Abstract
Lactation is a highly demanding lipid synthesis and transport process that is crucial for the development of newborn mammals. While PPARγ is known to promote adipogenesis and lipogenesis in adipose tissue, its role in the lactating mammary gland is unexplored. Here, we report that a targeted deletion of PPARγ in mice results in the production of “toxic milk” containing elevated levels of inflammatory lipids. Surprisingly, ingestion of this “toxic milk” causes inflammation, alopecia, and growth retardation in the nursing neonates. Genomic profiling reveals that PPARγ deficiency leads to increased expression of lipid oxidation enzymes in the lactating mammary gland. Consistently, metabolomic profiling detects increased levels of oxidized free fatty acids in the pups nursed by PPARγ-deficient mothers. Therefore, maternal PPARγ is pivotal for maintaining the quality of milk and protecting the nursing newborns by suppressing the production of inflammatory lipids in the lactating mammary gland.
Keywords: PPARg, mammary gland, lactation, inflammation, alopecia, lipid oxidation
Milk lipids provide not only calories, but also signaling molecules—such as essential fatty acids and derivatives—that regulate the development of the newborns. These unique demands make the lactating mammary gland one of the most active lipid synthesis and transport organs in the body. In humans, the lactating mammary glands secrete ∼32 g of fat daily or nearly 6 kg of fat during a typical 6-mo lactation period (Allen et al. 1991). The mammary glands of a lactating mouse perform an even more prodigious feat: Approximately 30 g of milk lipids are secreted over the course of 20 d, equivalent to the entire body weight of the mother (Schwertfeger et al. 2003). Triglycerides make up 98% of milk lipid content, and their composition depends on diet as well as intrinsic fatty acid synthesis and modification in the mammary gland (Neville and Picciano 1997). Dietary triglycerides are assembled into chylomicrons in the intestinal mucosa and delivered to adipose tissues such as subcutaneous fat. In the capillaries of adipose tissue, the free fatty acids (FFAs) of chylomicrons are released from the triacylglycerols by lipoprotein lipase (LPL) and absorbed by the tissues. Therefore, milk lipids influence tissue fatty acids composition and regulate multiple functions in the nursing newborns.
PPARγ is a member of the nuclear receptor superfamily of transcription factors that can be activated by lipophilic ligands. It enhances cellular processes involving lipid accumulation, such as adipogenesis and macrophage foam cell formation (Tontonoz et al. 1994, 1998). It also suppresses inflammation by inhibiting macrophage cytokine production (Jiang et al. 1998; Ricote et al. 1998). A versatile array of synthetic and natural substances can activate PPARγ, including thiazolidinediones (TZDs; e.g., BRL 49653) and 15-deoxy-Δ12,14-prostaglandin J2 (Forman et al. 1995; Lehmann et al. 1995; Willson et al. 1996), polyunsaturated fatty acids (Forman et al. 1997; Kliewer et al. 1997), and components of oxidized LDL particles such as 13-hydroxyoctadecadienoic acid (13-HODE) and 15-hydroxyeicosatetraenoic acid (15-HETE) (Nagy et al. 1998). The importance of PPARγ is accentuated by the widespread use of TZDs as drugs for the treatment of type II diabetes and insulin resistance.
Complete loss of PPARγ in mice leads to embryonic lethality due to defects in placental, cardiac, and adipose tissue development (Barak et al. 1999; Kubota et al. 1999; Rosen et al. 1999). Therefore, knowledge of the postnatal in vivo functions of PPARγ has been attained through the analyses of PPARγ heterozygous mice and flox/cre-mediated tissue-specific knockout mice (He et al. 2003; Hevener et al. 2003), as well as subtype-specific knockout mice (Zhang et al. 2004; Medina-Gomez et al. 2005), human mutation knock-in mice (Rangwala et al. 2003; Tsai et al. 2004), and hypomorphic mice (Koutnikova et al. 2003). In this study, we have generated hematopoietic and endothelial cell-specific PPARγ knockout mice using the Tie2cre transgenic mice. Unexpectedly, the consequences of maternal PPARγ deletion were reflected in the growth retardation and hair cycling defects in wild-type nursing pups, as the result of ingestion of “toxic milk” containing elevated levels of inflammatory lipids. These findings reveal a novel role for PPARγ in the postnatal maternal–offspring interaction via the mammary gland, in addition to the previously described prenatal maternal–fetal interaction via the placenta.
Results
Maternal PPARγ deficiency results in growth retardation and hair cycling defects in nursing neonates
We generated hematopoietic and endothelial cell-specific PPARγ knockout mice by crossing PPARγflox/flox mice (gf/f) (He et al. 2003) with Tie2cre transgenic mice (Constien et al. 2001; Kisanuki et al. 2001; Koni et al. 2001). In the process of breeding and expanding the colonies, we observed an unexpected phenotype: Pups of female gf/fTie2cre mice exhibited growth retardation and a hair cycling defect. At the time of weaning (postnatal day 21–28 [P21–P28]), these pups exhibited low body weights and near complete hair loss on their trunks (Fig. 1A). Approximately 2–3 wk after weaning to a standard chow diet, the pups regained a new coat of hair and remained phenotypically normal afterward. This phenotype was strictly dependent on maternal genotype (PPARγf/fTie2cre) and independent of paternal genotype as well as the genotype or sex of the progeny. Pups from female Tie2cre mice or PPARδf/fTie2cre mice, which were studied in parallel, never displayed this phenotype, indicating that this was not a Tie2cre-mediated effect. Furthermore, pups from female PPARγ heterozygous mice, such as PPARγ+/lacZ and PPARγf/+Tie2cre, exhibited a similar phenotype but at lower penetrance, confirming that PPARγ deficiency was the cause of the defects.
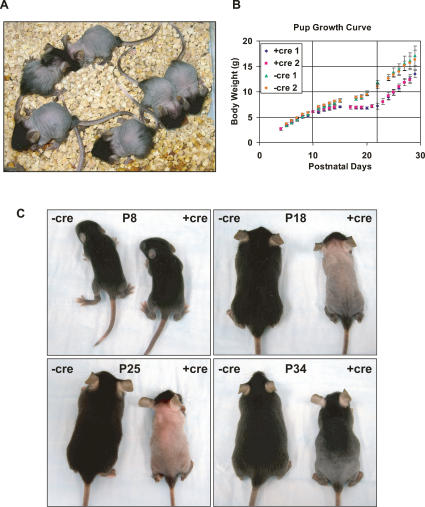
Figure 1.
Maternal PPARγ deletion by Tie2cre results in growth retardation and hair cycling defect in the nursing neonates. (A) gf/fTie2cre mothers raise defective pups. At weaning on P21, pups nursed by mutant mothers (+cre) exhibited growth retardation and nearly complete hair loss over the trunk, with sparing of the head, extremities, rump, and tail. (B) The developmental time line is depicted as the average body weight of pups from each of four independent litters on each postnatal day. During the first postnatal week, weight gain was similar for all pups. Around P10, weight gain in pups from +cre mothers stopped completely until after weaning at P21, when it resumed at a pace similar to the control pups. Error bars represent SD (n = 6). (C) The progression of hair cycling defect during lactation and pelage recovery after weaning at P21.
To carefully examine the time line of this phenotype, female gf/fTie2cre mutants (+cre) and littermate gf/f wild-type controls (−cre) were bred with wild-type C57BL/6J male mice. On P2, the litter size was normalized to six pups. Pup body weights were measured daily. During the first postnatal week, pups from mutant mothers gained weight at rates similar to pups from wild-type mothers (Fig. 1B). Also, the first postnatal pelage appeared normal and comparable to the controls (Fig. 1C). At about P10, weight gain in pups from mutant mothers ceased completely until weaning at P21 (Fig. 1B). At about P16, initial hair loss was observed around the neck and proceeded caudally. By about P18, the hair loss was complete on the trunk, but often with the head, rump, tail, and extremities spared (Fig. 1C). Interestingly, however, weight gain resumed immediately after pups were placed on a standard chow diet at P21 and continued at a pace similar to control pups (Fig. 1B). Also, the hairless skin gradually became pigmented, and a new pelage of hair grew back caudally 2–3 wk after weaning (Fig. 1C).
The dramatic rescue of the phenotype at weaning suggested that a maternal component, either nutritional or behavioral, was the cause of the defect. To distinguish between these two possibilities, we tested if separation of the pups from the mother at weaning was necessary for the rescue. Pups left with the mother until P35 also showed weight gain and hair recovery once they were able to eat the standard chow diet (data not shown), suggesting that a nutritional defect in the milk associated with lactation was the cause.
