-PCR assays in MCF cells that were infected with hZimp10 shRNA-2, hZimp7 shRNA-3, or control lentiviruses. The experimental details were described in the Materials and Methods section. (I) Mouse embryo fibroblasts isolated from either wild type or −/− animals were transfected with 100 ng of either MDM2-Luc or PG13-Luc either 3TP-luc and 25 ng of CMV-β-galactosidase in the presence or absence of 20 ng of p53 expression vector. Forty-eight hours after transfection, luciferase and β-galactosidase values were measured, and relative luciferase/β-galactosidase units were calculated. Bars represent the mean and SD of triplicate determinations. Similar results were obtained from two independent MEF isolations.
To further demonstrate the role of hZimp10 in a more biologically relevant setting, we have recently generated mice in which the Zimp10 gene locus has been disrupted by replacing exons 8–10 with a neomycin resistance cassette. The phenotype of this disruption is embryonic lethality at approximately E10.5 (Beliakoff et al., unpublished data). To determine whether endogenous Zimp10 regulates p53-mediated transcription, we generated mouse embryo fibroblasts (MEFs) from E9.5 day embryos and transfected them with the p53-responsive MDM2-Luc and pG13-Luc reporters (45). As shown in Figure 4I, ∼4-fold induction of luciferase activity by ectopically expressed p53 on either MDM2 or pG13 promoters, respectively, was observed in MEFs prepared from wild-type embryos (Zimp10+/+). In contrast, no activity was observed in MEFs where both Zimp10 alleles were disrupted (Zimp10−/−). These data demonstrate a crucial role for endogenous Zimp10 in the regulation of p53-mediated transcription in vivo.
p53 and hZimp10 co-localize in the nucleus
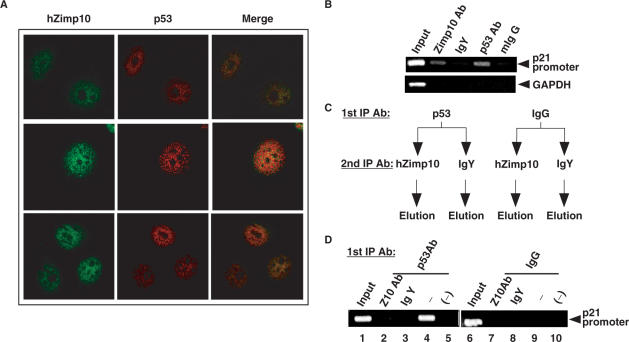
To examine whether a dynamic interaction between p53 and hZimp10 exists in cells, we detected the cellular distribution of both endogenous p53 and hZimp10 proteins using specific antibodies. HEK293 cells were synchronized by adding 0.5 mM mimosine overnight. Cells were then washed three times with PBS and stimulated with full medium for 12 h (42). As shown in Figure 5A, both p53 and Zimp10 proteins show a strong nuclear distribution in human HEK293 cells, which is consistent with previous reports (26,46,47). Intriguingly, a significant amount of overlay between endogenous p53 and hZimp10 proteins was observed in these cells. Based on these observations, we conclude that hZimp10 can co-localize with p53 in the nucleus, where the proteins may form a ternary transcriptional complex.
Figure 5.
p53 and hZimp10 co-localize with each other in the nucleus and form a ternary transcriptional complex. (A) HEK293 cells were synchronized with 0.5 mM mimosine in culture media for 24 h, washed three times with PBS and incubated with fresh media for 12 additional hours. Cells are immunostained with either a chicken antihuman Zimp10 or anti-p53 antibody followed by incubation with species-specific Alexafluor 488 and 594-conjugated secondary antibodies, respectively. Images were analyzed by confocal microscopy with 60× and 40× objectives. (B) HEK293 cells were seeded on three 10 cm dishes at 85% confluency and allowed to adhere overnight. The cells were UV irradiated at 40 J/m2 UV, cross-linked with DSP (1.6 mM), then cross-linked with 1% formaldehyde before chicken anti-human Zimp10, IgY, mouse anti-human p53, or mouse IgG antibody were added to immunoprecipitate chromatin. Primers specific to the distal p53 responsive element on the p21 promoter (−2301) were used to PCR amplify the eluted chromatin. Primers specific to GAPDH were used as a control to monitor immunoprecipitation specificity. (C) Schematic representation of ChIP and re-ChIP procedure. (D) HEK293 cells were prepared as described in (A) and mouse anti-human p53 or mouse IgG antibody was added to immunoprecipitate chromatin. The elution was subjected to a second immunoprecipitation by chicken antihuman Zimp10 or IgY antibody. The final elution was analyzed by p21 promoter-specific primers as described in (B).
