Short abstract
The recent sequencing of a large chunk of euchromatin from the human Y chromosome is a technical tour de force. It answers some evolutionary questions about this unusual chromosome while raising others.
Abstract
The recent sequencing of a large chunk of euchromatin from the human Y chromosome is a technical tour de force. It answers some evolutionary questions about this unusual chromosome while raising others.
The Y chromosome is probably the most bizarre part of the human genome, reflecting its unique status as a huge block of largely non-recombining DNA, maintained in a permanently heterozygous state, and transmitted solely through males [1]. Early in the history of genetics, it was realized that the pattern of sex-linked inheritance, seen in species with chromosomal sex determination, implies that the Y chromosome lacks most of the genes carried on its pairing partner, the X chromosome. HJ Muller inferred that the X and Y were originally homologous chromosomes, and that the Y had lost most of the genes that it once contained, in response to its lack of recombinational exchange with the X and its permanent heterozygosity [2].
Later work has largely validated this inference, although the details of the evolutionary mechanisms leading to Y chromosome degeneration are now thought to differ substantially from that proposed by Muller [3,4]. It is also now known that Y chromosomes have evolved independently in many different groups of animals and plants that have sex chromosomes; the Y chromosomes of mammals share a common origin with each other, but have nothing in common with their counterparts in birds or Drosophila [1,5]. In addition, Y chromosomes tend to be unusually rich in repetitive DNA, which is contributed both by transposable elements and by tandem arrays of satellite DNA sequences, as is often the case for regions of the genome that have low frequencies of crossing over [1]. Much of the human Y chromosome consists of heterochromatin, made up entirely of such repeats (Figure 1) [6].
Figure 1.
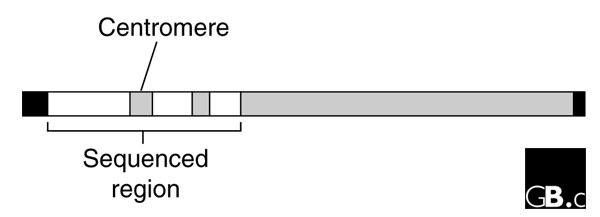
The structure of the human Y chromosome (features are shown approximately to scale). The black regions at each end of the Y chromosome are the pseudoautosomal regions, which cross over with homologous sequences on the X chromosome. The gray regions are heterochromatin. The white region contains the 23 megabase portion of the Y chromosome sequenced by Skaletsky et al. [7] (modified from Figure 1 of [7]).
The sequence of the human Y chromosome
The first complete sequence of a 23 megabase (Mb) euchromatic (non-heterochromatic) portion of a human Y chromosome, from a single male, has recently been published by David Page's group [7]. This is a considerable achievement, given the difficulties of sequencing chromosomes that are rich in repeats, as is the case even for the euchromatin of the human Y. The researchers exploited the fact that a single Y chromosome provided the material for sequencing: this allowed them to attribute slight sequence differences between similar tracts of DNA to within-chromosome rather than between-individual variation, and hence facilitated the identification of repeats.
The results provide a uniquely detailed picture of the organization of a Y chromosome - not yet available for other species - which confirms many previous findings but modifies others. The sequenced region is nearly the entire portion of the euchromatic Y chromosome that does not cross over with the X, together with some of the repeat-rich heterochromatic part of the Y chromosome. Skaletsky et al. [7] call this the 'male-specific Y' (MSY), because the small part of the Y that does cross over is essentially the same as the corresponding part of the X and is common to males and females. We now have a list of putative transcription units within the MSY that is considerably larger in number (158) than was previously thought, but this is still a small fraction of the number of the 1,000 or so on the 160 Mb of the X chromosome [6]. Many of the ones on the Y probably do not code for proteins.
