Abstract
We report the complete genome sequence of the model bacterial pathogen Pseudomonas syringae pathovar tomato DC3000 (DC3000), which is pathogenic on tomato and Arabidopsis thaliana. The DC3000 genome (6.5 megabases) contains a circular chromosome and two plasmids, which collectively encode 5,763 ORFs. We identified 298 established and putative virulence genes, including several clusters of genes encoding 31 confirmed and 19 predicted type III secretion system effector proteins. Many of the virulence genes were members of paralogous families and also were proximal to mobile elements, which collectively comprise 7% of the DC3000 genome. The bacterium possesses a large repertoire of transporters for the acquisition of nutrients, particularly sugars, as well as genes implicated in attachment to plant surfaces. Over 12% of the genes are dedicated to regulation, which may reflect the need for rapid adaptation to the diverse environments encountered during epiphytic growth and pathogenesis. Comparative analyses confirmed a high degree of similarity with two sequenced pseudomonads, Pseudomonas putida and Pseudomonas aeruginosa, yet revealed 1,159 genes unique to DC3000, of which 811 lack a known function.
The gamma subgroup of the Proteobacteria contains a number of important pathogens, including animal pathogenic Escherichia, Salmonella, Shigella, and Yersinia spp. and plant pathogenic Erwinia, Pantoea, Xanthomonas, Xylella, and Pseudomonas spp. (1). The genus Pseudomonas is notable because it contains the clinically important human pathogen Pseudomonas aeruginosa, the agriculturally important plant pathogen Pseudomonas syringae, and the nonpathogenic bioremediation agent Pseudomonas putida. Strains of P. syringae are noted for their diverse and host-specific interactions with plants and may be assigned to one of at least 50 pathovars based on host specificity (2). As a pathogen, P. syringae typically enters plant leaves through stomata, multiplies in the intercellular space (apoplast), and eventually produces necrotic lesions that are often surrounded by chlorotic halos (2).
P. syringae pv. tomato DC3000 (DC3000) causes bacterial speck of tomato (3), a worldwide, economically significant disease that is representative of numerous bacterial plant diseases for which effective controls are still needed (4). DC3000 is also a pathogen of the model plant Arabidopsis thaliana (5). Importantly, the pathogenicity of DC3000 resembles that of most animal and plant pathogens in the gamma Proteobacteria, which rely on a type III secretion system (TTSS) to inject virulence effector proteins into host cells (6). Highlighting the role of DC3000 as a model for plant-pathogen interactions, >35 genes encoding TTSS substrates have been experimentally identified in this strain (7-12), which exceeds the number demonstrated for any other bacterial pathogen to date. Furthermore, the pathogenicity of DC3000 resembles that of many important bacterial and fungal pathogens of plants where host specificity is controlled by “gene-for-gene” interactions in which a dominant allele in the host and a dominant allele in the pathogen condition the outcome of the plant-pathogen interaction (13). In this study, we report the complete genome sequence of DC3000, highlight genes with a likely role in virulence, and compare the virulence gene arsenal of DC3000 with that of other plant and animal pathogens.
Materials and Methods
Sequencing and Annotation Methods. Strain DC3000 was originally isolated in 1960 from tomato in the Channel Islands in Guernsey, United Kingdom (ref. 3; D. Cuppels, personal communication). The culture was confirmed as pathogenic on susceptible host plants before high-throughput sequencing. The complete genome was sequenced as described previously (14) and detailed information is available as Supporting Sequencing Methods, which is published as supporting information on the PNAS web site, www.pnas.org. ORFs were identified by using the GLIMMER algorithm (15). Predicted proteins were searched against a nonredundant amino acid database and searched for domains by using HMMER with the Pfam (16) and TIGRfam databases (17). The ORFs were manually curated and assigned to role categories adapted from Riley (18). Paralogous families were constructed as described previously (19). Insertion-sequence (IS) elements were classified according to transposase gene similarity by using BLAST analysis with the ISFinder database (www-is.biotoul.fr/).
Comparative Genomics. The presence of putative orthologs of DC3000 virulence factors in nine completely sequenced genomes of pathogenic proteobacteria was determined by BLASTP analysis by using a cutoff criterion of E < 10-5 (20). The reference genomes included the plant pathogens Ralstonia solanacearum (21), Xylella fastidiosa (22), Agrobacterium tumefaciens (23, 24), Xanthomonas campestris pv. campestris (25), and Xanthomonas axonopodis pv. citri (25) and the animal pathogens P. aeruginosa PAO1 (26), Salmonella typhimurium LT2 (27), Yersinia pestis CO92 (28), and Escherichia coli O157:H7 EDL933 (29).