To demonstrate the direct involvement of hZimp10 to coordinate p53-mediated transcription, chromatin immunoprecipitation (ChIP) assays were performed to detect the occupancy of hZimp10 on p53-regulated promoters. HEK293 cells were grown in DMEM with 5% FBS. Soluble chromatin was prepared after formaldehyde treatment of the cell cultures, and specific antibodies against hZimp10 were used to immunoprecipitate hZimp10-bound genomic DNA fragments. The genomic DNA was analyzed by PCR using specific pairs of primers spanning the p53-binding sites in the p21 promoter (Figure 5B). Both p53 and hZimp10 recruitment was detected within the region of the p21 promoter that contains a functional p53-binding site (40) in HEK293 cells after UV treatment. As the average length of the genomic DNA fragments produced in these experiments was ∼800 bp (data not shown), we could not distinguish with certainty whether the occupancy of hZimp10 on the p21 promoter is through an interaction with p53 or through interactions with other DNA-binding proteins. Therefore, we performed re-ChIP assays to assess the relationship between p53 and hZimp10 on the p21 promoter (Figure 5C). Using the hZimp10-specific antibody, we re-immunoprecipitated the elutions from the immunoprecipitates with a p53 antibody or normal IgG. As shown in Figure 5D, the presence of hZimp10 on the p21 promoter was selectively detected in the immunoprecipitates using the p53 antibody but not the IgG control. In addition, the samples re-immunoprecipitated with anti-hZimp10 showed a more intense p21 promoter PCR fragment than the ones using normal IgY. These data suggest that the recruitment of hZimp10 onto the p21 promoter is mediated through p53.
DISCUSSION
The p53 tumor suppressor is a DNA sequence-specific transcriptional factor that is mutated in ∼50% of human tumors (3). In response to a variety of cellular signals, perhaps the most well studied is its DNA damage function. p53 regulates the transcription of numerous genes involved in different cellular processes, including cell cycle arrest and cell death. Like other transcriptional factors, the transcriptional activity of p53 is largely dependent on its ability to recognize and bind specific DNA sequences and to recruit other necessary transcriptional co-regulators. In recent years, numerous transcription co-regulators have been shown to either directly or indirectly interact with p53 to modulate its transcriptional activity. For instance, physical and functional interactions between p53, p300 and HAT proteins have been well documented (48,49). The involvement of PRMT1 and CARM1 methyltransferases has also been demonstrated in previous studies (50). Importantly, p53 has been shown to facilitate formation of a preinitiation complex via direct interactions with the components of the general transcription complex (51). The experiments reported here demonstrate a specific protein–protein interaction between p53 and hZimp10, a novel PIAS-like protein (26). The interaction was first identified by a modified yeast two-hybrid screen. Using GST pull-down and immunoprecipitation assays, we then show that p53 binds to hZimp10 both in vitro and in intact cells. Moreover, immunofluorescence assays demonstrated that p53 co-localizes with hZimp10 within cell nuclei. Furthermore, analysis of the interaction by ChIP (chromatin immunoprecipitation assay) on the promoter of the p21 gene, a downstream target of p53, showed that hZimp10 is involved in the p53-mediated transcriptional complex. Taken together, these multiple lines of evidence clearly indicate that p53 and hZimp10 can specifically interact in a biologically relevant manner.