Hair loss results from follicular cyst (FC) formation mediated by increased inflammation
In mice, hair follicle morphogenesis as well as the first round of postnatal catagen (regression), telogen (quiescence), exogen (shedding), and anagen (growth) development follows a rather precise time scale (Paus and Cotsarelis 1999; Muller-Rover et al. 2001). To pinpoint the hair cycling defect, we examined the histology of affected skins throughout the postnatal time course. The hair follicles exhibited normal morphology until P16–P20. At P20, in pups from −cre control mothers, catagen started when the follicles regressed, followed by telogen when the follicles became quiescent. Around P32, the follicles underwent exogen, ejecting the hair shaft as the next anagen began (Fig. 2A). In contrast, in pups from +cre mutant mothers, a precocious formation of FCs was observed around P20 with hair shaft ejection but without the concurrent initiation of anagen (Fig. 2A). These aberrant follicular structures persisted until around P32, when the first postnatal anagen started in which all hair layers showed inward proliferation comparable to controls. The dermal papillae were well developed, and new hair shafts were present in the follicles (Fig. 2A). Therefore, the major defect was in the first postnatal catagen/telogen. Furthermore, there appeared to be profound leukocyte infiltration in the interfollicular region and subcutaneous fat in the affected skin (Fig. 2B), suggesting the presence of inflammation. Indeed, immunofluorescence staining for CD11b/Mac-1α demonstrated a marked increase in macrophage accumulation in the skin of the pups nursed by +cre mutant mothers as early as P10 (Fig. 2C). Together, the histological evidence suggests that the hair loss was due to FC formation during the first postnatal catagen/telogen, possibly due to increased inflammation.
Figure 2.
Hair loss results from precocious FC formation. (A) H&E-stained sagittal sections of the dorsal skin of pups from selected time points during postnatal development (10×). During P20–P28, catagen/telogen hair follicles with the hair shaft (H) protruding predominated the skin of control pups (−cre, top), but was absent in the skin of pups nursed by mutant mothers (+cre, bottom). Instead, the hair follicles formed cyst-like structures (FC and arrows). By P32, anagen hair follicles with large dermal papilla (DP) developed in the skin of pups nursed by both wild-type and mutant mothers. (B) H&E-stained sagittal sections of dorsal skin at higher magnification (20× for both −cre and +cre) showing the infiltration of leukocytes (marked by arrows) in the interfollicular region and subcutaneous fat (sf) in the defective skin (+cre, bottom) compared with the control skin (−cre, top). (C) Immunofluorescence-stained (anti-CD11b/Mac-1) sagittal sections of the dorsal skin of pups from P10 showing increased macrophage infiltration in the skin of pups nursed by mutant mother (+cre) compared with wild-type mother (−cre). (D) Expression of cyclin D1 and GATA3 was decreased in the skin of pups nursed by mutant mothers (+cre) during hair loss, measured by RT-QPCR and illustrated as relative expression normalized by L19. Error bar represents SD (n = 4).
We performed gene expression analyses of the skins from pups nursed by mutant or control mothers using real-time quantitative PCR (RT-QPCR). First, we tested two genes that are important for hair follicle development. Cyclin D1 and GATA3 are critical for stem cell differentiation in the outer root sheath (ORS) and inner root sheath (IRS), respectively (Kaufman et al. 2003; Xu et al. 2003). The expression of both genes was decreased in the affected skin (+cre) during the stages of hair loss (P20–P31) (Fig. 2D), but by P34, these differences resolved along with the recovery of pelage. Reduced expression of both genes indicated the lack of stem cell proliferation and differentiation required for anagen.
To determine if there was increased inflammation in the affected skin at the transcriptional level, the expression of a set of inflammatory markers was measured (Fig. 3A). Expression of cytokines IL-1β and TNFα, chemokines MCP-1 and MCP-3, and the matrix metalloproteinase MMP9 were all dramatically increased in the affected skin compared with the control skin. IL-1β and TNFα are important mediators of the inflammatory response that are mainly secreted by macrophages. MCP-1 and MCP-3 are chemotactic factors that attract monocytes, basophils, or eosinophils. MMP9 plays an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. Therefore, the affected skin exhibited an inflammatory response hallmarked by leukocyte infiltration and cytokine/chemokine secretion.
Figure 3.
Hair loss is mediated by increased inflammation and COX signaling, and can be rescued by COX inhibitor or foster mothers. (A) The expression of inflammatory markers and cyclooxygenases was elevated in the skin of pups nursed by mutant mothers (+cre). Error bar represents SD (n = 4). All RNA expression was measured by RT-QPCR and illustrated as relative expression normalized by L19. (B) Increased inflammatory marker expression started between P7 and P13 before hair loss. Error bar represents SD (n = 4). (C) Protein levels of selected genes in the skin of pups were determined by Western blot analyses. (pAb) Polyclonal antibody; (mAb) monoclonal antibody. (D) Oil Red O-stained liver sections (P8, 40×) demonstrated increased lipid content in the livers of pups nursed by mutant mothers (+cre). (E) The expression of inflammatory markers and PGT was also increased in the livers of pups nursed by mutant mothers. Error bars represent SEM (n = 6). (*) P < 0.05. (F,G) The hair cycling defect can be rescued by topical application of COX inhibitor. (F) The expression of inflammatory markers in the skin of pups nursed by +cre mutant mothers was inhibited by topical aspirin treatment. Error bar represents SD (n = 6). (**) P < 0.01. (G) By P29, the DMSO-treated pups exhibited complete hair loss over the trunk (left), while the aspirin-treated pups retained fairly normal pelage on the back (right), but with some hair loss observed on the scalp (red arrow) that was not directly treated. This is the representative result from two independent litters (n = 6 for each treatment). (H) Wild-type foster mothers (−cre) fully rescue the hair loss of pups from mutant mothers (+cre). Pups born and nursed by mutant mothers exhibited alopecia by P20 (left), while pups born from mutant but nursed by wild-type mothers displayed normal pelage (right). This is the representative result from two independent fostering pairs.
The arachidonic acid/COX/prostaglandin pathway has been shown to be an important regulator of inflammatory responses. Interestingly, this pathway is also involved in the regulation of hair cycling. Two independent studies have demonstrated that overexpression of COX-2 in mouse skin leads to alopecia (Bol et al. 2002; Muller-Decker et al. 2003). Therefore, we examined the expression of several genes in this pathway. The expression of both COX-1 and COX-2 genes were markedly increased in the affected skin (Fig. 3A), as was that of the prostaglandin transporter (PGT), which mediates the release of newly synthesized prostaglandins from cells. Western blot analyses further confirmed these gene expression changes at the protein level (Fig. 3C). As shown in Figure 3,A and C, COX-2 expression in the control skin declined in catagen (P17–P20) and was barely detectable in early telogen (P21), but increased again during anagen (P28–P40) (Muller-Decker et al. 2003). In contrast, COX-2 expression in the affected skin was significantly elevated during catagen/telogen (P16–P25) (Fig. 3A,C). This was also the case in the K5-COX2 and K14-COX2 transgenic mice that developed alopecia (Bol et al. 2002; Muller-Decker et al. 2003). Together with the increased expression of COX-1, this pattern of COX expression was unscheduled compared with the normal controls and correlated with the precocious FC formation and hair loss. The increased expression of these inflammatory genes and macrophage infiltration began between P7 and P13, preceding the hair loss (Figs. 3B, 2C), suggesting the inflammation was likely the cause rather than the consequence of the hair loss. Furthermore, the expression of most inflammatory markers in the affected skin decreased by P48, indicating that the inflammation slowly resolved after the pups were weaned to a standard diet (Fig. 3A).
Detailed examination of the pups nursed by gf/fTie2cre mothers revealed that their livers often appeared paler compared with control pups. Oil Red O staining of liver sections indicated increased lipid accumulation (Fig. 3D). Hepatic steatosis is often linked to inflammation, and indeed, expression of several inflammatory markers were also elevated in liver (Fig. 3E), suggesting the presence of inflammation at multiple tissues and manifested in the skin as hair loss.
Hair loss can be rescued by COX inhibitors or foster mothers
To test if the increased inflammation and COX signaling was the cause of the hair cycling defect, we performed rescue experiments by topical treatment with the COX-1/2 inhibitor aspirin. Gene expression analyses indicated that aspirin treatment resulted in the inhibition of inflammation in the skin of pups from gf/fTie2cre mothers (Fig. 3F). By P29, all DMSO-treated pups exhibited complete hair loss over the trunk, while all the aspirin-treated pups retained fairly normal pelage on the back (Fig. 3G), although some hair loss was observed on the scalp (Fig. 3G, red arrow) and the chest (data not shown) that was not directly treated. Aspirin treatment did not have any visible effect on the skin of pups from the control mothers. This demonstrated that the hair loss was the result of COX-mediated inflammation.
