Abstract
Our intestine is the site of an extraordinarily complex and dynamic environmentally transmitted consortial symbiosis. The molecular foundations of beneficial symbiotic host-bacterial relationships in the gut are being revealed in part from studies of simplified models of this ecosystem, where germ-free mice are colonized with specified members of the microbial community, and in part from comparisons of the genomes of members of the intestinal microbiota. The results emphasize the contributions of symbionts to postnatal gut development and host physiology, as well as the remarkable strategies these microorganisms have evolved to sustain their alliances. These points are illustrated by the human-Bacteroides thetaiotaomicron symbiosis. Interdisciplinary studies of the effects of the intestinal environment on genome structure and function should provide important new insights about how microbes and humans have coevolved mutually beneficial relationships and new perspectives about the foundations of our health.
Keywords: gut microbial ecology, gnotobiotic mice, glycobiome, ecogenomics, environmental sensing
Microorganisms represent the largest component of biodiversity in our world. For example, an estimated 1010 bacterial genes are distributed throughout the biosphere (1). For the past ≈109 years, members of the Bacteria superkingdom have functioned as a major selective force shaping eukaryotic evolution (2). Coevolved symbiotic relationships between bacteria and multicellular organisms are a prominent feature of life on Earth. These alliances can be broadly classified according to (i) their means of transmission from generation to generation (acquisition from the environment vs. transfer via gametes from the female parent, as in bacteriocyte-insect symbioses); (ii) the physical relationship between symbiont and host (intracellular vs. extracellular); and (iii) whether the association is binary (one host and one microbial species), as is typical in invertebrates, or consortial, as in the cow rumen and human gut (2). Remarkably, there are only a few cases where the mechanisms underlying these relationships have been analyzed experimentally. Notable examples are the changes in structure and function of root tissues in leguminous plants through interactions with nitrogen-fixing rhizobia (3) and the recruitment of Vibrio fischeri from bacterioplankton so that the light organ of the sepiolid squid, Euprymna scolopes, can develop and use bacterial light (2).
We Are Not Alone
As inhabitants of this microbial world, we need to take a comprehensive view of ourselves as a life form by understanding that we are host to a remarkable variety and number of environmentally transmitted extracellular microorganisms. Acquisition of our microbial nation begins at birth (4). As adults, our total microbial population is thought to exceed our total number of somatic and germ cells by at least an order of magnitude (5, 6). Our largest collection of microorganisms resides in the intestine. As with most natural ecosystems, the true extent of biodiversity in the adult gastrointestinal tract remains to be defined. A current view is that the intestinal microbiota is composed of 500-1,000 different species, with an aggregate biomass of ≈1.5 kg. Most are refractory to cultivation, although new methods are being developed to help overcome this problem (e.g., ref. 7). Although most species are members of Bacteria, the microbiota contains representatives from Archaea (8) and Eukarya. Assuming 1,000 bacterial species, and using Escherichia coli as an arbitrarily selected representative of the community, the aggregate size of all intestinal microbial genomes may be equivalent to our own genome, and the number of genes in this “microbiome” may exceed the total number of human genes by a factor of ≈100.
The human intestinal ecosystem is remarkably dynamic. The host organ is lined with a perpetually and rapidly renewing epithelium: ≈20-50 million cells are shed per minute in the small intestine and 2-5 million per minute in the colon (9, 10). This epithelium is able to maintain marked regional differences in the differentiation programs of its component cell lineages as it undergoes continuous replacement. In a healthy individual, all available intestinal niches are presumably occupied by members of the microbiota (6). Within a given niche, some microbial members function as committed “residents” (autochthonous components, as defined by D. C. Savage; ref. 5), whereas others are transient tourists who are just “passing through” (allochthonous members). At any given position along the proximal-distal axis of the intestine, or at its luminal-mucosal interface, the tourists may represent autochthonous members of more proximal niches that have been dislodged (shed), or they can be derived from ingested food and water.
These considerations, together with reports that host development (4, 11, 12), host genotype (13), and environmental factors (14) influence the composition of the microbiota, emphasize how challenging it is to define and compare microbial community structures within and between specified intestinal niches of a given individual at a particular point in his or her life history, let alone to compare the microbiota among groups of individuals living in a particular geographic locale or among more broadly distributed populations.
Nonetheless, some general features of the human intestinal microbiota are apparent. First, in adults, >99.9% of the cultivatable bacterial population are obligate anaerobes (15). Prominently represented genera typically include Bacteroides, Clostridium, Lactobacillus, Fusobacterium, Bifidobacterium, Eubacterium, Peptococcus, Peptostreptococcus, Escherichia, and Veillonella (5). Second, population density increases by ≈8 orders of magnitude from the proximal small intestine (103 organisms per milliliter luminal contents) to the colon (1011 per g of contents) (5). Biodiversity also appears to increase along this axis, although the extent of diversification has yet to be systematically defined with secure region-specific sampling methods and enumeration methods that do not require cultivation (e.g., sequencing libraries of 16S rDNA amplicons). Third, the microbiota functions as a multifunctional organ whose component cell lineages provide metabolic traits that we have not fully evolved in our own genome. These traits include the ability to break down otherwise indigestable plant polysaccharides (16, 17), biotransformation of conjugated bile acids (18), degradation of dietary oxalates (19), and synthesis of certain vitamins (20). Fourth, postnatal colonization of our intestine educates our immune system, so we become tolerant of a wide variety of microbial immunodeterminants. This education appears to reduce allergic responses to food or environmental antigens (21).
The relationship between the microbiota and gut-associated lymphoid tissue (GALT) is reciprocal: for example, the GALT plays a key role in shaping the microbiota, although details of the mechanisms that underlie this reciprocity are just emerging (22).
Benefits of Studying Symbiotic Relationships in the Human Gut
The word “commensal” is often used to describe the relationship between vertebrate hosts and most members of their indigenous microbial communities. The implication is that these microbes have no discernible effect on the fitness of their host (23). However, this view is generally a reflection of our lack of knowledge about the specific contributions of community members, rather than representing an evidence-based conclusion that benefit is truly restricted to one partner (23).
The current revolution in genomics provides an unprecedented opportunity to analyze whether and how components of the intestinal microbiota modulate features of our postnatal development and adult physiology. One notion that motivates such an analysis is that our coevolved microbial partners have developed the capacity to synthesize novel chemical entities that help establish and sustain beneficial symbioses. Prospecting for these chemicals and characterizing the signaling pathways through which they operate may provide new strategies and reagents for manipulating our biology in ways that enforce health or that correct or at least ameliorate certain pathophysiologic states.
The potential rewards extend beyond identification of new therapeutic agents and their targets. In a dynamic densely populated ecosystem such as the gut, horizontal gene transfer between bacterial species can have important effects on organismal gene content and physiology. Thus, the intestinal ecosystem, or simplified derivative models, provides an opportunity to address general questions related to the field of ecogenomics (1). For example, how do symbionts sense and respond to variations in their environments? How does a given intestinal environment shape the evolution of its component microbial species? What are the functions of genetic diversity and apparent redundancy within or between niches? If there is significant microevolution of a given species and redistribution of genetic traits to other members of the consortium, what are genome-based definitions of speciation and extinction? What is the genomic basis for nutrient cycling and syntrophy (the cooperative interactions that take place between organisms so that they can consume a substrate that neither one alone can process)?
This article focuses on two complementary approaches for examining the molecular foundations of symbiosis in the human gut, from the perspectives of both host and microbe. One approach involves functional genomics studies of a simplified in vivo model of the intestinal ecosystem consisting of germ-free mice colonized with a Gram-negative anaerobe prominently represented in our distal gut microbiota, Bacteroides thetaiotaomicron. The other approach consists of a comparison of this organism's recently decoded genome (24) with the genomes of other members of the human intestinal microbiota.