Results and Discussion
Genome Features. The DC3000 genome is composed of one circular chromosome of 6,397,126 bp and two plasmids of 73,661 bp (pDC3000A) and 67,473 bp (pDC3000B) (Fig. 1 and Table 1). A total of 5,763 ORFs were identified within the DC3000 genome. Among the predicted genes, 3,510 (61%) have been assigned a putative function, with the remaining designated as encoding a protein with unknown function, a conserved hypothetical protein, or a hypothetical protein. Classification of the known proteins into biological role categories (18) revealed that the largest category was transport and binding, followed by cellular processes (see Fig. 4, which is published as supporting information on the PNAS web site). DC3000 is representative of pathogenic bacteria that have diverse relationships with plants as epiphytes and pathogens and thus likely requires an array of regulatory genes to cope with diverse niches, host responses, and environmental conditions. Indeed, 12% (711) of the DC3000 genes could be assigned to role categories associated with gene regulation, signal transduction, and transcription. A high degree of gene duplication exists in DC3000, and 2,735 genes (48%) were assigned to 687 paralogous families. Although the majority of paralogous families contained two members (see Fig. 5, which is published as supporting information on the PNAS web site), 12 families have >25 members, and many of these large families encode either transporters or regulatory proteins. However, this group also includes families encoding methyl-accepting chemotaxis proteins, oxidoreductases of largely unknown specificities, and transposases of the IS4 family. A total of 5 rRNA operons, 5 structural RNAs, and 63 tRNA genes were present, all on the chromosome.
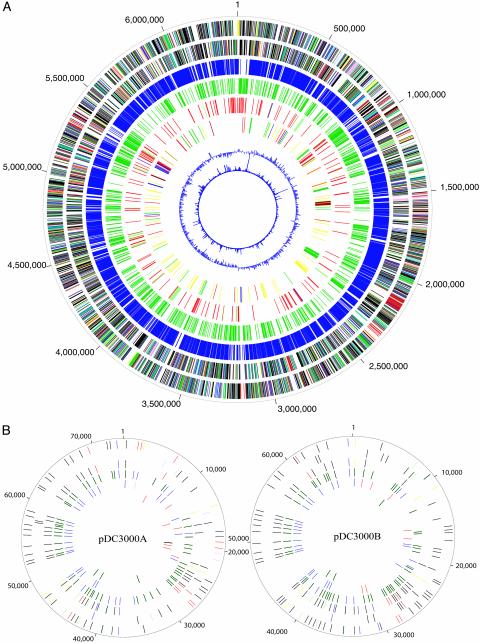
Fig. 1.
Features of the DC3000 genome. (A) The chromosome, with the outermost circle depicting ORFs on the positive strand and the second circle depicting ORFs on the negative strand. The ORFs are color-coded based on the major grouping of role categories as follows: amino acid biosynthesis (salmon); biosynthesis of cofactors, prosthetic groups, and carriers (light blue); cell envelope (light green); cellular processes (red); central intermediary metabolism (brown); DNA metabolism (yellow); energy metabolism (green); fatty acid and phospholipid metabolism (purple); protein fate/synthesis (pink); purines, pyrimidines, nucleosides, and nucleotides (orange); regulatory functions (navy blue); transcription (gray); transport and binding proteins (teal); and unknown function, conserved hypothetical, and hypothetical (black). The third circle represents the ORFs with homologs present in all three Pseudomonas spp. (E < 10-5). The fourth circle depicts the ORFs that are unique to DC3000 compared with P. aeruginosa and P. putida (E ≥ 10-5). The fifth circle represents mobile genetic elements, including IS elements (red), phage-related genes (light green), and plasmid-related functions (blue). The sixth circle depicts ORFs associated with virulence, which are color coded as follows: adhesins and other cell surface-associated factors (dark green), miscellaneous virulence factors (yellow), TTSS effectors, candidates, and helpers (red), TTSS regulatory and secretory proteins (brown), and low molecular weight diffusible factors (blue). The seventh circle is the GC skew, and the eighth circle represents atypical nucleotides. (B) The plasmids with the outermost circle representing ORFs on the positive strand and the second circle depicting ORFs on the negative strand, with the same color coding as A. The third circle represents ORFs shared by the two plasmids (E < 10-5). The fourth circle depicts the ORFs with homologs present in all three Pseudomonas spp. (E < 10-5) in navy blue and the ORFs that are unique to DC3000 compared with P. aeruginosa and P. putida (E ≥ 10-5) in green. The fifth circle represents mobile genetic elements (color-coded as in A). The sixth circle depicts ORFs associated with virulence (color-coded as in A).
Table 1. Features of the DC3000 genome.
| Feature | Chromosome | pDC3000A | pDC3000B |
|---|---|---|---|
| Size, bp | 6,397,126 | 73,661 | 67,473 |
| G + C, % | 58.4 | 55.1 | 56.1 |
| Number of ORFs | 5,615 | 71 | 77 |
| Percent coding | 86.8 | 81.9 | 84.7 |
| Average ORF length, nt | 988 | 849 | 742 |
| No. of ORFs encoding | |||
| Protein with a known function | 3,402 (61%) | 52 (73%) | 46 (60%) |
| Hypothetical protein | 610 (11%) | 10 (14%) | 16 (21%) |
| Conserved hypothetical protein | 961 (17%) | 8 (11%) | 12 (16%) |
| Protein of unknown function | 642 (11%) | 1 (1%) | 3 (4%) |
Comparative Genomics with Fluorescent Pseudomonads. The complete genome sequences for two other pseudomonads, P. aeruginosa PAO1 (26), an opportunistic animal pathogen, and P. putida KT2440, a saprophytic soil bacterium (30), are available. Of the 5,608 ORFs in DC3000 that predict a functional protein, 4,449 (79%) have homologs (BLASTP E < 10-5) in either P. aeruginosa or P. putida and 3,797 (68%) are present in all three genomes (Fig. 1). The 1,159 (21% of the total) genes that are unique to DC3000 are heavily enriched in hypothetical proteins (551), conserved hypothetical proteins (203), mobile genetic elements (116), and proteins of unknown function (57). Other genes unique to DC3000 include 96 that are implicated in virulence (see below for more details).