The identifiable coding sequences among the 158 predicted genes on the Y chromosome can be divided into two categories. The first comprises 27 genes with clear signs of homology to genes on the X chromosome, betraying the common origin of these two chromosomes; 13 of these have degenerated into pseudogenes. The remaining 14 active Y-linked genes within this category tend to have a broad range of expression in many different tissues, with the notable exception of the male-determining gene, Sry, which is expressed early in development in the germ cells of males, and has an X-linked counterpart, Sox3. There is considerable variation among these 14 genes in the extent of their silent-site sequence divergence (Ks) [8] between the copies on the X and Y chromosomes, although it is not clear that they can be sharply divided into four 'evolutionary strata', representing discrete phases in the differentiation of the X and Y chromosomes, as was previously proposed for a subset of these genes [9].
There are two possible explanations for the range of differences in divergence: one is that the more highly diverged genes have been isolated from recombination with the X chromosome for longer than the less highly diverged genes; and the other is that there are different rates of genetic exchange via gene conversion between X and Y for different genes. The first hypothesis is consistent with the fact that genes with different Ks values tend to cluster together on the X chromosome in contiguous blocks, with Ks values increasing from the distal short arm to the distal long arm of the X. The order of these genes differs greatly between X and Y chromosomes, suggesting that there have been rearrangements involving chromosomal inversions that would have helped to suppress crossing-over between the evolving X and Y chromosomes. The evolutionary advantages to such suppression of crossing-over have long been discussed [1,3,10]. Sry and Sox3 have the highest Ks value, as would be predicted from such evolutionary considerations for descendants of a gene that must have been involved in early stages of the evolution of the Y chromosome, equivalent to a time of isolation of over 250 million years ago. The second hypothesis - that different genes have different rates of genetic exchange between X and Y - is supported by the fact that gene conversion between genes on the X and Y chromosomes has been detected in the cat family [11], and seems also to be occurring at a high rate within the human Y chromosome (discussed below), but this does not explain the ordering of Ks values along the X chromosome and the very ancient origin of several Y chromosome genes.
The second category of Y-linked genes consists of nine gene families that mostly have no resemblance to genes on the X chromosome. These are organized into repeats of two or more units, and show testis-specific expression. They seem to comprise genes whose functions are important for males but possibly deleterious for females; seven out of the nine have originated by transposition from an autosome, and two come from the X chromosome. It is interesting that a parallel case of a transposition of a gene from an autosome onto a Y chromosome has been found in the plant Silene latifolia; in this case the Y-linked copy is specifically expressed in male reproductive tissue [12]. These observations suggest that there may be an ongoing evolutionary process of acquisition of genes with male-specific functions by the Y chromosome; if higher levels of expression of these genes are advantageous for males but disadvantageous for females, this would be favored by selection [1,10].
Another unusual feature of the human Y chromosome is the presence of a 3.4 Mb tract of predominantly non-coding sequence derived from the X chromosome by a transposition event of some kind. The extent of sequence divergence from the corresponding region of the X chromosome suggests that the transposition occurred about 3-4 million years ago. An inversion of part of the short arm of the Y subsequently split the transposition into two non-contiguous blocks. Only two genes are present in this part of the Y chromosome [7].
Palindromes on the Y chromosome
A startling finding is that, with one exception, the Y chromosome repeats are organized as palindromes, with two very similar long sequences pointing in opposite directions, connected by a 'spacer' (Figure 2) [7]. A companion paper [13] shows that at least some of the palindromes are present in chimpanzees and gorillas, so their origin predates the split between humans and their closest relatives. There is very little sequence divergence between the arms of the palindromes, compared to the divergence between human and chimpanzee sequences. This strongly suggests that there is ongoing gene conversion between the arms, maintaining their sequence similarity. A simple population genetics argument yields an estimate of the rate of conversion as 2.2 × 10-4 per nucleotide per generation, comparable with values from other sources [13]. The MSY therefore experiences recombination, although of a type that does not affect its divergence from the X.
Figure 2.
