Short abstract
An informatic procedure has been used to analyze the euchromatic sequences of Drosophila melanogaster and D. pseudoobscura for conserved sequences that adopt an extended stem-loop structure and display other characteristics of known miRNAs.
Abstract
Background
MicroRNAs (miRNAs) are a large family of 21-22 nucleotide non-coding RNAs with presumed post-transcriptional regulatory activity. Most miRNAs were identified by direct cloning of small RNAs, an approach that favors detection of abundant miRNAs. Three observations suggested that miRNA genes might be identified using a computational approach. First, miRNAs generally derive from precursor transcripts of 70-100 nucleotides with extended stem-loop structure. Second, miRNAs are usually highly conserved between the genomes of related species. Third, miRNAs display a characteristic pattern of evolutionary divergence.
Results
We developed an informatic procedure called 'miRseeker', which analyzed the completed euchromatic sequences of Drosophila melanogaster and D. pseudoobscura for conserved sequences that adopt an extended stem-loop structure and display a pattern of nucleotide divergence characteristic of known miRNAs. The sensitivity of this computational procedure was demonstrated by the presence of 75% (18/24) of previously identified Drosophila miRNAs within the top 124 candidates. In total, we identified 48 novel miRNA candidates that were strongly conserved in more distant insect, nematode, or vertebrate genomes. We verified expression for a total of 24 novel miRNA genes, including 20 of 27 candidates conserved in a third species and 4 of 11 high-scoring, Drosophila-specific candidates. Our analyses lead us to estimate that drosophilid genomes contain around 110 miRNA genes.
Conclusions
Our computational strategy succeeded in identifying bona fide miRNA genes and suggests that miRNAs constitute nearly 1% of predicted protein-coding genes in Drosophila, a percentage similar to the percentage of miRNAs recently attributed to other metazoan genomes.
Background
Although the analysis of sequenced genomes to date has focused most heavily on the protein-coding set of genes, all genomes also contain a constellation of non-coding RNA genes. With the exception of certain classes of RNAs with strongly conserved sequences and/or structures, such as ribosomal and transfer RNAs, identification of most non-coding RNAs has historically been a relatively serendipitous affair. Only very recently have there been concerted efforts to identify such genes systematically, using both experimental and computational approaches [1].
Our collective ignorance of the totality of non-coding RNA genes was laid bare by recent work on microRNAs (miRNAs), an abundant family of 21-22 nucleotide non-coding RNAs [2,3]. The founding members of this family, lin-4 and let-7, were identified through forward analysis of extant Caenorhabditis elegans mutants [4,5]. Both of these RNAs are post-transcriptional regulators of developmental timing that function by binding to the 3' untranslated regions (3' UTRs) of target genes [5-8]. Although they were long regarded as genetic curiosities possibly specific to nematodes, let-7 was subsequently found to be broadly conserved across bilaterian evolution [9] and miRNA genes are now recognized as a pervasive and widespread feature of animal and plant genomes [10-16].
In general, it is thought that miRNA biogenesis proceeds via intermediate precursor transcripts of more than 70 nucleotides that have the capacity to form an extended stem-loop structure (pre-miRNA), although at least some pre-miRNAs are further derived from even longer transcripts (primary miRNA transcripts, or pri-miRNAs). These can exist as long individual pre-miRNA precursor transcripts, as operon-like multiple pre-miRNA precursors, or even as part of primary mRNA transcripts. Processing of pri-miRNA into the pre-miRNA stem-loop occurs in the nucleus, while subsequent processing of pre-miRNA into 21-22 mers is a cytoplasmic event mediated by the RNAse III enzyme Dicer [17-20]; Dicer is also responsible for cleavage of long perfectly double-stranded RNA into 21-22 nucleotide fragments during RNA interference (RNAi) [2,21]. These latter molecules, known as silencing RNA (siRNA), bind to and trigger the degradation of perfectly homologous mRNA molecules via RISC, a double-strand RNA-induced silencing complex containing nuclease activity [22,23].
Although the in vivo function of only a few miRNAs is known so far, it is believed that the vast majority are likely to participate in post-transcriptional gene regulation of complementary mRNA targets. Interestingly, perfect or near-perfect target complementarity is associated with mRNA degradation [24-26], similar to the effects of siRNA, whereas imperfect base-pairing is associated with regulation by translational inhibition [6,27]. Recently, siRNAs with imperfect match to target mRNA were observed to function as translational inhibitors [28], suggesting that the type of 21-22 nucleotide RNA-mediated regulation may be largely determined by the quality of target complementarity.
The vast majority of the approximately 300 miRNAs currently known were identified through direct cloning of short RNA molecules. Although this method has been quite successful thus far, its practicality is limited by the necessity for a considerable amount of RNA as raw material for cloning, and cloned products are often dominated by a few highly expressed miRNAs. For example, 41% of miRNAs cloned from HeLa cells are variants of let-7, 28% of human brain miRNAs are variants of miR-124, and 45% of miRNAs cloned from human heart and 32% of those cloned from early Drosophila embryos are miR-1 [10,29]. In fact, it has been opined that few additional mammalian miRNAs will be easily identified by the direct cloning method [30].
As a complementary approach to miRNA identification, we developed an informatic strategy ('miRseeker') and applied it to the completed genomes of Drosophila melanogaster and D. pseudoobscura, which are some 30 million years diverged. miRseeker subjects conserved intronic and intergenic sequences to an RNA folding and evaluation procedure to identify evolutionarily constrained hairpin structures with features characteristic of known miRNAs. The specificity of this computational procedure was shown by the presence of 18 out of 24 reference miRNAs within the top 124 candidates. We identified a total of 48 novel miRNA candidates whose existence was strongly supported by conservation in other insect, nematode or vertebrate genomes. Expression of 24 novel miRNA genes was verified by northern analysis (including 20 out of 27 candidates that were supported by third-species conservation and 4 out of 11 high-scoring predictions specific to Drosophila), demonstrating that the bioinformatic screen was successful. As might be expected, the newly verified miRNA genes vary tremendously with respect to abundance and developmental expression profile, suggesting diverse roles for these genes. Inference of our false-positive prediction and false-negative verification rates (based on our ability to identify known miRNAs and detect the expression of highly conserved, and thus presumed genuine, novel miRNAs) leads us to estimate that drosophilid genomes contain around 110 miRNA genes, or nearly 1% of the number of predicted protein-coding genes. In combination with other concurrent genomic analyses [31-34], it is likely that most miRNAs in completed animal genomes have now been identified. Collectively, this sets the stage for both genome-wide and targeted studies of this functionally elusive family of regulators.
Results
Evolutionarily conserved characteristics of miRNA genes
The starting point for our studies was a reference set of 24 Drosophila pre-miRNA sequences (let-7, the 21 originally identified by Lagos-Quintana and colleagues, mir-125, and a previously undescribed paralog of mir-2 that we named mir-2c [9,10,29]). We analyzed this set to derive rules and determine parameters that specifically describe known miRNA genes within anonymous genomic sequence.
Examination of the genomic sequence of D. melanogaster and D. pseudoobscura showed that all 24 members of the reference set are highly conserved along the entirety of the predicted precursor transcripts, which typically range between 70-100 nucleotides. When viewed in VISTA plot alignments [35], miRNA genes reside in short regions of exceptional conservation, easily seen as local 'peaks' (Figure 1). As is the case for many other non-coding RNA genes, their degree of conservation usually exceeds that of coding regions, due to their lack of third-position wobble. This suggested that miRNA genes might be found by folding fixed lengths of conserved sequence to identify ones that display the high degree of relatively continuous helicity characteristic of known pre-miRNAs. However, pilot studies identified a very large number of conserved stem-loops in genomic sequence, suggesting that additional criteria were necessary to make effective miRNA gene predictions.
Figure 1.
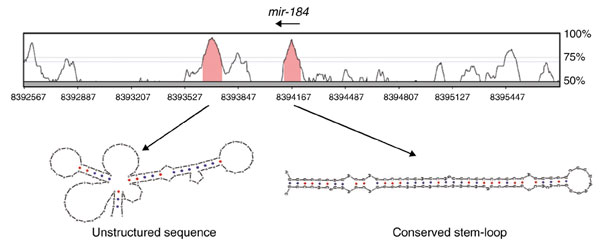
miRNA genes are isolated, evolutionarily conserved genomic sequences that have the capacity to form extended stem-loop structures as RNA. Shown are VISTA plots of globally aligned sequence from D. melanogaster and D. pseudoobscura, in which the degree of conservation is represented by the height of the peak. This particular region contains a conserved sequence identified in this study that adopts a stem-loop structure characteristic of known miRNAs. Expression of this sequence was confirmed by northern analysis (Table 2), and it was subsequently determined to be the fly ortholog of mammalian mir-184. Most conserved sequences do not have the ability to form extended stem-loops, as evidenced by the fold adopted by the sequence in the neighboring peak.
We next aligned the 24 pairs of orthologous Drosophila pre-miRNA sequences and assessed their pattern of nucleotide divergence. There are only three pairs that have been completely conserved (Figure 2a, class 1), indicating that most pre-miRNAs have diverged to some extent within Drosophila. Unexpectedly, we detected mild selective pressure on the precise sequence of the non-miRNA-encoding arm. This attribute is not self-evident. It might have been the case that point mutations along an arm would be neutral as long as the status of base-pairing was maintained; this is possible due to the acceptability of G-U base-pairing in RNA. Instead, we observed preferential divergence within the loop sequence. Ten members of the reference set have diverged exclusively within their loop sequence (Figure 2a, class 2), whereas there are no members that have diverged exclusively along an arm (Figure 2a, class 5). This is well illustrated by the Drosophila let-7 orthologs, which have accumulated four mismatches and gaps in the loop sequence but maintain perfectly conserved arms (Figure 2b). Thus, the terminal loop is the most evolutionary labile portion of pre-miRNA.