Gnotobiotics: Creating a Simplified in Vivo Model of the Intestinal Ecosystem
The intestinal microbiota operates through a complex network of interspecies communications and an elaborate web of nutrient sharing/cycling. The complexity of the system presents a seemingly overwhelming experimental challenge when envisioning how to (i) identify the principles that govern establishment of these environmentally transmitted communities; (ii) characterize the spectrum of contributions that community members make to postnatal gut development and adult physiology; (iii) dissect bacterial-host and bacterial-bacterial communications pathways; and (iv) understand the forces that direct coevolution and coadaptation of bacteria and host in specified intestinal niches.
Next year marks the 50th anniversary of the first announcements of successful propagation of strains of mice raised under germ-free conditions (25, 26). We have taken advantage of this “old” technology to simplify the intestinal ecosystem. Gnotobiotic (“known-life”) mice can be viewed as having a complete ablation of their multilineage microbial “organ.” They can be colonized with a recognized or candidate intestinal symbiont (“cell lineage”), during or after completion of postnatal gut development (27). The gene expression profiles of age-matched germ-free and monoassociated animals are then compared in designated regions of their intestines by using genome-based tools, such as DNA microarrays. The cellular origins of selected host transcriptional responses to colonization can then be characterized by quantitative analysis (e.g., real-time RT-PCR) of laser capture microdissected (LCM) cell populations retrieved from intestinal cryosections (LCM allows all cellular responses to input signals from neighboring cells and from the gut lumen to be preserved during their harvest; ref. 28). The impact of colonization with one symbiont can be compared and contrasted to another species, to defined collections of symbionts, or to an unfractionated microbiota harvested from specified regions of the intestines of mice that have acquired a microbiota from birth (conventionally raised animals).
There were a number of reasons why B. thetaiotaomicron was selected as the model symbiont for these gnotobiotic experiments. It provides a key metabolic capability to humans: degradation of plant polysaccharides (17, 29). It is genetically manipulatable (30), easy to culture, and a predominant member of the distal intestinal microbiota of both mice and humans (15). Finally, it becomes prominent during a critical postnatal transition: the switch from mother's milk to a diet rich in plant polysaccharides (31). Some findings from this binary model of the intestinal ecosystem are summarized below.
Postnatal Developmental Phenomena as Manifestations of Underlying Host-Bacterial Symbioses
B. thetaiotaomicron has multiple effects on postnatal gut development. For example, it is able to direct expanded synthesis of glycans containing terminal α-linked fucose in members of the principal intestinal epithelial cell lineage (enterocytes) positioned in the distal immature small bowel (32). α-Fucosidases secreted by B. thetaiotaomicron (24, 33) allow the bacterium to harvest the pentose sugar from enterocytic glycans and use it as a carbon source. These observations suggest that structurally diverse outer chain segments of epithelial glycans may function as sign posts to direct colonization of distinct intestinal niches, and that this process is governed, at least in part, by members of the microbiota through their modulation of expression of components of the host glycobiome [for a list of known components of the human and mouse glycobiomes, see the Carbohydrate Active Enzymes (CAZy) database at http://afmb.cnrs-mrs.fr/CAZY].
This organism also affects development of the intestine's elaborate submucosal network of interconnected capillaries (34). In adult germ-free mice, the complexity of this capillary network is quite primitive compared with their age-matched conventionally raised counterparts. However, when adult germ-free mice are colonized by B. thetaiotaomicron, the angiogenic program is abruptly restarted, and construction of the network is completed within 10 d (34). The microbial signal that stimulates angiogenesis is processed via bacteria-sensing Paneth cells, a small intestinal epithelial lineage that is a key component of the gut's innate immune system (34). In this manifestation of the symbiotic relationship, B. thetaiotaomicron benefits its host by ensuring there is an adequate absorptive capacity for nutrients that the microbe processes [or that are processed through host metabolic pathways regulated by the bacterium: for example, B. thetaiotaomicron induces mediators of dietary triacylglycerol absorption and represses expression of an angiopoietin family member (ANGPLT4) that is secreted from the epithelium and functions as an inhibitor of lipoprotein lipase, the rate-limiting enzyme for import and storage of triacylglycerol-derived fatty acids in adipocytes (35-37). This finding suggests that components of the microbiota may also function as an environmental factor affecting acquisition and storage of lipids].
B. thetaiotaomicron influences another essential developmental process: establishment of the intestinal mucosal barrier. Colonization of germ-free mice with B. thetaiotaomicron alone or with an unfractionated distal gut microbiota harvested from conventionally raised mice induces expression of a Paneth cell protein, Ang4, that is secreted into the gut lumen (38). Ang4 is bactericidal for several Gram-positive gut pathogens, including Listeria monocytogenes (but not its more benign relative, Listeria innocua). It has little effect on B. thetaiotaomicron (38). Ang4 expression is normally induced as the microbiota of conventionally raised mice undergoes a pronounced shift in composition from facultative to obligate anaerobes during the suckling-weaning transition (38). In this embodiment of the symbiosis, components of the microbiota protect the host against invasion by pathogens by regulating expression of a species-selective endogenous protein antibiotic (Ang4), while presumably benefiting themselves by obtaining a degree of control over the composition of their microbial neighborhood.
Together, these findings illustrate the principle that certain postnatal developmental phenomena in mammals are manifestations and consequences of coevolved beneficial symbioses (39).
The Triumph of a Glycophile: A Comparative Microbial Genomics View of the Human-B. thetaiotaomicron Symbiosis
Members of the genus Bacteroides account for ≈25% of the total bacterial population in the adult human intestine (40). According to the current phylogenetic classification, Bacteroides belongs to the Bacteroidaceae family, Bacteroidales order, Bacteroides class, Bacteroidetes phylum, and Bacteroidetes/Chlorobi group (Superphylum) (www.ncbi.nlm.nih.gov/Taxonomy). Bacteroidetes diverged early in the evolution of Bacteria and is not closely related to either the Proteobacteria Phylum or the Firmicutes Phylum (low GC content Gram-positive bacteria) (Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org). The genome sequence of the B. thetaiotaomicron type strain VPI-5482 (ATCC 29148; originally isolated from the feces of a healthy human) provides the first view of the genetic features of a member of Bacteroidetes and of the proteome of a major member of our adult intestinal microbiota (24).
Among reported sequenced prokaryotes, B. thetaiotaomicron, two members of Archaea (Methanosarcina acetivorans and Methanosarcina mazei), Mycobacterium leprae, and two Xanthomonas species have an unusually low gene content for their genome size (Fig. 4, which is published as supporting information on the PNAS web site). The two Archaeons have low coding potential (75% vs. 85%-90% for most members of Bacteria; refs. 41, 42). Xanthomonas campestris pv. campestris and Xanthomonas axonopodis pv. citri also have relatively low coding potential plus a slightly larger than average coding sequence (CDS) size (43). M. leprae contains a large number of pseudogenes (”a genome in decay”; ref. 44). In contrast, the low gene content of B. thetaiotaomicron is explained by its CDS size: 1,170 bp, the largest value reported to date. This reflects a marked increase in the number of proteins containing >1,000 aa. For example, in the 1,000- to 1,160-aa size range, there are 83 homologs of SusC, an outer membrane protein (OMP) involved in acquisition of starch (see below), 14 glycosylhydrolases, and 17 membrane-associated transporters.
Polysaccharides are the most abundant biological polymer on Earth. Fermentation of polysaccharides is an important activity in many bacterial communities and contributes to a number of ecologically important processes, including the recycling of carbon (45). Bacterial metabolism of otherwise indigestible plant polysaccharides in the distal human intestine generates short chain fatty acids that account for ≥10% of our daily absorbed calories (17).
