Abstract
To detect and characterize autoreactive T cells in diabetes-prone NOD mice, we have developed a multimeric MHC reagent with high affinity for the BDC-2.5 T cell receptor, which is reactive against a pancreatic autoantigen. A distinct population of T cells is detected in NOD mice that recognizes the same MHC/peptide target. These T cells are positively selected in the thymus at a surprisingly high frequency and exported to the periphery. They are activated specifically in the pancreatic LNs, demonstrating an autoimmune specificity that recapitulates that of the BDC-2.5 cell. These phenomena are also observed in mouse lines that share with NOD the H-2g7 MHC haplotype but carry diabetes-resistance background genes. Thus, a susceptible haplotype at the MHC seems to be the only element required for the selection and emergence of autoreactive T cells, without requiring other diabetogenic loci from the NOD genome.
Introduction
Type 1 diabetes, or IDDM, results from the destruction of pancreatic islet β cells by a complex autoimmune process to which both genetic and environmental factors contribute (1–3). In humans, as in model NOD mice, genetic control is complex: the main genetic contribution to susceptibility resides in class II loci of the MHC, with particular sequences and structures in haplotypes that confer susceptibility in NOD and in diabetic patients (4–8). In mice, as in humans, a complex assortment of other susceptibility “background” genes that affect immune function also contribute to diabetes risk (3, 9, 10). IDDM is marked by the presence and activation of autoreactive T cells, whose origin remains a puzzle. It is not clear whether they result from a failure of central or of peripheral tolerance, and whether MHC susceptibility alleles or background genes play a determining role in their selection. It has been suggested, for example, that the particular MHC-II alleles of the NOD mouse, because they bind peptides with peculiar kinetics or specificity, may inefficiently affect negative selection of autoreactive cells (11–13). Others have suggested that the background genes of the NOD mouse lead to poor negative selection and central-tolerance induction (14, 15). Finally, it is not clear whether the disease in NOD mice depends on a central-tolerance defect that lets a large number of high-affinity cells escape, or on a peripheral incapacity to control their aggressiveness (15–17). Transgenic models have shown that, for the most part, T cells reactive against peripheral antigens are selected in the thymus much like other T cells are (18–21), without the imprints of negative selection that mark T cell populations reactive against antigens expressed in the thymus (22–25). Yet these analyses only yield examples of the fates of individual T cell receptors (TCRs), reflecting a single affinity for self. It would be desirable to track broader populations, but the analysis has suffered from the absence of tools that would allow, in nontransgenic animals, side-by-side comparison of T cell repertoires between susceptible and resistant individuals. The advent of MHC multimer reagents, with which TCRs of particular specificities can be detected, now offers this possibility (26, 27).
Several autoreactive T cell clones isolated from diabetic NOD mice have been shown to be pathogenic in transfer experiments (28–33). BDC-2.5 is the best characterized of these clones. BDC-2.5 was derived from a diabetic female NOD mouse and was shown to greatly accelerate disease when transferred to young animals (30–32). This CD4+ Th1 clone displays a Vβ4/Vα1 (AV1S5) TCR heterodimer (34). BDC-2.5 is restricted by Ag7, the lone MHC class II molecule of NOD mice (4, 5). The nature of the presented antigen is still unknown, although it has been shown to be associated with the membrane fraction of β granules (35). Mice transgenic for the BDC-2.5 TCR show a robust positive selection of CD4+ cells in the thymus. These are then exported, naive and fully reactive, to peripheral lymphoid organs. Infiltration of the pancreatic islets is precocious and synchronized (18). It seems, in this transgenic mouse, that the regulatory genes (36), molecules (37), cells (36), or intercellular milieu (38, 39) that modulate the progression to diabetes all act at a peripheral level, affecting the damage wrought there by the BDC-2.5 T cells, rather than the thymic control of their maturation.
We decided to use the BDC-2.5 system to test directly for the presence of autoreactive T cells, in nontransgenic animals, that might share the same antigenic specificity. Thus, an MHC-mimetic peptide with high agonistic activity for BDC-2.5 T cells (40, 41) was complexed to the Ag7 molecule in an MHC multimer molecule (26, 27). Tetramers of this Ag7/mimotope complex did prove specific for BDC-2.5 T cells. Strikingly, in NOD mice, they labeled a “natural” Ag7/BDC-2.5 mimotope–reactive population, whose fate we could track. This report demonstrates the unique role of the MHC susceptibility genes in selecting this specificity.
Methods
Antibodies, hybridomas, and T cell lines.
T cell hybridomas BDC-2.5 and R28 have been described elsewhere and were maintained in RPMI-1640/10% FCS (30, 42). BDC T cell clones were maintained in culture by successive rounds of stimulation with β cell granule membranes from β cell tumors as a source of antigen and γ-irradiated NOD splenocytes as APCs as described previously (31, 35). T cell clones were restimulated with β cell granule membrane protein for 4 days and expanded in the presence of IL-2 prior to staining. All antibodies mentioned in this study were obtained from BD Pharmingen (San Diego, California, USA). Vβ TCR-specific antibodies were directed against Vβ’s 2, 3, 4, 5.1, 5.2, 6, 7, 8.1, 8.2, 8.3, 9, 10, 11, 12, 13, 14, and 17, respectively.
Mouse strains and immunization.
NOD/LtJ, NOR/LtJ, C57BL/6, BALB/c, BDC-2.5/N, RAGo/o/NOD (18, 43), and KRN TCR transgenic mice that recognize glucose-6-phosphate isomerase peptide 282–292 (hereafter referred to as GPI) in the context of Ag7 MHC molecules (23, 44) were either purchased from The Jackson Laboratory (Bar Harbor, Maine, USA) or bred in our special pathogen-free animal facility (Joslin Diabetes Center and The Scripps Research Institute). For in vivo T cell response studies, mice were immunized in the base of the tail and in footpads with 10 μg of BDC-2.5 mimotope peptide reconstituted in PBS and emulsified in CFA. Animals were sacrificed 8 days after immunization.
Construction, expression, and purification of BDC-2.5 TCR and Ag7/peptide molecules; generation of tetrameric MHC molecules.
