Summary
The SUMO ubiquitin-like proteins play regulatory roles in cell division, transcription, DNA repair, and protein subcellular localization. Paralleling other ubiquitin-like proteins, SUMO proteins are proteolytically processed to maturity, conjugated to targets by E1-E2-E3 cascades, and subsequently recognized by specific downstream effectors containing a SUMO-binding motif (SBM). SUMO and its E2 from the budding yeast S. cerevisiae, Smt3p and Ubc9p, are encoded by essential genes. Here we describe the 1.9 Å resolution crystal structure of a noncovalent Smt3p–Ubc9p complex. Unexpectedly, a heterologous portion of the crystallized complex derived from the expression construct mimics an SBM, and binds Smt3p in a manner resembling SBM binding to human SUMO family members. In the complex, Smt3p binds a surface distal from Ubc9's catalytic cysteine. The structure implies that a single molecule of Smt3p cannot bind concurrently to both the noncovalent binding site and the catalytic cysteine of a single Ubc9p molecule. However, formation of higher-order complexes can occur, where a single Smt3p covalently linked to one Ubc9p's catalytic cysteine also binds noncovalently to another molecule of Ubc9p. Comparison with other structures from the SUMO pathway suggests that formation of the noncovalent Smt3p–Ubc9p complex occurs mutually exclusively with many other Smt3p and Ubc9p interactions in the conjugation cascade. By contrast, high-resolution insights into how Smt3p–Ubc9p can also interact with downstream recognition machineries come from contacts with the SBM mimic. Interestingly, the overall architecture of the Smt3p–Ubc9p complex is strikingly similar to recent structures from the ubiquitin pathway. The results imply that noncovalent ubiquitin-like protein–E2 complexes are conserved platforms, which function as parts of larger assemblies involved many protein post-translational regulatory pathways.
Keywords: SUMO, ubiquitin, E2, ubiquitin conjugating enzyme, Ubc9
Covalent attachment of ubiquitin-like proteins (Ubls) is emerging as a predominant mechanism for regulating protein function in eukaryotes.1; 2 In this process, the C-terminus of a Ubl becomes covalently attached to the target.3 Protein modification by the prototypic Ubl, ubiquitin, is often associated with altered target half-life, subcellular localization, or protein–protein interactions. In addition to ubiquitin, several structurally related Ubls, such as ISG15, NEDD8, and members of the SUMO family have been found to modify a vast number of targets to alter protein functions in a variety of ways. For example, SUMO, or Smt3p (S. cerevisiae SUMO) modifications have been associated with altering target protein subcellular localization, enzymatic activity, and ability to interact with protein or DNA partners.4; 5; 6; 7 In general, Ubls, including ubiquitin and SUMO, are conjugated by their own enzymatic cascades, which share many common features.3 Ubls are initially translated with extended C-termini, which become proteolytically processed to generate the functional C-terminal Gly-Gly motif. Once processed, a Ubl's C-terminus becomes covalently attached to its targets via an enzymatic cascade involving Ubl-specific E1, E2, and E3 enzymes. The E1 first binds noncovalently to the Ubl and catalyzes adenylation of the Ubl's C-terminus in a MgATP-dependent reaction. The E1 subsequently forms a covalent thioester intermediate between its catalytic cysteine and the Ubl's C-terminus. The E1 binds E2 for a transthiolation reaction, where the Ubl is transferred from the E1's catalytic cysteine to that of E2. The product of this reaction is a covalent E2∼Ubl complex (∼ denotes covalent interaction), linked by a thioester bond between the E2's catalytic cysteine and the Ubl's C-terminus. The covalent E2∼Ubl complex is released from E1, and then generally interacts with an E3, which facilitates Ubl transfer, most often to a target's lysine's ε-amino group. The modified target interacts with Ubl recognition machinery, resulting in altered function of the target. Finally, the dynamic nature of this process is maintained by Ubl deconjugation, carried out by Ubl specific proteases.
In addition to many conserved features of Ubl cascades, there are several unique features distinguishing the SUMO pathway. First, relative to the structure of ubiquitin, SUMO contains a ∼20 residue disordered N-terminal extension.8 Second, whereas the ubiquitin cascade is significantly expanded at the level of E2, a single E2 (Ubc9) is responsible for all SUMO modification.9; 10 Third, in some cases, Ubc9 directly recognizes a target's SUMO modification consensus motif (ϕ-K-X-D/E, ϕ, hydrophobic; K, target lysine; and D/E, acidic) either in the absence or presence of an E3.11 Fourth, whereas the sequence of ubiquitin is highly conserved across phyla, in mammals there are four reported SUMO family members, with the best-studied being SUMO-1, -2, and -3.7 These have distinct features, but all share ∼45% sequence identity with the single S. cerevisiae SUMO, Smt3p. S. cerevisiae Smt3p, and mammalian SUMO-2 and SUMO-3, can form polymeric chains, in which a lysine from ϕ-K-X-D/E consensus sites found in the unstructured N-terminal extension from one SUMO is linked to the C-terminus of the next in the chain.12; 13 However, unlike ubiquitin, chain formation is not an essential feature of Smt3p, as neither the entire N-terminal extension nor any Smt3p lysines are essential for S. cerevisiae viability.14 A fifth unique feature of the SUMO pathway is a distinct set of E3s: all eukaryotes appear to have SUMO E3s of the Siz/PIAS class.15; 16; 17 These contain a Siz/PIAS-RING (SP-RING) domain, which resemble RING and U-box sequences found in ubiquitin E3s. SP-RINGs are thought to interact with Ubc9 in a manner resembling RING/U-box interactions with ubiquitin E2s. Higher eukaryotes are also known to have additional families of SUMO E3s, such as Nup358/RanBP2 and PC2,18; 19 which do not resemble any known ubiquitin E3s.