Figure 2.
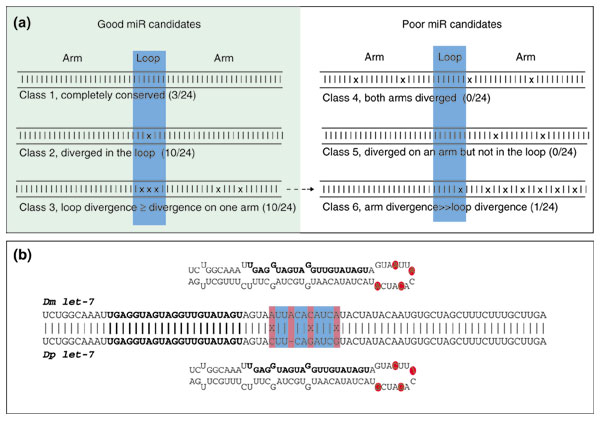
Classification of conserved stem-loop sequences. (a) Patterns of Drosophila pre-miRNA nucleotide divergence patterns imply a canonical progression in miRNA evolution. The Drosophila orthologs of 23/24 previously described miRNAs are either completely conserved (class 1), contain one or more mismatches or gaps located exclusively in the loop (class 2) or contain an equal or greater number of mutations within the loop compared to the non-miRNA-encoding arm (class 3). We consider these to represent successive steps in the normal evolution of miRNAs and therefore connect them with arrows. Members of classes 1-3 are considered as equally good candidates while members of classes 4-6 are poor candidates. As we expect class 3 candidates to eventually evolve into class 6 candidates (broken arrow), these evolutionary considerations are most relevant to species separated by an evolutionary distance comparable to D. melanogaster and D. pseudoobscura. (b) Preferential divergence of miRNAs within their loop sequences is illustrated by let-7. The Drosophila orthologs of let-7 contain three mismatches and one gap within the loop, whereas both arms have been completely conserved.
Mutations do eventually accumulate on the non-miRNA-encoding arm, and orthologous pre-miRNAs from more diverged species will often preserve only the 21-24 nucleotide mature miRNA itself. However, because of preferential divergence within the loop, orthologous miRNAs from species of an appropriate evolutionary distance (such as D. melanogaster and D. pseudoobscura) show an equal or greater amount of change within the loop than on the non-miRNA-encoding arm (Figure 2a, class 3). This is the case despite the fact that the loop is typically only a third to a quarter the length of each arm. Of the eleven members of the reference set that show divergence on both an arm and the loop, seven show more changes in the loop than on the non-miRNA-encoding arm and three have an equal number of changes on the loop and non-miRNA-encoding arm (Figure 2a, class 3); only one member of the reference set (miR-2b-1) shows a greater number of changes on the non-miRNA arm than the loop (Figure 2a, class 6). Finally, there are no cases where both arms have diverged (irrespective of loop status), a situation that would imply that the miRNA sequence itself had diverged (Figure 2a, class 4).
We propose then that classes 1-3 represent the normal pattern of evolutionary divergence of miRNAs, and consider Drosophila candidates that fall into these classes to represent 'good' miRNA candidates. Conversely, we consider classes 4-6 to have a low probability of reflecting a genuine miRNA in Drosophila, regardless of how 'impressive' the helical nature of the conserved stem-loop is. Indeed, we tested one class 4 and seven class 5 candidates by northern blot and failed to observe expression for any of them, despite extensive, conserved stem-loop structure (data not shown). We emphasize that these evolutionary considerations are most relevant to relatively closely related species: since the loop is much shorter than the arms, we expect class 3 candidates to eventually evolve into class 6 candidates in species separated by greater evolutionary distance than the two Drosophila species analyzed in the present study (Figure 2a).
Computational prediction of Drosophila miRNA genes using miRseeker
Overall, the collected observations indicated that miRNAs are phylogenetically conserved, extended stem-loop structures that display a characteristic pattern of nucleotide divergence. We proceeded to identify such sequences in the completed drosophilid genomes using the following three-part computational pipeline that we call miRseeker (Figure 3).
Figure 3.
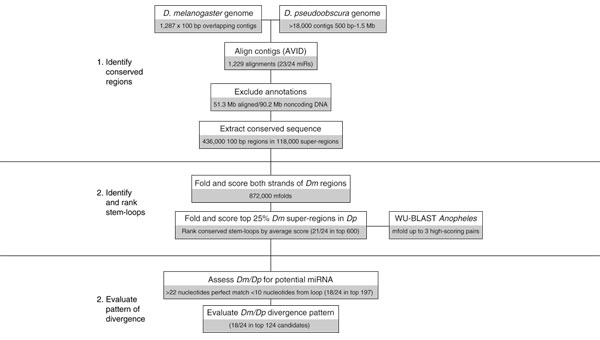
Overview of miRseeker, a computational strategy for identifying Drosophila miRNAs. See text for details.
Extraction of candidate, conserved, 'nongenic' Drosophila sequences
We first subdivided Release 3 of the D. melanogaster genome [36] into 1,287 contigs of 100,500 nucleotides each, with 500 nucleotides of overlap at either end. These were matched to the approximately 18,000 contigs in the first assembly of the D. pseudoobscura genome produced by the Human Genome Sequencing Center at the Baylor College of Medicine [37], using a dataset provided by the Berkeley Genome Pipeline [38]. We then aligned the repeat-masked D. melanogaster sequence to corresponding D. pseudoobscura sequence using the global alignment tool AVID [35,39]. We subsequently eliminated from the alignment all sequences associated with the following Release 3.1 annotations [40]: exons, transposable elements, snRNA, snoRNA, tRNA and rRNA genes. In total, we were able to align 51.3 out of 90.2 megabases of intronic and intergenic D. melanogaster sequence by this procedure.
Using the reference set, we empirically determined parameters for extracting conserved sequences that could contain miRNA genes (designated as 'regions'). We found that a 100-unit segment of the alignment (where a unit is either a single paired or gapped nucleotide) containing no more than 13% gaps or 15% mismatches, was sufficient to identify all known miRNA genes within their respective genomic neighborhoods, with a minimum of additionally selected sequences.
Conserved intergenic or intronic sequences are frequently longer than 100 nucleotides. We chose to evaluate these as a series of 'regions' that overlap each other by around 10 nucleotides, and grouped these regions into a single 'super-region'. The rationale for defining 'regions' and 'super-regions' came from our observation that RNA folding algorithms would not necessarily identify characteristic pre-miRNA structures if they were folded within the context of longer RNAs, owing to base-pairing with non-miRNA sequence. Thus, region-folding should offer optimal structures, while tracking of super-regions would ensure that multiple overlapping regions were evaluated as a single candidate gene. We took care to verify that our windowing parameters segregated individual miRNAs within known miRNA clusters (the mir-2, mir-13 and mir-3 → mir-6 clusters [10]) into distinct super-regions. This analysis identified 436,000 conserved regions that originate from 118,000 super-regions.
Identification of conserved stem-loops and evaluation of their quality
We analyzed conserved regions with mfold 3.1, an RNA-folding algorithm [41]. As miRNA genes can be located on either strand, and the quality of predicted hairpin structures can vary significantly between the respective strands (depending on the amount of G-U base-pairing), we folded each region as both the forward and the reverse complement of the extracted sequence (436,000 regions × 2 = 872,000 mfolds). Evaluation of candidate structures took into account the length of the longest helical arm (with a minimum cutoff of 23 base-pairs) and the free energy of this isolated arm (with a minimum cutoff of ΔG ≤-23.0 kcal/mol). The physical resemblance to the canonical stem-loop precursor was also assessed with metrics that reward continuous helical pairing and progressively penalize internal loops of increasing size. Since unpaired nucleotides in known miRNAs have a tendency to be found in symmetric loops, the presence of asymmetric loops and bulged nucleotides was further penalized. The size of the hairpin loop was not specifically evaluated as it appears to be variable in known pre-miRNAs; however it was indirectly assessed, as maximization of stem length concomitantly minimizes terminal loop size.
Given a 100-nucleotide input sequence, mfold 3.1 typically returns one to six alternate structures, each containing one to four helical arms; thus, the structure containing the highest-scoring helical arm had to be determined for each folded sequence. The highest-scoring region in each super-region (which could be located on either strand) was then saved as its representative, and these were ordered. For the top 25,000 D. melanogaster super-regions, we repeated this analysis on all regions in the corresponding D. pseudoobscura super-regions. We averaged the scores obtained for each aligned pair of Dm/Dp regions (termed a region-pair) and selected the highest-scoring region-pair within each super-region as its most probable miRNA candidate sequence. We then searched for homologs of these selected regions in Anopheles gambiae using WU-BLAST of the Dm sequence [42], extending the returned sequences as necessary on their 5' and 3' ends to make 100-nucleotide windows equivalent to the queried sequence. The top three mosquito hits were then folded and scored as before. However, as a large fraction of known fly miRNAs lack mosquito counterparts (Table 1), we decided to rank the candidates solely on their average Dm/Dp score.
Table 1.
