Abstract
The biotic consequences of climate change have attracted considerable attention. In particular, the “refugial debate” centers on the possible retraction of habitats to limited areas that may have served as refuges for many associated species, especially during glaciations of the Quaternary. One prediction of such scenarios is that populations must have experienced substantial growth accompanying climatic amelioration and the occupation of newly expanded habitats. We used coalescence theory to examine the genetic evidence, or lack thereof, for late Pleistocene refugia of boreal North American and tropical Amazonian mammals. We found substantial and concordant evidence of demographic expansion in North American mammals, particularly at higher latitudes. In contrast, small mammals from western Amazonia appear to have experienced limited or no demographic expansion after the Late Pleistocene. Thus, demographic responses to climate change can be tracked genetically and appear to vary substantially across the latitudinal gradient of biotic diversity.
The Pleistocene was characterized by worldwide climatic perturbations that resulted in the development of large ice sheets and changes in the landscape and ecology of high-latitude continental areas of the northern hemisphere. There is widespread agreement that glacial cycles dramatically affected continental biota in those areas. Indeed, numerous distributional and phylogeographic patterns of the fauna and flora of northern Europe, Asia, and North America are thought to reflect the processes of colonization of newly available land subsequent to the last glaciation (1, 2). In Europe in particular, patterns of molecular variation across a variety of organisms are consistent with population expansion into deglaciated regions after the Pleistocene (2-4). During full glacial advances, many boreal species persisted at lower latitudes in North America south of the ice sheets (5), although some species may have been found along the north Pacific coast or in eastern Beringia (central Alaska and far western Yukon). Controversy and uncertainty remain regarding the number, locations, and significance of biotic refugia as sources for the colonization of the higher latitudes of North America after the Pleistocene (6, 7). However, we can predict that (i) populations that have colonized habitat made available by the retreat of ice sheets should bear the genetic footprint of a history of rapid expansion, and (ii) different species that share such history should have broadly concordant footprints.
The consequences of Pleistocene glacial cycles at low latitudes have also attracted substantial attention, particularly since Haffer (8) and Vanzolini and Williams (9) proposed that the retraction of Amazonian forests and accompanying fauna to distinct refugia might have promoted geographical speciation and, therefore, may account for the high biological diversity that characterizes tropical rainforests. The causes (10), significance for speciation (11-13), and even the reality (14-17) of such Pleistocene refugia have been the focus of sharp controversy. Alternative models for diversification have been proposed, including the hypothesis that the Andes served as a “species pump” for Amazonia (18).
Although conflicting points of view (19, 20) have been expressed since Haffer's proposal, the “refugial debate” has gained intensity in recent years (2, 10-13, 21, 22). The task of testing refugial hypotheses is a difficult one. For instance, the locations of proposed refugia may vary among taxa, or they may be defined without sufficient precision to allow phylogenetic predictions and tests (23). Fortunately, genetic tests of population expansions are not affected by such concerns. If populations from an area are part of an historical expansion from a refuge, a genetic footprint of that history should remain. Furthermore, consistent genetic signals of historical expansion should be found across species that have taken advantage of the expansion of their habitat from the more restricted refuge.
Here, we present analyses of boreal North American and tropical Amazonian mammals in an attempt to examine the genetic evidence, or lack thereof, for late Pleistocene refugia. We focus on the possible consequences of the last glacial cycle for several reasons. First, the last glaciation lasted some 60,000-80,000 years with a maximum ≈18,000 years ago (1). A prolonged glacial period followed by a rapid and recent climatic amelioration provides optimal conditions for detecting its biotic consequences under the refugial hypothesis, i.e., long-lasting refugia from which dramatic expansions must have occurred. Second, coalescence theory makes specific predictions about patterns of genetic variation in populations that have undergone significant demographic expansions (24-28). These predictions can be examined in samples of extant taxa from the areas of interest. Importantly, analyses can be carried out without explicit reference to predefined refugial areas, and without making assumptions about common sources, times, and patterns of expansion across taxa. Units for analyses can be clades previously identified by phylogenetic methods, or they may even represent particular localities if sample sizes are sufficiently large. Finally, a common methodology can be used for both boreal North American and Amazonian organisms, thus allowing a broad comparative assessment of postglacial demographic processes in distinctive regions.