SUMO also has its own distinct recognition machineries, which contain a “SUMO Binding Motif” (SBM, also called a “SUMO Interaction Motif” (SIM)) of sequence ϕ-X-ϕ-ϕ or ϕ-ϕ-X-ϕ, where ϕ is hydrophobic and generally Leu, Ile, or Val.20; 21; 22 SBMs adopt a β-strand structure, and bind a groove in SUMO with orientation depending on whether the pair of hydrophobic side-chains is at the N- or C-terminus of the motif.21 SBM binding by SUMO can exert either negative or positive effects on intermolecular interactions, depending on context. For example, SUMO binding to the SBM of its covalently attached thymine DNA glycosylase disrupts DNA binding,23 whereas SUMO-1 modification of RanGAP1 promotes interaction with the SBM of the nuclear pore component Nup358/RanBP2.24; 25 Notably, Nup358/RanBP2 is also a SUMO E3, and for this activity the SBM can position SUMO-1 relative to Ubc9 for the ligation reaction.26 After the reaction, the Nup358/RanBP2 SBM remains bound to SUMO-1, allowing recruitment of modified RanGAP1 to the nuclear pore.
Several high-resolution structures have provided insights into both general characteristics of Ubl conjugation, and specialized features of the SUMO cascade. These include multiple structures of Ulp/SENP proteases bound to Smt3p/SUMO representing different steps in Smt3p/SUMO processing and deconjugation (for examples, see references 27; 28; 29), the human SAE1–SAE2 E1 complexed with SUMO-1 at the adenylation active site,30 and several structures of Ubc9 bound to the ϕ-K-X-D/E-sequence-containing target RanGAP-1, including SUMO-1-modified RanGAP-1 in complex with Ubc9 and Nup358/RanBP2.26; 31; 32 SUMO recognition by SBMs was revealed by this structure, and also by structures of SUMO-bound to thymine DNA glycosylase, and by the structure of a SUMO–PIASX peptide complex.21; 26; 33 Further insights into SUMO conjugation can been inferred from structures of complexes from the ubiquitin or NEDD8 pathways that represent interactions common among Ubl cascades. Nonetheless, an intriguing complex remains less characterized: SUMO and its E2, Ubc9, bind each other, noncovalently, with high affinity.13; 34; 35 The functions of noncovalent SUMO–Ubc9 interactions remain poorly understood. To gain high-resolution insights into this complex, and to understand the potential for SUMO and Ubc9 in the noncovalent complex to interact with their other binding partners in the cascade, we determined the structure of a noncovalent Smt3p–Ubc9p complex from S. cerevisiae.
Protein expression, purification, crystallization, data collection, and structure determination
Our cloning, expression, and purification of Smt3p, Ubc9p, and Smt3p's heterodimeric E1 complex (Aos1p–Uba2p) have been described previously,13 although additions to protocols for proteins in the crystallized complex are described here. For the crystallized complex, Smt3p and Ubc9p were generated by coexpression in BL21(DE3), from the pRSF-Duet vector, where a thrombin cleavage site-containing linker sequence (DPLVPRGS) was inserted between the His6SQ-tag and Smt3p's residue 13 (residue 13 was based on previous findings that residues 1-13 are disordered, and that Smt3p starting at residue 13 or 20 is functional14; 36), and full-length Ubc9p was expressed untagged. The complex was purified by Ni2+-affinity chromatography (His-Select resin, Sigma) and by gel filtration chromatography (SD200 column, GE Healthcare) in 25 mM Tris-Cl, 150 mM NaCl, 2 mM β-mercaptoethanol, pH 7.6, and concentrated to 28 mg/ml by ultrafiltration (Amicon). Crystals were obtained only for the uncleaved His6SQ/thrombin site linker/Smt3p–Ubc9p complex, by the hanging drop vapor diffusion method at 4°C in 18% PEG 3350, 0.2M MgCl2, 0.1 Bis-Tris pH 5.5. Crystals were cryoprotected in mother liquor supplemented with 4% PEG 3350 and 30% glycerol prior to flash-freezing in liquid nitrogen and data collection. The structure was refined to a resolution of 1.9 Å (Table 1, Fig. 1). In an unexpected coincidence, the thrombin cleavage site-containing linker sequence loosely mimics an SBM sequence. Crystal packing explains the requirement for the uncleaved thrombin-site linker for forming these crystals: the linker from one Smt3p forms a β-strand that docks in the groove from the adjacent symmetry-related Smt3p, in a manner resembling SBM binding to human SUMO family members.21; 26; 33 The model contains two complexes per asymmetric unit. One contains all residues except the His6SQ-tag, Smt3p residues 96-98, and Ubc9 residues 1-2 and 157, which were not visible in the electron density. These and Smt3p residues 15-18 and 95 are not visible in the other complex.
Table 1.
Crystallographic Data and Refinement Statistics
| Data Collection | |
| Wavelength (λ) | 0.97625 |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 120.89, 84.58, 80.14 |
| α, β, γ (°) | 90, 124.31, 90 |
| Resolution (Å) | 50.0−1.87 (1.94−1.87) |
| Total reflections | 1,491,311 |
| Unique reflections | 52503 |
| Overall Rsym (%) | 7.0 (19.9) |
| Overall I/σI | 56.0 (10.1) |
| Completeness (%) | 96.3 (78.5) |
| Mean Redundancy | 6.5 (5.3) |
| Refinement | |
| Rwork/Rfree | 0.225/0.250 |
| Number of atoms | |
| Protein | 3919 |
| Water | 386 |
| B-factors | |
| Protein | 30.4 |
| Water | 34.1 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.642 |
| Ramachandran Plot Statistics | |
| Residues in most favored regions | 92.6 % |
| Residues in additional allowed regions | 6.9 % |
| Residues in generously allowed regions | 0.5 % |
| Residues in disallowed regions | 0 |
Highest resolution shell is shown in parenthesis. Rwork = Σ|Fo−Fc|/ΣFo. Rfree is the cross-validation of R-factor, with 5% of the total reflections omitted in model refinement.
Figure 1.