List of Drosophila miRNAs and additional unverified candidates supported by third-species conservation
| Reference set miRNAs | ||||||||||
| Rank | Score | miR name | miR position | Cytological position | Sequence | Ano | Apis | Other | Nearest gene | Comment |
| 2 | 26.15 | miR-2a-2 (1) | 2L:19547562 | 37E | UAUCACAGCCAAGCUUUGAUGAGC | - | - | In the intron of spi (sense) | [10]; 3 miR cluster | |
| 3 | 26.00 | miR-2a-1 (2) | 2L:19547974 | 37E | UAUCACAGCCAGCUUUGAUGAGC | + | + | Worm | In the intron of spi (sense) | [10]; 3 miR cluster |
| 6 | 24.16 | miR-2b-2 (3) | 2L:19548259 | 37E | UAUCACAGCCAGCUUUGAGGAGC | + | + | In the intron of spi (sense) | [10]; 3 miR cluster | |
| (8) | 24.01* | miR-2b-1 | 2L:8250840 | 28B | UAUCACAGCCAGCUUUGAGGAGC | - | - | 895 upstream of Btk29A | [10]; failed conservation filter | |
| 8 | 23.52 | miR-13b-2 (4) | X:8830202 | 8C | UAUCACAGCCAUUUUGACGAGU | - | - | In the intron of CG7033 (sense) | [10] | |
| 9 | 23.45 | miR6-3 (5) | 2R:14724424 | 56E | UAUCACAGUGGCUGUUCUUUUU | - | - | 1732 upstream of CG11018 | [10]; 7 miR cluster | |
| 12 | 22.89 | miR-12 (6) | X:15240478 | 13D | UGAGUAUUACAUCAGGUACUGGU | + | + | 1986 upstream of Ac13E | [10]; 2 miR cluster | |
| 13 | 22.84 | miR-7 (7) | 2R:15669777 | 57A | UGGAAGACUAGUGAUUUUGUUGU | + | + | Vertebrate | 816 upstream of CG30147 | [10] |
| 17 | 22.45 | miR-14 (8) | 2R:4614375 | 45E | UCAGUCUUUUUCUCUCUCCUA | + | + | 5855 upstream of Or45b | [10] | |
| 27 | 20.94 | miR-9 (9) | 3L:19515075 | 76C | UCUUUGGUUAUCUAGCUGUAUGA | + | + | 6462 upstream of Shal | [10] | |
| 29 | 20.77 | miR6-2 (10) | 2R:14724582 | 56E | UAUCACAGUGGCUGUUCUUUUU | - | - | 1574 upstream of CG11018 | [10]; 7 miR cluster | |
| 31 | 20.65 | miR6-1 (11) | 2R:14724711 | 56E | UAUCACAGUGGCUGUUCUUUUU | - | - | 1445 upstream of CG11018 | [10]; 7 miR cluster | |
| 33 | 20.38 | miR-13a (12) | 3R:11243269 | 88F | UAUCACAGCCAUUUUGAUGAGU | + | + | 4626 upstream of CG6118 | [10]; 3 miR cluster | |
| 36 | 20.08 | miR-5 (13) | 2R:14724858 | 56E | AAAGGAACGAUCGUUGUGAUAUG | - | - | 1298 upstream of CG11018 | [10]; 7 miR cluster | |
| 62 | 18.86 | let-7 (14) | 2L:18450101 | 36E | UGAGGUAGUAGGUUGUAUAGU | + | + | Vertebrate/ worm | 932 upstream of CG10283 | [9]; 3 miR cluster |
| 18.45* | miR-10 | 3R:2635277 | 84B | ACCCUGUAGAUCCGAAUUUGU | + | + | Vertebrate | 13566 upstream of Scr | [10]; (not on aligned contig) | |
| 74 | 18.42 | miR-1 (15) | 2L:20457182 | 38D | UGGAAUGUAAAGAAGUAUGGAG | + | - | Vertebrate/ worm | 14444 upstream of CG15476 | [10] |
| 96 | 17.79 | miR-3 (16) | 2R:14725313 | 56E | UCACUGGGCAAAGUGUGUCUCA | - | - | 843 upstream of CG11018 | [10]; 7 miR cluster | |
| 114 | 17.49 | miR-11 (17) | 3R:17439181 | 93E | CAUCACAGUCUGAGUUCUUGC | - | - | In the intron of E2f (sense) | [10] | |
| 124 | 17.36 | miR-4 (18) | 2R:14724998 | 56E | AUAAAGCUAGACAACCAUUGA | - | - | 1158 upstream of CG11018 | [10]; 7 miR cluster | |
| 172 | 16.54 | miR-13b-1 (19) | 3R:11243135 | 88F | UAUCACAGCCAUUUUGACGAGU | + | + | 4760 upstream of CG6118 | [10]; 3 miR cluster | |
| 192 | 16.27 | miR-8 (20) | 2R:11895154 | 53D | UAAUACUGUCAGGUAAAGAUGUC | + | + | 3783 downstream of CG6301 | [10] | |
| 14.20 | miR-125 | 2L:18450405 | 36E | UCCCUGAGACCCUAACUUGUGA | + | + | Vertebrate/ worm | 628 upstream of CG10283 | [29]: low score; 3 miR cluster | |
| miR-2c | 3R:11243493 | 88F | UAUCACAGCCAGCUUUGAUGGGC | - | - | 4402 upstream of CG6118 | Hom; score n/a; 3 miR cluster | |||
| Newly verified miRNAs | ||||||||||
| Rank | Score | miR name | miR position | Cytological position | Sequence | Anos | Apis | Other | Nearest gene | Comment |
| 4 | 24.67 | miR-184 | 2R:8394117 | 50A | UGGACGGAGAACUGAUAAGGG | + | + | Vertebrate | 24406 upstream of CG17048 | Expression verified |
| 7 | 24.15 | miR-274 | 3L:11614451 | 68C | UUUUGUGACCGACACUAACGGGUAAU | - | - | In the intron of CG32085 (antisense) | Expression verified | |
| 10 | 23.10 | miR-275 | 2L:7418027 | 27F | cAGUCAGGUACCUGAAGUAGCGCGCG | + | + | 1070 upstream of CG5261 | 2miR cluster (+Ano and Apis); expression verified | |
| 16 | 22.57 | miR-92a | 3R:21461594 | 96E | CAUUGCACUUGUCCCGGCCUG | + | + | Vertebrate | 6578 upstream of BcDNA:LD22548 | 2miR cluster |
| 21 | 21.72 | miR-219 | 3L:17263886 | 74A | UGAUUGUCCAAACGCAAUUCUUG | + | + | Vertebrate | 4955 upstream of CG6485 | Expression not seen |
| 25 | 21.12 | miR-276a | 3L:10322758 | 67E | CAGCGAGGUAUAGAGUUCCUACG | + | + | 47587 upstream of CG12362 | Duplicated, 45 kb apart; expression verified; 1copy in Ano and Apis | |
| 28 | 20.88 | miR-277 | 3R:5925763 | 85F | UGUAAAUGCACUAUCUGGUACGACAU | + | + | 1391 upstream of Fmr1 | 2 miR cluster; expression verified | |
| 30 | 20.73 | miR-278 | 2R:10720792 | 52B | ggUGGGACUUUCGUCCGUUUGUAA | + | - | 386 upstream of fus | Expression verified | |
| 34 | 20.27 | miR-133 | 2L:20586360 | 38D | UUGGUCCCCUUCAACCAGCUGU | + | + | Vertebrate | 1059 downstream of CG15475 | 3 miR cluster; expression verified; [45] |
| 37 | 20.03 | miR-279 | 3R:25030674 | 99A | UGUGACUAGAUCCACACUCAU | + | + | 1328 upstream of CG31044 | Related to miR-286; expression verified | |
| 38 | 19.90 | miR-33 | 3L:19716503 | 76C | AGGUGCAUUGUAGUCGCAUUG | - | - | Vertebrate | In the intron of HLH106 (sense) | |
| 39 | 19.77 | miR-280 | 2R:3358854 | 44C | UGUAUUUACGUUGCAUAUGAAAUGAUA | - | - | 21740 upstream of CG30358 | Expression verified | |
| 41 | 19.73 | miR-281a | 2R:7235078 | 48E | ACUGUCGACGGACAGCUCUCUU | - | - | 356 downstream of SmD3 | Duplicated cluster; expression verified; 1 copy in Ano | |
| 43 | 19.64 | miR-282 | 3L:3231652 | 63C | aaucUAGCCUCUACUAGGCUUUGUCUGU | + | - | 7132 upstream of CG14959 | Expression verified | |
| 44 | 19.55 | miR-283 | X:15238971 | 13D | AAAUAUCAGCUGGUAAUUCUGGG | + | + | 3493 upstream of Ac13E | 2 miR cluster; expression verified | |
| 46 | 19.52 | miR-284 | 3R:8377257 | 87C | UGAAGUCAGCAACUUGAUUCCAGCAAUUG | - | - | 1128 upstream of CG6989 | Expression verified | |
| 47 | 19.47 | miR-281b | 2R:7234866 | 48E | ACUGUCGACGGAUAGCUCUCUU | + | - | 144 downstream of SmD3 | Duplicated cluster; expression verified | |
| 49 | 19.35 | miR-34 | 3R:5926677 | 85F | UGGCAGUGUGGUUAGCUGGUUG | + | + | Vertebrate/ worm | 477 upstream of Fmr1 | 2 miR cluster; expression verified; [45] |
| 50 | 19.27 | miR-263a | 2L:11942273 | 33B | aAUGGCACUGGAAGAAUUCACg | + | + | Vertebrate | 4764 downstream of CG16964 | Expression verified; [34] |
| 59 | 18.89 | miR-124 | 2L:17544454 | 36D | AUAAGGCACGCGGUGAAUGCCA | + | + | Vertebrate/ worm | 10606 downstream of CG7094 | 2 miR cluster; [45] |
| 66 | 18.58 | miR-79 | 2L:16676639 | 36A | AUAAAGCUAGAUUACCAAAGC | + | + | Worm | 822 upstream of CG31782 | 3 miR cluster; expression verified; [45] |
| 67 | 18.57 | miR-276b | 3L:10277315 | 67E | CAGCGAGGUAUAGAGUUCCUACG | - | - | Vertebrate | 7073 downstream of CG6559 | Duplicated, 45 kb apart; expression verified; 1copy in Ano and Apis |
| 77 | 18.36 | miR-210 | X:17859179 | 16F | UUGUGCGUGUGACAGCGGCUA | + | + | Vertebrate | 1193 downstream of CG32553 | |
| 83 | 18.11 | miR-285 | 3L:11903642 | 68E | UAGCACCAUUCGAAAUCAGUGCU | - | - | Vertebrate | 1592 upstream of CG7252 | Similar to miR-29 |
| 18.08* | miR-100 | 2L:18449518 | 36E | AACCCGUAAAUCCGAACUUGUG | + | - | Vertebrate | 1515 upstream of CG10283 | Failed conservation filter; 3 miR cluster; expression verified; [45] | |
| 91 | 17.93 | miR-92b | 3R:21466486 | 96E | AAUUGCACUAGUCCCGGCCU | + | - | Vertebrate | 1686 upstream of BcDNA:LD22548 | Expression verified; 2 miR cluster; [45] |
| 145 | 17.12 | miR-286 | 2R:14724858 | 56E | AGUGACUAGACCGAACACUCG | + | - | 1013 upstream of CG11018 | Expression verified; 7 miR cluster; related to miR-279 | |
| 146 | 17.11 | bantam | 3L:622845 | 61C | AGUGAGAUCAUUUUGAAAGCUG | + | - | Worm | 6301 upstream of CG12030 | [44] |
| 208 | 16.09 | miR-289 | 3L:13578391 | 70C | UAAAUAUUUAAGUGGAGCCUGCGACU | - | - | In the intron of bru-3 (antisense) | Expression verified | |