Metabolism. The key metabolic pathways in DC3000 (glycolysis, gluconeogenesis, tricarboxylic acid, and pentose phosphate cycles) are shared with P. putida and P. aeruginosa. However, the absence of lactate dehydrogenase in DC3000 results in an inability to convert pyruvate to lactate. Taxonomic classification based on the arginine dihydrolase and oxidase tests also distinguishes DC3000 from P. putida and P. aeruginosa, which are positive in both tests (31). DC3000 lacks arginine dihydrolase and subunits of the aa3-type cytochrome c oxidase.
The utilization of γ-aminobutyric acid (GABA) occurs by means of the GABA transaminase-catalyzed deamination of GABA to succinic semialdehyde, which is then oxidized to succinate by succinate semialdehyde dehydrogenase (SSADH) before entering the tricarboxylic acid cycle. In DC3000, there are three copies each of GABA transaminase and SSADH, as well as a GABA permease. Although the composition of the apoplast during a compatible interaction with DC3000 is unknown, GABA is present at milli-molar concentration in the apoplast during infection of tomato with the biotrophic fungal pathogen Cladosporium fulvum (32). Thus, if GABA is available in the apoplast during infection of tomato, DC3000 appears amply equipped to use this as a carbon and nitrogen source.
General Transport Systems. Like the other sequenced pseudomonads, DC3000 possesses very extensive transport capabilities. However, it differs significantly from P. putida and P. aeruginosa in having a greater capacity for sugar transport and a correspondingly decreased number of amino acid transporters. Importantly, DC3000 has 15 ABC transporter systems with predicted specificities for arabinose, xylose, ribose, and other plant-derived sugars, whereas P. putida and P. aeruginosa each encode only 4 ABC sugar transporters. In contrast, DC3000 encodes only 4 APC-family transporters for amino acids and other compounds, compared with the 21 APC transporters present in both P. putida and P. aeruginosa. DC3000 also encodes 9 RND-type efflux systems for drugs or cations, including probable orthologs of the Mex systems of P. aeruginosa, as well as a broad range of other predicted efflux systems. Thus, broad export capabilities appear to be a common feature of the pseudomonads.
Type I secretion is one-step process exemplified by E. coli hemolysin secretion (33), and homologs of the essential components of the pathway are present in DC3000. The type II secretion pathway is an extension of the general secretory pathway (34), and 14 genes predicting components of this pathway were identified in DC3000. Genes similar to gspC and gspO were not found; however, gspC is not present in all systems, and the function of GspO can be replaced by the prepilin peptidase PilD, which is present in the genome. A functional type IV secretion system was not identified in DC3000.
Virulence Mechanisms. DC3000 encodes a wide range of proteins that are implicated in virulence (35). We are interpreting virulence here to include processes such as motility, adhesion, injection of effector proteins into host cells, degradation of host cell walls, production of phytotoxic compounds, iron acquisition, and interference with host defenses, which may promote epiphytic growth as well as actual disease expression (Fig. 2). We identified 298 genes (5% of the total) in the virulence category (Table 2, which is published as supporting information on the PNAS web site). The virulence genes are addressed below.
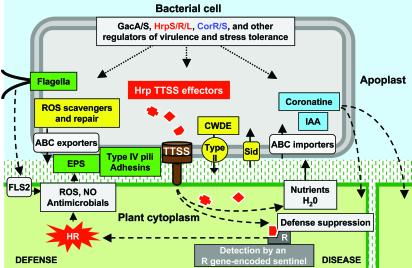
Fig. 2.
Diagrammatic overview of P. syringae-plant interactions, with virulence factors color-coded as in Fig. 1. Host-pathogen factors associated with defense and disease are separately grouped on the left and right of the diagram, respectively. The central pathogenic process is the injection of multiple effector proteins into plant cells by the TTSS, which is depicted as the brown structure traversing the bacterial inner and outer membranes, plant cell wall, and plasma membrane. The effectors may suppress defenses and promote nutrient and water accumulation in the apoplast unless any one of them is detected by a resistance (R) gene-encoded sentinel, in which case strong defenses associated with the hypersensitive response (HR) are triggered. Motility favors bacterial epiphytic survival and entry into leaves, but flagellin is also recognized by means of the FLS2 receptor-like kinase as an elicitor of general defenses (62), which are not as strong as HR-associated defenses and may be suppressed by Hrp effectors. Exclusion or tolerance of reactive oxygen species (ROS) and other antimicrobials is likely promoted by extracellular polysaccharides (EPS), scavengers, and ABC exporters. Favoring virulence are type IV pili (Tfp) and possibly adhesins, coronatine synthesis and regulation, plant cell-wall-degrading enzymes (CWDE) and other variously secreted proteins, iron-scavenging siderophores (Sid), indoleacetic acid (IAA), and probably numerous ABC transporters and other nutrient uptake systems.