To further determine if the phenotype was maternally dependent, we performed foster mother experiments. All pups born and nursed by gf/fTie2cre mothers exhibited hair loss, while the pups born from gf/fTie2cre mothers but fostered by gf/f mothers were rescued at 100% penetrance (eight out of eight pups, Fig. 3H). Reciprocally, all pups born and nursed by gf/f mothers showed normal pelage, while the pups born from gf/f mothers but fostered by gf/fTie2cre mothers developed hair loss at 75% penetrance (six out of eight pups). Thus, the phenotype was indeed maternally dependent. The fact that the hair cycling defect could be reversed by fostering demonstrates that it was due to postnatal factors such as milk, rather than prenatal factors such as the placenta. In addition, fostering (Fig. 3H), but not topical aspirin treatment (Fig. 3G), also exhibited a rescuing effect on the growth retardation. Together, these results suggest that components in the milk of gf/fTie2cre mothers were able to elicit inflammatory responses in the pups.
PPARγ deficiency increases lipid accumulation in the lactating mammary gland
We next assessed if there was any difference in the lactating mammary gland between gf/f and gf/fTie2cre mice. Mammary glands were collected on different lactation days for histological analyses. H&E staining indicated that the mutant mammary glands had fewer lobular alveolar structures and more stroma fat cells (adipocytes) compared with the controls on lactation day 4 (L4) (Fig. 4A). The size of the adipocytes was also larger in the mutant mammary gland (Fig. 4A). In addition, the epithelial cells in the mutant glands were filled with many large lipid droplets throughout lactation (L4–L16) (Fig. 4A shows examples from L4 and L10), indicating increased lipid synthesis and milk fat globule fusion. In contrast, in the wild-type mammary gland, intracellular lipid droplets were much smaller, and only the biggest globules in the process of being secreted were visible (Fig. 4A). Consistently, Oil Red O staining further demonstrated that there was increased lipid content in both adipocytes and epithelial cells in the mutant mammary glands on both L4 and L10 (Fig. 4A).
Figure 4.
Tie2cre-mediated PPARγ deletion increases lipid accumulation in the lactating mammary gland. (A) Histological analyses of lactating mammary glands. (Panels a–d) In the H&E-stained sections (20×), there was an increase in the number and size of the adipocytes (Ad), and a decrease in lobular alveolar epithelial cells (Ep), in the +cre glands (panel b) compared with the −cre control glands (panel a) on L4. On both L4 and L10, there was an increase in the number and size of the intracellular lipid droplets in the epithelial cells in +cre glands (panels b,d) compared with −cre control glands (panels a,c). Arrows mark the milk fat globules in the process of being secreted into the lumen. (Panels e–h) In the Oil Red O-stained sections, there was an increase in lipid content in both adipocytes (Ad) and epithelial cells in +cre glands (panels f,h) compared with −cre control glands (panels e,g) on both L4 and L10. (B) Expression of genes regulating mammary gland morphogenesis was diminished throughout lactation in the mutants (+cre). Error bars represent SD (n = 3).
To determine the transcriptional consequences of Tie2cre-mediated PPARγ deletion, we compared the gene expression profiles in the mutant and control mammary glands from L4 and L10 using Affymetrix microarray analysis. Genes that were differentially expressed by an average fold change of more than two across all four comparison pairs from L10 are listed in Supplementary Table 1. Several genes were selected for further examination throughout the time course of lactation using RT-QPCR (Figs. 4B, 5D).
Figure 5.
Tie2cre-mediated PPARγ deletion results in production of inflammatory lipids in milk. (A) Milk from mutant mothers (+cre) activated macrophage inflammation only after LPL digestion. The mouse macrophage RAW264.7 cells were treated with milk in the absence or presence of LPL (10 μg/mL) for 6 h. RNA was harvested and expression of inflammatory markers was determined by RT-QPCR. Representative result of three independent experiments; error bars indicate SEM. (*) P < 0.05 (n = 4). (B,C) Biochemical fractionation of hydrolyzed milk lipids enriched the inflammatory mediators from +cre milk. (B) A schematic diagram for the fractionation procedure. (EA/H) Ethyl acetate/hexanes. (C) Compared with −cre milk, HPLC fraction 4 from +cre milk increased the expression of both IL-1β and COX-2 in RAW cells by 110-fold and 67-fold, respectively. (D) Expression of two lipid oxidation enzymes was elevated throughout lactation in the mutant mammary glands (+cre). Error bars indicate SD (n = 3).
One group of genes whose expression was found to change throughout lactation includes modulators of mammary gland morphogenesis (Fig. 4B). The expression of Hoxd9, Hoxd10, and Hoxd8 were severely diminished in mutant mammary glands compared with controls. These three homeobox genes are localized adjacently in the mouse HoxD cluster and are likely regulated by the same enhancer elements (Spitz et al. 2001). Deletion of Hoxd9 in female mice, especially combined with the deletion of Hoxa9 and Hoxb9, leads to early postnatal death of newborns due to decreased milk production, as the result of fewer lobular alveolar structures and more fat droplets in the lactating mammary gland (Chen and Capecchi 1999). Also, both Timp4 and Timp2 were down-regulated in the mutant mammary glands (Fig. 4B; Supplementary Table 1). TIMPs are a family of matrix metalloproteinases inhibitors. Timp3-deficient mice show increased adipogenesis and accelerated involution in the mammary gland (Fata et al. 2001). Therefore, decreased expression of Hoxd9 and Timps could be part of the underlying mechanisms for the increased lipid content in the mutant mammary glands. Together, these results suggest that the lipid metabolism become deregulated in the lactating mammary gland of the gf/fTie2cre mice.
PPARγ deficiency leads to production of inflammatory lipids in milk
To determine if milk from gf/fTie2cre mothers has increased inflammatory mediators, we exploited an in vitro assay using the mouse macrophage cell line RAW264.7. The cells were treated with milk for 6 h in the presence or absence of LPL, and the expression of inflammatory markers was measured. In the absence of LPL digestion, there was no significant difference in the expression of IL-1β or COX-2 between −cre and +cre milk treatments. However, in the presence of LPL digestion, there was an activation of both genes by the +cre milk (Fig. 5A). This suggested that, indeed, there were increased inflammatory mediators in the +cre milk lipid, but they were released only after LPL digestion and possibly in the form of FFAs. To enrich the inflammatory mediators in the +cre milk, we extensively fractionated the hydrolyzed milk lipids to separate the active components from the bulk of the inactive FFAs, using silica gel chromatography followed by reverse phase HPLC (Fig. 5B). Compared with −cre control milk, HPLC fraction 4 from the +cre milk robustly induced the expression of IL-1β and COX-2 by 110-fold and 67-fold, respectively (Fig. 5C).
Interestingly, the expression of two lipid oxidation enzymes, 12-lipoxygenase (12-LO) and microsomal epoxide hydrolase (Ephx1), were elevated throughout lactation in the mutant mammary glands (Fig. 5D). Fatty acid oxidative metabolism produces potent inflammatory mediators. The lipoxygenases convert arachidonic acid into leukotrienes, which are eicosanoid lipid mediators of inflammation (Funk 2001). Thus, elevated 12-LO expression may lead to an increased production and secretion of leukotrienes. In addition, the cytochrome P450 epoxygenases transform arachidonic and linoleic acids to epoxyeicosatrienoic acids (EETs) and epoxyoctadecenoic acids (EpOMEs, leukotoxins), respectively. Although EETs are anti-inflammatory (Node et al. 1999), further metabolism of EETs and EpOMEs by Ephx1s generates the corresponding diols that can cause cytotoxicity and inflammation (Moghaddam et al. 1997; Morisseau et al. 1999). Thus, elevated Ephx1 expression indicates a likely increase in the conversion of fatty acid epoxides into their corresponding proinflammatory diols. Collectively, these results provide compelling evidence that PPARγ deficiency leads to the secretion of “toxic milk” containing inflammatory lipids, likely comprised of a combination of oxidized FFAs generated by lipoxygenase and Ephx1.