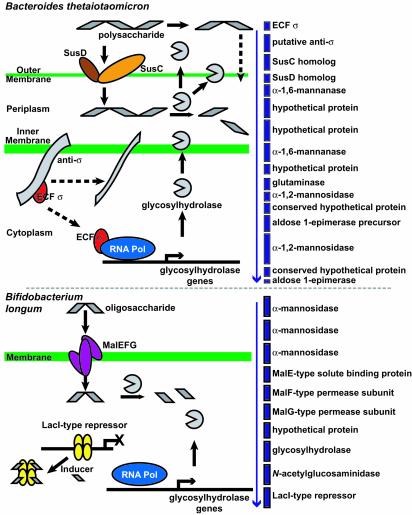
B. thetaiotaomicron has a very well developed glycobiome. The largest paralogous group in its genome contains 106 members with homology to SusC, whereas another 57-member group of paralogs has homology to SusD. SusC and SusD are components of an eight-component starch utilization system (Sus). SusC, together with SusD, is a member of a protein complex involved in binding starch and intermediate-size maltooligosaccharides (46) so that they can be subsequently broken down by outer membrane and periplasmic glycosylhydrolases (Fig. 1). The N-terminal domains of these SusC homologs have weak homology to TonB-dependent receptor family proteins, such as the E. coli outer membrane ferric transporter, FepA (47). The SusD homologs have no detectable sequence homology to other known proteins. Of the 57 SusD homologs, all but one is positioned immediately downstream of a SusC homolog (the exception, BT4085, has the weakest similarity to SusD among the group).
Fig. 1.
Strategies used by B. thetaiotaomicron and Bifidobacterium longum to regulate expression of their glycobiomes. B. thetaiotaomicron has an elaborate apparatus for retrieving polysaccharides from the luminal environment (SusC/D outer membrane proteins) and hydrolyzing them to oligosaccharides (secreted extracellular and periplasmic glycosylhydrolases). A highly developed environmental sensing apparatus, composed of ECF-type σ factors and hybrid two-component systems, is postulated to play a key role in regulating expression of components of its glycobiome and providing the means to behave as an adaptive forager of polysaccharides. ECF-typeσ factors are components of 12 gene clusters that contain downstream SusC/D homologs and glycosylhydrolases (one cluster is presented for illustration with the arrow indicating the direction of transcription). The Gram-positive bacterium, Bifidobacterium longum, has no SusC/D homologs but contains transporters that allow it to recover oligosaccharides generated from dietary polysaccharides by species such as B. thetaiotaomicron. An example is shown of one of the organism's seven gene clusters, each encoding MalEFG subunits of an oligosaccharide transporter, and glycosylhydrolases under the control of a LacI-type transcriptional repressor (51).
Little is known about the structural and functional organization of outer membrane transport systems in anaerobes. A high molecular weight porin complex, Omp200, composed of two proteins, Omp121 and Omp71, has been purified recently from Bacteroides fragilis (48). B. fragilis Omp121 has 26% amino acid sequence identity to B. thetaiotaomicron SusC. Although Omp71 has no detectable similarity to SusD, it has 15 homologs in the B. thetaiotaomicron genome: all but three are positioned next to a SusC homolog. Another group of 38 SusC homologs are located immediately upstream of ORFs encoding hypothetical proteins predicted to be associated with the outer membrane with sizes similar to that of SusD and Omp71 but without detectable sequence similarity to either. These findings invite speculation that many of the organism's SusC homologs are conserved components of a series of multifunctional outer membrane porins. The genes immediately downstream of SusC homologs may encode specificity components of these porins and can be divided into at least two groups: one with 56 SusD homologs that may effect acquisition/utilization of polysaccharides, the other with 12 homologs of Omp71, a protein whose function remains to be defined (48).
Nearly half of the genes encoding SusC homologs (47 of 106) are located next to glycosylhydrolases. B. thetaiotaomicron contains more glycosylhydrolases than any sequenced prokaryote, both in terms of absolute number (172) and when corrected for genome size (Table 1). The majority (61%) of these glycosylhydrolases are noncytosolic according to psort predictions (49). Together, the 172 enzymes appear to be capable of cleaving most glycosidic bonds identified in nature (based on GO term classifications of glycosidases; ref. 50). B. thetaiotaomicron also has at least 24 specific sugar transporters, including eight fucose permeases.
Table 1. Glycosylhydrolases encoded by the genomes of selected sequenced members of the adult human distal intestinal microbiota.
| Gene | B. thetaiotaomicron VPI 5482 | E. coli K12, MG1655 | Bifidobacterium longum NCC2705 | C. perfringens strain 13 | Enterococcus faecalis V583 | P. aeruginosa PAO1 |
|---|---|---|---|---|---|---|
| Amylase | 8 | 2 | 0 | 2 | 1 | 0 |
| Arabinase | 2 | 0 | 0 | 0 | 0 | 0 |
| α-Arabinofuranosidase | 4 | 0 | 5 | 0 | 0 | 0 |
| α-Arabinosidase | 7 | 0 | 5 | 0 | 0 | 0 |
| Chitinase | 3 | 0 | 0 | 0 | 0 | 1 |
| β-Fructofuranosidase (levanase) | 2 | 0 | 1 | 0 | 0 | 0 |
| α-Fucosidase | 3 | 0 | 0 | 1 | 0 | 0 |
| α-Galactosidase | 8 | 1 | 2 | 2 | 0 | 0 |
| β-Galactosidase | 31 | 3 | 6 | 5 | 4 | 0 |
| α-Glucosidase | 14 | 0 | 3 | 4 | 3 | 0 |
| β-Glucosidase | 10 | 8 | 7 | 1 | 10 | 1 |
| α-Glucuronidase | 1 | 0 | 0 | 0 | 0/1 | 0 |
| β-Glucuronidase | 2 | 1 | 1 | 1 | 1/0 | 0 |
| β-Hexosaminidase | 14 | 0 | 2 | 3 | 0 | 1 |
| α-Mannanase | 8 | 0 | 0 | 0 | 0 | 0 |
| α-Mannosidase | 14 | 1 | 3 | 2 | 0 | 0 |
| β-Mannosidase | 5 | 0 | 0 | 0 | 0 | 0 |
| α-N-Acetylglucosaminidase | 3 | 0 | 0 | 1 | 0 | 0 |
| β-N-Acetylglucosaminidase | 6 | 0 | 1 | 2 | 1 | 0 |
| α-Rhamnosidase | 5 | 0 | 0 | 0 | 0 | 0 |
| α-Xylosidase | 11 | 1 | 2 | 5 | 0 | 0 |
| β-Xylanase | 3 | 0 | 1 | 0 | 1 | 0 |
| β-Xylosidase | 8 | 1 | 0 | 0 | 0 | 1 |
| Total | 172 | 18 | 39 | 29 | 21 | 4 |
| Genome size, Mb | 6.26 | 4.64 | 2.26 | 3.03 | 3.22 | 6.26 |
Estimated numbers of genes for each category are based on genome annotation files in GenBank, as well as analysis of functional domains by using interpro. P. aeruginosa, a member of the Gamma branch of Proteobacteria whose genome size and coding potential are similar to those of B. thetaiotaomicron, is included to illustrate features in a Gram-negative bacterium with considerable ecological versatility.