To search for the biological consequence of the interaction between p53 and hZimp10, we performed a series of experiments to assess the effect of hZimp10 on p53-mediated transcription. As shown in this article, hZimp10 acts as a transcriptional co-activator to augment p53-mediated transcription. We observed that expression of exogenous hZimp10 or knockdown of endogenous hZimp10 affects p53-mediated transcription on both the p21 and Mdm2 promoters. Introducing exogenous hZimp10 into MCF7 cells also augments endogenous p53-mediated transcription by increasing p21 transcript levels. Interestingly, hZimp10 consistently up-regulates p53-mediated transcriptional activity in all cell contexts examined to date. This result is consistent with the observation that hZimp10 harbors a strong intrinsic transactivation domain within its C-terminus (26). It appears that through this domain hZimp10 can act as a transcriptional co-activator to augment p53-mediated transcription, which is consistent with previous observations showing that hZimp10 functions as a transcriptional co-activator of the androgen receptor and Smad3 (26,34).
It has been shown that the transcriptional activity of p53 can be regulated by multiple post-translational modifications, including phosphorylation, ubiquitination and acetylation (52). In addition, p53 can also be covalently modified by sumoylation, which is mainly regulated through SUMO-1 (Small Ubiquitin-related Modifier 1) (53,54). Recent studies have shown that PIAS proteins can bind to, sumoylate, and influence the activity of p53 (22,25). In particular, PIAS1 and PIASxβ act as E3 ligases to enhance sumoylation of p53 in vivo and in vitro (24). Although it has been shown that PIAS proteins negatively regulate the transcriptional activity of p53 through sumoylation, recent data indicated that PIAS1 and PIAS3 may function as activators of p53-dependent gene expression (22). Previously, we have shown that Zimp10 co-localizes with the AR and SUMO-1 at replication foci and enhances AR sumoylation. However, the mechanism for hZimp10-mediated enhancement of p53 activity appears to be through a sumoylation-independent pathway because over-expression of hZimp10 and SUMO-1 in HEK293 cells showed no effect on sumoylation of the p53 protein (Supplementary Data). In addition, our results indicate that the hZimp10-mediated enhancement of p53 activity may be at least partially Miz domain-independent because the strongest interaction was observed with a hZimp10 region containing only a portion of the Miz sequence (Figure 2A).
Recently, we have demonstrated that both hZimp7 and hZimp10 enhance the transcriptional activity of several transcriptional factors (26,27,34,44). However, the precise mechanism(s) for these Zimp proteins in transcriptional regulation still remains unclear. Our previous data showing that hZimp10 co-localizes with newly synthesized DNA at replication foci throughout S phase suggest that hZimp10 may play an important role in both chromatin assembly and maintenance of chromatin (26). Intriguingly, a homolog of human Zimp proteins, termed tonalli (tna), has been identified in Drosophila and was shown to genetically interact with SWI2/SNF2 and the Mediator complexes in complementation studies (28). In addition, we have shown previously that the C-terminal proline-rich domains of hZimp7 and 10 possess significant intrinsic transcriptional activity (26,27), and through these domains, the Zimp proteins can enhance transcription both in trans and in cis. The finding that hZimp10 augments p53-mediated transcription is consistent with our previous studies showing that hZimp10 functions as a transcriptional co-activator of the androgen receptor and Smad3/Smad4 (26,34). Interestingly, the C-terminal proline-rich region is not found in other PIAS or PIAS-like proteins, suggesting that the Miz domain family may consist of distinct groups of proteins that contain unique structures and play distinct roles in regulating transcription and other cellular processes. Therefore, it is conceivable that although hZimp10 and other PIAS proteins interact with p53 physically, they may regulate the function of p53 through different mechanisms. Indeed, this is in agreement with our results suggesting that the Miz domain is generally dispensable for the hZimp10–p53 interaction while published reports suggest that the Miz domain is important for PIAS–p53 interactions (25).
hZimp7 and hZimp10 share significant sequence similarity, particularly within their C-terminal regions (27). Both proteins contain an intrinsic transactivation domain and function as transcriptional co-activators (26,27). These two Zimp proteins show different tissue distribution profiles, which may suggest unique roles for these proteins in regulating different target genes. Our recent data showing that disruption of the Zimp10 gene in mice results in embryonic lethality at approximately E10.5 suggest that hZimp7 and 10 are not functionally redundant. In this study, even though we observed that hZimp7 interacts with p53 in a yeast two-hybrid assay, it showed no interaction between the two intact proteins in immunoprecipitation assays (see Supplementary Data). In addition, we only observed a very weak effect of hZimp7 on p53-mediated transcription. These data suggest that hZimp7 and hZimp10 proteins, although structurally similar, likely play a different role in p53-mediated transcription.