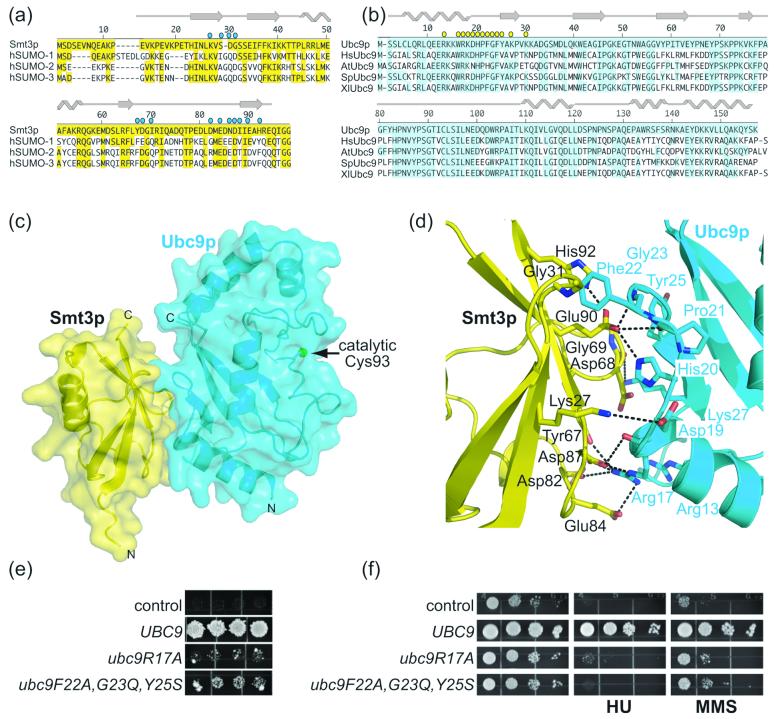
Structure of a Smt3p–Ubc9p complex. a) Sequence alignment of Smt3p from S. cerevisiae and human SUMO-1, SUMO-2, and SUMO-3, with secondary structure elements indicated above, and residues identical to Smt3p highlighted in yellow. Residues in Smt3p that contact Ubc9p are denoted with cyan circles. b) Sequence alignment of Ubc9p from S. cerevisiae with human, A. thaliana, S. pombe, and X. laevis Ubc9. Secondary structure elements are indicated above, and residues identical to S. cerevisiae Ubc9p are highlighted in cyan. Residues in Ubc9p that contact Smt3p are denoted with yellow circles. c) Overall structure of the complex, with Smt3p in yellow and Ubc9p in cyan. Secondary structures are shown overlaid with a semi-transparent surface. Ubc9p's catalytic cysteine (Cys 93) is represented by a green sphere. The crystals form in C2 with a = 120.89 Å, b = 84.58 Å, c = 80.14 Å, and β = 124.31, and two complexes per asymmetric unit. Data were collected using the mail-in program at the 22-BM (SER-CAT, Southeast Regional Collaborative Access Team) beamline at the Advanced Photon Source. Reflection data were indexed, integrated and scaled using HKL2000.50 Initial phases were obtained by molecular replacement using the coordinates of Ubc9p37 as a searchmodel in CNS.51 Electron density for Smt3p was readily visible in initial maps. The model was built manually using O,52 using the previous structures of Smt3p as guides,27; 36 and refined using CNS alternating with cycles of rebuilding relying on simulated-annealing omit and composite omit maps.51 The structure was refined from 50.0 Å to 1.9 Å. The final model has excellent geometry, with no Ramachandran outliers in disallowed regions. Statistics from data collection and refinement are given in Table 1. This and other figures representing structures were generated with Pymol.53 d) Close-up view of the interface between Smt3p and Ubc9p, oriented as in panel c. Smt3p is shown in yellow, with specific residues labeled in black. Ubc9p is shown in cyan, with specific residues labeled cyan. Oxygen atoms are colored red, and nitrogen atoms blue. Hydrogen bonds and salt bridges are represented with dashes. e) ubc9 bearing mutations in interface residues with Smt3p do not complement ubc9Δ cells. In a plasmid shuffle assay, ubc9Δ (PTY30 or PTY34) cells, transformed with a LEU2 vector containing an intron-less cDNA sequence for wild-type UBC9 or the indicated mutant allele expressed from the UBC9 promoter, were spotted onto selective medium supplemented with dextrose, as described previously.37; 54 Individual transformants were successively replica plated onto 5-FOA (US Biological) plates to cure cells of YCpUBC9·U and incubated at 26°C, 30°C, and 36°C.37 Similar results were obtained at all three temperatures, and representative results are shown for the experiment at 30°C. Plasmids were isolated from viable YCpUBC9·U-cured strains and sequenced to verify the identity of the ubc9 allele. f) ubc9 bearing mutations in interface residues with Smt3p do not restore resistance to genotoxic stress. The ubc9-10 mutant strain of S. cerevisiae, bearing a ubc9P123L conditional mutation, was shown previously to display increased sensitivity to a wide range of DNA damaging agents including hydroxyurea (HU) and methyl methane sulfonate (MMS).54 Exponentially growing cultures of ubc9-10 cells (A595=0.3) transformed with the indicated plasmids were serially 10-fold diluted and 5 μl aliquots were spotted onto the appropriate selective media supplemented with dextrose and incubated at 36°C. To assay cell sensitivity to HU or MMS, plates were supplemented with 5 mg/ml HU or 0.0125% MMS, respectively.37
In order to precisely characterize the association state of a Smt3p and Ubc9p mixture, we performed analytical ultracentrifugation analysis. Sedimentation equilibrium and velocity experiments provide experimentally determined molecular weights of 29.75 and 29.88 kDa, respectively, which agree well with the calculated molecular weight of 30.08 kDa for a 1:1 Ubc9p–Smt3p complex (Supplementary Fig. 1). Therefore, as both complexes in the asymmetric unit superimpose well (rmsd 0.738 Å), only one is discussed below.
Overall structure and Smt3p–Ubc9p interactions
Smt3p residues 21-94 adopt the canonical, globular ubiquitin-like β-grasp fold, with a twisted β-sheet forming one face, and an α-helix forming the other, resembling the previous solution structure of free Smt3p (1.572 Å rmsd)36 and the previous crystal structure of Smt3p in complex with the processing/deconjugating enzyme Ulp1p (0.658 Å rmsd) (Fig. 1).27 Ubc9p from the complex superimposes well with the previous crystal structure of free Ubc9p (0.406 Å rmsd),37 adopting the canonical E2 domain fold comprising a central anti-parallel 4-stranded β-sheet surrounded by 4 α-helices (Fig. 1).