| 13.73 | miR-287 | 2L:17552694 | 36D | UGUGUUGAAAAUCGUUUGCAC | + | - | 14896 upstream of Oli | Very low score;found by proximity to miR-124; expression verified | ||
| 13.35 | miR-87 | 2L:9942828 | 30D | UGAGCAAAAUUUCAGGUGUG | - | - | Worm | 2009 upstream of CG13126 | Hom: very low score | |
| miR-263b | 3L:15666960 | 72D | cuUGGCACUGGGAGAAUUCACa | + | - | Vertebrate | 4243 upstream of comm | Hom; score n/a | ||
| miR-288 | 2L:20588106 | 38D | UUUCAUGUCGAUUUCAUUUCAUG | + | - | 2805 downstream of CG15475 | Score n/a; found by proximity to miR-133; expression verified | |||
| Unverified Ano-conserved candidates | ||||||||||
| Rank | Score | miR name | miR position | Cytological position | Sequence | Ano | Apis | Other | Nearest gene | Comment |
| 1 | 26.76 | 2R:4681879 | 46A | CAUCACACCCAGGUUGAGUGAGU | + | + | In the intron of Mmp2 (antisense) | NT | ||
| 5 | 24.35 | 3R:121090 | 82A | AAAUUGACUCUAGUAGGGAGUCC | + | + | 533 downstream of CG9780 | NT | ||
| 14 | 22.63 | X:1545630 | 2B | UGCAGGUUUCGUCGACAACGA | + | - | 732 upstream of CG32806 | NT | ||
| 19 | 22.13 | 3L:21585985 | 79A | CGAUUUGUCUUUUUCCGCUUACUG | + | - | 1727 downstream of CG7160 | NT | ||
| 20 | 21.95 | 3L:18809845 | 75E | UUUUGAUUGUUGCUCAGAAAGCC | + | + | 3283 upstream of CG6865 | No expression seen either strand | ||
| 23 | 21.38 | 3L:8530512 | 66D | GUGAGAUAUGUUUGAUAUUCUUGGUUGUU | + | + | 2374 upstream of CG6638 | NT | ||
| 40 | 19.75 | X:12366993 | 11B | UAUCAUAAGACACACGCGGCUAU | + | - | in the intron of tomosyn (sense) | NT | ||
| 54 | 19.06 | 2R:11128979 | 52E | guUAUUGCUUGAGAAUACACGUAGUU | + | + | 15915 upstream of Dg | No expression seen either strand | ||
| 61 | 18.86 | 2L:859210 | 21D | AGUUUGUUCGUUUGGCUCGAGUUAU | + | - | 2208 downstream of CG13949 | NT | ||
| 104 | 17.64 | 2L:16676008 | 36A | UCUUUGGUAUUCUAGCUGUAGA | + | - | 1453 upstream of CG31782 | No expression seen; miR-79 cluster | ||
| 117 | 17.44 | 3R:21403955 | 96E | UGAUAUUGUCCUGUCACAGCAGUA | + | - | 3265 upstream of CG12250 | No expression seen | ||
| 123 | 17.36 | 2L:7418192 | 27F | AUUGUACUUCAUCAGGUGCUCUGGUG | + | + | 905 upstream of CG5261 | NT | ||
| 126 | 17.31 | 3R:16621175 | 92F | UUUGUUUUGCAAUUUUCGCUUU | + | - | In the intron of CG17838 (sense) | NT | ||
| 130 | 17.24 | 2L:16676828 | 36A | CUUUGGUGAUUUUAGCUGUAUG | + | - | 633 upstream of CG31782 | No expression seen; miR-79 cluster | ||
| 183 | 16.39 | 2R:7223583 | 48E | UCAUCCCCUUGUUGCAAACCUCACGC | + | - | In the intron of CG8877 (sense) | NT | ||
| 190 | 16.28 | 3R:5916861 | 85F | UGGGAUACACCCUGUGCUCGCU | + | - | 17107 upstream of CG5361 | NT | ||
| 195 | 16.24 | 2L:243049 | 21B | CAUAAGCGUAUAGCUUUUCCC | + | + | In the intron of kis (sense) | NT | ||
These sequences were identified as high-scoring candidates through miRseeker analysis of drosophilid genomes (except as noted) and are ordered by their rank and score. The first part of the table includes members of the reference set, whose rank within the reference set is given in parentheses after the gene name; thus miR-4 ranked 18th among the reference set and 124th overall. The second part of the table includes miRNAs newly identified in this study. In general, we defined a candidate miRNA sequence on the basis of the bounds of conserved sequence; this is often longer than the presumed 21-22 nucleotide mature product. The third part of the table includes unverified gene predictions supported by conservation in Anopheles and/or Apis. Drosophila-specific predictions without confirming expression data may be viewed on the web [43]. References in the comments are to miRNAs that have been independently identified in previous or concurrent studies. n/a, score not available; NT, expression not tested; Hom, miRNA identified solely by homology to other miRNAs. The following miRs were not identifed by miRseeker: miR-10 was not aligned using the first release of the D. pseudoobscura genome while miR-2b-1 and miR-100 failed the conservation filters. These three received very high miRseeker scores, however, and they have been placed into the list for the sake of comparison, although they are not ranked. Six additional miRNAs scored poorly but are genuine. These include two members of the reference set (miR-125 and miR-2c), two that were identified by homology to miRNAs cloned from other species (miR-87 and miR-263b), and two that were identified as Anopheles-conserved stem-loops located in proximity to other Drosophila miRNAs whose expression was verified by northern analysis (miR-287 and miR-288). Most miRNAs are located in intragenic regions, and there is an apparent bias for intronic miRNAs to be located on the transcribed strand.
miR-125 and miR-2c received exceptionally low scores in this analysis, while miR-10 was absent because it was not located in an alignable contig in the first available assembly of D. pseudoobscura (although it was identifiable by BLAST search). The other 21 members of the reference set fell into the top 600 or so in the initial round of scoring, indicating that our method of scoring stem-loops effectively identified genuine miRNAs from among the 118,000 conserved super-regions.
Evaluation of the divergence pattern in conserved stem-loops
As discussed earlier, 23/24 members of the Drosophila reference set are described by one of three patterns of divergence (Figure 2a, classes 1-3). In the final set of tests, we implemented a series of Boolean filters to eliminate high-scoring, conserved stem-loops whose patterns of nucleotide divergence are incompatible with a high likelihood of representing genuine miRNAs (Figure 2a, classes 4-6).
We began this analysis by trimming the 100-unit region to exclude sequences at the ends of the windowed sequence that lie outside of the main helical arm. We then defined a potential miRNA candidate sequence as being a perfectly conserved block of sequence greater than 22 nucleotides in length located less than 10 nucleotides from the end of the terminal loop, and eliminated those candidates that did not contain a potential miRNA (Figure 2a, class 4). If both arms passed this test, it was kept as a miRNA candidate regardless of loop status (Figure 2a, classes 1 and 2) as either arm could potentially contain a miRNA. The remaining candidates contain only a single conserved candidate miRNA arm. We defined the nonconserved arm as the non-miRNA-encoding arm and eliminated the candidate if it displayed a perfectly conserved loop (Figure 2a, class 5). The remaining candidates contain divergent positions in both the loop and the non-miRNA-encoding arm. We eliminated those that contained more than four additional non-conserved positions in the non-miRNA-encoding arm compared to within the loop (Figure 2a, class 6), leaving behind class 3 candidates as potential miRNAs. Of the reference set miRNAs, only mir-2b-1 failed these filters (as a class 6 miRNA), even though it received the eighth highest score of all super-regions in the entire euchromatic sequence of the drosophilid genomes.
Only about one-third of the highest-scoring conserved stem-loops passed these filters (with an even greater fraction of lower-scoring candidates failing these filters), leaving behind around 200 candidates from the initial top 600. Of the reference set, 18/24 (75%) reside in the first 124 candidates, demonstrating that the overall procedure robustly selected for genuine miRNAs (Figure 4, Table 1). A second measure of the utility of assessing patterns of nucleotide divergence is their ability to select against self-complementary repeats. Many high-scoring candidates were previously noticed to be rich in complementary nucleotide repeats (such as CAG, UUG and CUG, AUAU, or poly(A)-poly(U) repeats) and were presumed to be poor candidates in spite of occasionally remarkable helical structure: nearly all of them were eliminated by the step 3 filters. We have generated a web interface where folded structures and evaluation of the top 208 miRseeker candidates may be accessed [43].