Global metabolite profiling reveals increased oxidized FFAs in the affected skin
The hair loss was a progressive process as the result of digestion of “toxic milk” and accumulation of inflammatory lipids at the skin over time. Since the skin was the site of effectors as well as a major fat storage tissue, we next investigated if there was any difference in the lipid metabolome in the skin of pups nursed by mutant versus control mothers using discovery metabolite profiling (DMP) (Fig. 6). The untargeted liquid chromatography-mass spectrometry (LC-MS) approach employed in this strategy allowed the simultaneous comparison of known metabolites and the identification of unknown metabolites that were differentially regulated between wild-type and mutant tissues (Saghatelian et al. 2004). As shown by the pinnacles in Figure 6A, four groups of unknown metabolites were present at increased levels in the skin of pups nursed by mutant mothers. We performed further characterization on the metabolites in group 1 (Fig. 6B, top red box), since the m/z ratio and retention time indicated that they were a series of FFAs. Exact mass analyses revealed that they comprise monohydroxy FFAs; for example, the molecular formula of the most abundant member of this group (m/z ratio 271) is C16H32O3 (monohydroxy palmitic acid, OH-C16:0) (Fig. 6B; Supplementary Table 2). The paired increased levels of this group of oxidized FFAs in the affected skin (sixfold to 12-fold) along the elevated expression of lipid oxidation enzymes in the mutant mammary glands suggest these hydroxy FFAs represent stable downstream metabolites of the oxidized inflammatory lipids from the +cre milk. Furthermore, there were no significant differences in the levels of the majority of the unmodified fatty acids such as palmitic (C16:0), stearic (C18:0), and oleic (C18:1) acids (Fig. 6B). In contrast, there was a statistically significant reduction in the level of arachidonic acid (C20:4) (P = 0.00002, n = 18) (Fig. 6B, bottom red box). This observation is consistent with the increased COX expression and enhanced arachidonic acid catabolism in the affected pup skin. Together, these results provide further evidence that maternal PPARγ deficiency results in increased production of oxidized lipids in milk that, when released by digestion and accumulated in the skin, trigger inflammation-mediated hair loss in the nursing neonates.
Figure 6.
Global metabolite profiling revealed increased oxidized FFAs in the skin of pups nursed by mutant mothers. (A) DMP of skin samples from pups nursed by mutant mothers (+cre) versus by wild-type mothers (−cre). Organic soluble skin lipid was extracted and subjected to untargeted LC-MS metabolite profiling analyses in the negative ionization mode. Mass ion intensity ratios (+cre/−cre) of metabolites are presented on a three-dimensional surface plot, over a mass range of 200–1000 Da and liquid chromatography retention time of 20–60 min. The skin of pups nursed by +cre mothers possessed elevated levels of oxidized FFAs (group 1) and three unknown classes of metabolites (groups 2–4), while the majority of metabolites (∼1100 ions) were unaltered. Data represent the result of a group of n = 18 pups from eight independent mothers for each genotype. (B) Increased levels of hydroxyl fatty acids (top red box, group 1 in Fig. 7A) and decreased level of arachidonic acid (bottom red box) in the skin of the pups nursed by mutant mothers.
PPARγ suppresses 12-LO expression via a negative regulatory loop
X-gal staining of the lactating mammary gland from Tie2creROSAlacZ indicator mice demonstrated that Tie2cre specifically deleted the flox allele in hematopoietic and vascular endothelial cells, but not in mammary epithelial cells or adipocytes (Fig. 7A). Macrophages constitute a major hematopoietic cellular component in the lactating mammary gland (Gouon-Evans et al. 2002). Factors required for macrophage differentiation, such as MCSF, are also essential for normal mammary gland development and function (Van Nguyen and Pollard 2002). Immunofluorescence staining for CD11b/Mac-1α revealed that there was an increased population of macrophages in the mutant lactating mammary glands (Fig. 7B). Since PPARγ deletion by Tie2cre resulted in the production of inflammatory milk and increased expression of 12-LO and Ephx1 in the lactating mammary gland (Fig. 5), we examined gene expression in macrophages derived from mutant or wild-type mice. As shown in Figure 7C, PPARγ deletion in the mutants (+cre) resulted in elevated expression of 12-LO, MCP-1, and MCP-3 compared with wild-type (−cre). Moreover, the PPARγ agonist BRL inhibited the expression of both 12-LO and Ephx1, as well as MCP-1 and MCP-3. This inhibition was only observed in the wild-type but not the mutant cells, and thus was PPARγ dependent (Fig. 7C). Interestingly, saturated fatty acids such as palmitic acid (PA) that are abundant in the lactating mammary gland further induced COX-2 and iNOS expression in macrophages (Fig. 7D; Shi et al. 2006). However, BRL robustly suppressed this induction in wild-type cells, but not mutant cells (Fig. 7D). Together, these results demonstrate that PPARγ and its ligand suppress inflammation in a lipid-rich environment such as the lactating mammary gland. Loss of PPARγ results in elevated expression of lipid oxidation enzymes, chemokines, and saturated fatty acid-induced inflammatory genes, as well as resistance to the anti-inflammatory effect of endogenous PPARγ agonists.
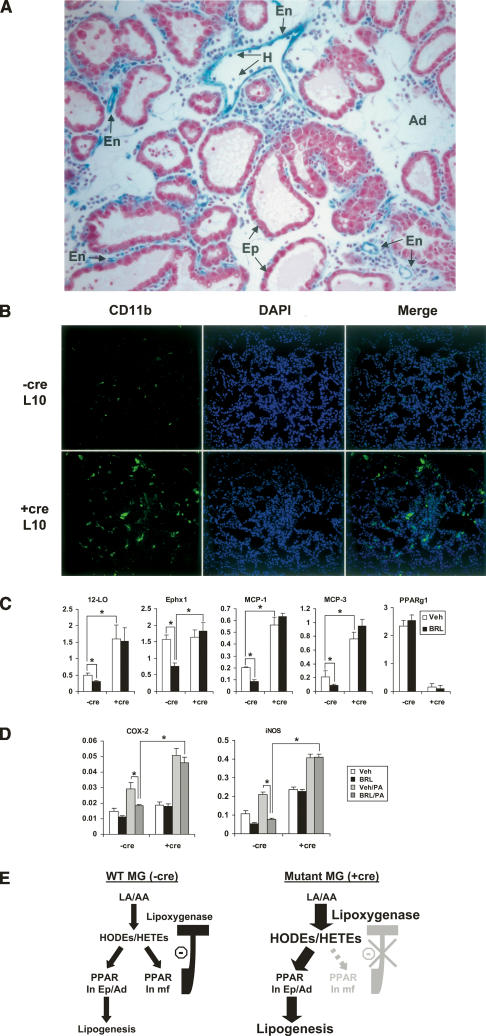
Figure 7.
PPARγ suppresses lipoxygenase expression via a negative regulatory loop. (A) X-gal staining of the mammary gland collected from lactating Tie2cre/ROSAlacZ mice. Tie2cre-mediated flox deletion was specific for the hematopoietic (H) and vascular endothelial (En) cells, but not in the mammary epithelial cells (Ep) or adipocytes (Ad). (B) Immunofluorescence staining (anti-CD11b/Mac-1) of mammary gland from L10 showing increased macrophage population in the mutant (+cre) compared with wild-type control (−cre). (C) PPARγ and ligand suppress the expression of 12-LO and Ephx1 and chemokines MCP-1 and MCP-3 in wild-type (−cre) but not mutant (+cre) macrophages. (D) PPARγ and ligand suppress the induction of COX-2 and iNOS by PA in wild-type (−cre) but not mutant (+cre) macrophages. Macrophages were differentiated from spleen progenitor cells with 10 ng/mL MCSF in the presence of vehicle (DMSO) or BRL (1 μM) for 6 d. The macrophages were stimulated with PA (200 μM) or BSA control for 6 h. RNA was isolated and gene expression analyses were performed with RT-QPCR. (*) P < 0.05. (E) A model of negative feedback loop regulation of lipoxygenase expression by PPARγ. Lipoxygenase produces endogenous PPARγ ligands (HODEs/HETEs). (Left) In the wild-type mammary glands, ligand activation of PPARγ in macrophages (mf) inhibits the expression of lipoxygenase, thereby maintaining its steady levels. (Right) However, in the mutant mammary glands, PPARγ expression in macrophages is absent due to Tie2cre-mediated deletion. The lack of the regulatory feedback loop results in sustained expression of lipoxygenase that in turn causes an even higher level of endogenous PPARγ ligands, resulting in increased lipogenesis in the mammary epithelial cell and adipocytes.
The fact that the expression of lipoxygenase can be inhibited by PPARγ and its ligand (Fig. 7C) suggests that it is under the regulation of a negative feedback loop (Fig. 7E). Lipoxygenases can generate endogenous PPARγ ligands, 13-HODE and 15-HETE, from linoleic and arachidonic acids, respectively (Conrad et al. 1992; Nagy et al. 1998; Huang et al. 1999). In wild-type mammary glands, ligand activation of PPARγ in macrophages inhibits lipoxygenase expression, thereby maintaining its steady levels (Fig. 7C,E). However, in mutant mammary glands, PPARγ deletion in macrophages and thus the lack of the regulatory feedback loop result in sustained expression of lipoxygenase (Fig. 7C), which in turn causes an even higher level of endogenous PPARγ ligands (Fig. 7E). Since Tie2cre does not mediate PPARγ deletion in epithelial cells and adipocytes (Fig. 7A), activation of PPARγ in these cells by endogenous ligands could be, in part, the underlying cause of the increased accumulation of lipid droplets in the mutant glands (Fig. 7E). Collectively, the combination of increased lipid accumulation and oxidation in the mutant mammary glands results in the elevated inflammatory mediators in the milk.