Overall, the Smt3p–Ubc9p complex adopts a “V” shape (Fig. 1c). Smt3p contributes one side of the “V”, with Ubc9p comprising the base and the other side. A broad, concave surface involving Smt3p's conserved β-sheet cradles a complementary, relatively localized surface from Ubc9p's N-terminal α-helix and β-strand (Fig. 1c). This N-terminal Ubc9p face is completely separate from the surface containing the catalytic Cys93: Cys93 is on the opposite side, and >25 Å away (Fig. 1c). In total, Smt3p–Ubc9p interactions bury ∼1400 Å2. Several observations suggest that these noncovalent interactions are conserved between Ubc9 and SUMO from other organisms. Many interface side chains are conserved in SUMO and Ubc9 family members (Fig. 1a, b). Also, NMR studies of human SUMO-1–Ubc9 and SUMO-3–Ubc9 complexes revealed the greatest chemical shift perturbations upon complex formation for Ubc9 resonances from the N-terminal helix and strand, and from the SUMO β-sheet.34; 38; 39 Although an NMR-based docking model suggested that these regions of Ubc9 and SUMO-3 undergo significant structural rearrangement as compared to existing crystal structures,39 we find no evidence of such rearrangement in the corresponding proteins from S. cerevisiae. High-resolution insights into a corresponding complex between human orthologs is described by Lima and colleagues.
The Smt3p–Ubc9p complex is stabilized by a combination of hydrophobic and electrostatic elements. The interface is anchored by hydrophobic contacts between Ubc9p's His20, Phe22, Gly23, and Tyr25 associating with Smt3p's Asp30, Gly31, Asp68 and Gly69 backbone carbons, Glu90 side-chain carbons, and Ile88 and His92 side-chains (Fig. 1d). The complex is further secured by a network of electrostatic interactions. A basic patch from Ubc9p's Arg13, Arg17, Lys27, and Lys30 side-chains interacts with an acidic patch involving Smt3p's Tyr67 side-chain hydroxyl group, Asp68, Asp82, Glu84 and Asp87. Much of this electrostatic network is water-mediated (not shown), but there are also many direct interactions, including between side-chains from Ubc9p's Arg17 and Smt3p's Asp82 and Asp87 (Fig. 1d). There are additional electrostatic contacts between Ubc9's Asp19 and Smt3p's Lys27 side-chains, and also between Smt3p's Glu90 side-chain oxygens with Ubc9p's His20 side-chain and Gly23 backbone nitrogens (Fig. 1d).
The relevance of the structurally observed contacts is confirmed by a previous mutational analysis of Ubc9p, where Ubc9p surface residues were swapped with sequences found on ubiquitin E2s, with the goal of identifying protein binding sites without perturbing the E2-fold structure.13 In agreement with key roles indicated by our Smt3p–Ubc9p structure, an R17A mutant Ubc9p, and a triple mutant version of Ubc9p harboring F22A, G23Q, and Y25S substitutions, showed significant defects in noncovalent interactions with Smt3p.13 These residues are contained within a 38-residue N-terminal stretch of Ubc9p's sequence we found previously to be important for function in vivo.37 Indeed, we find that neither the R17A nor the F22A/G23Q/Y25S mutants were able to complement the viability defect in ubc9Δ cells (Fig. 1e), or the temperature-sensitivity of the ubc9-10 strain of S. cerevisiae in the presence of the genotoxic agents hydroxyurea (HU) or methyl methane sulfonate (MMS) (Fig. 1f).
Comparison of noncovalent and covalent complexes between Smt3p and Ubc9p
The structure reveals that interactions observed in the noncovalent Smt3p–Ubc9p complex are distinct from those in the covalent Ubc9p∼Smt3p thioester-linked conjugate. The large distance between Ubc9's noncovalent Smt3p binding site and its catalytic cysteine, which is the site of the thioester linkage with Smt3p's C-terminus in the conjugation intermediate, would preclude a thioester-bound Smt3p from simultaneously occupying the noncovalent binding site of the same molecule of Ubc9p.
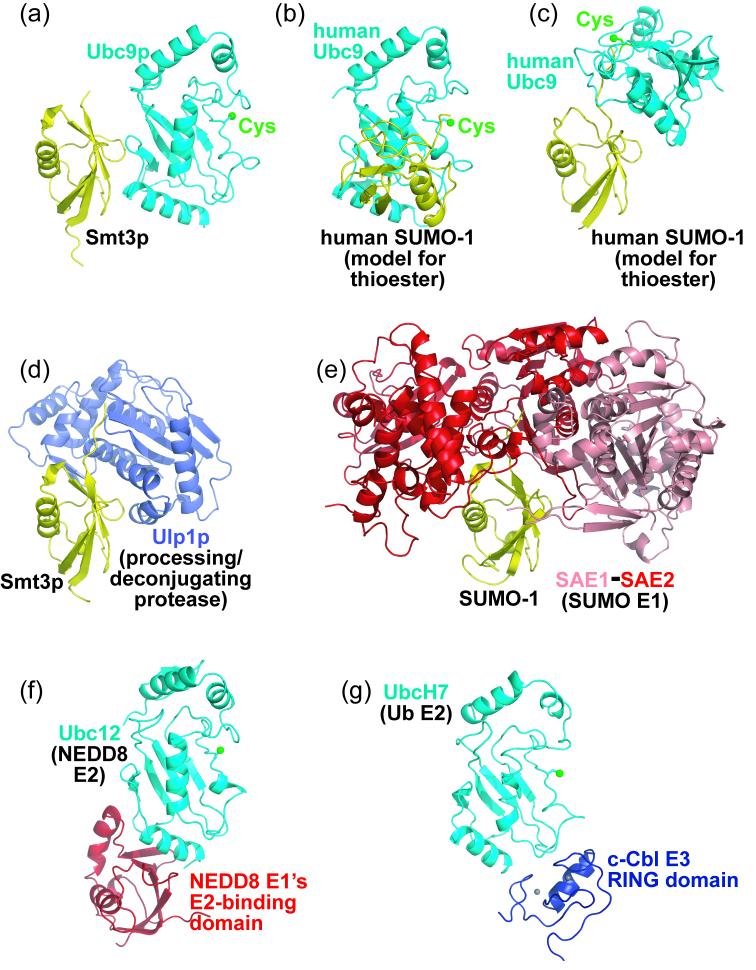