The cDNA for the α and β chains of the BDC-2.5 TCR was obtained by RT-PCR using Ready-To-Go RT-PCR beads (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) according to the manufacturer’s instructions, and RNA was isolated from BDC-2.5 T cells and R28 transgenic T cells. Oligo-dT primers and sequence-specific primers (Vα1 [AV1S5], 5′-AAAAAAGAATTCGGTACCGAAATGCATTCCTTACATGTTTCAC-3′; Vβ4, 5′-AAAAAATATAGGTACCGAATTCGGAATGGGCTCCATTTTCCTCAG-3′; Cα, 5′-AAAAAATAAGAATTCGGTACCGAAATGCATTCCTTACATGTTTCACTAGTG-3′; and Cβ, 3′-TTTATTTTGTCGACTCAACTGGACCACAGCCTCAGCGT-5′) were used for cDNA synthesis and PCR amplification, respectively. PCR products were subcloned into pCR2.1-TOPO cloning vector (Invitrogen Corp., Carlsbad, California, USA), sequenced, and subcloned into the metallothionein promoter–based fly expression vector pRMHa3 (45). Each of the final constructs coded for the α1α2 and the β1β2 domains, respectively, followed by a linker sequence (SSADL), a thrombin site (LVPRGS), a leucine zipper (acidic for the α chain, basic for the β chain) (46), and a hexahistidine tag. Vectors were transfected into SC2 cells, and stable cell lines and clones were established. Soluble TCRs were purified from culture supernatants as described previously (47). The generation of Ag7/peptide complexes has been previously reported in detail (6, 13). The Ag7/2.5mi and Ag7/GPI molecules bear the sequences AHHPIWARMDA and LSIALHVGFDH, respectively. The biotinylation sequence no. 85 from Schatz (48) follows the acidic zipper on the α chain. Biotinylation of purified molecules was performed according to the manufacturer’s instructions (13). Biotinylation was measured by immunodepletion on streptavidin-agarose beads and subsequent analysis by SDS-PAGE and scanning. Biotinylated molecules were tetramerized at 4°C overnight using phycoerythrin-labeled (PE-labeled) or APC-labeled streptavidin (BioSource International Inc., Camarillo, California, USA; and ProZyme Inc., San Leandro, California, USA), with a 5:1 molar ratio of biotinylated molecules to labeled streptavidin.
Cell preparation, cell staining, flow cytometry analysis, and adoptive transfer.
Single-cell suspensions were prepared by mechanical disruption of the corresponding organs in HBSS buffer, and erythrocytes were removed by lysis. Cells were washed in FACS buffer (PBS containing 2% FCS and 0.04% NaN3) and incubated with 0.5 mg/ml of streptavidin and Fc block in FACS buffer for 1 hour at room temperature. Cells were then washed once and stained with PE-labeled MHC/peptide tetramers at a final concentration of 10 μg/ml in FACS buffer for 1 hour at room temperature. For costaining of surface markers, APC–anti-CD4, FITC–anti-CD3, FITC–anti-CD44, PerCP–anti-B220, and PerCP–anti-CD8 from BD Pharmingen were used, as well as PE–Cy7–anti-CD8 and PE–Texas red–anti-CD45R (B220) from Caltag Laboratories Inc. (Burlingame, California, USA). Exclusion of dead cells was done by addition of 0.1 μg/ml of Hoechst 33342 (Molecular Probes Inc., Eugene, Oregon, USA) or propidium iodide. Flow cytometry was performed using a FACSCalibur instrument (Becton Dickinson Immunocytometry Systems, Mountain View, California, USA) at The Scripps Research Institute or a MoFlo instrument (Cytomation Inc., Fort Collins, Colorado, USA) at the Joslin Diabetes Center, and the data were analyzed using either CellQuest software (Becton Dickinson Immunocytometry Systems) or Summit software (Cytomation Inc.). Staining of T cell hybridomas and T cell clones with MHC tetramers was carried out in the same way. For cell transfer experiments, NOD splenocytes were stained the same way, sorted on a high-speed cell sorter, and injected intravenously into 4-week-old RAGo/o/NOD mice (with no irradiation).
Generation of T cell hybridomas.
Lymphocytes were isolated either from pancreatic lymph nodes (PLNs) of naive 8-week-old NOD females or from popliteal LNs 8 days after immunization in tail and footpads with 2.5mi peptide in CFA. Cells were stimulated with concanavalin A for 3 days and subsequently fused with BW5147αβ– lymphoma cells. Hybridomas were stained with 2.5mi tetramer, and single cells were cloned by FACS.
T cell activation assay.
Ninety-six-well flat-bottomed plates (Corning Inc., Corning, New York, USA) were coated with serially diluted MHC/peptide complexes in PBS overnight at 4°C and washed three times with PBS. T cell hybridomas were washed twice in PBS, resuspended in complete DMEM, and added at 5 × 104 cells per well in 200 μl, for 24 hours at 37°C. Alternatively, hybridomas were incubated with β cell granule membranes from β cell tumors as a source of antigen, or with purified pancreatic islets in the presence of γ-irradiated NOD splenocytes as APCs. Supernatants were harvested, and IL-2 production was determined using the IL-2–dependent cell line NK.
Surface plasmon resonance.
A Biacore 2000 instrument (Biacore Inc., Piscataway, New Jersey, USA) was used to determine interactions between purified MHC/peptide complexes and TCR molecules. BDC-2.5 TCR was immobilized by amine-coupling chemistry on a CM5 research-grade sensor chip. Surface densities for individual experiments are indicated in Table 1. Injections of MHC/peptide molecules at the appropriate concentrations were performed in filtered and degassed PBS at a flow rate of 20 μl/min. In all experiments, Ag7/GPI MHC molecules were used as negative control. Kd values as well as on and off rates were obtained by nonlinear curve fitting of subtracted curves using the Langmuir 1:1 binding model with the BIAevaluation program (version 3.0.2; Biacore Inc.) and the global-fitting software Clamp (version 3.3) (49). The equilibrium Kd, under steady-state conditions, was also determined using the BIAevaluation program.
Table 1.
