Abstract
Apoptosis is a regulated cell death program controlled by extrinsic and intrinsic signaling pathways. The intrinsic pathway involves stress signals that activate pro-apoptotic members of the Bcl-2 family, inducing permeabilization of mitochondria and release of apoptogenic factors. These proteins localize to the outer mitochondrial membrane. Ian4, a mitochondrial outer membrane protein with GTP-binding activity, is normally present in thymocytes, T cells, and B cells. We and others have recently discovered that a mutation in the rat Ian4 gene results in severe T cell lymphopenia that is associated with the expression of autoimmune diabetes. The mechanism by which Ian4 controls T cell homeostasis is unknown. Here we show that the absence of Ian4 in T cells causes mitochondrial dysfunction, increased mitochondrial levels of stress-inducible chaperonins and a leucine-rich protein, and T cell-specific spontaneous apoptosis. T cell activation and caspase 8 inhibition both prevented apoptosis, whereas transfection of T cells with Ian4-specific small interfering RNA recapitulated the apoptotic phenotype. The findings establish Ian4 as a tissue-specific regulator of mitochondrial integrity.
Apoptosis is a regulated cell death program that can be initiated by two different signaling pathways. The extrinsic pathway involves ligation of cell surface death receptors and recruitment of proteins to a death-inducing signaling complex (1). The intrinsic pathway is death receptor-independent and involves stress signals that activate pro-apoptotic members of the Bcl-2 family; this action in turn induces permeabilization of mitochondria and the release of apoptogenic factors (2). These proteins are localized to the outer mitochondrial membrane, but the mechanisms responsible for regulating mitochondrial homeostasis localize to the inner membrane (3). How the two systems interact is unknown.
Immune-associated nucleotide-binding protein 4 (Ian4) was originally identified as a highly expressed protein in Bcr/Abl-transformed 32D cells (4). It localized to the mitochondrial outer membrane and displayed GTP-binding activity in vitro (4). Cells transfected with mutated Bcr/Abl constructs lacked oncogenic potential and displayed lower Ian4 expression. Homologues of Ian4 are present in mouse, rat, and human (5). More recently, Ian4 was found to be disrupted in diabetes-prone BB (BBDP) rats (5, 6), which develop severe T lymphopenia due to apoptosis of recent thymic emigrants (7). We hypothesized that mitochondrial Ian4 (also known in the rat as Ian4l1 and Ian5) plays an important role in regulating T cell survival through control of T cell apoptosis.
Methods
Animals. Diabetes-resistant BB (BBDR) rats and BBDP rats were obtained from Biomedical Research Models (Worcester, MA). Approximately 90% of BBDP rats develop spontaneous autoimmune diabetes; they are Ian4-/- and congenitally lymphopenic (8). BBDR rats are Ian4+/+ and never become spontaneously diabetic (8). Wistar Furth (WF) rats were obtained from Harlan-Sprague-Dawley. A congenic, nondiabetic WF.ART2a rat bred in our laboratories was used in one experiment (9). Eight- to 10-week-old rats of either sex were used; all rats were nondiabetic at the time of study. Animals were housed in a viral-antibody-free facility and maintained in accordance with the Guide for the Care and Use of Laboratory Animals (10) and guidelines of our Institutional Animal Care Committee.
Antibodies. Polyclonal antiserum to Ian4 was generated against a keyhole limpet hemocyanin (KLH)-conjugated peptide translated from GenBank accession no. AC099444.2 that corresponds to residues 152-173 of rat Ian4S protein (AAL17698). This sequence would be absent from any truncated form of Ian4 that might have been translated in the BBDP rat (5). Purified and fluorochrome- or biotin-conjugated mAbs directed against CD3 (G4.18), CD28 (JJ319), and αβ T cell antigen receptor (TCR) (R7.3) were obtained from Pharmingen, as were isotype control mAb mouse IgG1 and allophycocyanin-conjugated streptavidin. Anti-actin and anti-cytochrome c antibodies were from Chemicon International and Pharmingen, respectively. Anti-rabbit and anti-mouse IgG horseradish peroxidase (HRP) conjugates were obtained from Santa Cruz Biotechnology or Promega.