TTSS and Effectors. The TTSS is key to the plant parasitism of P. syringae pathovars (Fig. 2). Most of the hrp (hypersensitive response and pathogenicity) and hrc (hrp conserved) genes encoding the TTSS system are essential for pathogenicity, which indicates the collective importance of the effector proteins that are injected into plant cells by the system (6). Hrp effectors are primarily known as Hop (Hrp outer protein) or Avr (avirulence) proteins, based on the phenotype by which they were discovered (11). Functional analysis of the DC3000 genome has yielded genes for 31 effectors and 7 additional proteins secreted by the Hrp system that likely have a role in translocating effectors across the plant cell wall and plasma membrane (8-12, 36). The associated hop, avr, and hol (Hop-like candidate) genes may provide markers to regions of the genome that are enriched in other virulence genes (Fig. 3). Over half of these TTSS effectors are encoded in one of several regions containing multiple effector genes (Fig. 1). One region with multiple effector genes is located on pDC3000A and contains avrPpiB2Pto, avrPphEPto, hopPtoS1, and hopPtoT1. Interestingly, an identical copy of avrPpiB2Pto is located on the chromosome, which could provide a site for integration of the plasmid in a manner similar to that observed with pFKN in P. syringae pv. maculicola M6 (37). Two other clusters of effector genes are found in the region encoding the phytotoxin coronatine (COR) (Fig. 3). Members of effector gene paralogous families are scattered around the genome and typically have an unusually low G+C content (Table 2), suggesting that such families may have grown by sequential horizontal acquisitions. Interestingly, at least four of the DC3000 effector genes are disrupted by mobile genetic elements (Tables 2 and 3, which are published as supporting information on the PNAS web site), as highlighted by comparison with orthologs in the draft genome of P. syringae pv. syringae B728a (12). Additional information on DC3000 TTSS effectors and an updated list can be found at http://pseudomonas-syringae.org.
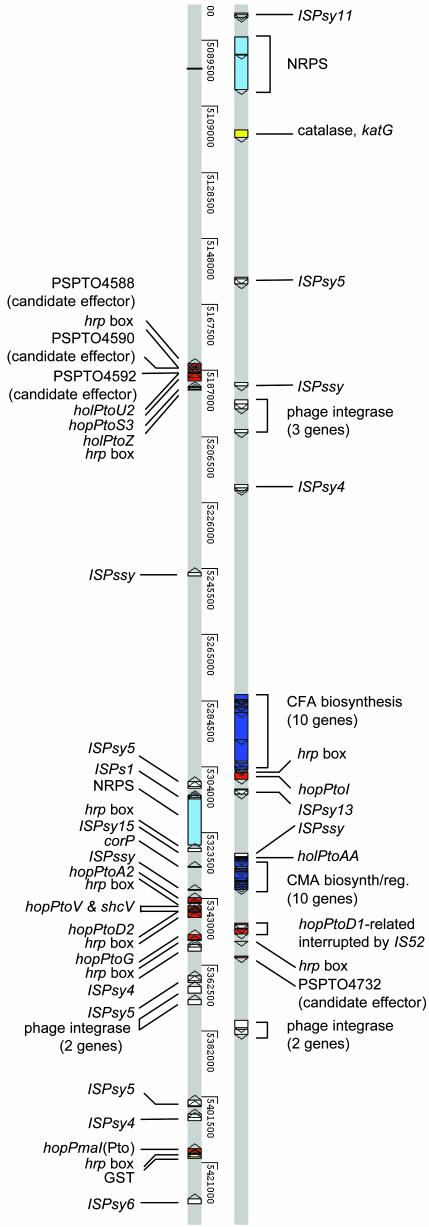
Fig. 3.
Detailed view of one region of the DC3000 genome that is enriched in virulence factors and mobile genetic elements. Only virulence-related genes, “hrp box” promoters, and mobile genetic elements are shown. Features are color-coded as in Fig. 1 (circle 6), with the exception that a lighter blue is used for the nonribosomal peptide synthase genes, and the mobile genetic elements are white. The genome display was generated by using artemis (www.sanger.ac.uk/Software/Artemis/) to view the chromosome (AE016853) with the aid of input files for a variety of virulence-related features that are available at http://pseudomonas-syringae.org and http://monod.cornell.edu.
Plasmid-Borne Virulence-Implicated Genes. Consistent with previous reports (38), DC3000 contains two plasmids (pDC3000A and pDC3000B) that are similar to the pPT23A family of P. syringae plasmids (39). There is substantial redundancy between the two plasmids, with 47 proteins sharing similarity (E < 10-5). The majority of the conserved proteins are involved in plasmid replication, conjugation, or transposition-related functions. There also are many genes unique to each of the plasmids, with a clear enrichment of genes implicated in virulence on pDC3000A. However, we found no evidence for a major role of the plasmids in virulence. Using previously described methods (40), we inserted sacB in pDC3000A and repA in pDC3000B to construct DC3000 derivatives lacking pDC3000A, pDC3000B, or both plasmids. Assays involving dipping of tomato cv. Moneymaker and A. thaliana Col-0 in inoculum (5 × 105 colony-forming units per ml) revealed no major difference in bacterial growth in planta or in symptom production (data not shown). This observation is consistent with the finding that most of the known plasmid-borne TTSS effector genes, which are all on pDC3000A, have paralogs on the chromosome.