Discussion
Milk is the sole food for young mammals and an important constituent of human diet. The quality of milk has great impact on both breast-fed infants and consumers of dairy products in general. In this study, by using integrated approaches of molecular genetics, biochemistry, genomic, and metabolomic profiling, we demonstrate that maternal PPARγ is pivotal for maintaining the quality of milk and protecting the nursing newborns by suppressing the production of inflammatory lipids in the lactating mammary gland. Maternal PPARγ deficiency results in increased expression of lipid oxidation enzymes in the lactating mammary gland, production of inflammatory milk, which consequently causes inflammation, alopecia, and growth retardation in the nursing pups.
There are other defects in milk that have been attributed to alopecia in infants, such as deficiencies in certain metals or vitamins. However, we did not detect any significant difference in the levels of zinc or iron between the milk from gf/fTie2cre mothers and gf/f controls. Also, feeding lactating mothers with a diet supplemented with Vitamins A or D had no rescuing effect on the hair cycling defect in the pups. Thus, the alopecia and growth retardation in the pups of gf/fTie2cre mothers were mainly due to the increased inflammatory signaling in the milk.
Our current study demonstrates that hematopoietic and endothelial cell-specific PPARγ deletion results in altered mammary gland morphogenesis and lipid metabolism during lactation. Interestingly, a previous study using a transgenic mouse model that overexpresses an active form of PPARγ (VpPPARγ) in the mammary epithelial cells did not observe any defect in mammary gland development (Saez et al. 2004). This suggests that PPARγ in hematopoietic and endothelial cells, rather than mammary epithelial cells, plays a critical role in the lactating mammary gland.
Hair cycling is sensitive to many genetic and metabolic factors (Nakamura et al. 2001; Tong and Coulombe 2003). In humans, alopecia often occurs due to nutritional, hormonal, or environmental factors in addition to genetic predisposition. For example, alopecia can result from essential fatty acid deficiency, androgen overproduction, chronic stress, or chemotherapy. Here, we have described a unique mouse model that links diet, inflammation, and hair cycling abnormalities in the absence of intrinsic genetic defects in the affected offspring. The alopecia in the pups results from the accumulation of inflammatory lipids transmitted from “toxic milk” and can be rescued by an anti-inflammatory agent. The hair loss was typically confined to the trunk; however, in severe cases, it could extend to the scalp (Fig. 3G, left). It is known that the duration of the hair cycle and the lengths of hair shafts are different among scalp, dorsal, and ventricle follicles, with the scalp hair having the longest cycle (Saitoh et al. 1970; Stenn and Paus 2001). Therefore, hair follicles from different skin regions have varying sensitivity to the inflammatory insult from the “toxic milk,” resulting in differences in the timing and severity of the hair loss. The acute onset and recovery after weaning suggests that this is a specific type of alopecia equivalent to “telogen effluvium” in human patients. Our findings will enhance the understanding of the roles for lipid metabolites and inflammation in the etiology of alopecia and other skin disorders, and facilitate the development of novel pharmacological strategies for the prevention and treatment of these diseases.
Materials and methods
Mice
gf/f mice (He et al. 2003) were backcrossed to C57BL/6J mice for four generations. Tie2cre transgenic mice were generously provided by the laboratory of Dr. M. Yanagisawa (University of Texas Southwestern Medical Center, Dallas, TX) (Kisanuki et al. 2001). gf/f mice were bred with Tie2cre mice to generate gf/f and gf/fTie2cre+/− mice, which were further backcrossed to C57BL/6J mice for three generations. To determine the tissue specificity of Tie2cre-mediated gene deletion in the lactating mammary gland, Tie2cre mice were bred to the ROSAlacZ indicator mice (Jackson Laboratory). Mice were maintained with regular rodent diet. For mammary gland and pup hair cycling analyses, 8- to 16-wk-old nulliparous female littermate mice were bred to wild-type C57BL/6J male mice. The litter sizes were normalized to six pups. For foster mother experiments, two pairs of gf/fTie2cre and control gf/f female mice that were bred with wild-type C57BL/6J male mice and delivered pups on the same day were used. On P1, the litter size was normalized to six pups. Four pups were switched between the gf/fTie2cre mother and the gf/f mother, and two pups were left with the original mother as controls. The fostered pups and original pups were distinguished by ear punching.
Histology and staining
For histological analysis, mammary glands and skin were collected and immediately fixed in 10% PBS-buffered formalin overnight. The tissues were then processed, paraffin-embedded, sectioned, and H&E-stained in the Histology Core Facility at the University of California at San Diego according to standard protocols. For Oil Red O staining, the tissue samples were immediately frozen in OCT compound in a dry ice/ethanol bath and stored at −80°C until sectioned. The frozen sections were fixed in formalin for 10 min. The sections were dipped in 60% isopropanol and then stained in Oil Red O working solution (60% 3 mg/mL Oil Red O in isopropanol) for 15 min. After rinsing in 60% isopropanol and H2O, the sections were counterstained with Mayer Hematoxylin for 3 min and mounted. For immunofluorescence staining, frozen OCT sections were fixed in acetone for 10 min and stained with FITC–anti-CD11b/Mac-1α (BD Pharmingen) at 1:250 for 1 h. The sections were washed twice and mounted with medium containing DAPI (Vector Laboratories).
Milk analyses
Milk was collected according to previously described procedures (Schwertfeger et al. 2003) with minor modifications. On L8–L12, lactating females were separated from their pups for 3–5 h to allow the mammary glands to refill. Before milking, the lactating female was anesthetized with an i.p. injection of avertin (0.6 mL of a 20 mg/mL stock), and then i.p. injected with 0.5 U of oxytocin (0.2 mL of a 2.5 U/mL stock) to induce milk ejection. Milk was immediately collected from mammary glands by gentle vacuum suction through a capillary tube. All 10 mammary glands were milked two rounds for a total of 20 min per mouse, allowing the collection of most of the milk. To assess for inflammatory mediators in the milk, mouse macrophage RAW264.7 cells were cultured in serum-free medium overnight, and then treated with milk at 1:2000 dilutions for 6 h in the presence or absence of LPL (10 μg/mL). RNA was isolated and the expression of inflammatory markers was determined by RT-QPCR.
To enrich the inflammatory lipids, 6 g of milk from each genotype was hydrolyzed with 2 M LiOH (10 mL) for 8 h at room temperature. The solution was then acidified using 3 M HCl and extracted with 15 mL of 2:1 chloroform/methanol. The organic layer was then isolated and concentrated to afford a waxy white solid. This material was fractionated by silica gel chromatography using a step gradient, with 30%, 60%, and 100% ethyl acetate/hexanes fractions being collected sequentially. Using the RAW264.7 cells in vitro assay, the inflammatory activity in +cre milk was determined to reside in the 60% fraction. This active fraction was further fractionated using reverse phase HPLC (Gemini C18 reversed phase column, Phenomenex, 5 μm, 50 × 10 mm) using a binary solvent system. The mobile phase A consisted of 95%/5% water/methanol and mobile phase B of 60%/35%/5% isopropanol/methanol/water, both containing 0.1% ammonium hydroxide. The gradient started at 0% B and then linearly increased to 100% B over 60 min, followed by an isocratic gradient of 100% B for 20 min at a flow rate of 2.5 mL/min. Fractions (one per minute) were collected using a Gilson FC 203B fraction collector. The resulting fractions were pooled to give seven samples (pool 1 included fractions 1–10, pool 2 included fractions 11–20, pool 3 included fractions 21–30, pool 4 included fractions 31–40, pool 5 included fractions 41–50, pool 6 included fractions 51–60, and pool 7 included fractions 61–80). The seven pooled fractions were concentrated under vacuum and dissolved in 50 μL of ethanol/chloroform (5:2) prior to testing for inflammatory activity using the RAW264.7 cells in vitro assay.
RNA extraction and RT-QPCR
Tissue samples were snap frozen in liquid nitrogen and stored at −80°C. RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s protocol. RNA was first treated with RNase-free DNase I using the DNA-free kit (Ambion) to remove all genomic DNA. Then the RNA was reverse-transcribed into first-strand cDNA using the SuperScript First-Strand Synthesis System (Invitrogen). RT-QPCR (SYBR Green) analyses of the cDNA were performed using an Applied Biosystems 7700 sequence Detection System. Primer sequences are available on request. Each reaction was performed in triplicate or quadruplicate in a 384-well format. The expression of each gene was normalized to L19 or β-actin expression.