Cell Isolation, Mitochondrial Isolation, and Western Blot Analysis. Single-cell suspensions were prepared as described (7). Purified T and B cells were prepared by using magnetic bead technology according to the manufacturer's directions (Miltenyi Biotec, Auburn, CA). Peritoneal exudate macrophages were obtained from animals injected 4 days previously with 3 ml of 3% thioglycollate (Sigma). Mitochondria were isolated from T lymphocytes and thymocytes as described (11). Western blot analyses of Ian4, actin, and cytochrome c were performed as described (12).
In Vitro T Cell Stimulation. Six-well culture plates (Falcon) were incubated with 10 μg of anti-CD3 mAb per well and 10 μg of anti-CD28 mAb per well in PBS at 4°C. After overnight incubation, plates were rinsed with PBS. Nylon wool-purified T cells were cultured in antibody-coated wells at 6 × 106 cells per well in 3 ml of complete AIM-V medium (AIM-V medium plus 55 μM 2-mercaptoethanol, Sigma; AIM-V medium was from Life Technologies, Grand Island, NY) for 17 h at 37°C in a humidified atmosphere of 95% air/5% CO2. Unstimulated cells were incubated in complete AIM-V medium alone.
Subdiploid DNA and Determination of Mitochondrial Membrane Potential (Δψm). The percentage of T cells with subdiploid DNA was measured as described (13). Δψm was measured by two- or three-color flow microfluorometry by using Mitolight (Chemicon) alone or in combination with biotin-conjugated anti-αβTCR mAb. Mitolight was used according to the manufacturer's instructions. As a positive control, cells were incubated for 15 min in AIM-V medium at 37°C with the uncoupling reagent carbonylcyanide m-chlorophenylhydrazone (50 μM, Sigma). In one experiment, cells were cultured with cyclosporin A (Sigma, 4 μM in DMSO) for 30 min at 37°C. Lymphoid cells were identified electronically by their forward and side light-scatter characteristics. For each analysis, a minimum of 10,000 events were analyzed.
Mass Spectrometry. Proteins resolved by SDS/PAGE were visualized by Sypro Ruby protein stain. Bands were excised from the gel and tryptically digested in the gel. Samples were analyzed by matrix-assisted laser desorption ionization-time of f light (MALDI-TOF) mass spectrometry. Digested samples were concentrated and desalted with Millipore Zip Tip C18 microtips. Peptide masses were determined by using a Kratos Analytical Axima curved field reflection MALDI-TOF mass spectrometer equipped with a curved field reflectron. Peptide masses were searched against the nonredundant protein database by using the protein prospector program (http://donatello.ucsf.edu). Fragmentation data from postsource decay analysis of individual peptides were searched against the nonredundant protein database by using the protein prospector MS-Tag routine.
RNA Interference Assays. RNA interference assays were performed by using subconfluent Jurkat cells (American Type Culture Collection). Cells were plated onto 12-well culture plates (Falcon 3043) (3 × 105 cells per well in 1 ml of 10% FBS in RPMI medium 1640). A predicted human Ian4 oligonucleotide (AAGGTGAAAGAGGTCTTTGGG) was synthesized, purified, and duplexed by Xeragon (Huntsville, AL), who also designed and synthesized a negative control duplex small interfering RNA (siRNA) (AATTCTCCGAACGTGTCACGT). Lyophilized siRNA was suspended in a supplied buffer according to the manufacturer's instructions. Transfection vehicle (TransTKO, 6 μl, Mirus, Madison, WI) was added to aliquots of RPMI medium 1640 (50 μl) and incubated for 15 min at 20°C, after which siRNA was added. After mixing gently for 20 min at 20°C, aliquots containing Ian4 siRNA, the control siRNA, or the transfection vehicle alone were added to each well. The final concentration of siRNA in each well was 250 or 500 nM. Cells were incubated for 48 h at 37°C in 95% air/5% CO2, after which the percentage of cells with subdiploid DNA was determined.