Despite the topological incompatibility of a single molecule of Smt3p interacting at both sites with a single molecule of Ubc9p, comparison of our noncovalent Smt3p– Ubc9p structure with a structural analog of Ubc9∼SUMO reveals that in the thioester intermediate, the noncovalent interaction surfaces are exposed in both Ubc9 and SUMO (Fig. 2a-c). A model for covalent interactions between SUMO and Ubc9's catalytic Cys93 face comes from the structure of the human SUMO-1∼RanGAP1–Ubc9– Nup358/RanBP2 complex.26 Although this contains a trapped-product complex where SUMO-1 is conjugated to a lysine from RanGAP1, the SUMO-1 C-terminus is adjacent to Ubc9's catalytic Cys93, suggesting a good structural model for a covalent Ubc9∼SUMO thioester complex,26 in which interactions primarily involve Ubc9's catalytic Cys93 face and SUMO's C-terminal tail and β1β2-loop (Fig. 2b-c), which do not overlap the noncovalent Smt3p–Ubc9p binding sites. Thus, it is possible that higher-order complexes could form, where a single Smt3p covalently linked to one Ubc9p's catalytic cysteine also binds noncovalently to another molecule of Ubc9p. Such higher-order complexes (Smt3p∼Ubc9p–Smt3p∼Ubc9p and so on in S. cerevisiae, SUMO∼Ubc9–SUMO∼Ubc9 in higher eukaryotes) could also interact with targets. Indeed, an analogous higher order complex has been found between ubiquitin and the ubiquitin E2, UbcH5: ubiquitin bound covalently via a thioester at the active site of one UbcH5 is also bound noncovalently to the “backside” of another UbcH5.40 In the case of ubiquitin and UbcH5, gel filtration analysis was used to confirm the nature of these interactions, where a higher-order assembly of the wild-type UbcH5∼ubiquitin thioester elutes earlier than the corresponding complex containing a UbcH5 mutant defective in noncovalent binding to ubiquitin.40 We performed such an analysis for the Smt3p pathway, taking advantage of the previously described F22A/G23Q/Y25S triple mutant version of Ubc9p, which does not bind noncovalently to Smt3p, but which maintains wild-type ability to form a covalent thioester complex with Smt3p.13 The F22A/G23Q/Y25S-Ubc9p∼Smt3p thioester complex elutes from the gel filtration as a compact, symmetric peak, consistent with one molecule of each F22A/G23Q/Y25SUbc9p and Smt3p in the conjugated complex (Supplementary Fig. 2). However, as with the higher-order UbcH5∼ubiquitin complexes,40 the wild-type Ubc9p∼Smt3p thioester complex elutes earlier, as a broad, asymmetric peak (Supplementary Fig. 2). These results are consistent with formation of larger molecular weight, higher order complexes.
Figure 2.
Recognition of common and distinct structural elements from Smt3p, Ubc9p, and their family members, by their multiple protein partners, with structural alignments performed by least-squares fitting in O.52 a) Ribbon diagram representation of the structure of the noncovalent Smt3p (yellow)–Ubc9p (cyan) complex. Ubc9p's catalytic Cys is shown as a green sphere. b) Model of a structure of human SUMO-1 (yellow)∼Ubc9 (cyan) covalent thioester intermediate, where SUMO-1's C-terminus would be thioester-linked to Ubc9's catalytic Cys. The model is based on the structure of a SUMO-1∼RanGAP1–Ubc9–Nup358/RanBP2 complex, where SUMO-1 is covalently linked to RanGAP1's target lysine, adjacent to Ubc9's catalytic Cys93.26 This structure is oriented with human Ubc9 in the same position as S. cerevisiae Ubc9p in panel a, after superposition. c) Model from (b), oriented with human SUMO-1 in the same position as S. cerevisiae Smt3p in panel a, after superposition. d) Structure of the Smt3p (yellow)–Ulp1p (slate) complex showing Smt3p recognition by the processing and deconjugating protease Ulp1p.27 This structure is oriented with Smt3p in the same position as in panel a, after superposition of Smt3p from both complexes. e) Structure of the human SUMO-1 (yellow)–SAE1 (pink)–SAE2 (red) complex showing SUMO-1 recognition by the heterodimeric SAE1–SAE2 E1 complex.30 This structure is oriented with SUMO-1 in the same position as Smt3p in panel a, after superposition of SUMO-1 from the complex with SAE1–SAE2 onto the structure of Smt3p from Smt3p–Ubc9p. f) Structure showing recognition of NEDD8's E2 by a domain conserved among E1s.41; 42 This structure is oriented with Ubc12 in the same position as Ubc9p in panel a, after superposition of Ubc12 from the complex with NEDD8's E1 onto the structure of Ubc9p from Smt3p–Ubc9p. g) Structure of the human UbcH7 (cyan)–c-Cbl ubiquitin E3 RING domain (blue) showing E2 recognition by a domain with sequence similarity conserved among a subset of E3s in the ubiquitin and SUMO pathways.44 This structure is oriented with UbcH7 in the same position as Ubc9p in panel a, after superposition of UbcH7 from the complex with the ubiquitin E3 c-Cbl onto the structure of Ubc9p from Smt3p–Ubc9p.