Methods and Materials
Mitochondrial cytochrome b gene (control region in the case of black bears) sequences of the following taxa [n = sample size, sequence length in base pairs (bp)] were used in the analyses: long-tailed vole, Microtus longicaudus (n = 69, 1,143 bp) (29); dusky shrew, Sorex monticolus (n = 29, 801 bp) (30); northern flying squirrel, Glaucomys sabrinus (n = 84, 1,140 bp) (31); black bear, Ursus americanus (n = 114, 329 bp) (32); Proechimys simonsi (n = 145, 390 bp) (33); Proechimys steerei (n = 185, 390 bp) (33); Proechimys brevicauda (n = 26, 801bp) (34); Proechimys cuvieri (n = 33, 798 bp) (34); Oryzomys perenensis (n = 37, 414 bp) (34); Oecomys bicolor (n = 32, 414 bp) (35); Isothrix bistrata (n = 18, 798 bp) (35); Oryzomys macconnelli (n = 12, 801 bp) (35); Mesomys hispidus (n = 38, 798 bp) (35); Neacomys spinosus (n = 17, 670 bp) (35); and Micoureus demerarae (n = 10, 630 bp) (36). We used a Markov chain Monte Carlo approach (25) to assess the evidence for postglacial population expansions in North American and Amazonian mammals. Possible genealogies and model parameters were sampled by using the Metropolis-Hastings algorithm as implemented in FLUCTUATE (25), Version 1.3. In each run, we used 10 short chains (sampling increments of 10; 1,000 steps per chain), 10 long chains (sampling increments of 10; 20,000 steps), and a random starting tree. Convergence was checked on a subset of the cases by carrying out replicate runs with different seed numbers. This method is most sensitive to demographic change because it exploits features of the genealogy not used in other tests (25). The method assesses the goodness of fit of a model of exponential growth (or decline), and generates maximum likelihood estimates of the growth parameter (g) and its standard deviation. The starting value for g was set to 1. Because these computations may show an upward bias and standard deviations are only approximate, we have conservatively used g to indicate population growth if g > 3 SD(g). Fu's Fs (28) test does not suffer from the aforementioned bias and is sensitive to demographic expansion, so it was used as an additional, independent assessment of historical changes in population sizes. Significance of Fs was evaluated by using 1,000 random permutations in ARLEQUIN (37).
Results and Discussion
Significant phylogeographic structure is the rule in both boreal and Amazonian mammals (6, 35). Nonetheless, northern species of mammals generally show limited molecular variation within populations, whereas Amazonian species of mammals often show high levels of variation within populations (6, 35). This contrast does not necessarily indicate the occurrence of late Pleistocene refugia in northern North America but not in Amazonia, because variation across species in mutation rates, selective regimes, and standing population sizes can all affect genetic polymorphism.
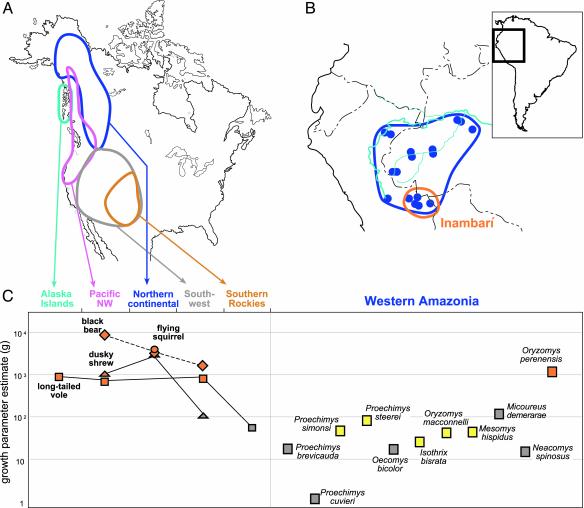
Further assessments are possible with coalescent-based approaches that allow estimation of historical demographic changes (24-28). All four species of North American mammals we examined using FLUCTUATE (25) show clear signs of rapid demographic expansion, especially, if not exclusively, in the northern parts of their ranges (Fig. 1 A and C). The long-tailed vole (Microtus longicaudus), for example, shows dramatic expansion (estimates of the exponential growth parameter g ≈ 103) in the islands of Alexander Archipelago in Alaska, in the Pacific Northwest, and in the Southwest. Three other species, from minute dusky shrews to black bears, show consistently high (g ≈ 103 to 104) growth parameter estimates in the northern parts of their distributions. This is precisely the expected pattern for populations that experienced dramatic growth from refugial areas, both locally and, especially, as they colonized newly available habitat. Selective sweeps can produce similar patterns, but the strong concordance of the observed patterns across northern populations of mammals as radically different in size and life history as shrews and bears suggests common demographic histories.