Caspase Inhibition. Lymph node T cells from BBDP rats were cultured in AIM-V medium in the presence or absence of various concentrations of the caspase inhibitors Z-VAD-fmk, ZVDVAD, Z-DEVD, D-IETD, and Z-LEHD (R & D Systems) according to the supplier's instructions. Cells were cultured for 17 h at 37°C in 95% air/5% CO2 and harvested; the percentage of cells with subdiploid DNA was determined as above.
Statistics. Parametric data are presented as arithmetic means ± 1 SD. Means were compared by ANOVA and the least significant differences method for a posteriori contrasts or by two-tailed Bonferroni-adjusted t tests.
Results
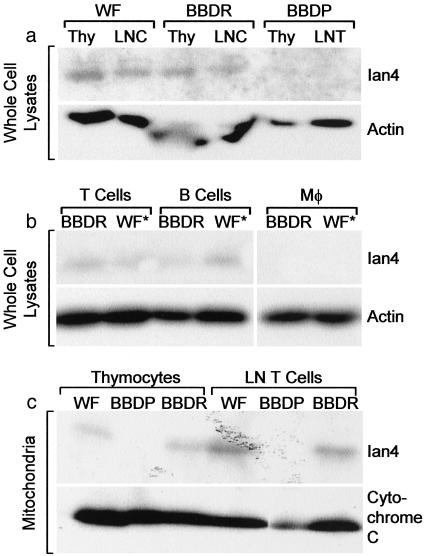
Ian4 Protein Is in Normal Rat Mitochondria, Absent from Ian4-/- Lymphopenic Rats. We compared lymphoid cells from nondiabetic Ian4-/- BBDP rats (5, 6, 8) to those isolated from control, nonlymphopenic WF and BBDR rats. The Ian4 frameshift mutation in the BBDP rat generates a truncated protein product that lacks the COOH-terminal transmembrane region required for mitochondrial membrane localization (5). Western blotting of Ian4+/+ BBDR and WF cell lysates with a polyclonal antibody to Ian4 revealed an ≈30-kDa band in thymocytes, lymph node T cells, and B cells but not macrophages (Fig. 1 a and b). No Ian4 was detected in lymphoid tissues of Ian4-/- BBDP rats. Western blots also demonstrated Ian4 protein in purified mitochondria obtained from thymocytes and T cells of Ian4+/+ but not Ian4-/- rats (Fig. 1c).
Fig. 1.
Western analyses of Ian4 protein. (a) Thymocyte (Thy) and lymph node cell (LNC) lysates from Ian4+/+ BBDR and WF and Ian4-/- BBDP rats. (b) Lymph node T cell, splenic B cell, and macrophage lysates from Ian4+/+ BBDR and WF.ART2a (WF*, see ref. 9) rats. (c) Purified mitochondrial lysates from T cells and thymocytes of Ian4+/+ BBDR and WF and Ian4-/- BBDP rats. Actin and cytochrome c were used as loading controls.