Siderophores. Siderophores are low molecular weight, high-affinity iron(III) chelators that function as important virulence factors in many bacterial pathogens because of their role in sustaining growth in iron-limited host environments. Synthesis of the siderophore pyoverdin is well documented among fluorescent pseudomonads (41), and the required genes are present in a single cluster in DC3000. Also, a cluster of genes homologous to those required for biosynthesis of the siderophore pyochelin by P. aeruginosa has been found. However, several observations suggest that the encoded proteins actually produce yersiniabactin, a siderophore that shares a common tricyclic core with pyochelin (42). First, DC3000 contains a gene encoding a large polyketide synthase (PKS) in the middle of the pyochelin gene cluster and this has no homolog in P. aeruginosa (or P. putida). This gene shows extremely high similarity with the PKS gene (irp1) found in the yersiniabactin gene cluster of Y. pestis (43). Second, there is no homolog of the pyochelin receptor gene of P. aeruginosa (fptA) in DC3000; however, a receptor gene is present that is highly similar to the pesticin/yersiniabactin receptor gene from Y. pestis (fyuA). Third, the structure of PSPTO2602, a nonribosomal peptide synthase in this cluster, differs from its P. aeruginosa counterpart in that a methylation domain and a terminal thioesterase domain are lacking, consistent with a yersiniabactin synthase.
Phytotoxins and Phytohormones. COR is a chlorosis-inducing phytotoxin produced by several P. syringae pathovars, including DC3000 (44). COR is hypothesized to function as a molecular mimic of methyl jasmonate and may promote colonization of host tissue by altering plant defense signaling pathways (45). COR is representative of biologically active small molecules that can diffuse in plant tissues (Fig. 2), and it is responsible for the chlorotic halos surrounding the necrotic lesions caused by DC3000 (44). The structure of COR consists of coronafacic acid and coronamic acid (CMA), joined by an amide linkage (46). Unlike many P. syringae pathovars where the COR genes are clustered and plasmid-encoded (46), the COR genes in DC3000 are chromosomally encoded, and the coronafacic acid and CMA structural genes are separated by 26 kb (Fig. 3). In DC3000, the CMA biosynthetic genes are linked with a modified two-component regulatory system, composed of corS, corR, and corP (46). Interestingly, analysis of the CMA region of DC3000 revealed four genes (PSPTO4707, 4708, 4711, and 4713) interspersed among those previously identified with roles in COR synthesis. Consistent with the annotation of the COR region, initial proteomic studies with DC3000 revealed that the HrpL-alternative σ factor, which regulates DC3000 hrp genes (8), induces production of a protein encoded by PSPTO4668 in the coronafacic acid region (D.J.S. and S.C., unpublished data; http://monod.cornell.edu). There is no evidence in the DC3000 genome for the biosynthesis of any of the known P. syringae lipodepsinonapeptide phytotoxins, including syringomycin, syringotoxin, syringostatin, and pseudomycin. Syringopeptin, a lipodepsipeptide produced by P. syringae pv. syringae (46), is also absent. However, DC3000 does contain five nonribosomal peptide synthases in three separate gene clusters (PSPTO2829, -2830, -4518, -4519, and -4699) that may direct the biosynthesis of undescribed peptide or lipodepsipeptide products.
Production of the phytohormone indole-3-acetic acid (IAA) by certain P. syringae pathovars has been shown to influence bacterial growth in planta and the expression of other virulence factors (47, 48). The two genes required for IAA production, iaaH and iaaM, are present in DC3000. Interestingly, iaaL (IAA-lysine ligase), which can convert IAA to a less active derivative and is widespread in P. syringae pathovars (47), is present in the DC3000 genome but lacking in the genomes of all of the other fully sequenced plant pathogens (Table 2). The presence of a functional Hrp promoter upstream of iaaL additionally suggests a role in P. syringae virulence (8).
Adhesins. Pathogenic bacteria rely on a variety of cell surface-associated virulence factors to facilitate adhesion to the host and provide protection during colonization. Type IV pili (Tfp) represent one of the major adhesins found in pseudomonads (49). Tfp were previously identified in P. syringae and appear to play a role in epiphytic fitness (50). Analysis of the 23 Tfp-associated genes suggests that regulation of Tfp production differs significantly between DC3000 and P. aeruginosa. Filamentous hemagglutinin (FHA) has also been shown to be an adhesin and a virulence factor in animal pathogenic bacteria and the plant pathogen Erwinia chrysanthemi (51). Three genes predicting FHA-like proteins were identified in DC3000 (PSPTO3210, 3214, and 3229).
Extracellular Polysaccharides. Components of the polysaccharide capsule, such as the exopolysaccharide alginate, play a role in both adhesion and protection of the bacterial cells from external stress (52). Among plant pathogenic pseudomonads, alginate production has been linked to epiphytic fitness and the production of water-soaked lesions (53). All of the genes required for alginate biosynthesis in P. aeruginosa are present in DC3000, although some of the alginate regulatory genes are absent, indicating that the regulation of alginate biosynthesis differs between P. aeruginosa and P. syringae, consistent with previous reports (54). Three genes encoding levansucrases, required for the biosynthesis of the polysaccharide levan, were also identified in DC3000.