Fig. 1.
(A) Map of North America indicating areas considered in the analyses. In the case of the long-tailed vole, the sample from the Southern Rockies was analyzed separately from those from the remaining Southwest. (B) Map of western Amazonia indicating the area and sampling localities considered in the analyses and the proposed Inambarí Refuge (8, 38). (C) Estimates of the exponential growth parameter g for boreal mammals, analyzed separately for each of the areas outlined in A and for mammals of western Amazonia. All estimates of g are color-coded as follows, regardless of symbol shape: gray, g < 3 standard deviations above 0; yellow, g between 3 and 6 standard deviations above 0; orange, g > 6 standard deviations above 0.
Signals of demographic expansion obtained with fluctuate (25) are more equivocal in lower-latitude populations of the North American species. Thus, high and significant estimates of g are found in black bears and long-tailed voles in the Southwest, where the corresponding estimate for dusky shrews is an order of magnitude smaller and not significant. Similarly, long-tailed voles in the Southern Rockies, an area that was not covered by ice sheets at the peak of the last glaciation, and where populations are restricted principally to relictual, alpine habitat, show the smallest value of g. Populations in that area are expected to have retracted and subdivided, not expanded, since the last glaciation. These results hold in analyses that treat samples gathered across large areas (Fig. 1 A and C). Clearly, geographic subdivision, which produces patterns opposite of those resulting from expansion, has not been sufficiently prolonged to override the effects of postglacial colonization. Analyses using Fu's Fs (28) are more conservative and are significant in all cases in the Alaska islands, Pacific northwest, and northern continental clades, but not in the southern Rockies and or the Southwest (Table 1).
Table 1.
Results of Fu's Fs test (28) for mitochondrial DNA sequences of North American (clades correspond to areas shown in Fig. 1) and Western Amazonian mammals
| Area/taxon | Clade | Fs | P |
|---|---|---|---|
| North America | |||
| Black bears | Pacific Northwest | −2.592 | 0.022 |
| Northern Continental | −3.242 | 0.008 | |
| Southwest | −0.475 | 0.223 | |
| Long-tailed voles | Alaska Islands | −12.600 | 0.001 |
| Pacific Northwest | −8.842 | 0.001 | |
| Southwest | −0.444 | 0.333 | |
| Southern Rockies | 0.788 | 0.570 | |
| Dusky shrews | Pacific Northwest | −9.915 | 0.001 |
| Northern Continental | −6.576 | 0.001 | |
| Southwest | −0.003 | 0.531 | |
| Flying squirrels | Northern Continental | −3.796 | 0.001 |
| Western Amazonia | |||
| Proechimys brevicauda | −4.608 | 0.071 | |
| P. cuvieri | 0.865 | 0.649 | |
| P. simonsi | −7.231 | 0.081 | |
| P. steerei | −14.326 | 0.004 | |
| P. bicolor | −2.722 | 0.161 | |
| Isothrix bistrata | −1.276 | 0.235 | |
| Oryzomys macconnelli | −2.478 | 0.075 | |
| Mesomys hispidus | −6.457 | 0.033 | |
| Micoureus demerarae | −1.364 | 0.181 | |
| Neacomys spinosus | −6.083 | 0.008 | |
| Oryzomys perenensis | −21.496 | 0.001 |
These geographical trends are concordant across several unrelated species of mammals, from large-bodied bears to diminutive shrews, suggesting that they have shared histories of northward expansion along coastal Pacific and continental paths. In some cases, populations have expanded onto islands off the North American continent at high latitudes (Fig. 1 A). The existing data, however, are insufficient to pinpoint the exact location and extent of the sources for such expansions. Coupled with paleoclimatic data and phylogeographic patterns, however, the population genetic data are suggestive of several common pathways from broadly shared sources at southern latitudes and in Beringia (30). Results also indicate areas where populations have remained stable, or areas supporting relicts of historically larger, more continuous habitats.