Figure 4.
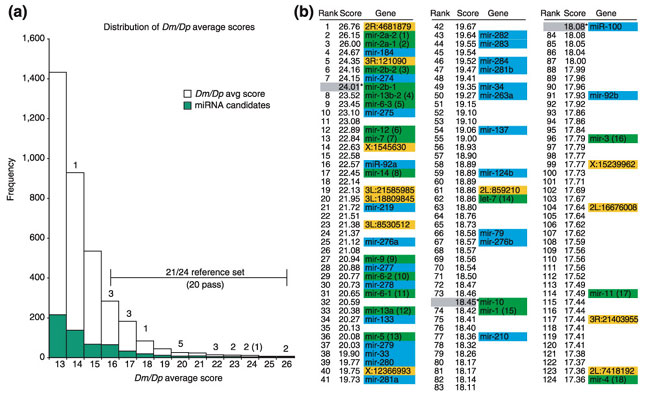
Efficient selection of genuine miRNAs by miRseeker. (a) Distribution of the top 2,996 candidates binned by helical/free energy score (white bars), of which 570 passed subsequent conservation filters (green bars). 21/24 members of the reference set received a score of 16 or higher, and 20 of these passed the conservation filters. Note that these figures do not include mir-10, which did not fall in an aligned contig and was thus not analyzed, even though its miRseeker score is 18.45 and it passes conservation filters. (b) List of the top 124 miRseeker candidates; members of the reference set are highlighted in green, newly identified miRNAs from this study in blue, and additional third-species-conserved candidates in orange. The vast majority of the highest-scoring candidates are bona fide.
Candidate mature miRNA sequences were defined by the bounds of the perfectly conserved sequence. A total of 42 novel candidates in the top 208 miRseeker predictions were supported by additional evidence of sequence and structural conservation in a third species (primarily Anopheles and Apis, with a smaller fraction in nematodes or vertebrates). In cases where candidate miRNAs were identifiable in non-insect species, a putative 21-24 nucleotide product was usually evident. A predicted miRNA produced from candidates whose only homologs were found in other insects could usually be inferred to within 5 nucleotides (Table 1).
Experimental verification of novel candidate miRNAs
We next sought to validate the predicted miRNAs by northern blot of total RNA isolated from 0-24 hour embryos, third instar larvae and early pupae, and adult males. The total number of genes authenticated by this method is an underestimate, for two main reasons. First, the mature miRNA can be derived from either arm of a given stem-loop, and many predicted pre-miRNAs fold well on either strand. In some cases (such as miR-4 and miR-100), the nontranscribed strand actually adopts a fold with longer continuous helices than does the transcribed strand. As we tested one or two probes for each candidate, a false-negative result will have been obtained if we tested either the incorrect arm or strand of a putative miRNA. Second, a significant fraction of miRNAs are likely to be expressed at extremely low levels or in a highly tissue-specific manner, and so may not be amenable to confirmation by these means. With these concerns in mind, we tested 38 candidate genes from two classes of predicted miRNAs: 27 that were conserved outside of Drosophila (25 of which were high-scoring candidates) and 11 high-scoring, Drosophila-specific candidates. This analysis confirmed 24 novel miRNA genes that give rise to processed 21-24 nucleotide RNAs (Figure 5, Table 2).
Figure 5.
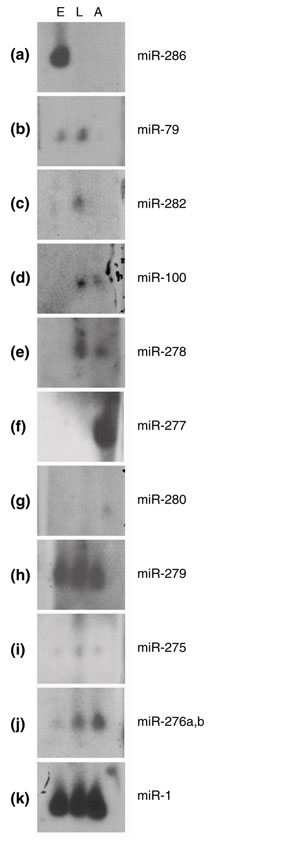
Diverse temporal and quantitative expression profiles of novel miRNAs by northern blotting. The three lanes represent 0-24-hour embryos (E), third instar larvae and 0-1-day pupae (L) and adult males (A), and hybridizing bands from the 21-24 nucleotide range are shown. (a-g) miRs with preferential expression at individual stages or a combination of two of these stages. (h-j) miRs that are expressed throughout development, either at uniform levels or in a graded fashion. (k) miR-1 was used as a control. Note that the blots shown were exposed for different lengths of time, so the relative levels of different miRNAs are not directly comparable; please refer to Table 2.
Table 2.
Summary of northern blot studies
| Gene | E | L | A |
| miR-286 | ++++ | ||
| miR-92b | ++ | ++ | |
| miR-79 | ++ | ++ | + |
| miR-275 | + | + | + |
| miR-287 | + | + | + |
| miR-283 | + | + | + |
| miR-281a, b | + | + | + |
| miR-279 | +++ | +++ | +++ |
| miR-263a | +++ | +++ | +++ |
| miR-276a, b | + | ++ | +++ |
| miR-288 | + | + | ++ |
| miR-184 | + | + | ++ |
| miR-282 | ++ | ||
| miR-289 | ++ | ||
| miR-133 | + | ++ | |
| miR-278 | + | + | |
| miR-284 | + | + | |
| miR-100 | ++ | ++ | |
| miR-34 | + | ++ | |
| miR-280 | + | ||
| miR-277 | ++++ | ||
| miR-274 | ++++ |
The relative abundance of a given miRNA at each stage is represented by the number of plus signs. miR-276a, b and miR-281a, b produce similar miRNAs that are not distinguished by northern analysis.
The expression profiles of computationally identifed miRNAs during development were much more heterogenous than those of the known set of embryonically derived miRNAs [10]. We identified miRNAs whose expression was highly restricted to individual developmental periods (embryogenesis, larval/pupal development, or adulthood), ones expressed in two of these developmental windows, and ones expressed throughout development, either at a relatively uniform level or in a progressive fashion. Selected hybridizations that illustrate the different temporal and quantitative profiles are shown in Figure 5 and the collected northern data are summarized in Table 2. We also note that these experimentally verified miRNAs vary tremendously in abundance, with several being two to three orders of magnitude less abundant than miRNAs discovered by direct cloning. Other undetected miRNAs orthologous to ones cloned in other species (that is, miR-137 and miR-219, Table 1) may be present at even lower levels. This suggests that their identification by sequencing miRNA cDNA libraries would have been unlikely.
In total, we observed expression for 20 out of 27 (74%) candidate genes that were conserved outside Drosophila and 4 out of 11 (36%) of high-scoring Drosophila-specific predictions (Figure 4b). Two of the former class were low-scoring candidates that were nonetheless conserved in Anopheles (miR-287 and miR-288), indicating that third-species conservation is in our hands a very strong indicator of a candidate gene's validity. Table 1 lists bona fide Drosophila miRNAs that scored in the top 208, grouped as members of the reference set followed by newly identified miRNAs that fulfill accepted criteria for miRNA biogenesis (that is, ones whose expression was confirmed here by northern blot and/or are homologous to miRNAs cloned in other species); each subset is rank-ordered by miRseeker score. The high-scoring, third-species-conserved candidates whose expression is unverified at present (either untested or negative by northern blot) are listed separately; they are provisionally referred to by their nucleotide position along the chromosome arm (that is, 2R:4681879). We note that while this work was in preparation, forward genetic analysis of bantam demonstrated that it encodes a miRNA [44] that was identified as a high-scoring candidate by miRseeker. In addition, a subset of the newly identified miRNAs (miR-34, miR-79, miR-87, miR-92a, miR-124b, miR-133 and miR-263a) were independently found by homology search or by informatic means and confirmed by northern analysis while this work was under review [34,45].
Conservation alone is an insufficient criterion for assessing the validity of a miRNA [46]. Indeed, one high-scoring candidate (number 78, 2L:13233310 that is strongly conserved in Anopheles and was identified by miRseeker appears to be an unannotated U2 snRNA [43]. Nevertheless, our high success rate (20/27) leads us to believe that failure to observe expression of a high-scoring, third-species-conserved miRseeker candidate could reflect a false-negative result. As an example, we show alignments and RNA folds of the four insect orthologs of 2R:11128979, which all adopt canonical, high-scoring stem-loop structures and collectively display patterns of nucleotide divergence characteristic of genuine miRNAs (preponderance of divergence within the loop, slightly less divergence along a nonconserved arm, and perfect conservation of a putative miRNA-encoding arm) (Figure 6). Although in this case evidence for expression of either strand by northern analysis was not obtained, it was subsequently found to be orthologous to miR-137, and thus accepted as a genuine miRNA. We hypothesize that other novel, high-scoring candidates with a similar level of third-species-conservation but which lack evidence of expression may in fact be genuine, thus implying up to 7/27 (26%) false-negative rate of northern analysis.
Figure 6.