Currently available sequenced genomes from other genera represented in the human intestinal microbiota include Bifidobacterium longum strain NCC2705 (51), Clostridium perfringens strain 13 (52), E. coli K12 MG1655 (53), and Enterococcus faecalis V583 (54). Like B. thetaiotaomicron, each of these species resides in the distal small intestine/colon. However, they are numerically far less prominent. Escherichia belongs to the Enterobacteriaceae family of the Gamma branch of Proteobacteria. All of the other species are Gram-positive. Bifidobacterium longum is an obligate anaerobe in the Actinomycetales branch of Bacteria that includes Corynebacteria, Mycobacteria, and Streptomycetes (55), and is a frequent component of over-the-counter probiotic dietary supplements. C. perfringens is a spore-forming anaerobe also found in soil (56). Enterococcus faecalis displays considerable ecologic versatility and is an important opportunistic pathogen (54, 57).
Table 1 summarizes the number and type of glycosylhydrolases encoded by the glycobiomes of these organisms (methods used for comparative microbial genome analyses are outlined in Supporting Text, which is published as supporting information on the PNAS web site). Several enzymes are unique to B. thetaiotaomicron (β-mannosidases, α-rhamnosidases, α-mannanases, arabinases, and chitinases). Other glycosylhyrolases are amplified in B. thetaiotaomicron to a degree that cannot be simply explained by the representation of particular glycan structures in the diet. For example, there are 31 β-galactosidases in B. thetaiotaomicron, a predominant member of the postweaning microbiota, vs. six in Bifidobacterium longum, a prominent member of the preweaning microbiota where lactose in mother's milk represents the principal ingested carbohydrate.
Further genomic evidence of B. thetaiotaomicron's versatility in harvesting polysaccharides is provided by its repertoire of mucin-degrading hydrolases and sulfatases. None of the other sequenced members of the human intestinal microbiota has predicted sulfatases. Studies of gnotobiotic mice simultaneously inoculated with two isogenic strains of B. thetaiotaomicron, one wild type, the other with a defect in its ability to harvest chondroitin sulfate, revealed that the capacity to use this mucopolysaccharide confers a competitive advantage (58). Unlike the other sequenced species, B. thetaiotaomicron lacks proteins with detectable homology to known adhesins or flagellar components (24). The presence of sulfatases and a large repertoire of outer membrane polysaccharide-binding proteins illustrates one way that B. thetaiotaomicron may be able to maintain residency in its niche in the absence of these components: it can interact with and harvest nutrients from the mucus layer that overlies the intestinal epithelium.
Herbivores are able to use ingested cellulose, a β-1,4-linked glucose polymer, due to the presence of cellulose-degrading microbes in their rumen, such as Ruminococcus and Fibrobacter spp. (59). These Gram-positive bacteria possess a cell surface-bound multienzyme complex known as the cellulosome. The best-studied cellulosome (from Clostridium cellulovorans) contains a scaffold protein for binding cellulose, and multiple endoand exo-glucanases that degrade this polysaccharide (60). Gram-negative bacteria have an outer membrane, and many of their glycosylhydrolases are presumably located in the periplasm. The outer membrane SusC/D complex and periplasmic glycosylhydrolases could be viewed as a functional equivalent of the cellulosome, evolved by Gram-negative Bacteroides to effectively capture and use other plant polysaccharides.
The distinctive features of the B. thetaiotaomicron glycobiome suggest that a hierarchical and collaborative nutritional network is embedded in the intestinal microbiota. For example, Bifidobacterium longum has no apparent homologs of SusC but possesses eight high-affinity MalEFG-type ABC transporters for importing oligosaccharides (more than any other published prokaryotic genome) plus a phosphotransferase system (51, 61). Phosphotransferase systems (PTSs) couple carbohydrate phosphorylation to transport, with the energy for transport provided by phosphoenolpyruvate (PEP). Two components are common to all PTSs: enzyme I (EI) and histidine protein (HPr). Carbohydrate-specific components (EII) consist of three domains (A, B, and C) that are either combined into a single protein or split into two or more proteins. EI, HPr, EIIA, and EIIC are responsible for the transfer of a phospho group from PEP to incoming carbohydrates, whereas EIIC forms membrane-associated translocation channels (62). Although PTSs are represented in many genera, B. thetaiotaomicron lacks a complete system. Thus, in the microbiota's nutrient network, B. thetaiotaomicron may function as a foundation species that breaks down a large variety of glycosidic bonds, thereby fashioning a nutrient broth for other members, such as Bifidobacterium longum, that are less capable of dealing with intact polysaccharides but more competent to import simpler sugars.
An ability to match expression of distinct subsets of outer membrane SucC/SucD homologs and periplasmic/extracellular glycosylhydrolases with carbohydrate availability would allow B. thetaiotaomicron to adaptively “graze” on glycans present in its niche. This feature would provide flexibility in the face of environment change and impart spatial (and temporal) characteristics to the microbial community. Moreover, ecological theory predicts that adaptive foraging promotes persistence of communities by stabilizing foodwebs (63). As described below, inspection of the B. thetaiotaomicron genome suggests that an environment-sensing transcriptional regulatory apparatus, coupled to the glycobiome, may allow for this type of foraging behavior.
B. thetaiotaomicron: Sense and Sensibility (and σ Factors)
Expression of B. thetaiotaomicron's elaborate repertoire of SusC/D homologs and glycosylhydrolases appears to be controlled by several types of transcriptional regulators. In the starch utilization system gene cluster, SusR binds maltose or larger oligosaccharides and activates expression of SusB-G (64). SusR appears to represent a distinct family of transcription factors: it has no detectable similarity to any family in the Pfam database (Ver. 8.0, February 2003). All four homologs (E value cutoff 10-10) deposited in GenBank (as of May 26, 2003) are found in B. thetaiotaomicron: one (BT3091) sits immediately upstream of a gene cluster containing SusC and SusD homologs, a cycloisomaltooligosaccharide glucanotransferase, and an α-glucosidase; another (BT3309) is positioned immediately upstream of SusC and SusD homologs, a glucosylceramidase, a hypothetical protein, and a β-glucosidase; another (BT4099) is located immediately upstream of an extracytoplasmic function (ECF)-type σ factor (see below for definition) followed by several glycosylhydrolases, whereas the fourth SusR homolog (BT2160) is positioned upstream of a putative oxidoreductase and a putative dehydrogenase.
B. thetaiotaomicron has an unprecedented expansion, among sequenced prokaryotes, of two classes of proteins involved in sensing environmental cues: ECF type σ factors and hybrid two-component systems (also known as one-component systems). σ factors are constituents of RNA polymerase complexes that are required for initiation of transcription and for coordinating cellular responses to various stimuli. They can be grouped into two families: σ-70 and σ-54 (65). The B. thetaiotaomicron genome encodes a total of 54 σ factors: two σ-70 family members (rpoD and rpoS homologs), two σ-54 related proteins, and 50 ECF-type σ factors. ECF-type σ factors belong to the Alternative Sigma Factors Group of the σ-70 family (66). ECF-type σ factors are often cotranscribed with and sequestered by anti-σ factors. Receipt of an environmental stimulus releases the ECF-type σ factor from its (membrane bound) anti-σ factor so that it can interact with RNA polymerase to regulate expression of target genes (66) (Fig. 1). Previously characterized ECF-type σ factors control a variety of functions including expression of heat-shock genes in E. coli (67); biosynthesis of alginates (exopolysaccharides containing d-mannuronic and l-guluronic acids) in Pseudomonas aeruginosa (68); iron uptake in E. coli (69); nickel and cobalt efflux in Alcaligenes europhus (70); outer membrane protein synthesis in response to variations in osmolarity, barometric pressure, and temperature in Photobacterium spp. (71); and cellular responses to disulphide stress in Streptomyces coelicolor (72).