In this study, we also assessed the interaction between hZimp10 and p53 in Zimp10 null cells. Using MEFs generated from Zimp10 knockout mice, we demonstrated that the disruption of Zimp10 inhibits p53-mediated transcription. In MEFs with an intact wild-type Zimp10 allele, a clear dose-dependent induction of p53 transcriptional activity was observed in cells transfected with increasing amounts of p53. In contrast, no enhancement was observed in cells where both Zimp10 alleles were disrupted. This perhaps provides the most convincing evidence that Zimp10 can indeed regulate p53 activity in an in vivo system. Further study using this in vivo system should help to elucidate the biological influence of Zimp10 on p53-mediated tumor repressive effects.
In conclusion, this study demonstrates for the first time that hZimp10, a novel PIAS-like protein, augments the transcriptional activity of the p53 tumor suppressor. This interaction provides an additional line of evidence to demonstrate that Zimp10 is involved in transcriptional regulation. Further studies into the molecular mechanisms by which hZimp10 and other PIAS proteins regulate p53-mediated transcription may provide new insight into the biological role of PIAS and PIAS-like proteins in cell growth, apoptosis, differentiation and tumorigenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grants CA070297, CA087767, DK061002 and 5T32 CA09302-27. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Harris N, Brill E, Shohat O, Prokocimer M, Wolf D, Arai N, Rotter V. Molecular basis for heterogeneity of the human p53 protein. Mol. Cell Biol. 1986;6:4650–4656. doi: 10.1128/mcb.6.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine AJ, Perry ME, Chang A, Silver A, Dittmer D, Wu M, Welsh D. The 1993 Walter Hubert Lecture: the role of the p53 tumour-suppressor gene in tumorigenesis. Br. J. Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathonnet G, Leger C, Desnoyers J, Drouin R, Therrien JP, Drobetsky EA. UV wavelength-dependent regulation of transcription-coupled nucleotide excision repair in p53-deficient human cells. Proc. Natl Acad. Sci. USA. 2003;100:7219–7224. doi: 10.1073/pnas.1232161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford JM, Hanawalt PC. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc. Natl Acad. Sci. USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 8.Gu W, Luo J, Brooks CL, Nikolaev AY, Li M. Dynamics of the p53 acetylation pathway. Novartis Found. Symp. 2004;259:197–205. discussion 205–197, 223–195. [PubMed] [Google Scholar]
- 9.Livengood JA, Scoggin KE, Van Orden K, McBryant SJ, Edayathumangalam RS, Laybourn PJ, Nyborg JK. p53 transcriptional activity is mediated through the SRC1-interacting domain of CBP/p300. J. Biol. Chem. 2002;277:9054–9061. doi: 10.1074/jbc.M108870200. [DOI] [PubMed] [Google Scholar]
- 10.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 11.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 12.Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl Acad. Sci. USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan J, Hall SH, Hamil KG, Grossman G, Petrusz P, Liao J, Shuai K, French FS. Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol. Endocrinol. 2000;14:14–26. doi: 10.1210/mend.14.1.0408. [DOI] [PubMed] [Google Scholar]
- 14.Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17:3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell. Mol. Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 17.Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 18.Kotaja N, Aittomaki S, Silvennoinen O, Palvimo JJ, Janne OA. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol. Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- 19.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 2002;277:30283–30288. doi: 10.1074/jbc.M204768200. [DOI] [PubMed] [Google Scholar]
- 21.Kotaja N, Vihinen M, Palvimo JJ, Janne OA. Androgen receptor-interacting protein 3 and other PIAS proteins cooperate with glucocorticoid receptor-interacting protein 1 in steroid receptor-dependent signaling. J. Biol. Chem. 2002;277:17781–17788. doi: 10.1074/jbc.M106354200. [DOI] [PubMed] [Google Scholar]
- 22.Megidish T, Xu JH, Xu CW. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1) J. Biol. Chem. 2002;277:8255–8259. doi: 10.1074/jbc.C200001200. [DOI] [PubMed] [Google Scholar]