The results obtained with FLUCTUATE (25) for western Amazonian mammals generally stand in sharp contrast with those for North American species. Only one species, the rice rat (O. perenensis), yielded an estimate of population growth that is both very high (g ≈ 103) and significant, and therefore comparable with expanding populations of boreal taxa at high latitudes (Fig. 1 B and C). Of the 10 remaining taxa, 5 have nonsignificant estimates of g, whereas the other 5 show values of g that are statistically significant but at least one order of magnitude lower than those of boreal taxa at high latitudes (<102). Analyses using Fu's Fs (28) also show mixed results. This test is highly significant for O. perenensis, and is significant for three other species (Table 1) that only partially coincide with those suggested to have undergone demographic expansion by FLUCTUATE (Fig. 1).
The pattern of rapid population expansion of O. perenensis appears to be an exception to a generally conservative pattern in the Amazon. This species may represent a recent and rapid colonizer of the region or, alternatively, a selective sweep recently may have affected the mitochondrial genome across the range of the species. More generally, and importantly for our purposes, an ad hoc explanation, rather than one based on shared history with codistributed Amazonian species, is required for this species of rice rat.
The signal shown by all other Amazonian species is mixed. Most have positive values of g, but only half of them have values that are significant. There are several not mutually exclusive factors that can account for this pattern. First, the Markov chain Monte Carlo approach tends to show upward bias in the estimation of g (21). Second, the method is designed to deal with panmictic, undivided populations that undergo exponential growth or reduction. This assumption is not met here, and it is unclear how the method will respond to more complex situations. In general, populations of small mammals in western Amazonia are highly structured geographically (34), a pattern that is not consistent with a history of dramatic expansion from a reduced source.
In sum, the genetic data at hand suggest that populations of western Amazonian small mammals, with the possible exception of O. perenensis, have either remained stable since the last glaciation, or, alternatively, have experienced only modest demographic expansion. Our results for western Amazonia thus stand in sharp contrast to the dramatic signals of expansion found for boreal North American taxa at high latitudes. These analyses are clearly consistent with those paleoclimatic reconstructions that attest to a history of stability of the Amazonian forest through the glacial cycles (16). Our analyses cover populations from the western and southwestern Amazon, an area bounded by the Amazon River on the north, by the Madeira River on the east and southeast, and by the Andes on the west. This area maps to the region presumed to have been subsequently colonized from the Inambarí Refuge (8, 38) located at the base of the Andes in northern Bolivia and southern Peru (Fig. 1B). It has been suggested (16) that the last glacial cycle resulted in changes in the composition of the Amazonian forest, but not in its retraction to small isolates. In accordance, populations of Amazonian mammals might have suffered only minor reductions at the peak of the last glaciation, allowing them to retain substantial geographical structure. The transition toward warmer, modern climates, in turn, thus would have resulted in modest or no population expansions. These conclusions may have to be altered for specific habitats that experienced more dramatic changes, or for areas at or near the edges of the forest that became forested only after the last glaciation.
Boreal North America and Amazonia represent extremes of the latitudinal gradient of biotic diversity. Genetic footprints reflect the variable demographic responses to climate change across this gradient. Greater demographic stability (and hence lower probability of extinction) may be a key factor in the accumulation of species diversity in the tropics. We foresee several avenues for further examination of some of the processes underlying the latitudinal gradient of biotic diversity with molecular genetic tools. First, studies should be undertaken across additional geographical areas along the gradient. For instance, a recent examination of high arctic lemmings (39) raises the possibility that multiple nonglaciated areas may have supported populations during the Pleistocene (see also ref. 40). A study of MacGillivray's warbler (41) shows greater genetic structure and variation in tropical than in temperate areas, suggesting that populations in temperate areas may have resulted from postglacial colonizations. Second, ancient DNA studies may reveal distributional changes that cannot be detected with recent samples (42). Third, multilocus studies will help discriminate demographic expansions from selective sweeps and, at the same time, eliminate the upward bias apparent in single-locus studies using FLUCTUATE (25). Finally, theoretical advances in modeling demographic change (43, 44) will allow more realistic assessments of population histories.