A hybrid two-component system consists of a single protein that contains features of sensor kinases and response regulators, specifically, a histidine kinase domain (HATPase_c in Pfam), a phosphoacceptor domain (HisKA in Pfam), and a response regulator receiver domain (response_reg in Pfam) (73). Our analysis of 102 bacterial proteins annotated as hybrid two-component systems in the 83 complete bacterial genomes published before August 2002 indicated that none had a detectable DNA-binding domain. Remarkably, B. thetaiotaomicron contains 33 hybrid two-component systems: 32 of these have an additional conserved DNA-binding domain, all of the AraC helix-turn-helix (HTH) type. The hybrid protein that lacks a HTH_AraC domain (BT1183) contains a glycosyltransferase domain (glycos_transf in Pfam) in addition to its HATPase_c, HisKA and response_reg domains. Thus, the B. thetaiotaomicron proteome contains 32 novel hybrid two-component proteins that combine all of the features of a classical two-component system needed to complete the process from sensing a given environmental stimulus to regulating a series of target genes.
Many of the genes encoding this elaborate collection of ECF-type σ factors and hybrid two-component systems are physically linked to B. thetaiotaomicron's glycobiome. There are 12 gene clusters with a conserved modular structure distributed throughout the genome. In each cluster, ORFs encoding an ECF-type σ factor and a putative anti-σ factor are positioned upstream of linked SusC and SusD homologs, glycosylhydrolases, plus other enzymes involved in carbohydrate metabolism (Fig. 1 and Fig. 5, which is published as supporting information on the PNAS web site). None of these 12 gene clusters are associated with an upstream ORF specifying other classes of transcriptional regulators. This arrangement suggests that B. thetaiotaomicron uses some of its ECF-type σ factors to link expression of distinct subsets of polysaccharide-binding OMPs and secreted glycosylhydrolases to polysaccharide availability. ORFs specifying 23 of its 33 hybrid two-component systems are also positioned just upstream of genes involved in polysaccharide or mucopolysaccharide utilization. Nineteen of the 23 ORFs are juxtaposed next to genes encoding oligo/polysaccharide hydrolases, whereas four are adjacent to sulfatases or a heparitin sulfate lyase.
This sensory apparatus is distinctive among sequenced members of the distal human intestinal microbiota. Bifidobacterium longum has only one ECF-type σ factor, seven classical two-component systems, and no hybrid two-component systems. Unlike B. thetaiotaomicron, Bifidobacterium longum relies predominantly on negative regulation: 62 of its 83 predicted transcriptional regulators are repressors, principally HTH-containing LacI and MarR family members (Fig. 6, which is published as supporting information on the PNAS web site). Bifidobacterium longum contains seven gene clusters, each with a LacI-type sugar-responsive repressor, an ABC-type MalEFG oligosaccharide transporter, and genes encoding various types of glycosylhydrolases (Fig. 1). Classical transcription factors (defined here as proteins that are not σ factors or classical or hybrid two-component regulators) appear to provide the major mechanism for regulating gene expression in the E. coli and C. perfringens genomes. Figs. 6 and 7, which are published as supporting information on the PNAS web site, illustrate how sequenced members of the distal human gut microbiota can be more readily distinguished by their divergent collections of transcriptional regulators than by clusters of orthologous groups (COG)-based (74) functional profiling of their proteomes.
Another perspective about the distinctiveness of the B. thetaiotaomicron glycobiome is provided by P. aeruginosa, a member of the Gamma branch of Proteobacteria (Fig. 7). Its genome size (6.26 Mb) and coding potential (89.4%) are virtually identical to those of B. thetaiotaomicron (75). Glycosylhydrolases are poorly represented (Table 1). There are no detectable SusC/D homologs, and most of its specific membrane transporters are dedicated to acquiring amino acids (40 vs. 4 in B. thetaiotaomicron) rather than sugars (3 vs. 24 in B. thetaiotaomicron). This organism, which can adapt to a wide range of ecologic niches, has a greater number of proteins predicted to be involved in regulating gene expression [468 or 8.4% of its proteome vs. 277 (5.8%) in B. thetaiotaomicron] and a greater representation of transcriptional regulators that are not σ factors or classical or hybrid two-component systems (335 vs. 112). Moreover, P. aeruginosa has a diversified portfolio of these transcription factors, whereas the B. thetaiotaomicron proteome is heavily skewed toward those of the AraC-type (76) [81 or 72% of the total (Fig. 6), some of which are positioned next to SusC/D homologs and glycosylhydrolases].
The extraordinary abundance of hybrid two-component regulators and ECF-type σ factors present in B. thetaiotaomicron suggests that its successful adaptation to intestinal niche required development of a complex set of sensors so that it could respond to what is presumably a very dynamic environment with significant natural variation. Its profusion of glycosylhydrolases raises intriguing questions about whether the seemingly redundant capacity to degrade carbohydrates is actually a manifestation of the organism's need to express enzymes with subtle differences in their substrate specificities and distinct cellular destinations and/or its need to possess an elaborate set of responses to potential changes in the glycan environment through evolution of gene clusters with different combinations of carbohydrate degrading enzymes and linked transcriptional regulators.
Evolving in the Intestinal Ecosystem
Analyzing the genomes of other members of Bacteroides with varying degrees of prominence in the human distal gut microbiota should yield testable hypotheses about how evolution of various paralogous groups determines representation within and functional contributions to the microbial community (and host).
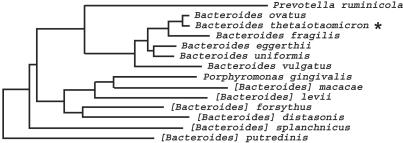
The B. fragilis type strain NCTC9343 provides a preview of the value of such comparisons. This species is phylogenetically close to B. thetaiotaomicron (Fig. 2) and an important opportunistic pathogen. However, it is only a minor component of the human intestinal microbiota. The 5.21-Mb B. fragilis genome has been sequenced at the Sanger Institute and can be obtained at ftp://ftp.sanger.ac.uk/pub/pathogens/bf. There are notable differences in its glycobiome and environmental sensing apparatus compared with B. thetaiotaomicron. The reduction in SusC homologs (69 vs. 106 in B. thetaiotaomicron) and SusD homologs (20 vs. 57) is disproportionate to the reduction in proteome size (≈4,200 vs. 4,779 members). There is one-half the number of glycosylhydrolases (89 vs. 172), although nearly all enzyme types (glycosidic bonds targeted) are present. Although there is proportional representation of ECF-type σ factors (41 vs. 50) and conservation of the modular structure of carbohydrate utilization gene clusters (10 vs. 12 with ECF-type σ factor linked to a putative anti-σ factor, SusC/D homologs, and glycosylhydrolases), there are far fewer hybrid two-component systems (8 vs. 33).
Fig. 2.
Phylogeny of Bacteroides and related species. Members of the Bacteroidales order are common inhabitants of the mammalian digestive tract. The order includes four established families: Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, and Rikenellaceae. Porphyromonas gingivalis (Porphyromonadaceae family) is associated with periodontal disease of humans. Its genome has been sequenced (www.tigr.org/tdb/mdb/mdbinprogress.html). Bacteroides forsythus, now renamed Tannerella forsythensis (Porphyromonadaceae family), is a human dental pathogen that has been partially sequenced (198 contigs, ≈3.6 Mb, unpublished work; www.tigr.org/tdb/mdb/mdbinprogress.html). Prevotella ruminicola (Prevotellaceae family) is a prominent member of the rumen and plays a central role in ruminal digestion of feed proteins. Brackets denote that for these species, initial assignment to this genus was based on biochemical phenotype: their 16S rDNA sequences indicate that their membership in Bacteroides should be viewed as tentative (adapted from ref. 82).