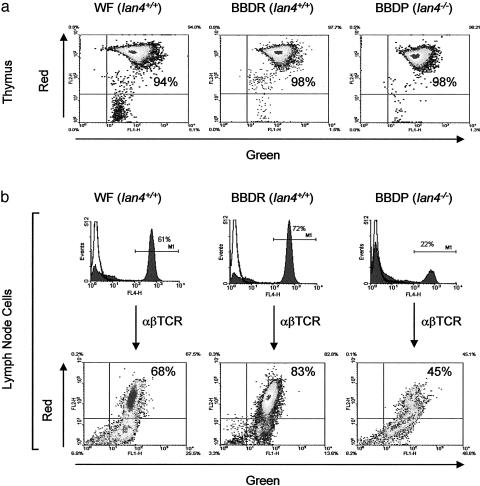
Mitochondrial Membrane Potential Is Decreased in T Cells, but Not Thymocytes, of Ian4-/- Lymphopenic Rats. We next compared mitochondrial membrane potential (Δψm) in WT and Ian4-/- T cells by using a mitochondrion-specific dye, Mitolight, which displays a green to red spectral shift proportional to Δψm (14). High-resolution digital fluorescence microscopy of cells from both Ian4-/- BBDP and Ian4+/+ BBDR rats confirmed that the dye labels mitochondrial structures, with no detectable staining of other organelles (data not shown). Mitochondrial structure appeared similar in T cells from Ian4-/- and Ian4+/+ rats. Quantitative analysis revealed that Δψm was similar and normal in freshly isolated thymocytes from all three rat strains (Fig. 2a) but substantially lower in freshly isolated resting T cells from Ian4-/- BBDP rats than in T cells from Ian4+/+ rats (Figs. 2b and 3a). The result suggests that loss of Δψm in Ian4-/- BBDP rat T cells occurs after thymic export.
Fig. 2.
Mitochondrial membrane potential (Δψm). (a) The Δψm of thymocytes from Ian4+/+ (WF and BBDR) and Ian4-/- (BBDP) rats; shown are the relative percentages of cells fluorescing red (vertical axis, high Δψm) and green (horizontal axis, low Δψm). The majority of thymocytes from both Ian4+/+ and Ian4-/- rats display high Δψm. Data are representative of two independent experiments. (b) The Δψm of lymph node T cells. Horizontal bars in histograms indicate gates used to identify αβTCR+ cells. The T lymphopenia of BBDP rats is evident. The dot plots show that the percentage of T cells with high Δψm was lower in Ian4-/- than in Ian4+/+ rats. Data are representative of three independent experiments; the complete dataset is shown in Fig. 3a. Control incubations with the mitochondrial uncoupler carbonylcyanide m-chlorophenylhydrazone uniformly reduced the percentage of T cells with high Δψm to ≈4% (data not shown).
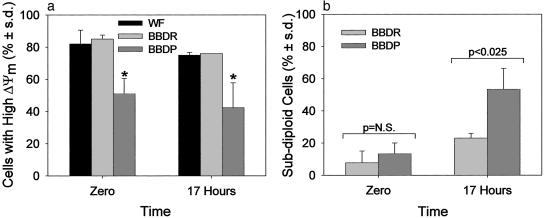
Fig. 3.
Kinetics of Δψm and DNA fragmentation. (a) Peripheral T lymphocytes from Ian4+/+ (WF and BBDR) rats and Ian4-/- (BBDP) rats were assayed for Δψm at time of isolation (time 0) and after 17 h of culture. Shown are results of three independent experiments analyzed by ANOVA. *, P < 0.003 at time 0 and P < 0.006 at 17 h vs. BBDR and WF. (b) The percentage of cells with subdiploid DNA in peripheral T lymphocytes from BBDR and BBDP rats was determined by propidium iodide staining and flow microfluorometry when isolated (time 0) and after 17 h of culture. Shown are results of three independent experiments.
Kinetics of Mitochondrial Membrane Potential and DNA Fragmentation. Kinetic analysis showed that the percentage of freshly isolated T cells with high Δψm did not change significantly in any of the strains during 17 h of culture (Fig. 3a). In contrast, the percentage of subdiploid (apoptotic) lymph node T cells immediately after isolation was similar in Ian4-/- and WT rats (Fig. 3b), but within 17 h became significantly higher in Ian4-/- animals.