Kinetic constants from the interaction of BDC-2.5 TCR with Ag7/2.5mi MHC complexes
Immunocytochemistry.
Immunocytochemistry was performed essentially as previously described (50). Briefly, organs of interest were removed, embedded in Tissue-Tek OCT compound (Sakura Finetek USA Inc., Torrance, California, USA), and immediately frozen on dry ice. Sections of 6 μm thickness were cut at –16°C on a microtome (Cryocut 1800; Reichert-Jung, Leica Microsystems, Bannockburn, Illinois, USA). Staining was performed at 4°C. Sections were incubated with APC-labeled Ag7 tetramers (5 μg/ml in PBS containing 2% FCS) and a rat anti-CD4 antibody (L3T4) for 15 hours. Sections were then washed briefly with PBS and fixed with paraformaldehyde. After 3 hours of incubation with a rabbit anti-APC antibody (Biomeda Corp., Foster City, California, USA), the signal was amplified by staining with a Rhodamine Red-X–conjugated goat anti-rat antibody and a Cy5-labeled goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA). Insulin was detected on paraformaldehyde-fixed sections by successive incubation with a guinea pig anti-insulin antibody (DAKO Corp., Carpinteria, California, USA) and a FITC-conjugated anti–guinea pig antibody. Staining was visualized using a Bio-Rad MRC1024 laser scanning confocal microscope (Bio-Rad, Hercules, California, USA) fitted with a krypton/argon mixed-gas laser (excitation at 488 nm, 568 nm, and 647 nm) using a ×40 oil objective.
Results
Production of functional BDC-2.5 TCR and Ag7/2.5 mimotope complexes.
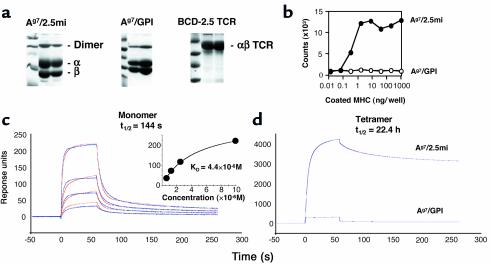
A surrogate ligand for BDC-2.5 T cells was isolated from a chemically synthesized random-peptide library based on its ability to stimulate BDC-2.5 T cells (41). We hypothesized that a strong agonistic peptide selected solely on the basis of its biological activity, without prior regard to the affinity of the peptide/MHC complex for the TCR, would be the optimum ligand to construct MHC class II tetramers. The peptide AHHPIWARMDA, hereafter referred to as BDC-2.5 mimotope (2.5mi), was isolated and optimized using this strategy. A fusion MHC β chain/2.5mi peptide was engineered as previously reported (6, 13). Similar molecules with Ag7-binding peptides from glucose-6-phosphate isomerase (282LSIALHVGFDH292, hereafter referred to as GPI) and hen-egg lysozyme (11AMKRHGLDNYRGYSL25, hereafter referred to as HEL) were also produced. The Ag7/GPI complex is recognized by the R28/KRN TCR (44, 51). The Ad α chain cDNA was modified at its 3′ end to harbor an acidic zipper sequence followed by the biotinylation sequence no. 85 from Schatz (48) and a histidine tag (46). Purified molecules were biotinylated using the BirA enzyme and repurified by size exclusion chromatography (Figure 1a). Ag7/2.5mi MHC molecules were functional when coated onto plates, stimulating BDC-2.5 hybridoma cells to secrete IL-2 (Figure 1b). Control Ag7/GPI molecules had no such effect (but did stimulate the R28 hybridoma; data not shown). In parallel, the α and β chains of the BDC-2.5 TCR were cloned by RT-PCR from thymocytes of BDC-2.5 transgenic mice and modified to produce zippered soluble recombinant TCR (47) (Figure 1a).
Figure 1.
Biophysical and functional characterization of Ag7/2.5mi and Ag7/GPI MHC molecules. (a) SDS-PAGE analysis of the various recombinant MHC and TCR molecules used in this study. Molecules were purified from culture supernatants of transfected Drosophila melanogaster cells. The peak fractions of the final size exclusion chromatography are shown. (b) Recombinant Ag7/2.5mi molecules can activate the BDC-2.5 T cell hybridoma. Ag7/2.5mi MHC monomers were coated at the indicated concentrations into 96-well plates. IL-2 production was measured from the supernatants after 24 hours of culture. (c) Surface plasmon resonance analysis of the Ag7/2.5mi MHC/BDC-2.5 TCR interaction. Left: BDC-2.5 TCR molecules were randomly immobilized on a CM5 chip, and Ag7/2.5mi or Ag7/GPI (negative control) MHC molecules were flown over the surface. Subtracted curves are shown in blue lines, calculated curves in red. A Langmuir 1:1 binding model was used for analysis. Concentrations of injected MHC molecules were 10 μM, 2.5 μM, 1.25 μM, and 0.625 μM. The inset shows a steady-state analysis of the same interaction. (d) Tetramers of Ag7 with either peptide were flown over a high-density BDC-2.5 TCR surface. Unsubtracted curves corresponding to a tetramer concentration of 1 μM are presented.
The interaction between the recombinant BDC-2.5 TCR and Ag7/2.5mi MHC molecules was measured by surface plasmon resonance using Ag7/GPI MHC molecules as a negative control. Association and dissociation rates were calculated by global fitting from subtracted sensorgrams (Figure 1c and Table 1). The calculated Kd was approximately 0.5 μM, whereas equilibrium data gave a value of 4.4 μM at 25°C. These values were relatively low compared with those for most TCR/MHC class II agonistic-peptide interactions (52), suggesting that the mimotope is a very strong agonistic peptide for the BDC-2.5 TCR in the context of Ag7. Measurements at 37°C showed a slight decrease of calculated Kd to approximately 1.5 μM (data not shown). The same MHC molecules were tetramerized with streptavidin and tetramers purified by gel filtration. Over a high-density surface (∼8,000 resonance units [RU]) of immobilized BDC-2.5 TCR, Ag7/GPI tetramers exhibited no binding, whereas Ag7/2.5mi tetramers showed specific binding and an increase of half-life to 22.4 hours compared with 144 seconds for the monomers (Figure 1, c and d). These results suggest that the interaction between the Ag7/2.5mi tetramer and the BDC-2.5 TCR is long lasting. The affinity of Ag7/GPI molecules for their cognate TCR was also measured by surface plasmon resonance using recombinant soluble TCR and was evaluated to be approximately 1 μM (T. Stratmann and L. Teyton, unpublished observations).