We have initiated sequencing of the Bacteroides distasonis and Bacteroides vulgatus genomes. Both are present at densities similar to that of B. thetaiotaomicron in the colonic microbiota (5) but are positioned at greater phylogenetic distances from B. thetaiotaomicron than B. fragilis (Fig. 2). Ex vivo studies indicate that B. distasonis is less capable of degrading polysaccharides than B. thetaiotaomicron (e.g., it is unable to ferment amylose, amylopectin, dextran, polygalacturonate, pectin, or larch arabinogalactan), whereas the substrate range of B. vulgatus is intermediate between that of B. distasonis and B. thetaiotaomicron. In contrast to B. thetaiotaomicron, neither of these two other Bacteroides spp. appears to be able to degrade mucopolysaccharides (77-79). Capsular polysaccharides contribute to the ability of Bacteroides spp. to cause human infections (80). Studies of clinical isolates indicate that B. distasonis does not produce a capsule, unlike B. thetaiotaomicron, B. fragilis, and B. vulgatus (81). B. distasonis has been placed at the junction between Bacteroides and Porphyromonas (82). A partial sequence of the human dental pathogen, Bacteroides forsythus (198 contigs, ≈3.6 Mb), originally isolated from pockets of periodontal disease, has been generated (www.tigr.org). Phylogenetic analyses based on 16S rDNA sequencing (81), initially placed B. forsythus close to B. distasonis (Fig. 2). A recent study identified important phenotypic differences between this organism and members of Bacteroides, leading to its renaming as Tannerella forsythensis (83). Thus, the B. distasonis genome sequence should allow further delineation of the evolution of Bacteroides, produce new insights about the contributions of polysaccharide metabolism to the human-Bacteroides symbioses, and permit comparison of the features of a predominant member of distal intestinal microbiota with those of a frequent member of the oral microbial community.
Symbiosis: Food for Many Disciplines
Future studies of the molecular foundations of human-bacterial symbioses in the intestine will require tools and concepts from many disciplines. In turn, the results of such studies should have broad implications that cross traditional disciplinary boundaries.
For example, genome scientists and microbial ecologists are now being confronted with the challenge of designing costeffective strategies for defining the microbiome in selected regions of the gastrointestinal tract. One approach is to take a community view and determine gene content without regard to function or species of origin (84). This could be accomplished by sequencing various types of shotgun libraries of the microbiome, although some type of kinetic fractionation/iterative normalization would presumably be required to facilitate new gene discovery, because a relatively few species dominate the microbiota. A complementary and more conservative approach is to focus on cultivatable members of dominant genera and systematically obtain improved draft (8×) coverage of their genomes by whole-genome shotgun sequencing. This effort could then progress to encompass less abundant components of the consortium, especially as new methods are developed for cultivating organisms that were previously refractory to growth ex vivo (7). Ultimately, these efforts, accompanied by careful annotation and software tools that allow in silico prediction of metabolic capabilities [e.g., kegg (85); wit (86), ecocyc (87); metacyc (88); and pathway tools (89)] should provide a view of the degree of functional distinctiveness as well as apparent redundancy among subspecies, species, and genera within the microbiota. The resulting information could provide new molecular tools, beyond 16S rDNA, for enumeration of this and other ecosystems, new perspectives about the degree of interchange of genetic material among community members (and thus what defines a species or constitutes true extinction within a consortium), plus new genomic views of features that distinguish symbionts from pathogens.
Cellular microbiologists, biochemists, and systems biologists will be challenged to develop the means for hypothesis-directed tests of in silico predictions of metabolic capabilities. The marriage of DNA microarray-based profiling of transcription (e.g., ref. 90) with mass spectrometry-based analysis of metabolism (metabolomics) in wild-type and isogenic mutant strains of a given species of bacteria, or in defined consortia, during growth in chemostats should allow characterization of responses to defined environmental conditions and perturbations (e.g., the foraging capability of B. thetaiotaomicron). In vivo expression technology (IVET) has already been used to identify bacterial genes induced during colonization of the intestines of conventionally raised mice with members of a genus represented in the human intestinal microbiota (Lactobacillus; ref. 91). Introducing gnotobiotics into the mix and developing effective tools for coincident profiling of bacterial and host gene expression during colonization of the intestines of normal and genetically manipulated germ-free mice (or other model organisms) should allow the results of ex vivo studies to be examined in the context of a simplified in vivo model of the intestinal ecosystem.
Studies of the coevolution of humans and intestinal symbionts should help evolutionary biologists, anthropologists, and those interested in human nutrition gain new information about how the need for dietary versatility shaped, and shapes, our supraorganismal biology. The “expensive tissue hypothesis” has emphasized the importance of selecting a high-quality diet to support expansion of a metabolically expensive brain as humans evolved, without demanding an accompanying marked increase in our gut size (92). Coevolving an intestinal microbiota that provided metabolic traits that improved extraction of nutrients from various diets may have provided part of the energetic solution to this evolutionary challenge. A microbial anthropology that focuses on genome-based analysis of the microbiota in suitably preserved and procured samples of feces and/or intestines derived from ancient humans (93), from current, relatively isolated human populations living in ecologically distinctive niches of our planet, or from nonhuman primates that inhabit various locales, should provide new understanding of how our migrations, dietary transitions, and social innovations/interactions conspired to craft modern symbionts (and Homo sapiens), as well as how they influenced the birth and spread of pathogens.
Plant molecular biologists seeking to genetically engineer improvements in the nutrient value of crops should consider the nutrient-processing capacity of the gut microbiota in targeted human populations. For example, an integrated research program centered on polysaccharides would select a carbohydrate source for introduction into or enrichment within a crop based on the following “bench-to-bowel” research pipeline. There would be an initial in silico assessment of the glycobiomes of the host plant, the human consumer, and members of their intestinal microbiota to help select the carbohydrate. Utilization of the carbohydrate could then be tested in chemostats containing members of the human gut microbiota postulated to be capable of processing the nutrient. This would be followed by in vivo physiological studies in groups of gnotobiotic mice, colonized with the same organisms used for the chemostat studies, and fed a diet containing the carbohydrate. The results would then set the stage for hypothesis-based clinical trials using individuals representing the targeted population. These individuals would be phenotyped and genotyped, in part, by molecular enumeration studies of their fecal microbiota (94, 95). In addition to evaluating nutrient processing/absorption, these trials could include an assessment of the effects of the carbohydrate manipulation on the hosts' microbiota, as well as the effects of attempted manipulations of their intestinal microbial communities (e.g., through coadministration of specified carbohydrate-degrading bacterial species that are absent or poorly represented).
As final examples of the potential interdisciplinary impact of this field, studying the molecular strategies used by symbionts for defining scarcity in their environment, for managing access to crucial resources when they are limiting, and for making decisions about sharing goods with others to ensure societal stability (concepts of cooperation and reciprocity; refs. 96 and 97) could yield operating principles of interest to systems and environmental engineers, mathematicians (including those that study game theory), ecologists, economists, business managers, and perhaps those who study, organize, and even govern our human communities.
Supplementary Material
Acknowledgments
We are grateful to our colleagues, Lora Hooper, Magnus Bjursell, Herb Chiang, Jason Himrod, Su Deng, Lynn Carmichael, Lynn Bry, David O'Donnell, Maria Karlsson, Janaki Guruge, Justin Sonnenburg, Peter Kang, and John Rawls, for their many contributions to these studies; and to Tore Midtvedt, Per Falk, Margaret McFall-Ngai, Nadia Shoemaker, and Abigail Salyers for many helpful discussions.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
Abbreviations: OMP, outer membrane protein; Sus, starch utilization system; ECF, extra-cytoplasmic function.
References
- 1.Stahl, D. A. & Tiedje, J. (2002) in Microbial Ecology and Genomics: A Crossroads of Opportunity, Critical Issues Colloquia (Am. Soc. Microbiol., Washington, DC).