- 23.Nishida T, Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 24.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma M, Li X, Wang Y, Zarnegar M, Huang CY, Palvimo JJ, Lim B, Sun Z. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 2003;22:6101–6114. doi: 10.1093/emboj/cdg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CY, Beliakoff J, Li X, Lee J, Li X, Sharma M, Lim B, Sun Z. hZimp7, a novel PIAS-like protein, enhances androgen receptor-mediated transcription and interacts with SWI/SNF-like BAF complexes. Mol. Endocrinol. 2005;19:2915–2929. doi: 10.1210/me.2005-0097. [DOI] [PubMed] [Google Scholar]
- 28.Gutierrez L, Zurita M, Kennison JA, Vazquez M. The Drosophila trithorax group gene tonalli(tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development. 2003;130:343–354. doi: 10.1242/dev.00222. [DOI] [PubMed] [Google Scholar]
- 29.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z. Linking beta-catenin to androgen signaling pathway. J. Biol. Chem. 2002;277:11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 30.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delenda C. Lentiviral vectors: optimization of packaging, transduction and gene expression. J. Gene Med. 2004;6(Suppl. 1):S125–S138. doi: 10.1002/jgm.501. [DOI] [PubMed] [Google Scholar]
- 33.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Thyssen G, Beliakoff J, Sun Z. The novel PIAS-like protein hZimp10 enhances Smad transcriptional activity. J. Biol. Chem. 2006;281:23748–23756. doi: 10.1074/jbc.M508365200. [DOI] [PubMed] [Google Scholar]
- 35.Sun Z, Yergeau DA, Tuypens T, Tavernier J, Paul CC, Baumann MA, Tenen DG, Ackerman SJ. Identification and characterization of a functional promoter region in the human eosinophil IL-5 receptor alpha subunit gene. J. Biol. Chem. 1995;270:1462–1471. doi: 10.1074/jbc.270.3.1462. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Louie MC, Yang HQ, Ma AH, Xu W, Zou JX, Kung HJ, Chen HW. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl Acad. Sci. USA. 2003;100:2226–2230. doi: 10.1073/pnas.0437824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaeser MD, Iggo RD. Promoter-specific p53-dependent histone acetylation following DNA damage. Oncogene. 2004;23:4007–4013. doi: 10.1038/sj.onc.1207536. [DOI] [PubMed] [Google Scholar]
- 40.Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl Acad. Sci. USA. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24:543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krude T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 1999;247:148–159. doi: 10.1006/excr.1998.4342. [DOI] [PubMed] [Google Scholar]
- 43.Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, Yokoyama S, Tanaka H, Yasuda H, et al. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- 44.Beliakoff J, Sun Z. Zimp7 and Zimp10, two novel PIAS-like proteins, function as androgen receptor coregulators. Nucl. Recept. Signal. 2006;4:e017. doi: 10.1621/nrs.04017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 46.Jiang M, Axe T, Holgate R, Rubbi CP, Okorokov AL, Mee T, Milner J. p53 binds the nuclear matrix in normal cells: binding involves the proline-rich domain of p53 and increases following genotoxic stress. Oncogene. 2001;20:5449–5458. doi: 10.1038/sj.onc.1204705. [DOI] [PubMed] [Google Scholar]
- 47.Mao Y, Mehl IR, Muller MT. Subnuclear distribution of topoisomerase I is linked to ongoing transcription and p53 status. Proc. Natl Acad. Sci. USA. 2002;99:1235–1240. doi: 10.1073/pnas.022631899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 49.Watts GS, Oshiro MM, Junk DJ, Wozniak RJ, Watterson S, Domann FE, Futscher BW. The acetyltransferase p300/CBP-associated factor is a p53 target gene in breast tumor cells. Neoplasia. 2004;6:187–194. doi: 10.1593/neo.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Farmer G, Colgan J, Nakatani Y, Manley JL, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol. Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez MS, Dargemont C, Hay RT, Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting SUMO-1 modification activates the transcriptional response of p53. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 54.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.