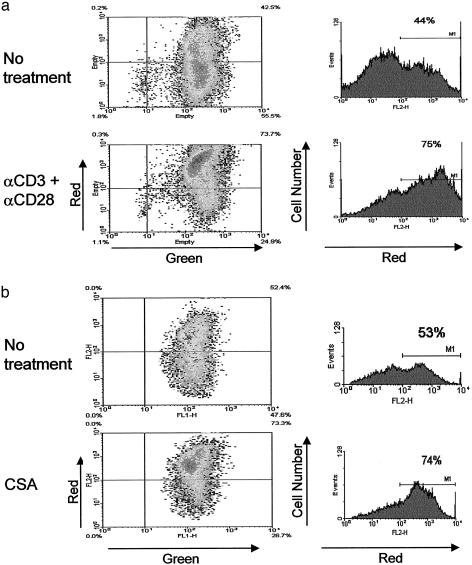
Rescue of Mitochondrial Membrane Potential. The apoptosis of T cells from BBDP rats is reportedly prevented by activation (15). We confirmed this observation (data not shown) and then found that activation in vitro normalized Δψm in these Ian4-/- T cells (Fig. 4a). The result suggests that loss of Δψm is an early event in the pathway leading to spontaneous apoptosis in Ian4-/- T cells.
Fig. 4.
Rescue of mitochondrial membrane potential. Ian4-/- BBDP T cells were incubated for 17 h without or with anti-CD3 plus anti-CD28 mAbs (a) or for 30 min without or with cyclosporin A (CSA) (b). Gates used to identify the percentage of cells with high Δψm are indicated by horizontal bars. Data are representative of three independent experiments. Both activation and cyclosporin A rescued Ian4-/- lymphocytes from loss of Δψm.
Maintenance of Δψm depends on mitochondrial membrane integrity and proper regulation of the permeability transition pore (16). We tested the hypothesis that Ian4 helps maintain normal permeability transition pore function. Ian4-/- BBDP T cells were incubated in the presence of cyclosporin A, which prevents mitochondrial membrane pore transition (16). Treatment of Ian4-/- T cells with cyclosporin A was associated with a rapid increase in the percentage of T cells with high Δψm (Fig. 4b), suggesting that Ian4 may be involved in the regulation of the permeability transition pore.
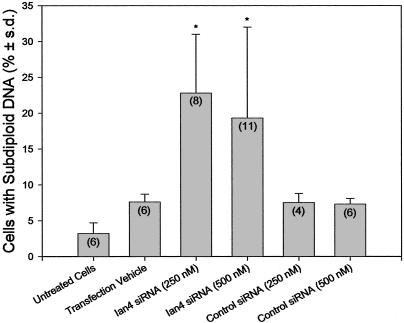
Ian4 RNA Interference Recapitulates Apoptosis. If Ian4 expression is required to prevent spontaneous apoptosis in T cells, its removal should induce apoptosis. This inference was confirmed by using human Jurkat T cells. Transfection with siRNA specific to Ian4 increased the percentage of T cells with subdiploid DNA, an effect not observed in nontransfected cells or in cells treated with transfection reagent alone or with nonspecific control siRNA (Fig. 5). Impairment of Ian4 translation in Jurkat T cells thus reproduces the spontaneous apoptosis of Ian4-/- BBDP T cells, the apoptotic phenotype of which is likely to be due specifically to loss of Ian4.
Fig. 5.
siRNA analyses. Transfection of Ian4-specific siRNA increased T cell apoptosis. Bars depict the percentage of cells with subdiploid DNA after 48 h of treatment. *, Statistically similar to each other and P < 0.025 vs. each of the other four groups. The number of independent measurements is shown in parentheses. Two other Ian4 siRNAs were less effective (data not shown).