- 2.McFall-Ngai, M. J. (2002) Dev. Biol. 242, 1-14. [DOI] [PubMed] [Google Scholar]
- 3.Schultze, M. & Kondorosi, A. (1998) Annu. Rev. Genet. 32, 33-57. [DOI] [PubMed] [Google Scholar]
- 4.Favier, C. F., Vaughan, E. E., De Vos, W. M. & Akkermans, A. D. (2002) Appl. Environ. Microbiol. 68, 219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage, D. C. (1977) Annu. Rev. Microbiol. 31, 107-133. [DOI] [PubMed] [Google Scholar]
- 6.Berg, R. D. (1996) Trends Microbiol. 4, 430-435. [DOI] [PubMed] [Google Scholar]
- 7.Zengler, K., Toledo, G., Rappe, M., Elkins, J., Mathur, E. J., Short, J. M. & Keller, M. (2002) Proc. Natl. Acad. Sci. USA 99, 15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckburg, P. B., Lepp, P. W. & Relman, D. A. (2003) Infect. Immun. 71, 591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croft, D. N. (1970) Proc. R. Soc. Med. 63, 1221-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croft, D. N. & Cotton, P. B. (1973) Digestion 8, 144-160. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, C. A. & Parrett, A. M. (2002) Br. J. Nutr. 88, Suppl. 1, S11-S18. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins, M. J., Sharp, R. & Macfarlane, G. T. (2001) Gut 48, 198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoetendal, E. G., Akkermans, A. D. L., Akkermans-van Vliet, W. M., de Visser, J. A. G. M. & De Vos, W. M. (2001) Microb. Ecol. Health Dis. 13, 129-134. [Google Scholar]
- 14.Sullivan, A., Edlund, C. & Nord, C. E. (2001) Lancet Infect. Dis. 1, 101-114. [DOI] [PubMed] [Google Scholar]
- 15.Moore, W. E. & Holdeman, L. V. (1974) Appl. Microbiol. 27, 961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, G. R. & Roberfroid, M. B. (1999) in Colonic Microbiota, Nutrition and Health (Chapman & Hall, London), pp. 37-53.
- 17.Hooper, L. V., Midtvedt, T. & Gordon, J. I. (2002) Annu. Rev. Nutr. 22, 283-307. [DOI] [PubMed] [Google Scholar]
- 18.Hylemon, P. B. & Harder, J. (1998) FEMS Microbiol. Rev. 22, 475-488. [DOI] [PubMed] [Google Scholar]
- 19.Duncan, S. H., Richardson, A. J., Kaul, P., Holmes, R. P., Allison, M. J. & Stewart, C. S. (2002) Appl. Environ. Microbiol. 68, 3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, M. J. (1997) Eur. J. Cancer Prev. 6, Suppl. 1, S43-S45. [DOI] [PubMed] [Google Scholar]
- 21.Braun-Fahrlander, C., Riedler, J., Herz, U., Eder, W., Waser, M., Grize, L., Maisch, S., Carr, D., Gerlach, F., Bufe, A., et al. (2002) N. Engl. J. Med. 347, 869-877. [DOI] [PubMed] [Google Scholar]
- 22.Fagarasan, S., Muramatsu, M., Suzuki, K., Nagaoka, H., Hiai, H. & Honjo, T. (2002) Science 298, 1424-1427. [DOI] [PubMed] [Google Scholar]
- 23.McFall-Ngai, M. J. & Gordon, J. I. (2003) in Evolution of Microbial Virulence, eds. Seifert, H. & DiRita, V. (Am. Soc. Microbiol., Washington, DC), in press.
- 24.Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V. & Gordon, J. I. (2003) Science 299, 2074-2076. [DOI] [PubMed] [Google Scholar]
- 25.Reyniers, J. A. (1956) in Proceedings Third International Congress of Biochemistry, ed. Lierbecq, C. (Academic, New York).
- 26.Pleasants, J. R. (1965) Proc. Indiana Acad. Sci. 75, 220-226. [Google Scholar]
- 27.Hooper, L. V., Mills, J. C., Roth, K. A., Stappenbeck, T. S., Wong, M. H. & Gordon, J. I. (2002) Methods Microbiol. 31, 559-589. [Google Scholar]
- 28.Stappenbeck, T. S., Hooper, L. V., Manchester, J. K., Wong, M. H. & Gordon, J. I. (2002) Methods Enzymol. 356, 168-196. [DOI] [PubMed] [Google Scholar]
- 29.Salyers, A. A., Valentine, P. & Hwa, V. (1993) in Genetics and Molecular Biology of Anaerobic Bacteria, ed. Sebald, M. (Springer, New York), pp. 505-516.
- 30.Salyers, A. A., Bonheyo, G. & Shoemaker, N. B. (2000) Methods 20, 35-46. [DOI] [PubMed] [Google Scholar]
- 31.Mackie, R. I., Sghir, A. & Gaskins, H. R. (1999) Am. J. Clin. Nutr. 69, 1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 32.Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1996) Science 273, 1380-1383. [DOI] [PubMed] [Google Scholar]
- 33.Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1999) Proc. Natl. Acad. Sci. USA 96, 9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stappenbeck, T. S., Hooper, L. V. & Gordon, J. I. (2002) Proc. Natl. Acad. Sci. USA 99, 15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hooper, L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G. & Gordon, J. I. (2001) Science 291, 881-884. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, K., Shimizugawa, T., Ono, M. & Furukawa, H. (2002) J. Lipid Res. 43, 1770-1772. [DOI] [PubMed] [Google Scholar]
- 37.Weinstock, P. H., Levak-Frank, S., Hudgins, L. C., Radner, H., Friedman, J. M., Zechner, R. & Breslow, J. L. (1997) Proc. Natl. Acad. Sci. USA 94, 10261-10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hooper, L. V., Stappenbeck, T. S., Hong, C. V. & Gordon, J. I. (2003) Nat. Immunol. 4, 269-273. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert, S. F. (2001) Dev. Biol. 233, 1-12. [DOI] [PubMed] [Google Scholar]
- 40.Moore, W. E., Cato, E. P. & Holdeman, L. V. (1978) Am. J. Clin. Nutr. 31, S33-S42. [DOI] [PubMed] [Google Scholar]
- 41.Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R. A., Martinez-Arias, R., Henne, A., Wiezer, A., Baumer, S., Jacobi, C., et al. (2002) J. Mol. Microbiol. Biotechnol. 4, 453-461. [PubMed] [Google Scholar]
- 42.Galagan, J. E., Nusbaum, C., Roy, A., Endrizzi, M. G., Macdonald, P., FitzHugh, W., Calvo, S., Engels, R., Smirnov, S., Atnoor, D., et al. (2002) Genome Res. 12, 532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M., et al. (2002) Nature 417, 459-463. [DOI] [PubMed] [Google Scholar]
- 44.Cole, S. T., Eiglmeier, K., Parkhill, J., James, K. D., Thomson, N. R., Wheeler, P. R., Honore, N., Garnier, T., Churcher, C., Harris, D., et al. (2001) Nature 409, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 45.Maier, R. M. (2000) in Environmental Microbiology, eds. Maier, R. M., Pepper, I. L. & Gerba, C. P. (Academic, San Diego), pp. 321-331.