Tolerance to Reactive Oxygen Species or Heavy Metals. P. syringae and other pathogens can trigger an oxidative burst in plants, which results in the production in the apoplast of reactive oxygen species such as superoxide anion and hydrogen peroxide (H2O2). Superoxide dismutases (SODs) convert superoxide to H2O2, which can be detoxified to water by catalase. In addition to its antimicrobial properties, H2O2 functions as a signaling molecule within plant cells in processes involving pathogen defense and programmed cell death (55). DC3000 has three catalases and three SODs. Other enzymes in DC3000 that may play a role in scavenging reactive oxygen species include six peroxidases, eight GSTs, and two glutathione reductases. In addition, DC3000 encodes a single peptide methionine sulfoxide reductase, which reduces methionine sulfoxide to methionine, thereby functioning as a repair enzyme for proteins inactivated by oxidation.
One strategy to control bacterial plant pathogens in the field is the use of copper-based bactericides, which are toxic to bacterial cells at high concentrations. For bacterial speck of tomato, this has led to the emergence of copper-resistant strains (56). In P. syringae pv. tomato strain PT23.2, resistance to copper is mediated by four structural genes, copA-D, as well as two regulatory genes, copR and copS, which encode a two-component regulatory system (57). DC3000 is not resistant to copper (C.L.B., unpublished data), and examination of the genome revealed that although copA and copB are present, DC3000 lacks the structural proteins CopC and CopD, as well as the regulatory proteins CopR and CopS. Interestingly, homologs of CopR and CopS (PSPTO3603, 1306) are present in DC3000 and may function in regulating responses to other heavy metals.
Other Proteins Relevant to Virulence. Pectic enzymes and cellulases were previously identified in other Pseudomonas spp., and pectate lyase has been shown to influence the final symptoms produced by P. syringae pv. lachrymans (58). Genes encoding cell-wall-degrading enzymes are present in DC3000 and include a pectin lyase, a polygalacturonase, and three enzymes predicted to have cellulolytic activity (PSPTO1029, PSPTO3534, and PSPTO0905). As previously noted, DC3000 has two insecticidal toxin gene clusters (9). Although P. syringae is not known to be an insect pathogen, insects may be casual vectors of epiphytic bacteria on wet leaves (2). It is also worth noting that DC3000 lacks a gene encoding the outer-membrane ice nucleation protein, which enables many strains of P. syringae to cause frost damage on plants (2).
Mobile Genetic Elements and Their Potential Impact on the Genome.
A total of 382 genes (7%) in the DC3000 genome were classified as mobile genetic elements, including genes encoding 270 proteins that function in transposition, 54 proteins with plasmid functions, and 58 proteins with prophage functions. Compared with other published plant pathogen genomes (21-23, 25), this is the highest percentage seen to date, resulting in 24 ORFs interrupted by an IS element or phage protein (Table 3). The transposable element class, which includes the IS elements, accounts for 5% of the DC3000 ORFs (Table 4, which is published as supporting information on the PNAS web site). The IS elements vary widely in their copy number, presumably reflecting a combination of transposition frequency and residence time in the genome. Multiple prophage regions also are present in DC3000, which collectively encompass ∼118 kbp of the genome (Table 5, which is published as supporting information on the PNAS web site). The impact of mobile genetic elements on the DC3000 genome is seen by their frequent association with regions that lack homologs in P. aeruginosa and/or P. putida, that are enriched in atypical nucleotides potentially indicative of horizontal acquisition, and/or that encode either TTSS effectors or proteins of unknown function. One such region, shown in Fig. 3, contains a potpourri of known and candidate virulence factors, including catalase and GST genes, COR biosynthetic genes, a nonribosomal peptide synthase preceded by a Hrp promoter, and two clusters of effector genes, along with multiple mobile genetic elements, one of which has disrupted a hopPtoD paralog.
Comparative Genomics of Virulence. We performed a comparative analysis involving 298 putative virulence genes in DC3000 and the complete genome sequences of nine plant and animal pathogenic proteobacteria, which revealed that 86 genes are common to all of these pathogens (Table 2 and Fig. 6, which is published as supporting information on the PNAS web site). The pathogen with the most similar genes was P. aeruginosa PAO1 (191 genes; 64% of the virulence factors), which reflects the relatedness of DC3000 and PAO1 and the large number of similar genes involved in flagellum, alginate, and Tfp production. Not surprisingly, among the 65 virulence genes that are unique to DC3000, 33 encode TTSS effectors, helpers, and candidates, which is consistent with the specialized nature of TTSS-based host-pathogen interactions.
Another promising source of virulence candidates is the set of genes unique to DC3000 that were identified in comparative analyses with P. putida and P. aeruginosa. A search of the 811 genes of unknown function in this category revealed that 635 also were unique to DC3000 in comparisons with all eight of the reference pathogen genomes. However, four of these genes were present in all of the plant pathogenic bacteria, and another set of four were present exclusively in all animal pathogenic bacteria (Table 6, which is published as supporting information on the PNAS web site).
In considering DC3000 virulence genomics, it is worth noting that pioneering genetic studies with P. syringae yielded a paradox that has long frustrated comprehensive exploration of the virulence of this pathogen (59, 60). That is, despite the many indications that P. syringae is a highly coevolved plant parasite, screens for mutants with reduced virulence have yielded primarily mutants disabled in a single system, the Hrp TTSS. The complete genome sequence of DC3000 clarifies the basis for this paradox in the redundancy of many of the candidate virulence factors. The sequence will now facilitate the construction of polymutants that can test the role of genes with overlapping function, and it will foster the identification of novel virulence factors through targeted investigation of genes that are unique or duplicated in comparison with P. aeruginosa and P. putida or in regions showing signatures of horizontal acquisition. In conclusion, P. syringae pathobiology has been described as a “highly dynamic, highly variable matrix of interactions of bacteria of widely variable genotypes with a broadly variable group of plants” (61), and the plethora of mobile genetic elements and associated virulence factors that we found provides a genomic basis for such a dynamic association with plants.