Ag7/2.5mi tetramers are specific for the BDC-2.5 TCR.
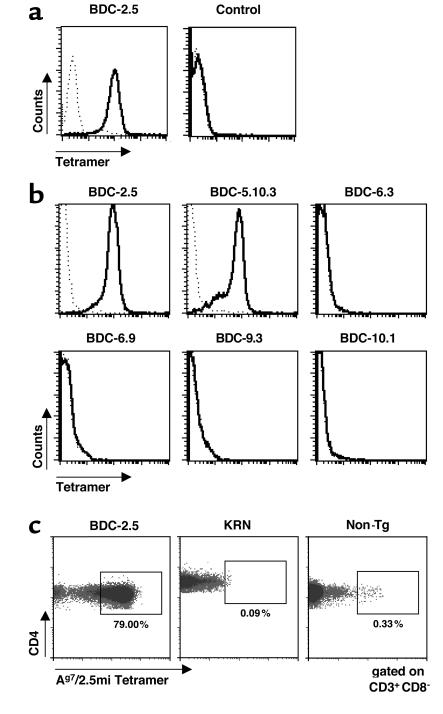
Ag7/2.5mi and Ag7/GPI molecules were then tetramerized with streptavidin-phycoerythrin conjugate and the specificity of these reagents was tested against a panel of T cells. First, we stained T cell hybridomas for flow cytometry. As expected, Ag7/2.5mi multimers stained the BDC-2.5 but not the control hybridoma (Figure 2a), whereas Ag7/GPI multimers stained only R28 cells (data not shown).
Figure 2.
Specific recognition of 2.5mi+ T cells by Ag7/2.5mi multimers. (a) BDC-2.5 T cell hybridoma and 19.2B T cell hybridoma (specific for glutamic acid decarboxylase peptide 282–292) were labeled with either Ag7/2.5mi tetramers (solid line) or Ag7/GPI tetramers (dotted line). (b) Staining of T cell clones specific for β cell granule membranes with Ag7/2.5mi tetramers. After 4 days of stimulation in vitro with islet-membrane preparations, BDC clones were stained with Ag7/2.5mi tetramers. (c) Inguinal LN cells from young adult BDC-2.5/NOD, KRN, and NOD (Non-Tg) mice were stained for CD4 and Ag7/2.5mi tetramers.
Second, we tested the original BDC-2.5 T cell clone, as well as five other NOD-derived clones that are all specific for β cell antigens (31, 35) (Figure 2b). Ag7/2.5mi multimers stained specifically both BDC-2.5 and BDC-5.10 T cells, but not any of the other clones. This observation was consistent with the ability of the BDC-2.5 mimotope peptide to stimulate both these clones to produce IFN-γ (data not shown). Finally, we tested our reagent with primary T cells from transgenic BDC-2.5 mice or another Ag7-restricted TCR transgenic mouse, KRN (23). The staining of naive T cells with MHC class II multimers has been a consistent hurdle for these reagents, and, in our hands at least, most of them failed even though they could stain hybridomas of similar specificity. When splenocytes from a BDC-2.5 transgenic animal were stained with Ag7/2.5mi multimers and analyzed by FACS, 79.0% of the CD4+ T cells were positive for the Ag7/2.5mi multimers (Figure 2c), consistent with the known frequency of clonotype expression in these mice (N. Martin-Orozco, unpublished observations). The same reagent was unable to stain T cells from the KRN transgenic mouse used as a control (Figure 2c). With the nontransgenic NOD control, the staining was also largely absent, but we were intrigued by a small, but clear, population of CD4+ T cells that stained with the same intensity as in the BDC-2.5 transgenic mouse (Figure 2c).
BDC-2.5–like T cells in the NOD mouse.
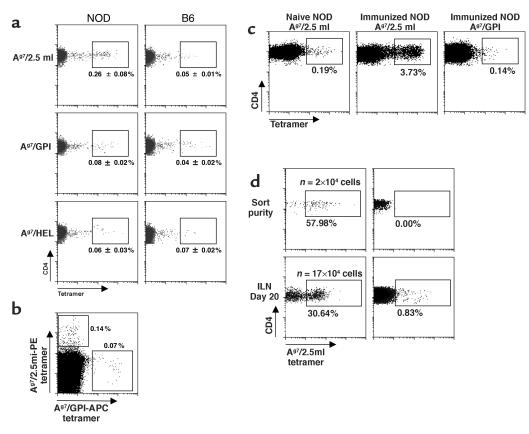
Several experiments were performed to ascertain the relevance of the small population of CD4+ T cells that stained with the Ag7/2.5mi multimer (hereafter referred to as 2.5mi+ T cells). The 2.5mi+ T cells were alive, judging from scatter and vital dye exclusion, and expressed normal levels of CD4 and CD3 (not shown). The staining appeared specific, since it was not observed with control multimers loaded with GPI or HEL peptides (Figure 3a), which gave frequencies close to the background level (around 0.05% in most of our experiments). No CD8+ T cells of similar specificity were identified (not shown). The genetic control provided by B6 mice (Ab haplotype) showed that the 2.5mi+ T cells were indeed specific to NOD mice (Figure 3a). Dual-label experiments were also performed, with two tetramer reagents added together. The 2.5mi+ T cells did not bind the GPI tetramer simultaneously, arguing against nonspecific “sticking” of the reagents (Figure 3b). To verify the responsiveness of these cells, NOD mice were injected subcutaneously with the mimotope peptide in adjuvant. Eight days after immunization, 2.5mi+ T cells increased in frequency from about 0.2% to 3.7% in the draining LNs (Figure 3c), demonstrating that the 2.5mi+ T cell population was not anergized but responsive to antigenic challenge. To further demonstrate the specificity of Ag7/2.5mi tetramer staining, we tested the capacity of 2.5mi+CD4+ cells to proliferate in response to homeostatic cues, after transfer into lymphopenic RAGo/o/NOD mice. As shown in Figure 3d, 2.5mi+ T cells expanded significantly in these recipients, forming a very distinct population among LN cells 20 days after transfer. Insulitic attack was noted in one of the recipients, but not in all (data not shown), consistent with the requirement for CD8+ cells, and conversely the blockade by other CD4+ cells, in the development of autoimmunity in BDC-2.5 transgenic mice (18, 36).