Acknowledgments
We thank Mary Kuhner, John Wakeley, and two anonymous reviewers for comments on earlier drafts. E.P.L. gratefully acknowledges the support of a Guggenheim Fellowship and the expert guidance of John Wakeley (Harvard University, Cambridge, MA) in the study of coalescence theory. J.A.C. was supported by the National Science Foundation and the U.S. Fish and Wildlife Service and thanks John Demboski, Chris Conroy, Karen Stone, and Amy Runck for their contributions to this work. J.L.P. was supported by the National Science Foundation, the Wildlife Conservation Society, and the National Geographic Society and thanks Maria Nazareth da Silva, Yuri R. Leite, Leonora P. Costa, Jay R. Malcolm, Louise H. Emmons, and the governments of Perú and Brazil.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pielou, E. C. (1991) After the Ice Age: The Return of Life to Glaciated North America (Univ. of Chicago Press, Chicago).
- 2.Hewitt, G. (2000) Nature 405, 907-913. [DOI] [PubMed] [Google Scholar]
- 3.Hewitt, G. (1996) Biol. J. Linn. Soc. 58, 247-276. [Google Scholar]
- 4.Taberlet, P., Fumagalli, L., West-Saucy, A. G. & Cossons, J.-F. (1998) Mol. Ecol. 7, 453-464. [DOI] [PubMed] [Google Scholar]
- 5.Graham, R. W., Lundelius, Jr., E. L., Graham, M. A., Schroeder, E. K., Toomey, R. S., III, Anderson, E., Barnosky, A. D., Burns, J. A., Churcher, C. S., Grayson, D. K., et al. (1996) Science 272, 1601-1606. [DOI] [PubMed] [Google Scholar]
- 6.Demboski, J. R., Stone, K. D. & Cook, J. A. (1999) Evolution (Lawrence, Kans.) 53, 2008-2012. [DOI] [PubMed] [Google Scholar]
- 7.Byun, S. A., Koop, B. F. & Reimchen, T. E. (1997) Evolution (Lawrence, Kans.) 51, 1647-1653. [DOI] [PubMed] [Google Scholar]
- 8.Haffer, J. (1969) Science 165, 131-137. [DOI] [PubMed] [Google Scholar]
- 9.Vanzolini, P. E. & Williams, E. E. (1970) Arq. Zool. (São Paulo) 19, 1-298. [Google Scholar]
- 10.Nores, M. (1999) J. Biogeog. 26, 475-486. [Google Scholar]
- 11.Klicka, J. & Zink, R. M. (1997) Science 277, 1666-1669. [Google Scholar]
- 12.Avise, J. C., Walker, D. & Johns, G. C. (1998) Proc. R. Soc. London Ser. B 265, 1707-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chek, A. A., Lougheed, S. C., Bogart, J. P. & Boag, P. T. (2001) Mol. Phylogenet. Evol. 18, 370-385. [DOI] [PubMed] [Google Scholar]
- 14.Clapperton, C. M. (1993) Quaternary Geology and Geomorphology of South America (Elsevier, Amsterdam).
- 15.Colinvaux, P. A. (1996) in Evolution and Environment in Tropical America, eds. Jackson, J. B. C., Budd, A. F. & Coates, A. G. (Univ. of Chicago Press, Chicago), pp. 359-405.
- 16.Colinvaux, P. A. & De Oliveira, P. E. (2001) Palaeogeogr. Palaeoclimat. Palaeoecol. 166, 51-63. [Google Scholar]
- 17.Burnham, R. J. & Graham, A. (1999) Ann. Mo. Bot. Gard. 86, 546-589. [Google Scholar]
- 18.Fjeldså, J. (1994) Biol. Cons. 3, 207-226. [Google Scholar]
- 19.Endler, J. A. (1982) in Biological Diversification of the Tropics, ed. Prance, G. T. (Columbia Univ. Press, New York), pp. 641-657.