Supplementary Material
Acknowledgments
We thank the members of The Institute for Genome Research Sequencing Facility, Informatics Department, and Information Technology Support Group who contributed to the work. Funding for the work described in this study was provided by National Science Foundation Grants DBI-0077622 (to A.C., J.R.A., C.R.B., S.C., A.K.C., T.P.D., S.G.L., G.B.M., and X.T.) and IBN-0135286 (to C.L.B.) and National Institutes of Health Grant AI 43311 (to C.L.B.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CMA, coronamic acid; COR, coronatine; DC3000, Pseudomonas syringae pv. tomato DC3000; GABA, γ-aminobutyric acid; IS, insertion sequence; Tfp, type IV pili; TTSS, type III secretion system.
Data deposition: The sequence and annotation of P. syringae pv. tomato DC3000 have been deposited in the GenBank database (accession nos. AE016853-AE016855).
References
- 1.Gupta, R. S. (2000) FEMS Microbiol. Rev. 24, 367-402. [DOI] [PubMed] [Google Scholar]
- 2.Hirano, S. S. & Upper, C. D. (2000) Microbiol. Mol. Biol. Rev. 64, 624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuppels, D. A. (1986) Appl. Environ. Microbiol. 51, 323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson, M., Campbell, H. L., Ji, P., Jones, J. B. & Cuppels, D. A. (2002) Phytopathology 92, 1284-1292. [DOI] [PubMed] [Google Scholar]
- 5.Whalen, M. C., Innes, R. W., Bent, A. F. & Staskawicz, B. J. (1991) Plant Cell 3, 49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis, G. R. & Van Gijsegem, F. (2000) Annu. Rev. Microbiol. 54, 735-774. [DOI] [PubMed] [Google Scholar]
- 7.Boch, J., Joardar, V., Gao, L., Robertson, T. L., Lim, M. & Kunkel, B. N. (2002) Mol. Microbiol. 44, 73-88. [DOI] [PubMed] [Google Scholar]
- 8.Fouts, D. E., Abramovitch, R. B., Alfano, J. R., Baldo, A. M., Buell, C. R., Cartinhour, S., Chatterjee, A. K., D'Ascenzo, M., Gwinn, M. L., Lazarowitz, S. G., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G. & Greenberg, J. T. (2002) Science 295, 1722-1726. [DOI] [PubMed] [Google Scholar]
- 10.Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., Schechter, L. M., Buell, C. R., Tang, X., Collmer, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collmer, A., Lindeberg, M., Petnicki-Ocwieja, T., Schneider, D. J. & Alfano, J. R. (2002) Trends Microbiol. 10, 462-469. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, J. T. & Vinatzer, B. A. (2003) Curr. Opin. Microbiol. 6, 20-28. [DOI] [PubMed] [Google Scholar]
- 13.Keen, N. T. (1990) Annu. Rev. Genet. 24, 447-463. [DOI] [PubMed] [Google Scholar]
- 14.Tettelin, H., Nelson, K. E., Paulsen, I. T., Eisen, J. A., Read, T. D., Peterson, S., Heidelberg, J., DeBoy, R. T., Haft, D. H., Dodson, R. J., et al. (2001) Science 293, 498-506. [DOI] [PubMed] [Google Scholar]
- 15.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haft, D. H., Selengut, J. D. & White, O. (2003) Nucleic Acids Res. 31, 371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley, M. (1993) Microbiol. Rev. 57, 862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nierman, W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen, J. A., Heidelberg, J. F., Alley, M. R., Ohta, N., Maddock, J. R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., Billault, A., Brottier, P., Camus, J. C., Cattolico, L., et al. (2002) Nature 415, 497-502. [DOI] [PubMed] [Google Scholar]
- 22.Simpson, A. J. G., Reinach, F. C., Arruda, P., Abreu, F. A., Acencio, M., Avarenga, R., Alves, L. M., Araya, J. E., Baia, G. S., Baptista, C. S., et al. (2000) Nature 406, 151-157. [DOI] [PubMed] [Google Scholar]
- 23.Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., Zhou, Y., Chen, L., Wood, G. E., Almeida, N. F., Jr., et al. (2001) Science 294, 2317-2323. [DOI] [PubMed] [Google Scholar]
- 24.Goodner, B., Hinkle, G., Gattung, S., Miller, N., Blanchard, M., Qurollo, B., Goldman, B. S., Cao, Y., Askenazi, M., Halling, C., et al. (2001) Science 294, 2323-2328. [DOI] [PubMed] [Google Scholar]
- 25.Da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Sluys, M. A., Almeida, N. F., Alves, L. M., et al. (2002) Nature 417, 459-463. [DOI] [PubMed] [Google Scholar]
- 26.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 27.McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. (2001) Nature 413, 852-856. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 29.Perna, N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, K. E. (2002) Environ. Microbiol. 4, 777. [DOI] [PubMed] [Google Scholar]
- 31.Palleroni, N. J. (1984) in Bergey's Manual of Systematic Bacteriology, eds. Krieg, N. R. & Holt, J. G. (Williams & Wilkins, Baltimore), pp. 141-199.