Figure 3.
Presence of 2.5mi+ T cells in NOD mice. (a) Specific 2.5mi+CD4+ T cells were identified in subcutaneous LNs from young adult NOD but not from B6 mice. Control stains for either Ag7/GPI or I-Ag7/HEL tetramers are shown in the lower panels. (b) Lack of cross-reactivity of Ag7/2.5mi tetramers with nonspecific T cells. Splenocytes from a NOD mouse were stained with PE-labeled Ag7/2.5mi tetramers, followed by incubation with APC-labeled Ag7/GPI tetramers. T cells were only stained by either of the tetramers, not by both simultaneously, indicating specific recognition. (c) 2.5mi+CD4+ T cells expansion after immunization with the 2.5 mi+ peptide. Popliteal LN cells from nonimmunized and immunized NOD mice were stained with either Ag7/2.5mi or Ag7/GPI tetramers. (d) Expansion of 2.5mi+CD4+ cells in a lymphopenic mouse reconstituted with 2.5mi+CD4+ cells (or 2.5mi–CD4+ cells as control), sorted from splenocytes of 5-week-old NOD mice and immediately analyzed for sort purity. Twenty thousand sorted cells were injected into RAGo/o/NOD mice. Inguinal LN cells were stained 20 days after transfer for the presence of 2.5mi+CD4+ cells. The number of injected cells was not corrected for the presence of nonviable cells and is therefore likely to be overestimated. The number of 2.5mi+CD4+ cells present in the whole mouse (n) was extrapolated to a full lymphoid compartment from the number of 2.5mi+CD4+ cells observed in the inguinal LN by flow cytometry (taking into account multiple losses during the staining procedures and the flow cytometry acquisition itself).
To more directly visualize these results, in situ staining was carried out with MHC multimers and anti-CD4 antibodies. To date, only a few reports have mentioned using MHC class I tetramers for immunocytochemical staining (50, 53–55), and, to our knowledge, none has used MHC class II multimers. A technique for frozen nonfixed tissues was developed (50), and antibody- and tetramer-stained sections were analyzed by confocal microscopy (costained with anti-CD4 to identify T cells). In spleen or pancreas sections from BDC-2.5 transgenic mice, clear staining with the Ag7/2.5mi multimer could be identified, coincident with anti-CD4 staining (Supplemental Figure 1, http://www.jci.org/cgi/content/full/112/6/902/DC1). In sections from nontransgenic NOD mice, 2.5mi+ T cells were also identified in situ by immunocytochemistry (Figure 4). Typically, we found between one and two 2.5mi+ T cells per T cell zone in the spleen (not shown). Interestingly, they were also seen in PLNs and in islets in the infiltrating CD4+ T cell population (Figure 4). Thus, by all these criteria, the prediabetic NOD mouse contains in its lymphoid organs a small but clearly identifiable population of 2.5mi+ T cells that stains specifically with the Ag7/2.5mi reagent, is responsive to antigenic stimulation, and localizes to the expected organs.
Figure 4.
Histological detection of 2.5mi+CD4+ T cells. In situ staining of 2.5mi+CD4+ T cells in PLNs (a and b) and pancreatic islets (c–e). I-Ag7/2.5mi MHC tetramer stains are shown in green (a and c). Staining with an anti-CD4 antibody is shown in red and overlaid by the tetramer staining (b and d). Insulin staining is shown in e.
Life history of the 2.5mi+ T cell population.
The dynamics of CD4+/2.5mi+ T cells were characterized further by FACS analysis. A representative experiment is shown in Figure 5, a and b, and compiled data are shown in Figure 5c. 2.5mi+ T cells were clearly detectable among mature CD4+CD8–CD3hi thymocytes (Figure 5a) but not among CD4–CD8+CD3hi cells, as might be expected. These cells colonize the peripheral lymphoid organs: CD4+ T cells in the spleen and subcutaneous LNs also showed clear staining above background, indicating that 2.5mi+ T cells are exported and survive efficiently in the peripheral immune system (indeed, expand slightly, when thymus and LN frequencies are compared; Figure 5c). This proportion increased further in the LNs of the pancreatic group (PLNs) and among pancreas-infiltrating cells, suggesting that they were expanding in response to antigen. The expansion of BDC-2.5–like CD4+ T cells in the pancreas has also been recently observed by You and colleagues using a very similar mimotope/Ag7 molecule to identify those cells (56). This expansion and accumulation in the PLN and pancreas was suggestive of activation of the 2.5mi+ T cell population by pancreatic antigens, similar to that of BDC-2.5 transgenic mice. This was confirmed by examination of T cell activation markers in the CD4 population and its 2.5mi+ T cell subset. As shown in Figure 5d, an increased proportion of CD44hi cells was found among 2.5mi+ T cells in the PLNs of young NOD mice, mirroring BDC-2.5 transgenics. This difference was not observed in bulk CD4+ T cells of the same mice (10% ± 2% and 12% ± 2% of CD44hi cells in inguinal LN and PLN, respectively). This hallmark of activation was detected, to a lesser extent, in the neighboring superior mesenteric LNs, into which some drainage of the pancreas likewise occurs, but not in the spleen or other LNs unrelated to the pancreas (axillary, iliac, renal, and lower mesenteric), whereas almost no 2.5mi+ cells were detected in Peyer’s patches (data not shown). These data indicate that the 2.5mi+ T cells do share the autoimmune specificity of the BDC-2.5 clonotype.