- 20.Mayr, E. & O'Hara, R. J. (1986) Evolution (Lawrence, Kans.) 40, 55-67. [DOI] [PubMed] [Google Scholar]
- 21.Willis, K. J. & Whittaker, R. J. (2000) Science 287, 1406-1407. [DOI] [PubMed] [Google Scholar]
- 22.Wilf, P., Cúneo, N. R., Johnson, K. R., Hicks, J. F., Wing, S. L. & Obradovich, J. D. (2003) Science 300, 122-125. [DOI] [PubMed] [Google Scholar]
- 23.Moritz, C., Patton, J. L., Schneider, C. J. & Smith, T. B. (2000) Annu. Rev. Ecol. Syst. 31, 533-563. [Google Scholar]
- 24.Wakeley, J. & Hey, J. (1997) Genetics 145, 847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhner, M. K., Yamato, J. & Felsenstein, J. (1998) Genetics 149, 429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slatkin, M. & Hudson, R. R. (1991) Genetics 129, 555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu, Y.-X. (1997) Genetics 147, 915-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conroy, C. J. & Cook, J. A. (2000) Mol. Ecol. 9, 165-175. [DOI] [PubMed] [Google Scholar]
- 30.Demboski, J. R. & Cook, J. A. (2001) Mol. Ecol. 10, 1227-1240. [DOI] [PubMed] [Google Scholar]
- 31.Demboski, J. R., Jacobsen, B. K. & Cook, J. A. (1998) Can. J. Zool. 76, 1771. [Google Scholar]
- 32.Wooding, S. & Ward, R. (1997) Mol. Biol. Evol. 14, 1096-1105. [DOI] [PubMed] [Google Scholar]
- 33.Matocq, M. D., Patton, J. L. & da Silva, M. N. F. (2000) Evolution (Lawrence, Kans.) 54, 1423-1432. [DOI] [PubMed] [Google Scholar]
- 34.Patton, J. L., da Silva, M. N. F. & Malcolm, J. R. (2000) Bull. Am. Mus. Nat. Hist. 244, 1-306. [Google Scholar]
- 35.Patton, J. L., da Silva, M. N. F. & Malcolm, J. R. (1994) Evolution (Lawrence, Kans.) 48, 1314-1327. [DOI] [PubMed] [Google Scholar]
- 36.Patton, J. L. & Costa, L. P. (2003) in Predators with Pouches: The Biology of Carnivorous Marsupials, eds. Jones, M., Dickman, C. & Archer, M. (CSIRO, Canberra, Australia), pp. 63-81.
- 37.Schneider, S., Roessli, D. & Excoffier, L. (2000) arlequin: A Software for Population Genetics Data Analysis (Univ. of Geneva, Geneva), Version 2000.
- 38.Brown, Jr., K. S. (1987) in Biogeography and Quaternary History of Tropical America, eds. Whitmore, T. C. & Prance, G. T. (Oxford Univ. Press, New York), pp. 175-196.
- 39.Fedorov, V. B., Goropashnaya, A. V., Jaarola, M. & Cook, J. A. (2003) Mol. Ecol. 12, 725-732. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, J. R. & Lister, A. M. (2001) Trends Ecol. Evol. 16, 608-613. [Google Scholar]
- 41.Milá, B., Girman, D., Kimura, M. & Smith, T. (2000) Proc. R. Soc. London Ser. B 26, 1033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes, I., Matheus, P., Shapiro, B., Jensen, D. & Cooper, A. (2002) Science 295, 2267-2270. [DOI] [PubMed] [Google Scholar]
- 43.Fay, J. C. & Wu, C. I. (1999) Mol. Biol. Evol. 16, 1003-1005. [DOI] [PubMed] [Google Scholar]
- 44.Hey, J. & Harris, E. (1999) Mol. Biol. Evol. 16, 1423-1426. [DOI] [PubMed] [Google Scholar]