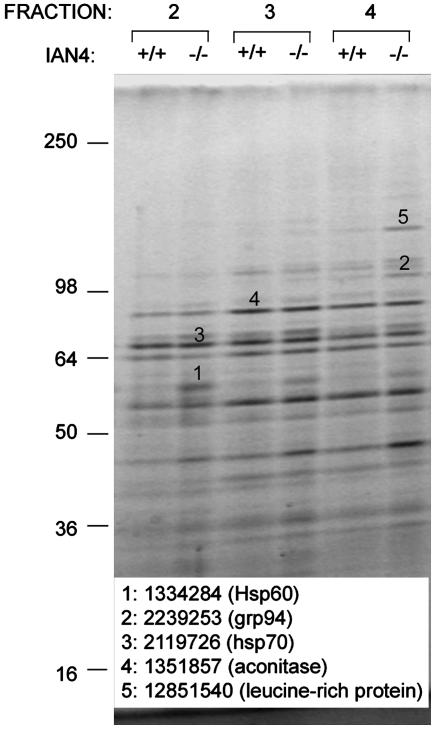
Mitochondrial Proteins. We next compared the protein profiles of purified mitochondria from BBDP, BBDR, and WF rat cells. Analysis by velocity gradient centrifugation (11) revealed proteins of ≈60, 100, and 130 kDa that were overrepresented in mitochondrial fractions from Ian4-/- T cells (Fig. 6). No proteins were overrepresented in Ian4-/- thymus or liver mitochondrial extracts (data not shown). MALDI-TOF mass spectrometric analysis identified the ≈60- and ≈100-kDa proteins as the stress-inducible proteins hsp60 and GRP94, respectively (Fig. 6). The ≈130-kDa protein was very similar to a putative mouse protein of unknown function with a predicted mass of 130 kDa that is highly homologous to a human 130-kDa leucine-rich protein up-regulated in the HepG2 hepatoblastoma cell line (17). Mitochondrial proteins unaffected by the presence or absence of Ian4 included aconitase and hsp70 (Fig. 6).
Fig. 6.
Proteomic analyses. Purified mitochondrial proteins from Ian4+/+ WF and Ian4-/- BBDP rat T cells resolved by SDS/PAGE. Bands 1, 2, and 5 were differentially expressed in Ian4+/+ vs. Ian4-/- T cells, excised, and identified by MALDI-TOF mass spectrometry. Not all differentially expressed bands were examined. Bands 3 and 4, which were similarly expressed, were excised to verify the purity of the mitochondrial isolation. Identities and gene identifiers for excised bands are indicated. Shown is one of two similar gels.
Caspase Inhibitors Prevent Apoptosis of Ian4-/- T Cells. These data suggest that absence of an Ian4-dependent signal in T cell mitochondria leads to indolent activation of intrinsic apoptosis pathways after thymic emigration. As many apoptotic pathways involve caspase activity, we tested a general inhibitor of caspases, Z-VAD-fmk, and specific inhibitors of caspases 2, 3, 8, and 9 in Ian4-/- T cells. These four caspases reportedly exist as proenzymes in the mitochondrial intermembrane space (18) and are released and activated by apoptotic stimuli (19). Both the general caspase inhibitor and inhibitors of caspases 3 and 8 prevented apoptosis of Ian4-/- T cells after 17 h of culture (data not shown). Inhibitors of caspases 2 and 9 were much less effective. The apparent lack of involvement of caspase 9 suggests that Ian4-mediated apoptosis is different from the classical intrinsic pathway or possibly that Ian4 is an unrecognized component of the extrinsic pathway, consistent with the ability of caspase 8 inhibitors to prevent apoptosis.
Discussion
The mitochondrion-dependent pathway for apoptosis is regulated principally by members of the Bcl-2 family (2). Like Ian4, these localize to the outer membrane of the mitochondrion and alter membrane potential, possibly by interacting with the permeability transition pore (3). The differential effect of Ian4 on T and B cells is paralleled by different roles that Bcl-2 plays in B and T cell development and homeostasis (20). Unlike Bcl-2, however, Ian4 is a GTP-binding protein (4) and in this respect may be unique among proteins involved in apoptotic pathways. How its function is restricted to T cells remains to be determined. The inhibition of spontaneous apoptosis in Ian4-/- BBDP T cells by activation (15) suggests that at least two independent pathways participate in T cell survival. One requires Ian4 in quiescent recent thymic emigrants and a second is Ian4-independent and can be activated by TCR ligation in the presence of costimulation. Apoptosis may also play a role in the senescence of the adaptive immune system (21), and it will be of interest to learn if Ian4 participates in this process.