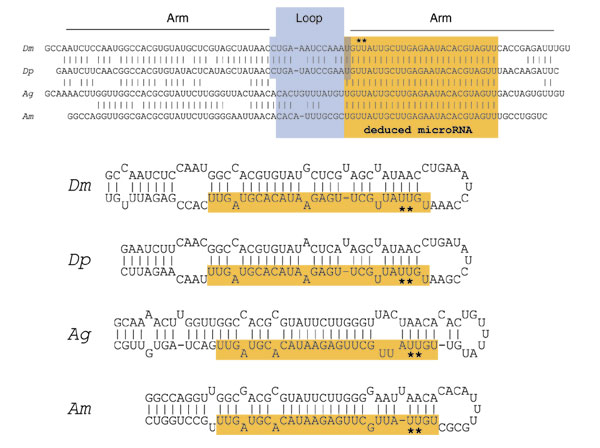
Example of a miRNA with false-negative evidence by northern blot (2R:11128979 = miR-137). In this example, four related sequences from four species of insects (Dm, D. melanogaster; Dp, D. pseudoobscura; Ag, A. gambiae; Am, A. mellifera) all adopt a phylogenetically conserved stem-loop structure. One arm has been perfectly preserved among all four species, and we presume that a miRNA is processed from within the conserved sequence (orange). Patterns of nucleotide divergence characteristic of miRNAs are seen, with more related sequences (Dm/Dp and Ag/Am) showing approximately equal amounts of divergence within the loop and along one arm, whereas the Dm/Dp vs Ag/Am comparison shows complete divergence within the loop (blue), with slightly less overall divergence along the putative non-miRNA-encoding arms. We deduce that a mature miRNA may initiate at one of the U residues that are highlighted by asterisks, as the first residue of the conserved region is found in the loop of the drosophilid hairpins and the second G residue is unfavored as the 5' residue of a miRNA. Northern analysis was negative using a probe complementary to the conserved region as well as with a probe identical to the conserved region (in the event that a miRNA is transcribed from the other strand). This sequence was only subsequently discovered to be orthologous to vertebrate miR-137 (which initiates at the second highlighted U). We consider other unverified predicted genes conserved in other insect species with similar characteristics to be potential candidates (see also Table 1).
Together, these data allow us to estimate the number of Drosophila miRNA genes. In the first 124 candidates, there are 18 members of the reference set and 25 novel miRNAs that meet accepted criteria for representing genuine miRNAs. Of the remaining candidates, around 36% may be genuine, although this rises to a maximum of approximately 62% if one considers the inferred false-negative rate of northern analysis. Thus, there may be between 81 × (0.36 to 0.62) = 29 to 50 additional miRNAs in this list of unverified and/or untested candidates. Therefore, we estimate 72-93 miRNAs (18 reference + 25 novel verified genes + 29-50 additional unverified candidates) at a cutoff that includes 18/24 (75%) members of the reference set, allowing us to extrapolate that drosophilid genomes may contain 96-124, or around 110 miRNA genes. This suggests that nearly 1% of Drosophila genes are miRNAs, a figure that is in relative accord with the percentage recently ascribed to vertebrate genomes [31].
miRNA genes: clusters and paralogs
A subset of miRNA genes are known to reside in local genomic clusters with possible operon-like organization. The largest cluster of miRNA genes in Drosophila includes six that were previously identified as the mir-3 → mir-6-3 cluster [10]. We identified two additional conserved stem-loop structures that flank mir-3 (Figure 7a), one of which (mir-286) was confirmed by northern blot (Figure 5a). Surprisingly, mir-286 is the only member of this seven-miRNA cluster that is conserved in Anopheles, indicating tremendous flux in the miRNA content of this region even within the Diptera. We also observed that miR-286 is related at its 5' end to another experimentally verified miRNA (miR-279, Figure 5h), suggesting that they may have related functions.
Figure 7.
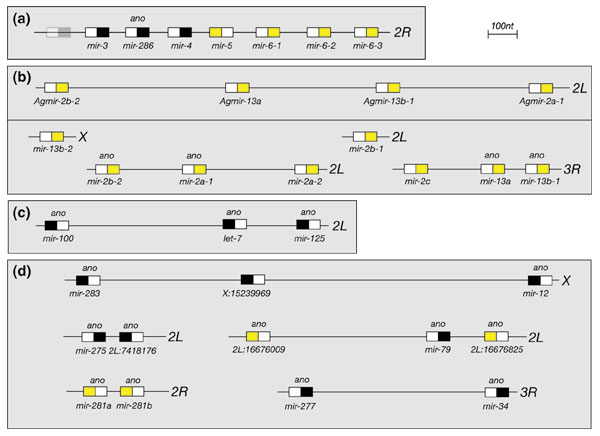
Examples of Drosophila miRNA gene clusters. In this figure, pre-miRNAs are represented by rectangles and the arm that gives rise to the mature miRNA is colored. (a) The largest miRNA cluster was previously identified by Tuschl and colleagues [10]; we identified and experimentally verified a new member of this cluster, mir-286. A second conserved hairpin was found (light gray box), but its expression was not seen. Of the seven genes in this cluster, only mir-286 is conserved in Anopheles (ano). Note also that this cluster contains both related miRNA genes (mir-6-1, -2, -3 and the K-box antisense gene mir-5, yellow), as well as unrelated miRNA genes (black). (b) A second example of rapid miRNA gene evolution. The Anopheles genome contains four members of the mir-2/mir-13 family, which are all located in a single cluster. In contrast, drosophilid genomes contain eight members of this family, located at four distinct genomic locations on three different chromosomes. (c) A cluster of putative developmental regulators. let-7 and mir-125 are orthologous to the genetically characterized genes let-7 and lin-4 in C. elegans. A similar gene cluster exists in Anopheles, although mir-100 is separated from the other two by several kilobases (not shown). (d) Other examples of miRNA clusters. Note that, as is the case for the other clusters shown, miRNA clusters can contain related genes (yellow), but appear to be as likely to contain unrelated genes (black).
The volatility of miRNA genes is further indicated by members of the miR-2/miR-13 K box subfamily [47]. Seven members were previously reported that are scattered in four genomic locations on three different chromosome arms, including a cluster of three mir-2 genes on 2L and a cluster of two mir-13 genes on 3R; we also identified an additional paralog of mir-2 (mir-2c) that is located in the mir-13 cluster. Unexpectedly, the Anopheles genome contains only four members of this subfamily, and all are located in a single cluster on 2L (Figure 7b); the same four members were identifiable in the unassembled genomic sequence of Apis [48]. The simplest scenario is that members of the K box-family have undergone radical duplication and dispersal about the genome in Drosophila. This is consistent with the finding that the remaining members of the K-box family [47], including mir-11, the three mir-6 paralogs and the K-box-antisense gene mir-5, are similarly absent from both Anopheles and Apis. However, one new putative member of the K-box family (2R:4681879) was identified by miRseeker, and it has been conserved in all four sequenced insect genomes.
Another notable cluster that includes previously identified miRNAs is one containing mir-100, let-7 and mir-125 (Figure 7c). miR-125 is an apparent homolog of C. elegans lin-4 [29], which functions with let-7 to regulate developmental timing in nematodes. Although complementary sites for both miRNAs are found in the 3' UTRs of several putative nematode targets, these miRNAs are not physically linked in the C. elegans genome and their mutant phenotypes are principally due to misregulation of different transcripts [4-6,8]. The function of these miRNAs has not yet been explicitly demonstrated in other species, but Drosophila let-7 is regulated at the larval-pupal transition by ecdysone [49], and all three members of this cluster are present in other insects and in vertebrates, thus implying broadly conserved functions. Their existence in a gene cluster in Drosophila and Anopheles (data not shown) may imply that their functions overlap to a greater extent in insects than in nematodes. This could explain why individual let-7 or miR-125 mutants with strong developmental defects have not yet been identified in Drosophila by genetic means. Our observation that the temporal expression profile of miR-100 (Figure 5d) is similar to that described for let-7 and miR-125 (that is, expression is initiated during larval/pupal development and continues through adulthood [9,29]) is consistent with probable coordinate expression of all three as a single pri-miRNA transcript, and may further implicate miR-100 in developmental timing. While this work was under review, the clustering of miR-100, let-7 and miR-125 was independently reported; these researchers also provide evidence for coordinate expression of these miRNAs as a single pri-miRNA [45,50].
The characteristics of miR-100 are highly unusual and thus serve as a useful caution. First, although miRseeker correctly identified it as a high-scoring conserved stem-loop, the incorrect strand was identified as its nontranscribed strand adopts greater helical structure than does its transcribed strand. Second, it is the only confirmed Drosophila miRNA that deviates from the expected pattern of divergence in two fundamental ways: it not only contains a mismatch on the arm while maintaining a perfectly conserved loop (and thus failed the conservation filter as a class 5 candidate), but the mismatch actually resides within the mature miRNA sequence itself. Therefore, the identification of miR-100 in flies relied upon its conservation in other species.
We identified several additional examples of closely linked miRNAs that are transcribed from the same strand (Figure 7d). These include clusters containing paralogous miRNA genes (such as mir-281a and mir-281b) others that contain unrelated genes (mir-12/mir-283, mir-34/mir-277 and mir-275/2L:7418176 clusters), and some that are a mixture of both (mir-79 cluster). Unexpectedly, we did not find that clusters of paralogous miRNA genes were more prevalent, as might have been predicted a priori if gene clusters generally result from local gene duplications. This is apparently corroborated by the identification of several miRNA clusters whose processed products derive from opposing arms of their respective precursors (Figure 7) [10,29]. A second curious observation is that relatively few Drosophila miRNAs are members of paralogous gene sets, irrespective of whether they are physically linked or not. In fact, we identified only four new examples (miR-281a+b,miR-276a+b, miR-92a+b and miR-263a+b) to add to the previously described sets of miR-6 and miR-2/miR-13 genes. This contrasts with observations of miRNAs in vertebrates, where the majority of known miRNAs are members of paralogous gene sets [31]. The most extreme examples of this disparity are let-7 and mir-29, which are present in single copies in Drosophila, but are represented by thirteen and six distinct human homologs, respectively.