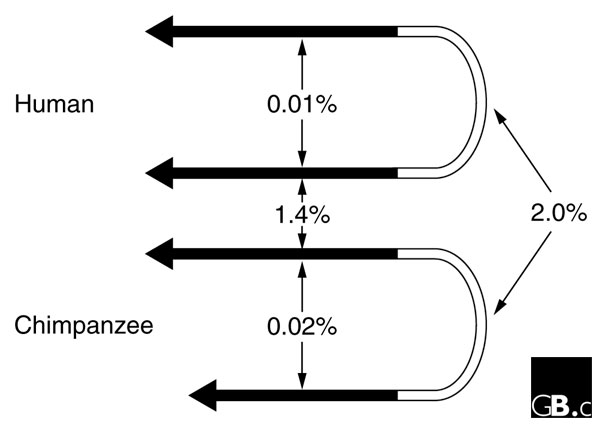
Portions of the Y chromosome palindrome P6, sequenced in both human and chimpanzee. The white region is the 'spacer' separating the arms of the palindrome (the spacer is about 46 kb in length). The black regions are the arms of the palindrome. The numbers indicate the percentage sequence divergence, either between the arms of the palindrome within a species, or between homologous regions in humans and chimpanzees. (Modified from Figure 1 of [13].)
A consequence, though probably not a cause (given that evolution has no foresight), of the existence of the palindromes is that ongoing gene conversion will inhibit the spread by genetic drift of a deleterious mutation through just one of the two arms, allowing selection to preserve the functionality of both copies [14]. It is possible that the unusual abundance of palindromes among functional duplications on the Y reflects the fact that unequal crossovers between sister chromatids cannot occur among palindromes without causing disruptive chromosome rearrangements; this will prevent their loss through the generation of single-copy units by unequal crossing over, as can happen with tandem duplications [15].
The organization of the human Y chromosome thus reflects a combination of several evolutionary processes: the degeneration of the bulk of the genes, which were originally common to the primeval X/Y chromosome pair; the accumulation of genes with male-specific functions by sporadic transpositions from the X chromosome and autosomes; and a unique recent transposition of a large piece of X chromosomal material. It is therefore probably premature to predict the ultimate demise of the human Y chromosome, as has recently been done [16]. Some species have indeed lost all trace of the Y chromosome, so that males are Xo, not XY [1]. But this can happen only if the male-determining function of the Y chromosome is abolished and is replaced by an X/autosome-balance sex-determination system of the kind found in Drosophila and Caenorhabditis, which is probably rather hard to pull off [17]. Together with the presence of several genes required for male fertility on the human Y chromosome, these considerations imply that human males are likely to be stuck with their bizarre genetic make-up for the foreseeable evolutionary future.
References
- Bull JJ. Evolution of Sex Determining Mechanisms. Menlo Park: Benjamin Cummings; 1983. [Google Scholar]
- Muller HJ. Genetic variability, twin hybrids and constant hybrids in a case of balanced lethal factors. Genetics. 1918;3:422–499. doi: 10.1093/genetics/3.5.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Evolution of the Y sex chromosome in animals. Biosciences. 1999;46:331–343. [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, Zend-Ajusch E, Shan Z, Grutzner F, Schartl M, Burt DW, Koehler M, Fowler VM, Goodwin G, Schneider WJ, et al. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re)view on avian sex determination. Cytogenet Cell Genet. 2000;89:67–78. doi: 10.1159/000015567. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntnikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Li W-H. Molecular Evolution. Sunderland: Sinauer; 1997. [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Slattery JP, Sanner-Wachter L, O'Brien SJ. Novel gene conversion between X-Y homologues located in the nonrecombining region of the Y chromosome in Felidae (Mammalia). Proc Natl Acad Sci USA. 2000;97:5307–5312. doi: 10.1073/pnas.97.10.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S, Isono E, Kejnovsky E, Vyskot B, Kawano S, Charlesworth D. Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol Biol Evol. 2003;20:1062–1069. doi: 10.1093/molbev/msg114. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marzalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Ohta T. The mutational load of a multigene family with uniform members. Genet Res. 1989;53:141–145. doi: 10.1017/s0016672300028020. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The rise and fall of SRY. Trends Genet. 2002;18:259–264. doi: 10.1016/s0168-9525(02)02666-5. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]