Comparison to other Smt3p/SUMO and Ubc9p/Ubc9/E2 interactions involved in conjugation
Smt3p and Ubc9p interact with numerous other proteins in the SUMO conjugation cascade. We sought to understand which other proteins and complexes in the pathway might also involve Smt3p–Ubc9p noncovalent interactions, and which assemblies cannot. Comparison with existing structures shows that residues at the Smt3p–Ubc9p interface also play critical roles in forming many other Smt3p and Ubc9p complexes with roles early in the cascade. During Smt3p conjugation, Smt3p is first processed by a Ulp protease, and then Smt3p is activated by Aos1p–Uba2p, Smt3p's dedicated heterodimeric E1 enzyme. The noncovalent Ubc9p binding site overlaps with 44% of the Smt3p surface that interacts with the Ulp1p protease (Fig. 2a, d). Although there is no structure of Smt3p in complex with Aos1p–Uba2p, a high degree of sequence identity suggests that this will resemble the corresponding structure containing human SUMO-1 (45% identical to Smt3p) bound to the human SUMO E1 (the heterodimeric SAE1–SAE2 complex, 36% identical to Aos1p–Uba2p).30 50% of Smt3p's Ubc9 binding surface overlaps the corresponding SAE1–SAE2 binding surface of human SUMO-1 (Fig. 2a, e). Together, Ulp1 and E1 bind the key Smt3p residues involved in the noncovalent interaction with Ubc9p.
During Smt3p activation, the Aos1p–Uba2p E1 also interacts with Ubc9p, to generate the covalent Smt3p∼Ubc9p thioester-linked intermediate, which is distinct from the noncovalent Smt3p–Ubc9p complex. Although there is no structure of Ubc9p in complex with Aos1p–Uba2p, structures of E1–E2 complexes are available from the pathway for the Ubl NEDD8,41; 42 and the E2-binding domains from the SUMO/Smt3p and NEDD8 E1s share related folds.30; 43 The structures show N-terminal helix residues from NEDD8's E2 (Ubc12) binding to the E1.41; 42 Some of these E1-binding residues from Ubc12 correspond to Smt3p-binding residues from Ubc9p (Fig. 2a, f). This suggests that Smt3p and Aos1p–Uba2p bind partially overlapping surfaces on Ubc9p. Indeed, a previous study showed that mutation of Ubc9p's N-terminal helix residue Arg17 reduces binding to both Smt3p and Aos1p–Uba2p.13 Interestingly, there may be subtle species-specific differences in SUMO E1-Ubc9p interactions, because mutation of basic residues, including Arg17, from human Ubc9's N-terminal helix reduces binding to SUMO-1 without significantly affecting binding to SAE1-SAE2,35 and Aos1p-Uba2p charges human Ubc9 very poorly37. To further probe whether S. cerevisiae Smt3p and Aos1p–Uba2p bind overlapping, but non-identical surfaces on Ubc9p, we used an in vitro gel mobility shift binding assay. We found previously that in such an assay, titration of excess free Smt3p leads to displacement of Ubc9p from an Aos1p–Uba2p–Ubc9p complex into a Smt3p–Ubc9p complex.13 To test whether this comes from Smt3p and Aos1p–Uba2p competing to bind overlapping sites on Ubc9p, we used the F22A/G23Q/Y25S triple mutant version of Ubc9p. Phe22, Gly23, and Tyr25 are all located at the Smt3p–Ubc9p interface, but are distal from Ubc9p's N-terminal helix implicated in binding to both Smt3p and Aos1p–Uba2p. Therefore, if Smt3p and Aos1p–Uba2p compete for binding to Ubc9p's N-terminal helix, then the F22A/G23Q/Y25S triple mutant version of Ubc9p, which does not bind noncovalently to Smt3p, would be able to bind Aos1p–Uba2p equally well in the absence or presence of Smt3p. In competition experiments, we found that unlike the case with wild-type Ubc9p, excess free Smt3p is unable to displace the F22A/G23Q/Y25S triple mutant Ubc9p from the complex with Aos1p–Uba2p (Supplementary Fig. 3). These results support the notion that parts of the Smt3p and Aos1–Uba2p binding surfaces on Ubc9p overlap.
Our structure suggests that noncovalent Smt3p–Ubc9p interactions may coexist in some complexes involved in latter steps conjugation cascades. During conjugation, following formation of the covalent Ubc9p∼Smt3p thioester intermediate, Ubc9p can interact with an E3, which promotes Smt3p transfer from Ubc9p's catalytic Cys93 to the target's lysine. All presently characterized S. cerevisiae Smt3p E3s (Siz1p, Siz2p, Mms21p, and Zip3p) are members of the SP-RING family.7 Although there are no reported structures of any Ubc9–SP-RING E3 complexes, there are related structures of E2–RING E3 complexes from the ubiquitin pathway.44; 45 There is no structural overlap upon superposition of Ubc9p–Smt3p onto structures of ubiquitin E2s in complex with RING and RING-related domains (Fig. 2g). By contrast, many human Ubc9 residues involved in binding the mammalian E3 Nup358/RanBP226 overlap with the corresponding noncovalent Smt3p-binding site on S. cerevisiae Smt3p. Thus, there may be variation in the abilities of different E3s to interact with a noncovalent SUMO–Ubc9 complex.
Insights into ternary SBM (SUMO Binding Motif)–Smt3p–Ubc9p interactions
Following conjugation, SUMO binds specific recognition machineries, resulting in altered function of the SUMOylated target. Structural insights into interactions of Smt3p with endogenous recognition machineries remain poorly characterized, although detailed binding and structural studies are available for some human SBM–SUMO complexes.21; 26; 33 Like human SUMO-1, Smt3p was found to bind an SBM peptide derived from human PIASX with high affinity.21 Our structure suggests that the noncovalent Smt3p–Ubc9p complex can interact with Smt3p recognition machineries, and provides insight into how Smt3p recruits its S. cerevisiae protein partners like mammalian SUMOs recruit SBMs. Interestingly, a portion of the uncleaved thrombin-site linker sequence (DPLVPRGS) inserted between the His6SQ-tag and Smt3p loosely resembles the ϕ–X-ϕ-ϕ sequence of an SBM, with Leu, Pro, and the substantial aliphatic portion of an arginine side-chain all hydrophobic. This engineered sequence attached to one Smt3p binds the symmetry-related Smt3p in the crystal lattice, in a manner resembling SBM binding to human SUMO family members (Fig. 3a-e). The Leu and Pro, and the hydrophobic portion of the Arg side-chain, bind in a conserved hydrophobic groove between Smt3p's β2-strand α-helix comprising Smt3p's Ile35, Phe36, Phe37, Ile39, Arg47, Leu48, Ile51, and Arg55. The hydrophobic regions of corresponding side-chains in mammalian SUMO family members also play critical roles in recruiting bonafide mammalian SBMs (Fig. 3a-e).20; 21; 22; 26; 33
Figure 3.