Tissue protein extraction and Western blot
Tissue samples were homogenized in RIPA buffer supplemented with protease and phosphatase inhibitors. After centrifugation, the clear homogenate was collected. Protein concentration was determined using BCA Protein Assay Reagents (Pierce). For Western blot analyses, 15 μg of each tissue protein extract were separated by SDS-PAGE and transferred to nitrocellulose membrane. Primary antibodies anti-COX-1, anti-COX-2 (monoclonal and polyclonal), anti-PGT, and anti-12-LO were from Cayman Chemicals. The anti-GATA3 antibody was from Santa Cruz Biotechnologies. The anti-β-actin antibody was from Sigma.
Affymetrix microarray analysis
Total RNA was isolated from the fourth inguinal mammary glands of littermate gf/f or gf/fTie2cre mice on L4 or L10. Each condition was sampled in biological duplicates. RNA (5 μg) was subjected to Affymetrix microarray gene expression profiling using the Affymetrix Gene Chip Mouse Expression Set 430 2.0A according to the procedures suggested by Affymetrix. This protocol resulted in four measurements for each analyzed gene at L4 and L10; i.e., two measurements each for “L4 − cre” and “L4 + cre” and two each for “L10 − cre” and “L10 + cre.” Probe hybridization and data scanning were performed by the Salk microarray core facility. Differentially expressed genes were assigned by the MICROSUITE software for each possible pairwise comparison. Genes differentially expressed in all four pairwise comparisons between two −cre and two +cre samples were filtered by the BULLFROG analysis software.
Topical treatment
Nulliparous female littermate mice (10 wk old) were bred to wild-type C57BL/6J male mice in individual cages. Litter sizes were normalized to six pups on P2. The six pups were divided into two groups by ear punching. One group was topically treated by spreading 20 μL (P2–P14) or 50 μL (P15–P29) of aspirin (40 mg/mL or 222 mM in DMSO, Cayman Chemicals) with parafilm on dorsal skin daily, while the control group was treated with an equal volume of DMSO.
LC-MS of tissue metabolomes
Dorsal skin samples were collected from pups after hair was removed with a clipper, snap frozen on dry ice, and stored at −80°C. Organic soluble metabolites were extracted with a 2:1:1 CHCl3/MeOH/H2O solution and dounce homogenization. Each sample was then centrifuged at 2500 rpm for 10 min at 4°C in a glass vial. After centrifugation, the organic (bottom) layer was carefully removed and transferred to another vial and concentrated under a stream of nitrogen. Samples were stored at −80°C and dissolved in CHCl3 prior to analysis by LC-MS (Saghatelian et al. 2004). Untargeted LC-MS analysis was performed using an Agilent 1100 LC-MSD SL instrument under negative ionization mode as previously described (Saghatelian et al. 2004). Data were analyzed using the XCMS software for peak alignment and quantification (Smith et al. 2006). The peak ratio between +cre and −cre samples (n = 18) provided a quantitative measurement of the relative metabolite levels. To determine the molecular formula of unknown metabolites that were elevated in the +cre samples, the metabolite extracts from 15 samples were combined for a single LC purification as described (Saghatelian et al. 2004). Fractions containing the m/z 271 metabolite were identified by MS and used for exact mass analysis. The molecular formulas of several other metabolites were confirmed by exact mass measured with ESI-TOF to be within 5 parts per million (PPM) from the calculated mass.
Acknowledgments
We thank Drs. Dennis Volker (National Jewish Center for Immunology and Respiratory Medicine), Anthony Oro (Stanford University), Elaine Fuchs (Rockefeller University), and Peggy Neville (University of Colorado Health Sciences Center) for their helpful discussion; Dr. Kaylee Schwertfeger (Baylor College of Medicine) for technical advice on mouse milk collection; Drs. Johan Jonker, Jun Sonoda, Ruth Yu, and Grant Barish (Salk Institute) for discussion and critical reading of the manuscript; and Lita Ong and Sally Ganley for administrative assistance. R.M.E. is an investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. Y.W. is supported by a post-doctoral fellowship from American Cancer Society (PF-03-081-01-TBE). This work was supported by the funding from HHMI and NIH (SCOR/HL569898 and DK57978).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1567207
References
- Allen J.C., Keller R.P., Archer P., Neville M.C., Keller R.P., Archer P., Neville M.C., Archer P., Neville M.C., Neville M.C. Studies in human lactation: Milk composition and daily secretion rates of macronutrients in the first year of lactation. Am. J. Clin. Nutr. 1991;54:69–80. doi: 10.1093/ajcn/54.1.69. [DOI] [PubMed] [Google Scholar]
- Barak Y., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M., Chien K.R., Koder A., Evans R.M., Koder A., Evans R.M., Evans R.M. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Bol D.K., Rowley R.B., Ho C.P., Pilz B., Dell J., Swerdel M., Kiguchi K., Muga S., Klein R., Fischer S.M., Rowley R.B., Ho C.P., Pilz B., Dell J., Swerdel M., Kiguchi K., Muga S., Klein R., Fischer S.M., Ho C.P., Pilz B., Dell J., Swerdel M., Kiguchi K., Muga S., Klein R., Fischer S.M., Pilz B., Dell J., Swerdel M., Kiguchi K., Muga S., Klein R., Fischer S.M., Dell J., Swerdel M., Kiguchi K., Muga S., Klein R., Fischer S.M., Swerdel M., Kiguchi K., Muga S., Klein R., Fischer S.M., Kiguchi K., Muga S., Klein R., Fischer S.M., Muga S., Klein R., Fischer S.M., Klein R., Fischer S.M., Fischer S.M. Cyclooxygenase-2 overexpression in the skin of transgenic mice results in suppression of tumor development. Cancer Res. 2002;62:2516–2521. [PubMed] [Google Scholar]
- Chen F., Capecchi M.R., Capecchi M.R. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc. Natl. Acad. Sci. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D.J., Kuhn H., Mulkins M., Highland E., Sigal E., Kuhn H., Mulkins M., Highland E., Sigal E., Mulkins M., Highland E., Sigal E., Highland E., Sigal E., Sigal E. Specific inflammatory cytokines regulate the expression of human monocyte 15-lipoxygenase. Proc. Natl. Acad. Sci. 1992;89:217–221. doi: 10.1073/pnas.89.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constien R., Forde A., Liliensiek B., Grone H.J., Nawroth P., Hammerling G., Arnold B., Forde A., Liliensiek B., Grone H.J., Nawroth P., Hammerling G., Arnold B., Liliensiek B., Grone H.J., Nawroth P., Hammerling G., Arnold B., Grone H.J., Nawroth P., Hammerling G., Arnold B., Nawroth P., Hammerling G., Arnold B., Hammerling G., Arnold B., Arnold B. Characterization of a novel EGFP reporter mouse to monitor Cre recombination as demonstrated by a Tie2 Cre mouse line. Genesis. 2001;30:36–44. doi: 10.1002/gene.1030. [DOI] [PubMed] [Google Scholar]
- Fata J.E., Leco K.J., Voura E.B., Yu H.Y., Waterhouse P., Murphy G., Moorehead R.A., Khokha R., Leco K.J., Voura E.B., Yu H.Y., Waterhouse P., Murphy G., Moorehead R.A., Khokha R., Voura E.B., Yu H.Y., Waterhouse P., Murphy G., Moorehead R.A., Khokha R., Yu H.Y., Waterhouse P., Murphy G., Moorehead R.A., Khokha R., Waterhouse P., Murphy G., Moorehead R.A., Khokha R., Murphy G., Moorehead R.A., Khokha R., Moorehead R.A., Khokha R., Khokha R. Accelerated apoptosis in the Timp-3-deficient mammary gland. J. Clin. Invest. 2001;108:831–841. doi: 10.1172/JCI13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman B.M., Tontonoz P., Chen J., Brun R.P., Spiegelman B.M., Evans R.M., Tontonoz P., Chen J., Brun R.P., Spiegelman B.M., Evans R.M., Chen J., Brun R.P., Spiegelman B.M., Evans R.M., Brun R.P., Spiegelman B.M., Evans R.M., Spiegelman B.M., Evans R.M., Evans R.M. 15-Deoxy-Δ 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- Forman B.M., Chen J., Evans R.M., Chen J., Evans R.M., Evans R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C.D. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V., Lin E.Y., Pollard J.W., Lin E.Y., Pollard J.W., Pollard J.W. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002;4:155–164. doi: 10.1186/bcr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M., Nelson M., Ong E., Olefsky J.M., Evans R.M., Ong E., Olefsky J.M., Evans R.M., Olefsky J.M., Evans R.M., Evans R.M. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevener A.L., He W., Barak Y., Le J., Bandyopadhyay G., Olson P., Wilkes J., Evans R.M., Olefsky J., He W., Barak Y., Le J., Bandyopadhyay G., Olson P., Wilkes J., Evans R.M., Olefsky J., Barak Y., Le J., Bandyopadhyay G., Olson P., Wilkes J., Evans R.M., Olefsky J., Le J., Bandyopadhyay G., Olson P., Wilkes J., Evans R.M., Olefsky J., Bandyopadhyay G., Olson P., Wilkes J., Evans R.M., Olefsky J., Olson P., Wilkes J., Evans R.M., Olefsky J., Wilkes J., Evans R.M., Olefsky J., Evans R.M., Olefsky J., Olefsky J. Muscle-specific Pparγ deletion causes insulin resistance. Nat. Med. 2003;9:1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- Huang J.T., Welch J.S., Ricote M., Binder C.J., Willson T.M., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Welch J.S., Ricote M., Binder C.J., Willson T.M., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Ricote M., Binder C.J., Willson T.M., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Binder C.J., Willson T.M., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Willson T.M., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Witztum J.L., Funk C.D., Conrad D., Glass C.K., Funk C.D., Conrad D., Glass C.K., Conrad D., Glass C.K., Glass C.K. Interleukin-4-dependent production of PPAR-γ ligands in macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- Jiang C., Ting A.T., Seed B., Ting A.T., Seed B., Seed B. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- Kaufman C.K., Zhou P., Pasolli H.A., Rendl M., Bolotin D., Lim K.C., Dai X., Alegre M.L., Fuchs E., Zhou P., Pasolli H.A., Rendl M., Bolotin D., Lim K.C., Dai X., Alegre M.L., Fuchs E., Pasolli H.A., Rendl M., Bolotin D., Lim K.C., Dai X., Alegre M.L., Fuchs E., Rendl M., Bolotin D., Lim K.C., Dai X., Alegre M.L., Fuchs E., Bolotin D., Lim K.C., Dai X., Alegre M.L., Fuchs E., Lim K.C., Dai X., Alegre M.L., Fuchs E., Dai X., Alegre M.L., Fuchs E., Alegre M.L., Fuchs E., Fuchs E. GATA-3: An unexpected regulator of cell lineage determination in skin. Genes & Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki Y.Y., Hammer R.E., Miyazaki J., Williams S.C., Richardson J.A., Yanagisawa M., Hammer R.E., Miyazaki J., Williams S.C., Richardson J.A., Yanagisawa M., Miyazaki J., Williams S.C., Richardson J.A., Yanagisawa M., Williams S.C., Richardson J.A., Yanagisawa M., Richardson J.A., Yanagisawa M., Yanagisawa M. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Kliewer S.A., Sundseth S.S., Jones S.A., Brown P.J., Wisely G.B., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Sundseth S.S., Jones S.A., Brown P.J., Wisely G.B., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Jones S.A., Brown P.J., Wisely G.B., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Brown P.J., Wisely G.B., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Wisely G.B., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Devchand P., Wahli W., Willson T.M., Lenhard J.M., Wahli W., Willson T.M., Lenhard J.M., Willson T.M., Lenhard J.M., Lenhard J.M., et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koni P.A., Joshi S.K., Temann U.A., Olson D., Burkly L., Flavell R.A., Joshi S.K., Temann U.A., Olson D., Burkly L., Flavell R.A., Temann U.A., Olson D., Burkly L., Flavell R.A., Olson D., Burkly L., Flavell R.A., Burkly L., Flavell R.A., Flavell R.A. Conditional vascular cell adhesion molecule 1 deletion in mice: Impaired lymphocyte migration to bone marrow. J. Exp. Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutnikova H., Cock T.A., Watanabe M., Houten S.M., Champy M.F., Dierich A., Auwerx J., Cock T.A., Watanabe M., Houten S.M., Champy M.F., Dierich A., Auwerx J., Watanabe M., Houten S.M., Champy M.F., Dierich A., Auwerx J., Houten S.M., Champy M.F., Dierich A., Auwerx J., Champy M.F., Dierich A., Auwerx J., Dierich A., Auwerx J., Auwerx J. Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPARγ hypomorphic mice. Proc. Natl. Acad. Sci. 2003;100:14457–14462. doi: 10.1073/pnas.2336090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Satoh S., Nakano R., Ishii C., Sugiyama T., Nakano R., Ishii C., Sugiyama T., Ishii C., Sugiyama T., Sugiyama T., et al. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- Lehmann J.M., Moore L.B., Smith-Oliver T.A., Wilkison W.O., Willson T.M., Kliewer S.A., Moore L.B., Smith-Oliver T.A., Wilkison W.O., Willson T.M., Kliewer S.A., Smith-Oliver T.A., Wilkison W.O., Willson T.M., Kliewer S.A., Wilkison W.O., Willson T.M., Kliewer S.A., Willson T.M., Kliewer S.A., Kliewer S.A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ) J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Medina-Gomez G., Virtue S., Lelliott C., Boiani R., Campbell M., Christodoulides C., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Virtue S., Lelliott C., Boiani R., Campbell M., Christodoulides C., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Lelliott C., Boiani R., Campbell M., Christodoulides C., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Boiani R., Campbell M., Christodoulides C., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Campbell M., Christodoulides C., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Christodoulides C., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Perrin C., Jimenez-Linan M., Blount M., Dixon J., Jimenez-Linan M., Blount M., Dixon J., Blount M., Dixon J., Dixon J., et al. The link between nutritional status and insulin sensitivity is dependent on the adipocyte-specific peroxisome proliferator-activated receptor-γ2 isoform. Diabetes. 2005;54:1706–1716. doi: 10.2337/diabetes.54.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam M.F., Grant D.F., Cheek J.M., Greene J.F., Williamson K.C., Hammock B.D., Grant D.F., Cheek J.M., Greene J.F., Williamson K.C., Hammock B.D., Cheek J.M., Greene J.F., Williamson K.C., Hammock B.D., Greene J.F., Williamson K.C., Hammock B.D., Williamson K.C., Hammock B.D., Hammock B.D. Bioactivation of leukotoxins to their toxic diols by epoxide hydrolase. Nat. Med. 1997;3:562–566. doi: 10.1038/nm0597-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisseau C., Goodrow M.H., Dowdy D., Zheng J., Greene J.F., Sanborn J.R., Hammock B.D., Goodrow M.H., Dowdy D., Zheng J., Greene J.F., Sanborn J.R., Hammock B.D., Dowdy D., Zheng J., Greene J.F., Sanborn J.R., Hammock B.D., Zheng J., Greene J.F., Sanborn J.R., Hammock B.D., Greene J.F., Sanborn J.R., Hammock B.D., Sanborn J.R., Hammock B.D., Hammock B.D. Potent urea and carbamate inhibitors of soluble epoxide hydrolases. Proc. Natl. Acad. Sci. 1999;96:8849–8854. doi: 10.1073/pnas.96.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Decker K., Leder C., Neumann M., Neufang G., Bayerl C., Schweizer J., Marks F., Furstenberger G., Leder C., Neumann M., Neufang G., Bayerl C., Schweizer J., Marks F., Furstenberger G., Neumann M., Neufang G., Bayerl C., Schweizer J., Marks F., Furstenberger G., Neufang G., Bayerl C., Schweizer J., Marks F., Furstenberger G., Bayerl C., Schweizer J., Marks F., Furstenberger G., Schweizer J., Marks F., Furstenberger G., Marks F., Furstenberger G., Furstenberger G. Expression of cyclooxygenase isozymes during morphogenesis and cycling of pelage hair follicles in mouse skin: Precocious onset of the first catagen phase and alopecia upon cyclooxygenase-2 overexpression. J. Invest. Dermatol. 2003;121:661–668. doi: 10.1046/j.1523-1747.2003.12473.x. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S., Handjiski B., van der Veen C., Eichmuller S., Foitzik K., McKay I.A., Stenn K.S., Paus R., Handjiski B., van der Veen C., Eichmuller S., Foitzik K., McKay I.A., Stenn K.S., Paus R., van der Veen C., Eichmuller S., Foitzik K., McKay I.A., Stenn K.S., Paus R., Eichmuller S., Foitzik K., McKay I.A., Stenn K.S., Paus R., Foitzik K., McKay I.A., Stenn K.S., Paus R., McKay I.A., Stenn K.S., Paus R., Stenn K.S., Paus R., Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nagy L., Tontonoz P., Alvarez J.G., Chen H., Evans R.M., Tontonoz P., Alvarez J.G., Chen H., Evans R.M., Alvarez J.G., Chen H., Evans R.M., Chen H., Evans R.M., Evans R.M. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Sundberg J.P., Paus R., Sundberg J.P., Paus R., Paus R. Mutant laboratory mice with abnormalities in hair follicle morphogenesis, cycling, and/or structure: Annotated tables. Exp. Dermatol. 2001;10:369–390. doi: 10.1034/j.1600-0625.2001.100601.x. [DOI] [PubMed] [Google Scholar]