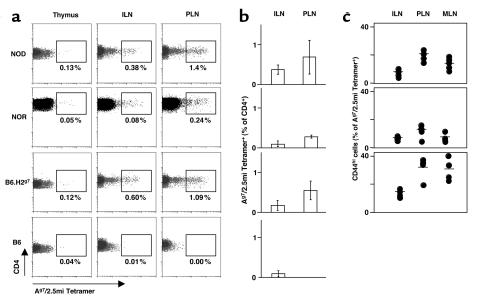
Figure 5.
T cells with the 2.5mi+ reactivity are positively selected in the thymus and expand in the periphery. (a and b) Thymocytes, splenocytes, inguinal LNs (ILNs), PLNs, and infiltrated pancreatic T cells were stained for CD4, CD8, CD3, and Ag7/2.5mi tetramer. The percentage of Ag7/2.5mi+CD4+CD3hi T cells was electronically calculated from the rectangle gate of each profile. (c) 2.5mi+CD4+ T cells expand in the periphery after leaving the thymus, and they accumulate in the PLNs. Average values represent independent experiments for seven mice analyzed. (d) Specific activation of 2.5mi+CD4+ T cells in PLNs. ILNs, PLNs, or superior mesenteric LNs (MLNs) from BDC-2.5 transgenic (Tg) mice (open squares) or from groups of 3-week-old (diamonds) or 4-week-old or older NOD mice (triangles) were costained with Ag7/2.5mi tetramers and anti-CD44.
We were curious whether the percentage of 2.5mi+ T cells in the CD4+ T cell population increases with age in NOD females, paralleling the development of insulitis and diabetes. PLNs and iliac LNs were removed from animals ranging from 2 to 16 weeks of age (four animals combined per time point), and the CD4 T cell population was analyzed for 2.5mi+ T cells. Frequencies of 2.5mi+ T cells varied from 0.05% to 0.15% over the course of the experiment (Figure 6). In the PLN, cells were found as early as week 2, and their frequency increased to peak at week 3 or 4. In the iliac LNs, 2.5mi+ cells appeared later and climbed to a steady level from week 4 to week 16. The early wave of expansion in the PLN suggests site-specific activation and is also reminiscent of the 3- to 4-week checkpoint 1 that has been described in the 2.5 transgenic mouse (57). The control GPI-reactive cells remained at background level in both locations for the entire period of the experiment (Figure 6).
Figure 6.
Time course analysis of 2.5mi+ T cells in naive NOD females. PLNs and iliac LNs (ILLNs) were removed and analyzed for 2.5mi+ T cells at the indicated ages. Harvested lymphocytes were pooled from four animals before analysis.
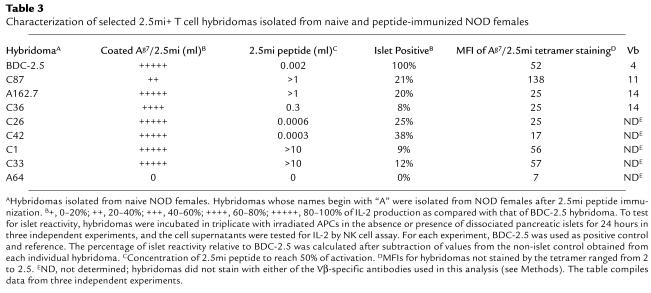
To further address the islet reactivity of these T cells, we immunized NOD female mice by injection of 2.5mi peptide into the footpads and generated hybridomas from T cells isolated from the draining LNs. A total of 120 hybridoma lines were generated, of which 80 were analyzed for their reactivity to coated peptide/MHC complexes and to free peptide in the presence of APCs, as well as by FACS analysis for staining with 2.5mi tetramers (Table 2, Table 3, and Figure 7a). About half of the lines were negative in both assays. Out of 39 and 38 lines, respectively, that tested positive for either reactivity toward coated peptide/MHC complexes or staining by FACS, 22 clones were positive in both assays. With three exceptions, there was a clear positive correlation between IL-2 secretion and intensity of tetramer staining (data not shown). Of the remaining clones, about half tested positive in the activation assay, whereas the other half were only positive in the FACS analysis. However, staining by FACS of these clones was very low (medium fluorescence intensity [MFI] < 10 vs. 7 for the negative control). Double-positive lines were tested for islet reactivity. Of those clones, only one (clone A162.7) tested clearly positive (Table 3 and Figure 7c).
Table 2.
Summary of the T cell hybridomas generated for this study from NOD female mice
Table 3.
Characterization of selected 2.5mi+ T cell hybridomas isolated from naive and peptide-immunized NOD females
Figure 7.
Reactivity of 2.5mi+ T cell hybridomas to 2.5mi peptide and pancreatic islet cells. T cell hybridoma clones obtained from NOD females after immunization with 2.5mi peptide (a and c) or from naive animals (b and c) were incubated in the presence of APCs with increasing concentrations of 2.5mi peptide (a and b) or pancreatic islet cells (c), and their IL-2 response was measured by incorporation of 3H-thymidine during an NK cell proliferation assay. Clone numbers were as follows: (a) A42, open squares; A162.7, filled circles; A202.2, filled squares; A64, open diamonds; A129, filled triangles; A129, filled diamonds; A72, open upward-pointing triangles; A86, open circles; and A166, open downward-pointing triangles; (b) C26, open squares; C20, open circles; C33, filled upward-pointing triangles; C87, filled circles; BDC-2.5, filled squares; C36, open upward-pointing triangles; C1, filled downward-pointing triangles; C26, open downward-pointing triangles.