Structure of a SUMO binding motif mimic bound to Smt3p–Ubc9p: structural conservation of noncovalent Ubiquitin-like protein–E2 complexes as platforms for interactions within Ubl pathways. a) Two complexes as in the crystal, where Smt3p (yellow)–Ubc9p (cyan) and Smt3p' (magenta)–Ubc9p' (blue) are related by crystallographic C2 symmetry, showing the uncleaved thrombin-site linker sequence from the adjacent, crystallographic symmetry-related Smt3p' packing in a groove in Smt3p. The linker from Smt3p extends five additional residues beyond the *. b) Structure of a SUMO-binding motif mimic bound to Smt3p–Ubc9p. A portion of the linker from the adjacent, crystallographic symmetry-related Smt3p' is shown in magenta sticks, as it interacts with the Smt3p (yellow)–Ubc9p (cyan) complex. The N- and C-terminal regions of the displayed portion of the linker are labeled to indicate directionality of the peptide-like interaction with Smt3p. Oxygen atoms are colored red, nitrogen atoms blue, and Ubc9p's catalytic Cys93 is marked with a green sphere. Hydrogen bonds and salt bridges are represented with dashes. c) Structures of human SUMO-1 (yellow) in complex with the SUMO-binding motif (SBM) regions (magenta) from Nup358/RanBP2,26 thymine DNA glycosylase,33 and PIASX21 are shown from left to right, oriented after superposition of SUMO-1 with Smt3p as in panel b. The N- and C-terminal regions of the displayed peptide-like regions of Nup358/RanBP2 and thymine DNA glycosylase, and the peptide from PIASX, to indicate directionality of the polypeptide interaction with SUMO-1. Oxygen atoms are colored red, and nitrogen atoms blue. Hydrogen bonds and salt bridges are represented with dashes. d) Close-up view of interactions between Smt3p and the uncleaved thrombin-site linker sequence from the adjacent, crystallographic symmetry-related Smt3p'. Smt3p is shown in yellow with black labels, and the SBM mimicking linker in magenta. Oxygen atoms are colored red, and nitrogen atoms blue. Hydrogen bonds are represented with dashes. e) Structure-based sequence alignments of the SUMO/Smt3p-binding sequences from Nup358/RanBP2, thymine DNA glycosylase (TDG), PIASX, and the SUMO-binding motif mimic from the uncleaved thrombin-site linker sequence upstream of Smt3p' residues used for crystallization (linker). Residues mediating key hydrophobic interactions are boxed. f) Structure of the human ubiquitin (yellow)–UbcH5 (cyan) noncovalent complex, showing noncovalent interactions between ubiquitin and a ubiquitin E2 involved in BRCA1-mediated polyubiquitin chain assembly.40 This complex is oriented with UbcH5 in the same position as Ubc9p in panel b, after superposition of UbcH5 from the complex with ubiquitin onto the structure of Ubc9p from Smt3p–Ubc9p. g) Structure of the human ubiquitin (yellow)–MMS2 (cyan) complex, showing noncovalent interactions between ubiquitin and a noncatalytic E2 variant involved in Lys63-linked polyubiquitin chain assembly.40 This complex is oriented with MMS2 in the same position as Ubc9p in panel b, after superposition of MMS2 from the complex with ubiquitin onto the structure of Ubc9p from Smt3p–Ubc9p.
Just as the three structurally characterized mammalian SBMs make unique contacts with their SUMO partners, there are distinguishing features of the linker–Smt3p interface (Fig. 3c-e). The proline in the linker sequence induces a kink in the strand not found in the existing SBM-SUMO structures. As a result, the proline backbone oxygen is able to form a hydrogen bond with Smt3p's Arg55. Critical electrostatic interactions are also mediated by the corresponding Arg conserved in human SUMO family members, but in some cases with SBM acidic side-chains in structural locations corresponding to the proline oxygen.21; 26; 33 Also, Smt3p's Ser33 side-chain forms a hydrogen bond with the side-chain from the Glu corresponding to the adjacent Smt3p's residue 13.
Additionally, Smt3p's Asn25 forms a hydrogen bond with the linker Arg side-chain (Fig. 3d). Such interactions might occur with human SUMO-2 and -3, which also contain an Asn at this position, but would likely be destabilized by the corresponding lysine in the sequence of SUMO-1 (Fig. 1a).
Conservation of noncovalent Ubl–E2 interactions
The Smt3p–Ubc9p structure globally resembles two recent high-resolution structures of noncovalent complexes for ubiquitin with (i) an active ubiquitin E2 (UbcH5),40 and (ii) a catalytically-inactive Ubiquitin E2 Variant protein (MMS2)46 (Fig. 3b,f, g). All three noncovalent E2 (or UEV)–ubiquitin-like protein complexes use parallel interaction surfaces (Fig. 3b,f, g). The functions of ubiquitin–E2 noncovalent interactions are better characterized, with clear roles in multiprotein complex formation. Higher-order (UbcH5∼ubiquitin-UbcH5∼ubiquitin)x self-assembled thioester complexes have been implicated in BRCA1 E3-mediated processive polyubiquitin chain assembly.40 Ubiquitin binding to the ubiquitin E2-variant (UEV) protein, MMS2, also plays a role in assembly of a complex for polyubiquitin chain assembly. UEV domains are structural homologs of E2s, but they lack the active site cysteine and C-terminal helices of bone fide E2 conjugating enzymes. MMS2 recruits ubiquitin, noncovalently, and directs ubiquitin's Lys63 toward the active site of the E2, Ubc13, to establish linkage-specificity of MMS2–Ubc13-catalyzed polyubiquitin chain assembly.46; 47; 48; 49 Although the mechanisms underlying polymeric SUMO chain formation, and the functions of polymeric SUMO chains themselves, remain poorly characterized, structural similarity with complexes involved in polyubiquitin chain assembly raise the possibility that the noncovalent Smt3p–Ubc9p complex will eventually be found to play a role in polymeric SUMO chain formation.