- 32.Solomon, P. S. & Oliver, R. P. (2001) Planta 213, 241-249. [DOI] [PubMed] [Google Scholar]
- 33.Binet, R., Letoffe, S., Ghigo, J. M., Delepelaire, P. & Wandersman, C. (1997) Gene 192, 7-11. [DOI] [PubMed] [Google Scholar]
- 34.Sandkvist, M. (2001) Mol Microbiol 40, 271-283. [DOI] [PubMed] [Google Scholar]
- 35.Preston, G. M. (2000) Mol. Plant Pathol. 1, 263-275. [DOI] [PubMed] [Google Scholar]
- 36.Zwiesler-Vollick, J., Plovanich-Jones, A. E., Nomura, K., Brandyopadhyay, S., Joardar, V., Kunkel, B. N. & He, S. Y. (2002) Mol. Microbiol. 45, 1207-1218. [DOI] [PubMed] [Google Scholar]
- 37.Rohmer, L., Kjemtrup, S., Marchesini, P. & Dangl, J. L. (2003) Mol. Microbiol. 47, 1545-1562. [DOI] [PubMed] [Google Scholar]
- 38.Cuppels, D. A. & Ainsworth, T. (1995) Appl. Environ. Microbiol. 61, 3530-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sesma, A., Sundin, G. W. & Murillo, J. (2000) Microbiology 146, 2375-2384. [DOI] [PubMed] [Google Scholar]
- 40.Jackson, R. W., Athanassopoulos, E., Tsiamis, G., Mansfield, J. W., Sesma, A., Arnold, D. L., Gibbon, M. J., Murillo, J., Taylor, J. D. & Vivian, A. (1999) Proc. Natl. Acad. Sci. USA 96, 10875-10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bultreys, A. & Gheysen, I. (2000) Appl. Environ. Microbiol. 66, 325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reimmann, C., Patel, H. M., Serino, L., Barone, M., Walsh, C. T. & Haas, D. (2001) J. Bacteriol. 183, 813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelludat, C., Rakin, A., Jacobi, C. A., Schubert, S. & Heesemann, J. (1998) J. Bacteriol. 180, 538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore, R. A., Starratt, A. N., Ma, S. W., Morris, V. L. & Cuppels, D. A. (1989) Can. J. Microbiol. 35, 910-917. [Google Scholar]
- 45.Kloek, A. P., Verbsky, M. L., Sharma, S. B., Schoelz, J. E., Vogel, J., Klessig, D. F. & Kunkel, B. N. (2001) Plant J. 26, 509-522. [DOI] [PubMed] [Google Scholar]
- 46.Bender, C. L., Alarcon-Chaidez, F. & Gross, D. C. (1999) Microbiol. Mol. Biol. Rev. 63, 266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glickmann, E., Gardan, L. & Dessaux, Y. (1998) Mol. Plant-Microbe Interact. 11, 156-162. [DOI] [PubMed] [Google Scholar]
- 48.Mazzola, M. & White, F. F. (1994) J. Bacteriol. 176, 1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wall, D. & Kaiser, D. (1999) Mol. Microbiol. 32, 1-10. [DOI] [PubMed] [Google Scholar]
- 50.Roine, E., Raineri, D. M., Romantschuk, M., Wilson, M. & Nunn, D. N. (1998) Mol. Plant-Microbe Interact. 11, 1048-1056. [DOI] [PubMed] [Google Scholar]
- 51.Rojas, C. M., Ham, J. H., Deng, W.-L., Doyle, J. J. & Collmer, A. (2002) Proc. Natl. Acad. Sci. USA 99, 13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gacesa, P. (1998) Microbiology 144, 1133-1143. [DOI] [PubMed] [Google Scholar]
- 53.Yu, J., Peñaloza-Vázquez, A., Chakrabarty, A. M. & Bender, C. L. (1999) Mol. Microbiol. 33, 712-720. [DOI] [PubMed] [Google Scholar]
- 54.Peñaloza-Vázquez, A., Kidambi, S. P., Chakrabarty, A. M. & Bender, C. L. (1997) J. Bacteriol. 179, 4464-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittler, R. (2002) Trends Plant Sci. 7, 405-410. [DOI] [PubMed] [Google Scholar]
- 56.Bender, C. L. & Cooksey, D. A. (1987) J. Bacteriol. 169, 470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silver, S. & Phung, L. T. (1996) Annu. Rev. Microbiol. 50, 753-789. [DOI] [PubMed] [Google Scholar]
- 58.Bauer, D. W. & Collmer, A. (1997) Mol. Plant-Microbe Interact. 10, 369-379. [DOI] [PubMed] [Google Scholar]
- 59.Niepold, F., Anderson, D. & Mills, D. (1985) Proc. Natl. Acad. Sci. USA 82, 406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindgren, P. B., Peet, R. C. & Panopoulos, N. J. (1986) J. Bacteriol. 168, 512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirano, S. S. & Upper, C. D. (1990) Annu. Rev. Phytopathol. 28, 155-177. [Google Scholar]
- 62.Gomez-Gomez, L. & Boller, T. (2000) Mol. Cell 5, 1003-1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.