However, immunization with peptide could lead to the preferential selection of T cell specificities distinct from the fine specificity inherent to the original BDC-2.5 clone. To better understand the natural 2.5mi+ T cell population, we generated another set of T cell hybridomas, from PLNs of naive animals. One hundred fifty-three lines were generated, and tetramer-positive cells were sorted by FACS and cloned. Ninety-nine clones were generated in such a way and reanalyzed by FACS for staining with the 2.5mi tetramer. Of these clones, 40 tested positive, with MFIs ranging from 17 to 138, indicating the heterogeneity of these clones in terms of avidity. (MFI > 10 defined positive clones, and MFI < 2 defined negative clones. TCR expression was identical for all clones.) Twenty-five of these clones were analyzed for IL-2 secretion in an activation assay using coated peptide/MHC complexes (Figure 7b). All clones positive for tetramer staining secreted robust amounts of IL-2 (<200,000 counts for 95% of the clones in an NK assay, 1,500 counts for the negative control; Table 3 and data not shown). A selection of clones (with different MFIs) was further analyzed for their reactivity toward peptide and islets in the presence of APCs (Figure 7b and Tables 2 and 3). Most of the clones responded toward the peptide, though some of them with a much lower functional avidity than others (IC50 ranging from 0.0003 to >10 μg/ml). We analyzed 14 clones for their reactivity to islets. Six of these clones showed reproducibly various degrees of IL-2 production (between 8% and 38% compared with BDC-2.5; Table 3 and Figure 7) as compared two five negative clones included in each experiment (a representative clone, A64, is shown in Table 3 and Figure 7). The Vβ TCR usage of the various clones, analyzed by FACS (Table 2 and Table 3), was as diverse as the staining and reactivity toward peptide, pMHC, and islet antigen.
The fact that some of these clones reacted to both peptide and islet antigen indicated strongly that the cross-reactivity between the 2.5mi peptide and a pancreatic antigen extended beyond the sole BDC-2.5 T cell to other 2.5mi-reactive T cells. These results confirmed a previous observation on the cross-reactivity of six islet-reactive, diabetogenic T cell clones with the same or similar mimotopes (41). As for the hybridomas, these clones were heterogeneous in terms of Vα/Vβ usage and dose-response curves. They also indicate that the anti–2.5 antigen CD4+ T cell response is diverse in terms of repertoire and range of functional avidity.
2.5mi+ T cells are present in H-2g7 mice with diabetes-resistant genetic backgrounds.
What is required for the selection of 2.5mi+ T cells? Do they represent defective negative selection, imparted by the diabetes-susceptibility alleles of the NOD background (15), or do they only require the presence of the Ag7 molecule for their selection? To test the relative contribution of MHC and background genes, we analyzed B6.H2g7 and NOR, two congenic strains that carry the same H-2g7 MHC haplotype as the NOD mouse but differ in other background loci. Both are fully resistant to diabetes. The 2.5mi+CD4+ T cells were easily detectable in lymphoid organs of these congenic mice. Their tissue distribution (Figure 8a) and numbers (Figure 8b) were very comparable in the three H-2g7 strains. Although frequencies in NOR mice tended to be slightly lower, the peripheral expansion in PLNs was always present, and, upon immunization, the expansion of 2.5mi+ cells was as important as the one seen on the NOD background (Supplemental Figure 2, http://www.jci.org/cgi/content/full/112/6/902/DC1). Furthermore, signs of antigen-specific activation were also present in the congenic mice: the increased frequency in PLNs (Figure 8b), and the increased frequency of CD44hi cells in the PLN (more pronounced in the B6.H2g7 animals) (Figure 8c). Again, the B6 mice used as a control showed no such populations. As for NOD mice, 2.5mi+ T cells expanded after peptide immunization with the 2.5 mimotope in NOR mice, arguing against anergy of this cell population in a diabetes-resistant background (Supplemental Figure 2, http://www.jci.org/cgi/content/full/112/6/902/DC1).
Figure 8.
Ag7 is sufficient to select 2.5mi+CD4+ T cells. (a) Thymocytes, inguinal LNs (ILNs), and PLNs from young adult NOD mice, NOR mice, C57BL/6 mice congenic for H-2g7 (B6.H2g7), and C57BL/6 (B6) mice were stained for CD4, CD8, CD3, and Ag7/2.5mi tetramer. The percentage of Ag7/2.5mi+CD4+CD3hi T cells was electronically calculated from the rectangle gate of each profile. (b) 2.5mi+ T cells accumulate in PLNs of mice that do not get insulitis or diabetes but share Ag7. Average representation (percent) of 2.5mi+CD4+ T cells from NOD, NOR, and B6.H2g7 mice (five to seven mice per group) was measured in four independent experiments. (c) 2.5mi+ T cells are activated in PLNs of mice that do not get insulitis or diabetes but share Ag7. The percentage of CD44hiCD4+Ag7/2.5mi+ T cells in ILNs, PLNs, and MLNs of NOD, NOR, and B6.H2g7 mice (five to seven mice per group) was measured in four independent experiments.
These observations allow three key conclusions: (a) MHC alleles of the H-2g7 haplotype are necessary and sufficient for the selection of the 2.5mi+ T cells; (b) negative selection of 2.5mi+ T cells is identical in susceptible or resistant mice; and (c) 2.5mi+ T cells are activated in pancreas-draining LNs regardless of the genetic background and diabetic status of the mouse.
Discussion
The MHC multimer reagent, with its highly efficient binding to the BDC-2.5 TCR, offers a glimpse of the development of a “normal” repertoire of T cells reactive against a peripheral antigen.
One question must be addressed at the outset. The BDC-2.5 TCR confers reactivity to a uniquely pancreatic antigen and cross-reacts with the AHHPIWARMDA mimotope analog. The 2.5mi+ population of CD4+ T cells binds the mimotope, but does that necessarily imply that it recognizes the original BDC-2.5 pancreatic antigen? This was not a foregone conclusion, as T cells are broadly cross-reactive but T cells of identical specificity need not share cross-reactive recognition (58). Here, the phenotype of 2.5mi+ T cells in the PLN provides a decisive clue: activation of a substantial proportion of the cells is seen quite exclusively in the PLN. We and others (57, 59) have shown that activation of pancreas-specific transgenic TCRs occurs solely in the draining PLN. That a substantial proportion of the 2.5mi+ T cells is also specifically activated in that LN shows these to be also reactive to a pancreas-specific antigen, and thus most likely to the BDC-2.5 antigen itself. We further confirmed this hypothesis by characterizing a series of tetramer-positive T cell hybridomas. All the cells that we tested were reactive toward the 2.5 peptide, the recombinant peptide/MHC molecules, and pancreatic islets cells, confirming the cross-reactivity between the mimotope peptide and the islet antigen seen in BDC-2.5. This cross-reactivity had been previously established by characterization of mimotopes for six diabetogenic T cell clones isolated in two separate laboratories (41). All clones reacted against mimotopes retaining the central P5 tryptophan residues and common P6–P9 segments. Optimization of reactivity was achieved by changing of P2 and P3 residues. All six clones had different Vα/Vβ pairing, and three of them were activated by the mimotope that we used in the present study. The BDC-2.5 mimotope published by Judkowski et al. (40) shares the same characteristics (similar P5–P9 segment, variation at P2 and P3) and was shown to be cross-reactive with our mimotope by induction of a similar 2.5mi+ cell expansion after immunization (data not shown). The relative variability of the N-terminal part of the mimotope suggests that the 2.5mi+ population will be more variable in its Vα usage than in its Vβ usage. This assumption is supported by the initial TCR usage characterization that we have done on the 2.5mi+ population (Supplemental Figure 3, http://www.jci.org/cgi/content/full/112/6/902/DC1). Altogether, these data strongly suggest that BDC-2.5 and 2.5mi+ T cells indeed recognize a common antigen, and that the usage of mimotope is warranted for the identification of T cell populations.