- Neville M.C., Picciano M.F., Picciano M.F. Regulation of milk lipid secretion and composition. Annu. Rev. Nutr. 1997;17:159–183. doi: 10.1146/annurev.nutr.17.1.159. [DOI] [PubMed] [Google Scholar]
- Node K., Huo Y., Ruan X., Yang B., Spiecker M., Ley K., Zeldin D.C., Liao J.K., Huo Y., Ruan X., Yang B., Spiecker M., Ley K., Zeldin D.C., Liao J.K., Ruan X., Yang B., Spiecker M., Ley K., Zeldin D.C., Liao J.K., Yang B., Spiecker M., Ley K., Zeldin D.C., Liao J.K., Spiecker M., Ley K., Zeldin D.C., Liao J.K., Ley K., Zeldin D.C., Liao J.K., Zeldin D.C., Liao J.K., Liao J.K. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R., Cotsarelis G., Cotsarelis G. The biology of hair follicles. N. Engl. J. Med. 1999;341:491–497. doi: 10.1056/NEJM199908123410706. [DOI] [PubMed] [Google Scholar]
- Rangwala S.M., Rhoades B., Shapiro J.S., Rich A.S., Kim J.K., Shulman G.I., Kaestner K.H., Lazar M.A., Rhoades B., Shapiro J.S., Rich A.S., Kim J.K., Shulman G.I., Kaestner K.H., Lazar M.A., Shapiro J.S., Rich A.S., Kim J.K., Shulman G.I., Kaestner K.H., Lazar M.A., Rich A.S., Kim J.K., Shulman G.I., Kaestner K.H., Lazar M.A., Kim J.K., Shulman G.I., Kaestner K.H., Lazar M.A., Shulman G.I., Kaestner K.H., Lazar M.A., Kaestner K.H., Lazar M.A., Lazar M.A. Genetic modulation of PPARγ phosphorylation regulates insulin sensitivity. Dev. Cell. 2003;5:657–663. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- Ricote M., Li A.C., Willson T.M., Kelly C.J., Glass C.K., Li A.C., Willson T.M., Kelly C.J., Glass C.K., Willson T.M., Kelly C.J., Glass C.K., Kelly C.J., Glass C.K., Glass C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M., Milstone D.S., Spiegelman B.M., Mortensen R.M., Spiegelman B.M., Mortensen R.M., Mortensen R.M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Saez E., Rosenfeld J., Livolsi A., Olson P., Lombardo E., Nelson M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Rosenfeld J., Livolsi A., Olson P., Lombardo E., Nelson M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Livolsi A., Olson P., Lombardo E., Nelson M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Olson P., Lombardo E., Nelson M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Lombardo E., Nelson M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Nelson M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Banayo E., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Cardiff R.D., Izpisua-Belmonte J.C., Evans R.M., Izpisua-Belmonte J.C., Evans R.M., Evans R.M. PPARγ signaling exacerbates mammary gland tumor development. Genes & Dev. 2004;18:528–540. doi: 10.1101/gad.1167804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A., Trauger S.A., Want E.J., Hawkins E.G., Siuzdak G., Cravatt B.F., Trauger S.A., Want E.J., Hawkins E.G., Siuzdak G., Cravatt B.F., Want E.J., Hawkins E.G., Siuzdak G., Cravatt B.F., Hawkins E.G., Siuzdak G., Cravatt B.F., Siuzdak G., Cravatt B.F., Cravatt B.F. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Saitoh M., Uzuka M., Sakamoto M., Uzuka M., Sakamoto M., Sakamoto M. Human hair cycle. J. Invest. Dermatol. 1970;54:65–81. doi: 10.1111/1523-1747.ep12551679. [DOI] [PubMed] [Google Scholar]
- Schwertfeger K.L., McManaman J.L., Palmer C.A., Neville M.C., Anderson S.M., McManaman J.L., Palmer C.A., Neville M.C., Anderson S.M., Palmer C.A., Neville M.C., Anderson S.M., Neville M.C., Anderson S.M., Anderson S.M. Expression of constitutively activated Akt in the mammary gland leads to excess lipid synthesis during pregnancy and lactation. J. Lipid Res. 2003;44:1100–1112. doi: 10.1194/jlr.M300045-JLR200. [DOI] [PubMed] [Google Scholar]
- Shi H., Kokoeva M.V., Inouye K., Tzameli I., Yin H., Flier J.S., Kokoeva M.V., Inouye K., Tzameli I., Yin H., Flier J.S., Inouye K., Tzameli I., Yin H., Flier J.S., Tzameli I., Yin H., Flier J.S., Yin H., Flier J.S., Flier J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Invest. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., Want E.J., O’Maille G., Abagyan R., Siuzdak G., Want E.J., O’Maille G., Abagyan R., Siuzdak G., O’Maille G., Abagyan R., Siuzdak G., Abagyan R., Siuzdak G., Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Peichel C., Vogt T.F., Duboule D., Zakany J., Gonzalez F., Peichel C., Vogt T.F., Duboule D., Zakany J., Peichel C., Vogt T.F., Duboule D., Zakany J., Vogt T.F., Duboule D., Zakany J., Duboule D., Zakany J., Zakany J. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes & Dev. 2001;15:2209–2214. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenn K.S., Paus R., Paus R. Controls of hair follicle cycling. Physiol. Rev. 2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- Tong X., Coulombe P.A., Coulombe P.A. Mouse models of alopecia: Identifying structural genes that are baldly needed. Trends Mol. Med. 2003;9:79–84. doi: 10.1016/s1471-4914(02)00025-4. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B.M., Hu E., Spiegelman B.M., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPARγ 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Nagy L., Alvarez J.G., Thomazy V.A., Evans R.M., Nagy L., Alvarez J.G., Thomazy V.A., Evans R.M., Alvarez J.G., Thomazy V.A., Evans R.M., Thomazy V.A., Evans R.M., Evans R.M. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- Tsai Y.S., Kim H.J., Takahashi N., Kim H.S., Hagaman J.R., Kim J.K., Maeda N., Kim H.J., Takahashi N., Kim H.S., Hagaman J.R., Kim J.K., Maeda N., Takahashi N., Kim H.S., Hagaman J.R., Kim J.K., Maeda N., Kim H.S., Hagaman J.R., Kim J.K., Maeda N., Hagaman J.R., Kim J.K., Maeda N., Kim J.K., Maeda N., Maeda N. Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARγ. J. Clin. Invest. 2004;114:240–249. doi: 10.1172/JCI20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nguyen A., Pollard J.W., Pollard J.W. Colony stimulating factor-1 is required to recruit macrophages into the mammary gland to facilitate mammary ductal outgrowth. Dev. Biol. 2002;247:11–25. doi: 10.1006/dbio.2002.0669. [DOI] [PubMed] [Google Scholar]
- Willson T.M., Cobb J.E., Cowan D.J., Wiethe R.W., Correa I.D., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Cobb J.E., Cowan D.J., Wiethe R.W., Correa I.D., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Cowan D.J., Wiethe R.W., Correa I.D., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Wiethe R.W., Correa I.D., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Correa I.D., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M., Moore L.B., Kliewer S.A., Lehmann J.M., Kliewer S.A., Lehmann J.M., Lehmann J.M. The structure-activity relationship between peroxisome proliferator-activated receptor γ agonism and the antihyperglycemic activity of thiazolidinediones. J. Med. Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- Xu X., Lyle S., Liu Y., Solky B., Cotsarelis G., Lyle S., Liu Y., Solky B., Cotsarelis G., Liu Y., Solky B., Cotsarelis G., Solky B., Cotsarelis G., Cotsarelis G. Differential expression of cyclin D1 in the human hair follicle. Am. J. Pathol. 2003;163:969–978. doi: 10.1016/S0002-9440(10)63456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fu M., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Fu M., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Zhong W., Xiao Y., Floyd D., Liang J., Li E., Xiao Y., Floyd D., Liang J., Li E., Floyd D., Liang J., Li E., Liang J., Li E., Li E., et al. Selective disruption of PPARγ 2 impairs the development of adipose tissue and insulin sensitivity. Proc. Natl. Acad. Sci. 2004;101:10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]