- 46.Cho, K. H. & Salyers, A. A. (2001) J. Bacteriol. 183, 7224-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchanan, S. K., Smith, B. S., Venkatramani, L., Xia, D., Esser, L., Palnitkar, M., Chakraborty, R., van der Helm, D. & Deisenhofer, J. (1999) Nat. Struct. Biol. 6, 56-63. [DOI] [PubMed] [Google Scholar]
- 48.Wexler, H. M., Read, E. K. & Tomzynski, T. J. (2002) Gene 283, 95-105. [DOI] [PubMed] [Google Scholar]
- 49.Nakai, K. & Kanehisa, M. (1991) Proteins 11, 95-110. [DOI] [PubMed] [Google Scholar]
- 50.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M. C., Desiere, F., Bork, P., Delley, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu, T., Ohtani, K., Hirakawa, H., Ohshima, K., Yamashita, A., Shiba, T., Ogasawara, N., Hattori, M., Kuhara, S. & Hayashi, H. (2002) Proc. Natl. Acad. Sci. USA 99, 996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blattner, F. R., Plunkett, G., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 54.Paulsen, I. T., Banerjei, L., Myers, G. S., Nelson, K. E., Seshadri, R., Read, T. D., Fouts, D. E., Eisen, J. A., Gill, S. R., Heidelberg, J. F., et al. (2003) Science 299, 2071-2074. [DOI] [PubMed] [Google Scholar]
- 55.Biavati, B. & Mattarelli, P. (2001) in The Prokaryotes, eds. Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H. & Stackebrandt, E. (Springer, New York), pp. 1-70.
- 56.Hatheway, C. L. (1990) Clin. Microbiol. Rev. 3, 66-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klare, I., Werner, G. & Witte, W. (2001) Contrib. Microbiol. 8, 108-122. [DOI] [PubMed] [Google Scholar]
- 58.Hwa, V. & Salyers, A. A. (1992) Appl. Environ. Microbiol. 58, 869-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynd, L. R., Weimer, P. J., van Zyl, W. H. & Pretorius, I. S. (2002) Microbiol. Mol. Biol. Rev. 66, 506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shoham, Y., Lamed, R. & Bayer, E. A. (1999) Trends Microbiol. 7, 275-281. [DOI] [PubMed] [Google Scholar]
- 61.Boos, W. & Shuman, H. (1998) Microbiol. Mol. Biol. Rev. 62, 204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moat, A. G., Foster, J. W. & Spector, M. P. (2002) in Microbial Physiology (Wiley-Liss, New York) pp. 390-392.
- 63.Kondoh, M. (2003) Science 299, 1388-1391. [DOI] [PubMed] [Google Scholar]
- 64.D'Elia, J. N. & Salyers, A. A. (1996) J. Bacteriol. 178, 7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wosten, M. M. (1998) FEMS Microbiol. Rev. 22, 127-150. [DOI] [PubMed] [Google Scholar]
- 66.Helmann, J. D. (2002) Adv. Microb. Physiol. 46, 47-110. [DOI] [PubMed] [Google Scholar]
- 67.Erickson, J. W. & Gross, C. A. (1989) Genes Dev. 3, 1462-1471. [DOI] [PubMed] [Google Scholar]
- 68.Hershberger, C. D., Ye, R. W., Parsek, M. R., Xie, Z. D. & Chakrabarty, A. M. (1995) Proc. Natl. Acad. Sci. USA 92, 7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Enz, S., Braun, V. & Crosa, J. H. (1995) Gene 163, 13-18. [DOI] [PubMed] [Google Scholar]
- 70.Tibazarwa, C., Wuertz, S., Mergeay, M., Wyns, L. & van Der Lelie, D. (2000) J. Bacteriol. 182, 1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chi, E. & Bartlett, D. H. (1995) Mol. Microbiol. 17, 713-726. [DOI] [PubMed] [Google Scholar]
- 72.Paget, M. S. B., Hong, H.-J., Bibb, M. J. & Buttner, M. J. (2002) in Control of Bacterial Gene Expression, eds. Hodgson, D. A. & Thomas, C. M. (Cambridge Univ. Press, Cambridge, U.K.), pp. 105-125.
- 73.Wolanin, P. M., Thoomason, P. A. & Stock, J. B. (2002) Genome Biol. 3, REVIEWS3013.1-3013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tatusov, R. L., Galperin, M. Y., Natale, D. A. & Koonin, E. V. (2000) Nucleic Acids Res. 28, 33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 76.Gallegos, M. T., Michan, C. & Ramos, J. L. (1993) Nucleic Acids Res. 21, 807-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCarthy, R. E., Pajeau, M. & Salyers, A. A. (1988) Appl. Environ. Microbiol. 54, 1911-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salyers, A. A., Vercellotti, J. R., West, S. E. & Wilkins, T. D. (1977) Appl. Environ. Microbiol. 33, 319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salyers, A. A., West, S. E., Vercellotti, J. R. & Wilkins, T. D. (1977) Appl. Environ. Microbiol. 34, 529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coyne, M. J., Tzianabos, A. O., Mallory, B. C., Carey, V. J., Kasper, D. L. & Comstock, L. E. (2001) Infect. Immun. 69, 4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Babb, J. L. & Cummins, C. S. (1978) Infect. Immun. 19, 1088-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paster, B. J., Dewhirst, F. E., Olsen, I. & Fraser, G. J. (1994) J. Bacteriol. 176, 725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakamoto, M., Suzuki, M., Umeda, M., Ishikawa, L. & Benno, Y. (2002) Int. J. Syst. Evol. Microbiol. 52, 841-849. [DOI] [PubMed] [Google Scholar]
- 84.Beja, O., Suzuki, M. T., Heidelberg, J. F., Nelson, W. C., Preston, C. M., Hamada, T., Eisen, J. A., Fraser, C. M. & DeLong, E. F. (2002) Nature 415, 630-633. [DOI] [PubMed] [Google Scholar]
- 85.Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. (2002) Nucleic Acids Res. 30, 42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Overbeek, R., Larsen, N., Pusch, G. D., D'Souza, M., Selkov, E., Jr., Kyrpides, N., Fonstein, M., Maltsev, N. & Selkov, E. (2000) Nucleic Acids Res. 28, 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karp, P. D., Riley, M., Saier, M., Paulsen, I. T., Collado-Vides, J., Paley, S. M., Pellegrini-Toole, A., Bonavides, C. & Gama-Castro, S. (2002) Nucleic Acids Res. 30, 56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karp, P. D., Riley, M., Paley, S. M. & Pellegrini-Toole, A. (2002) Nucleic Acids Res. 30, 59-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karp, P. D., Paley, S. & Romero, P. (2002) Bioinformatics 18, Suppl. 1, S225-S232. [DOI] [PubMed] [Google Scholar]
- 90.Conway, T. & Schoolnik, G. K. (2003) Mol. Microbiol. 47, 879-889. [DOI] [PubMed] [Google Scholar]
- 91.Walter, J., Heng, N. C., Hammes, W. P., Loach, D. M., Tannock, G. W. & Hertel, C. (2003) Appl. Environ. Microbiol. 69, 2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aiello, C. C. & Wheeler, P. (1995) Curr. Anthropol. 36, 199-221. [Google Scholar]
- 93.Cano, R. J., Tiefenbrunner, F., Ubaldi, M., del Cueto, C., Luciani, S., Cox, T., Orkand, P., Kunzel, K. H. & Rollo, F. (2000) Am. J. Phys. Anthropol. 112, 297-309. [DOI] [PubMed] [Google Scholar]
- 94.Harmsen, H. J., Raangs, G. C., He, T., Degener, J. E. & Welling, G. W. (2002) Appl. Environ. Microbiol. 68, 2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang, R. F., Beggs, M. L., Robertson, L. H. & Cerniglia, C. E. (2002) FEMS Microbiol. Lett. 213, 175-182. [DOI] [PubMed] [Google Scholar]
- 96.Hauert, C., De Monte, S., Hofbauer, J. & Sigmund, K. (2002) Science 296, 1129-1132. [DOI] [PubMed] [Google Scholar]
- 97.Stephens, D. W., McLinn, C. M. & Stevens, J. R. (2002) Science 298, 2216-2218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.