Our structure of the noncovalent Smt3p–Ubc9p complex, the structure of the human SUMO-1–Ubc9 complex described by Lima and colleagues, and the previous ubiquitin–E2/UEV structures40; 46 reveal conservation of noncovalent Ubl–E2 interactions. The striking structural similarity - for different Ubls, for divergent E2/E2-like proteins, and across phyla - implies important functions for these complexes. In the ubiquitin pathway, noncovalent ubiquitin–E2/UEV complexes interact with multiple additional partners, as parts of large assemblies involved in polyubiquitin chain formation. Our serendipitous finding of an SBM mimic also bound to Smt3p–Ubc9p suggests that the noncovalent Smt3p–Ubc9p complex (and mammalian SUMO–Ubc9) exists as a part of larger assemblies, linked to other proteins or complexes by simultaneous SBM–Smt3p and Smt3p–Ubc9p interactions. We anticipate that future studies will reveal how larger assemblies involving SBM–Smt3p–Ubc9p interactions fit into the broader context of the Smt3p cascade, to yield the precise post-translational regulation that make this pathway essential.
Supplementary Material
Acknowledgements
We thank K. Bencsath for initiating our studies of the noncovalent Smt3p–Ubc9p complex, D. Huang for the crystallized Smt3p–Ubc9p coexpression construct, and C. Ross for computer support. We are grateful to J. Chrzas and Z. Jin for performing mail-in data collection at the SER-CAT (22-BM) beamline at the Advanced Photon Source (APS). Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the American Lebanese Syrian Associated Charities (ALSAC), the National Institutes of Health (P30CA21765 for St. Jude Cancer Center support, R01GM069530 to BAS, R01CA111542 to MAB), a Beckman Young Investigator Award to BAS, an American Cancer Society Postdoctoral Fellowship to DMD, and the Howard Hughes Medical Institute. BAS is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein Data Bank accession numbers
Coordinates and structure factors have been deposited in the RCSB Protein Data Bank under the filenames 2EKE.
References
- 1.Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28:321–8. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 2.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–41. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Dohmen RJ. SUMO protein modification. Biochim Biophys Acta. 2004;1695:113–31. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 8.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–86. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 9.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci U S A. 1998;95:560–4. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–9. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 12.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–74. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 13.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem. 2002;277:47938–45. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 14.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–20. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–44. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 16.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–8. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi Y, Kahyo T, Toh EA, Yasuda H, Kikuchi Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem. 2001;276:48973–7. doi: 10.1074/jbc.M109295200. [DOI] [PubMed] [Google Scholar]
- 18.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–20. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 19.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–37. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 20.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–8. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–9. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 22.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–27. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 23.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. Embo J. 2002;21:1456–64. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc Natl Acad Sci U S A. 1997;94:3736–41. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–76. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, Chau SF, Lam KH, Chan HY, Ng TB, Au SW. Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease. Biochem J. 2006;398:345–52. doi: 10.1042/BJ20060526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol. 2006;13:1060–8. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- 30.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. Embo J. 2005;24:439–51. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 32.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–9. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 33.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–82. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, Jin C, Liao X, Shen Z, Chen DJ, Chen Y. The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1) J Biol Chem. 1999;274:16979–87. doi: 10.1074/jbc.274.24.16979. [DOI] [PubMed] [Google Scholar]
- 35.Tatham MH, Kim S, Yu B, Jaffray E, Song J, Zheng J, Rodriguez MS, Hay RT, Chen Y. Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry. 2003;42:9959–69. doi: 10.1021/bi0345283. [DOI] [PubMed] [Google Scholar]
- 36.Sheng W, Liao X. Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions. Protein Sci. 2002;11:1482–91. doi: 10.1110/ps.0201602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Waardenburg RC, Duda DM, Lancaster CS, Schulman BA, Bjornsti MA. Distinct functional domains of Ubc9 dictate cell survival and resistance to genotoxic stress. Mol Cell Biol. 2006;26:4958–69. doi: 10.1128/MCB.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding H, Xu Y, Chen Q, Dai H, Tang Y, Wu J, Shi Y. Solution structure of human SUMO-3 C47S and its binding surface for Ubc9. Biochemistry. 2005;44:2790–9. doi: 10.1021/bi0477586. [DOI] [PubMed] [Google Scholar]
- 39.Ding H, Yang Y, Zhang J, Wu J, Liu H, Shi Y. Structural basis for SUMO-E2 interaction revealed by a complex model using docking approach in combination with NMR data. Proteins. 2005;61:1050–8. doi: 10.1002/prot.20695. [DOI] [PubMed] [Google Scholar]
- 40.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol Cell. 2005;17:341–50. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 42.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445:394–98. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–4. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 44.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 46.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 47.McKenna S, Hu J, Moraes T, Xiao W, Ellison MJ, Spyracopoulos L. Energetics and specificity of interactions within Ub.Uev.Ubc13 human ubiquitin conjugation complexes. Biochemistry. 2003;42:7922–30. doi: 10.1021/bi034480t. [DOI] [PubMed] [Google Scholar]
- 48.McKenna S, Moraes T, Pastushok L, Ptak C, Xiao W, Spyracopoulos L, Ellison MJ. An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2.Ubc13. The structural basis for lysine 63 chain catalysis. J Biol Chem. 2003;278:13151–8. doi: 10.1074/jbc.M212353200. [DOI] [PubMed] [Google Scholar]
- 49.McKenna S, Spyracopoulos L, Moraes T, Pastushok L, Ptak C, Xiao W, Ellison MJ. Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J Biol Chem. 2001;276:40120–6. doi: 10.1074/jbc.M102858200. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 53.DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 54.Jacquiau HR, van Waardenburg RC, Reid RJ, Woo MH, Guo H, Johnson ES, Bjornsti MA. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J Biol Chem. 2005;280:23566–75. doi: 10.1074/jbc.M500947200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.