Ian4 is a member of a large family of poorly characterized immune-associated nucleotide-binding proteins (4). The mammalian Ian1 gene is highly homologous to Ian4 and is expressed during positive selection of thymocytes and in T cells and B cells, but not macrophages. An orthologous protein in plants, aig1, participates in responses to bacteria in a process that involves apoptosis (22).
The increase in the 130-kDa protein in Ian4-/- T cell mitochondria is intriguing because, like Ian4, it is reportedly up-regulated in a transformed cell line (17) but has not previously been associated with cell death pathways. Whether up-regulation of this protein or of heat shock proteins/chaperonins in Ian4-/- mitochondria is indicative of their participation in the cell death cascade or, rather, a consequence of apoptosis is unknown.
The natural knockout of Ian4 was discovered in the BBDP strain, which develops spontaneous autoimmune diabetes (8). The role of the mutation in disease pathogenesis is unclear, but loss of peripheral Ian4-/- regulatory T cells due to apoptosis combined with rescue of activated Ian4-/- autoreactive T cells is an attractive hypothesis (8). The up-regulation of heat shock proteins in apoptotic Ian4-/- T cells could also be important. Development of autoimmune diabetes in NOD (nonobese diabetic) mice is believed to involve Hsp60, and the disease can be prevented by vaccination with Hsp60, its peptides, and a DNA construct encoding human Hsp60 (23). Our thymocyte data suggest that Ian4 is unlikely to affect the generation of autoimmunity at the level of the thymus.
Our data do identify the mechanism by which the absence of Ian4 causes T cell lymphopenia in the BBDP rat, and suggest that Ian4 provides a T cell-specific survival signal that maintains mitochondrial integrity. The Ian4 mutation in the rat is of unusual interest because it leads to spontaneous intrinsic apoptosis in the absence of external stressors. In addition, it appears to subserve this function principally if not exclusively in T cells. Ian4-/- B cells appear to be relatively unaffected, their numbers in BBDP rats being only slightly lower than in WF rats (24), and their ability to generate T-independent antibody responses being normal.¶
The data reveal another level of complexity in the governance of the T and B cell apoptotic pathways that participate in adaptive immunity.
Acknowledgments
We thank Drs. Roger Davis, Michael Czech, and Eugene Handler for review of the manuscript, and Dr. John Leszyk (Proteomics Facility), Jeffrey Carmichael (Digital Imaging Facility), Marcia Woda, Elaine Norowski, Linda Leehy, and Eric Forsberg for assistance. This work was supported in part by Grants DK49106 (to D.L.G. and J.P.M.), DK36024 (to D.L.G.), DK25306 (to J.P.M.), and DK60837-01A1 (to S.C.) and Center Grant DK32520 from the National Institutes of Health, and Grant 1-2002-394 from the Juvenile Diabetes Research Foundation (to R.B.). Hagedorn Research Institute is an independent basic research component of Novo Nordisk A/S.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BBDP rat, diabetes-prone BB rat; BBDR rat, diabetes-resistant BB rat; Ian4, immune-associated nucleotide-binding protein 4; WF rat, Wistar Furth rat; TCR, T cell antigen receptor; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight; siRNA, small interfering RNA.
Footnotes
Gosselin, E., Woodland, R., Mordes, J. P., Pelletier, A. & Rossini, A. A. (1985) Diabetes 34, Suppl. 1, 66A (abstr.).