Discussion
Drosophilid genomes contain around 110 microRNA genes
Although the first two miRNA genes were identified through forward genetics, the vast majority of known miRNAs were identified by direct cloning of mature 21-22 nucleotide RNAs, either from size-selected total RNA or from purified miRNP complexes. The direct cloning method has the distinct advantages of being expression based and aimed at identifying the processed miRNA that is presumably the active regulatory species. While it is clear that cloning efforts in some organisms have been far from saturating, work in other organisms (mammals) suggests that efforts of this sort will give sharply diminishing returns. In any case, it is likely that many miRNAs will not be amenable to direct cloning, including those that are present at very low levels (be they poorly expressed, highly unstable, inefficiently processed, or expressed by small numbers of cells) or whose expression is otherwise restricted to times and/or locations for which the isolation of sufficient amounts of RNA for cloning is impractical.
In theory, a computational approach based on the structural features of known miRNA genes might permit the unbiased discovery of the remaining complement of miRNA genes in a given sequenced genome. However, in one test, around 5% of randomly selected C. elegans genomic sequences were found to have the capacity to fold into plausible miRNA precursor hairpins [11]. This suggests that computational prediction of miRNAs based on presumed structure alone would have an unacceptably high false-positive rate. This type of approach might be strongly aided by comparative genomics, whereby one confined the analysis to genomic regions subject to both evolutionary and structural constraint. The proof of principle behind this dual scheme was demonstrated by the identification of several miRNAs via a structural analysis of genomic sequence conserved over 50 million years of nematode evolution [12]. Indeed, the great majority of C. elegans miRNAs identified through the direct cloning approach are conserved in the genome of C. briggsae, with conservation typically extending across the length of the predicted miRNA precursor.
In this study, we describe miRseeker, a computational approach for the identification of Drosophila miRNA genes that ranks conserved stem-loop structures and assesses their pattern of nucleotide divergence. miRseeker successfully identified nearly all of the known Drosophila miRNA genes, and a strong majority of novel, high-scoring candidates were verified by northern analysis. In total, we catalogued 32 newly verified miRNAs (24 of which were confirmed here by northern blot), bringing the current Drosophila tally to 56. Our data allows us to estimate the presence of around 110 miRNA genes in Drosophila. The approach used in this study should be applicable to the analysis of other sets of sequenced genomes of related higher eukaryotic model organisms. While this manuscript was in preparation and in submission, several other computational predictions of miRNA genes by similar overall strategies were reported [31-34]. The results of our studies are most similar to analyses by Bartel and colleagues predicting 200-255 miRNA genes in vertebrates, or nearly 1% of the predicted genes in humans [31]; this is comparable to our estimate of flies.
A unique aspect of miRseeker amongst the recently described methodologies is its assessment of the pattern of nucleotide divergence within miRNA precursors. The existence of this pattern, which is reflected by initial divergence within loop sequence, was unexpected. Two conclusions may be drawn from this phenomenon. First, the loop appears to be the least critical feature of a pre-miRNA, an observation supported by the identification of orthologous insect miRNAs that vary quite significantly in terms of both loop size and sequence (data not shown). Second, there appears to be measurable selective pressure on the sequence of the non-miRNA arm, above and beyond the necessity to maintain a certain degree of helical structure that would make it a Dicer substrate. This is perhaps at odds with the nonspecific activity of Dicer, which efficiently processes virtually every input dsRNA ever tested in RNAi assays. It may be the case that specific sequence requirements become greater when processing imperfect stem-loops characteristic of pre-miRNA, or alternatively, that the non-miRNA-encoding product may in fact have some previously unappreciated function. Nonetheless, it is clear that the selective pressure on the non-miRNA-encoding arm is mild, and that it diverges long before the sequence of the mature miRNA does.
It is worth noting that the fraction of miRNAs conserved between mammals and fish, which are around 450 million years diverged, appears to be significantly higher than the fraction of miRNAs that are conserved between flies and mosquitoes, which are separated by only 250 million years of evolution (this paper and [31]). This indicates that insect miRNAs evolve much more rapidly than do vertebrate miRNAs. This is in general agreement with analyses of orthologous protein pairs, which showed that dipteran pairs had a higher rate of divergence than did fish/human pairs [51]. Since it may be argued that the selective environmental pressures are more different between these sequenced vertebrates, it may be that the relatively high rate of divergence in Diptera is a consequence of their shorter generation times.
Of false negatives and false positives
In these heady times of miRNA gene discovery, it is prudent to exercise caution in designating predicted genes as bona fide, just because they resemble known miRNA genes [46]. At the same time, while many thousands of annotated protein-coding genes are not associated with cDNA clones or other evidence of expression, computational methods of their identification are robust enough for them to be considered 'real' genes until proven otherwise. It is our hope that refinement of miRNA prediction algorithms may further reduce the false-positive rate and elevate confidence in their output to a comparable level. Consideration of nucleotide bias at the 5' end of miRNA (including a propensity to begin with U) may improve predictions [11], although we stress that our algorithms were designed to predict pre-miRNA genes and not mature miRNA sequences themselves. The extent of the precisely conserved arm (thus including a potential miRNA) was in most cases longer than the mature product, and it would have been arbitrary to select a candidate miRNA sequence just so that it began with a U. Notably, a recent study suggests that different chemical strategies for capturing miRNA molecules may differentially recover miRNAs according to their 5' ends [33], so reevaluation of miRNA 5' bias may be in order. Incorporation of promoter evidence in gene models may also help, given recent speculation that miRNA genes may be transcribed by RNA polymerase II. Our confidence in the high-scoring Drosophila-specific candidates, most of which we did not test in this study, will also undoubtedly be bolstered by their conservation in the genomes of additional species of Drosophila. We eagerly await the initiation of these sequencing projects.
Potential false negatives fall into several classes. First, our search was based on the currently sequenced and alignable portions of the D. melanogaster and D. pseudoobscura genomes. As mentioned earlier, one of the 24 reference miRNAs (miR-10) did not reside in an aligned region, even though it was readily identified in available D. pseudoobscura sequence by BLAST query. Next, as our strategy relies upon conservation of primary sequence, we will have missed exceptionally divergent miRNAs, including class 6 candidates. Comparisons with Anopheles demonstrate that dipteran miRNA genes evolve rapidly and although none of the reference set is absent from D. pseudoobscura, there may be examples that are specific to individual drosophilid species. The recent discovery of a large class of tiny non-coding RNA genes (tncRNAs) that share some features with miRNAs provides a precedent for exceptionally rapidly evolving small RNA genes; tncRNAs are not even conserved within Caenorhabditis [33]. We also masked exons from the search, so we will have excluded any miRNAs that might be processed from spliced mRNA, including untranslated regions. Finally, we will have missed those miRNAs that proceed through precursor transcripts of unusual structure. Although most known pre-miRNAs in Drosophila, Caenorhabditis, mice and humans form relatively canonical stem-loops of around 70 to 100 nucleotides in length, plant miRNA precursors are often processed through exceptionally long stem-loops (many 150 nucleotides or longer) [14-16]. miR-125 is a member of the Drosophila reference set that received a low score by our informatic procedure, possibly because it may derive from an unusually long stem-loop (120 nucleotides), only a portion of which was included in the queried window. In addition, other plant miRNAs may proceed through precursors of noncanonical structure, such as multiple stem-loops or stem-loops of poor quality [14]. The existence of these types of pre-miRNAs in metazoans is unknown at present.
Despite these potential sources of error, the robust ability of miRseeker to identify genuine miRNAs is clearly indicated by its ability to identify most of the previously known Drosophila miRNAs as very high-scoring candidates and by our experimental validation of a very large number of newly identified candidate miRNA genes.
Prospects
Perhaps the most outstanding current challenge regarding miRNAs lies in determining their regulatory targets. It is presumed that most of them will form RNA duplexes with complementary sequences in mRNA, but direct regulatory relationships are known for only a handful of miRNAs. Although many miRNAs in plants were found to be nearly or completely complementary to known or putative target mRNAs [24,52], the few cases of known or presumed animal miRNA targets involve imperfect and limited homology. Indeed, searches for complements to miRNAs in animal genomes to date have not succeeded in detecting matches more compelling than those identified in random sequence. Thus, matching miRNAs to their cognate targets in silico presents a daunting task.
The 5' ends of a large subset of experimentally derived Drosophila miRNAs were recently observed to be perfectly complementary to Brd boxes, K boxes and GY boxes, a set of 3' UTR sequence motifs involved in post-transcriptional regulation [47,53-55]. This suggests that, at least in Drosophila, a subset of relevant miRNA targets might be found by searching for complements to the 5'-most 8-10 nucleotides of miRNAs in 3' UTRs. This procedure certainly produces many candidate regulatory pairs (I. Holmes and E.C.L., unpublished observations), but so many matches are found that it is difficult to single out individual cases as being more likely to be genuine.
Comparative genomic analyses of orthologous drosophilid genes should aid in this endeavor, as evidenced by the recent demonstration that several conserved motifs in the 3' UTR of the pro-apoptotic gene hid are targets of the miRNA bantam [44]. Statistically significant pairings of conserved and/or overrepresented 3' UTR motifs can be assessed for complementarity to miRNAs. Comparisons with Anopheles orthologs may also prove useful in this regard. For example, as is the case for their drosophilid counterparts, Anopheles transcripts for two types of Enhancer of split genes (basic helix-loop-helix repressor and Brd-family) contain Brd, K and GY box motifs in their 3' UTRs (E.C.L., unpublished observations). We anticipate that other examples of sequence motifs that are conserved in orthologs of the three dipterans and that are strongly complementary to miRNAs will be prime candidates to test as new examples of miRNA-mediated post-transcriptional gene regulation.
Materials and methods
Genome analysis
We used the following genomic sequences and analyses in this work: D. melanogaster Release 3 sequence (Berkeley Drosophila Genome Project, BDGP) [36]; D. melanogaster Release 3.1 annotation (BDGP) [40]; D. pseudoobscura Release 1 sequence and assembly (Human Genome Sequencing Center at Baylor College of Medicine, HGSC at BCM) [37]; whole-genome alignment of D. melanogaster and D. pseudoobscura (Berkeley Genome Pipeline) [32]; Anopheles gambiae assembled genomic sequence (Anopheles Genome Project) [56]; Apis mellifera unassembled genomic trace sequence (HGSC at BCM) [48].
miRseeker computational pipeline
The computational screen for Drosophila miRNAs was executed with a pipeline of custom developed Perl scripts that we refer to as miRseeker. These integrate sequence inputs from flat files with parallel computation on a 55-node Beowulf Linux cluster [57], load results into a specialized MySQL database and produce web page summaries of miRNA candidates. The scripts are grouped into three general processes as schematized in Figure 3.
Extraction of conserved Drosophila sequences
Release 3 of the D. melanogaster genome [36] was divided into 1,287 contigs of 100,500 nucleotides each, with 500 nucleotides of overlap at either end. These contigs were aligned to the first assembly of the D. pseudoobscura euchromatic sequence [37] by running 1287 parallel AVID jobs on the Linux cluster. Aligned sequences with the following Release 3.1 annotations [40] were eliminated: exons, transposable elements, snRNA, snoRNA, tRNA and rRNA genes. Conserved miRNA candidate regions were extracted from the aligned nongenic drosophilid sequence as follows. A 100-unit window (where a unit is either an aligned or a gapped nucleotide) was advanced across the AVID alignment files by single units. Once a window that satisfied minimal conservation criteria (≤13% gaps and ≤15% mismatches) was identified, the corresponding Dm and Dp sequences (regions) were stored as a multiple fasta file. The window was then advanced by 10 units and reassessed. The window continued to advance by 10 units until it no longer satisfied our criteria for conservation, at which point it was advanced by single units until the next conserved region was identified. Around 436,000 conserved regions were identified in this way.
The forward and reverse complement sequences of the regions were loaded into a MySQL database. If two D. melanogaster regions initiated within less than 13 nucleotides of each other, they were grouped into a super-region; reiterated grouping sorted the 436,000 regions into 118,000 distinct super-regions.
RNA folding and scoring of conserved stem-loops
All Dm regions were folded with mfold 3.1 by submitting parallel batches of 500 mfolds to the cluster nodes and copying the .det and .out mfold output files to a final storage destination. All mfold outputs were parsed, and information about the number of structures, number of arms per structure, size of helices within arms and size and symmetry of the internal loops within arms was loaded into the MySQL database.
Two parameters for each individual arm in each output structure were evaluated: free energy (ΔG kcal/mol) and miRNA-like helicity. Helicity was calculated by awarding +1 for each paired nucleotide, -1 for each one-nucleotide symmetric loop and -2 for each two-nucleotide symmetric loop. A progressively increasing penalty was applied for symmetric loops greater than three nucleotides as well as for all bulged nucleotides and asymmetrically sized loops, as these are more rarely observed in genuine miRNAs. An overall score was calculated as (helical score + (ABS(ΔG)/2))/2. For each super-region a single region that includes the highest scoring arm was flagged.
All steps outlined above in the sub-section above were repeated for regions contained within Dp super-regions corresponding to the top 25% Dm super-regions. The average score of each corresponding Dm and Dp region (termed a region-pair) was determined, and the highest-scoring region-pair in each super-region was flagged as its best candidate miRNA.
Representative regions were then rank-ordered by average Dm/Dp score. The D. melanogaster sequences from the top 20% of Dm/Dp regions were blasted against the Anopheles genome. The top three Anopheles blast hits were subjected to the steps outlined in the previous sub-section and the best-scoring structure was determined and linked to rank-ordered Dm/Dp regions. In most cases, either no Anopheles blast hit or non-homologous sequence was returned. The blast hit was determined to be homologous if it folded into a similar structure as the drosophilid sequences and one or both helical arms were highly conserved; these candidates are flagged orange on the web output.
Evaluation of the divergence pattern in conserved stem-loops
The alignment of Dm/Dp sequences was processed to distinguish loop regions (colored blue on the web output) and arm regions (colored red on the web output). Since mfold tends to insert small helices within terminal loops that falsely reduce the size of the terminal loop, small terminal loops (3, 4 or 5 nucleotides in length) were extended to seven nucleotides. Potential miRNA-encoding arms were then identified as perfectly matched blocks of sequence more than 22 nucleotides long and less than 10 nucleotides from the end of either side of the terminal loop (colored yellow on the web output). If no such sequence was found, the region was eliminated. If both arms contained potential miRNA sequences, the region passed and it was considered a miRNA candidate (colored green on the web output). If only one potential miRNA-encoding arm was identified, then the other arm was defined as the non-miRNA-encoding arm. The terminus of the potential miRNA-encoding arm was provisionally defined as the end of the perfectly conserved sequence, and the non-miRNA-encoding arm was trimmed to an equal helical length. The number of mismatched and gapped nucleotides in the loop region and in the non-miRNA-encoding arm was evaluated. Candidates with only a single candidate miRNA-encoding arm were disqualified if they did not diverge in the loop or the number of mismatches in the non-miRNA-encoding arm was four or more mismatches greater than the number of mismatches within the loop. The candidates that pass the conservation filter are colored green on the web report. The first 208 candidates produced by miRseeker, along with information on structures, scores, alignments, and genomic locations, are accessible via the web [43].
miRseeker public interface
For the genome-wide search, several steps of the pipeline that do not require interaction with the database were run in parallel on the Linux cluster (including AVID alignments, extraction of conserved regions, and mfolds) and represent a significant amount of computational time. To allow public access to miRseeker, we developed a scaled-down version of our computational pipeline [58]. It allows a user to input two sets of homologous sequences (up to 100 kb) from any two species along with gene annotation for one of them, and performs all the steps of the miRNA-finding procedure described above, producing a list of miRNA candidates ordered by score. A cutoff score that reliably distinguishes genuine miRNAs will differ with sequences from different input genomes. Our experience with drosophilid genomes suggests that a cutoff of 16.00 produces genuine candidates but also a high fraction of probable false negatives, whereas a cutoff of 17.00 defines candidates of much higher overall quality. We stress that the conservation consideration described above applies only to appropriately related species (such as Dm/Dp) and should not be indiscriminately used to filter miRNA candidates derived from comparative analysis of other species. The miRseeker public interface currently supports only a single concurrent user.
Northern blot validation of predicted miRNA genes
Total RNA was isolated from embryos (made by combining equal amounts of 0-12-hour and 12-24-hour embryo RNA), larvae and pupae (made by combining equal amounts of third instar and 0-1-day-old pupal RNA), and 0-2-day-old adult males. Blots were prepared by electrophoresing 40 mg RNA from each time point per lane on 15% acrylamide gels, followed by electroblotting to ZetaProbe GT membranes (BioRad). These were then probed with radioactive DNA oligonucleotide probes end-labeled with the StarFire system (Integrated DNA Technologies, Coralville, IA).
Additional data file
A detailed description (Additional data file 1) of the miRseeker output data, including the folded structures and evaluation of the top 208 miRseeker candidates, that can be accessed at [43] is available.
Supplementary Material
A description of the miRseeker output data that are available at [43]
Acknowledgments
Acknowledgements
We would like to thank the Human Genome Sequencing Project at the Baylor College of Medicine for making the unpublished, assembled genomic sequence of D. pseudoobscura and the unassembled, whole genome shotgun sequence of A. mellifera publicly available, the groups of Inna Dubchak and Lior Pachter at the Lawrence Berkeley National Laboratory for providing a global drosophilid genome alignment, Erwin Frise for continual technical assistance with Beowolf cluster operations, and the two anonymous reviewers for useful comments on this manuscript. This work was supported by a grant from the Damon Runyon Cancer Research Fund (DRG 1632) to E.C.L. and by the Howard Hughes Medical Institute.
References
- Huttenhofer A, Brosius J, Bachellerie JP. RNomics: identification and function of small, non-messenger RNAs. Curr Opin Chem Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau N, Lim L, Weinstein E, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli A, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ketting R, Fischer S, Bernstein E, Sijen T, Hannon G, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNAs involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Bass BL. A role for the RNAse III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy A, Hammond S, Hannon G. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Doench J, Petersen C, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee R, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Grad Y, Aach J, Hayes G, Reinhart BJ, Church G, Ruvkun G, Kim J. Computational and experimental identification of C. elegans microRNAs. Mol Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:research0079.1–0079.9. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila pseudoobscura genome project http://hgsc.bcm.tmc.edu/projects/drosophila
- Berkeley Genome Pipeline http://pipeline.lbl.gov/pseudo
- Bray N, Dubchak I, Pachter L. AVID: A global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3:research0083.1–0083.3. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews D, Turner D. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark BFC, editor. In RNA Biochemistry and Biotechnology. Dordrecht: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
- WU-BLAST http://blast.wustl.edu
- Drosophila microRNA candidates http://www.fruitfly.org/seq_tools/flymiRcandidates.html
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol N, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Project http://hgsc.bcm.tmc.edu/projects/honeybee
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Pasquinelli A, Kiger A, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259:1–8. doi: 10.1016/s0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3' UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3' UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. Regulation of Drosophila neurogenesis by RNA:RNA duplexes?. Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Mungall C, Misra S, Berman B, Carlson J, Frise E, Harris N, Marshall B, Shu S, Kaminker J, Prochnik S, et al. An integrated computational pipeline and database to support whole-genome sequence annotation. Genome Biol. 2002;3:research0081.1–0081.11. doi: 10.1186/gb-2002-3-12-research0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- miRseeker public interface http://www.fruitfly.org/seq_tools/miRseeker.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A description of the miRseeker output data that are available at [43]