References
- 1.Zimmermann, K. C., Bonzon, C. & Green, D. R. (2001) Pharmacol. Ther. 92, 57-70. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer, G. & Reed, J. C. (2000) Nat. Med. 6, 513-519. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden, M. G., Chandel, N. S., Li, X. X., Schumacker, P. T., Colombini, M. & Thompson, C. B. (2000) Proc. Natl. Acad. Sci. USA 97, 4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daheron, L., Zenz, T., Siracusa, L. D., Brenner, C. & Calabretta, B. (2001) Nucleic Acids Res. 29, 1308-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornum, L., Rømer, J. & Markholst, H. (2002) Diabetes 51, 1972-1979. [DOI] [PubMed] [Google Scholar]
- 6.MacMurray, A. J., Moralejo, D. H., Kwitek, A. E., Rutledge, E. A., Van Yserloo, B., Gohlke, P., Speros, S. J., Snyder, B., Schaefer, J., Bieg, S., et al. (2002) Genome Res. 12, 1029-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwakoshi, N. N., Goldschneider, I., Tausche, F., Mordes, J. P., Rossini, A. A. & Greiner, D. L. (1998) J. Immunol. 160, 5838-5850. [PubMed] [Google Scholar]
- 8.Mordes, J. P., Bortell, R., Groen, H., Guberski, D. L., Rossini, A. A. & Greiner, D. L. (2001) in Animal Models of Diabetes: A Primer, eds. Sima, A. A. F. & Shafrir, E. (Harwood, London), pp. 1-41.
- 9.Mordes, J. P., Leif, J., Novak, S., DeScipio, C., Greiner, D. L. & Blankenhorn, E. P. (2002) Diabetes 51, 3254-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Institute of Laboratory Animal Resources (1996) Guide for the Care and Use of Laboratory Animals (Natl. Acad. Press, Washington, DC).
- 11.Wilson-Fritch, L., Burkart, A., Bell, G., Mendelson, K., Leszyk, J., Nicoloro, S., Czech, M. & Corvera, S. (2003) Mol. Cell. Biol. 23, 1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigby, M. R., Bortell, R., Greiner, D. L., Czech, M. P., Klarlund, J. K., Mordes, J. P. & Rossini, A. A. (1996) Diabetes 45, 1419-1426. [DOI] [PubMed] [Google Scholar]
- 13.Nicoletti, I., Migliorati, G., Pagliacci, M. C., Grignani, F. & Riccardi, C. (1991) J. Immunol. Methods 139, 271-279. [DOI] [PubMed] [Google Scholar]
- 14.Smiley, S. T., Reers, M., Mottola-Hartshorn, C., Lin, M., Chen, A., Smith, T. W., Steele, G. D., Jr., & Chen, L. B. (1991) Proc. Natl. Acad. Sci. USA 88, 3671-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanathan, S., Norwich, K. & Poussier, P. (1998) J. Immunol. 160, 5757-5764. [PubMed] [Google Scholar]
- 16.Scorrano, L., Nicolli, A., Basso, E., Petronilli, V. & Bernardi, P. (1997) Mol. Cell. Biochem. 174, 181-184. [PubMed] [Google Scholar]
- 17.Hou, J., Wang, F. & McKeehan, W. L. (1994) In Vitro Cell Dev. Biol. Anim. 30, 111-114. [DOI] [PubMed] [Google Scholar]
- 18.Parone, P. A., James, D. & Martinou, J. C. (2002) Biochimie 84, 105-111. [DOI] [PubMed] [Google Scholar]
- 19.Qin, Z. H., Wang, Y., Kikly, K. K., Sapp, E., Kegel, K. B., Aronin, N. & DiFiglia, M. (2001) J. Biol. Chem. 276, 8079-8086. [DOI] [PubMed] [Google Scholar]
- 20.Katsumata, M., Siegel, R. M., Louie, D. C., Miyashita, T., Tsujimoto, Y., Nowell, P. C., Greene, M. I. & Reed, J. C. (1992) Proc. Natl. Acad. Sci. USA 89, 11376-11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, S. (2000) J. Clin. Immunol. 20, 229-239. [DOI] [PubMed] [Google Scholar]
- 22.Reuber, T. L. & Ausubel, F. M. (1996) Plant Cell 8, 241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintana, F. J., Rotem, A., Carmi, P. & Cohen, I. R. (2000) J. Immunol. 165, 6148-6155. [DOI] [PubMed] [Google Scholar]
- 24.Guttmann, R. D., Colle, E., Michel, F. & Seemayer, T. (1983) J. Immunol. 130, 1732-1735. [PubMed] [Google Scholar]