That T cells with an autoimmune specificity can exist in a healthy individual has been observed previously (43, 60). On the other hand, the very number of these 2.5mi+ T cells was the first surprise. Even if more recent sequencing reports (61) have downgraded the size of the effective T cell repertoire from the 1013 originally predicted (62), the combinatorial arrangements that can make up a repertoire are such that one would not have expected nearly 0.5% of CD4+ cells to bind a single MHC/peptide complex. Indeed, for other tetramer analyses, the precursor frequency in the naive repertoire was considerably lower, often not detectable but estimated to be in the range of 1 or fewer in 20,000 (27, 63–66). The frequency is already high at the outcome of thymic selection (0.18% on average) and is reflected in the naive cells of peripheral LNs (0.25% on average). In the present instance, the overrepresentation is not due to any particular Vα/Vβ combination, as the αβ composition of the 2.5mi+ T cell repertoire is broad (T. Stratmann, unpublished observations). The overrepresentation is also reminiscent of “canonical” TCRs that dominate the response to particular antigens (67), which have been suggested to correspond to selection and maintenance on particular MHC/peptide combinations. The peculiarity, in the case of the 2.5mi+ T cell population, is that this overrepresented specificity corresponds to an autoimmune clonotype. One might hypothesize that the mimotope yields a “promiscuous” surface, able to bind a variety of TCRs.
From a practical standpoint, the present observation could also mean that the prediction or early detection of diabetes based on the identification of antigen-specific T cells in at-risk patients will be difficult. If, as for the 2.5mi+ specificity, frequencies are roughly the same in peripheral lymphoid organs of at-risk and healthy individuals (the human counterparts of high-risk NOD and resistant B6.H2g7 mice), the sheer number of islet-reactive T cells in blood or nonpancreatic LNs may not be a good indicator of disease activity. This would be consistent with the difficulty in reproducibly distinguishing patients from controls in antigen-specific proliferation assays.
The most far-reaching conclusion that can be drawn from the present study is that the diabetes-susceptible H-2g7 MHC haplotype, not the background genes, determines the selection of this autoimmune repertoire. Several lines of evidence have suggested that the genetic composition of the NOD mouse favors inefficient central tolerance, leading to the emergence of an autoimmune repertoire (14, 15). This is clearly not the case for this specificity, as the 2.5mi+ T cells share similar frequencies of thymic selection and peripheral representation on the NOD, B6, or NOR background. Only H-2g7 is required. On nonsusceptible B6 and NOR backgrounds, 2.5mi+ T cells are fully reactive, as shown by their specific activation in the PLN (Figure 8) or by peptide-injection experiments (not shown).
But then why are NOR or B6.H2g7 mice not insulitic and diabetic? Where do background genes come into play to control diabetogenesis? Clearly, these strains can select an autoreactive repertoire of competent 2.5mi+ T cells. We cannot rule out fine affinity differences in NOD versus other backgrounds, but the cells do appear similar in numbers, staining intensity, activation potential, and repertoire bias. Further, they are activated in the PLN in all strains, indicating that their presence and activation there is not pathogenic by itself. The implication is that NOD background genes are peculiar in how they allow the consequence of autoimmune selection and activation to play out after autoantigen recognition: subtle phenotypic differences imparted by APCs, which allow the activated T cells to become fully aggressive; differences in regulatory T cell populations that control the initial response; differences in tissue infiltration. That NOD background alleles play a role by perturbing secondary immunoregulation has been proposed previously (3, 17, 43), and the notion is consistent with the fact that the recognized idd regions contain immunoregulatory loci: the costimulation complex on chr1, and the IL-2 locus (10).
Indeed, the present observations with 2.5mi+ T cells reproduce, to a striking extent, the behavior of the T cells in the original BDC-2.5 transgenic mice: robust positive selection in the thymus unencumbered by any signs of negative selection, export to the periphery in large numbers, and PLN-specific activation, all occurring with a similar independence from background genes (18, 43, 57). The present observations thus reinforce the validity of the transgenic system in analyzing immunoregulatory pathways. There is one apparent paradox: while BDC-2.5/B6.H2g7 transgenic mice progress to diabetes quite readily (in fact more efficiently than mice on the NOD background) (43), B6.H2g7 mice never do. If they share, at least in part, the same specificity as BDC-2.5 cells, why do 2.5mi+ T cells not generate disease? The difference likely resides in the sheer numbers of clonotype-positive cells in the transgenic mouse, where allelic exclusion strongly limits the number of regulatory cells. Accordingly, the aggression of diabetes in the transgenic mouse is correlated with the number of nonclonotypic cells and is far greater if the mouse is crossed onto a RAG-deficient background (36). Here again, the difference between pathogenicity and nonpathogenicity may lie in the regulatory environment, rather than in autoimmune cells themselves.
MHC haplotypes that confer susceptibility to particular autoimmune diseases can be thought of in two different ways. First, they may favor an autoimmune repertoire by inefficient negative selection of unwanted specificities; because of poor or promiscuous binding of self-peptides imparted by the unusual structure of MHC-II molecules devoid of an Asp residue at β-57, specificities that are normally negatively selected would appear in the repertoire (12, 68, 69). On the other hand, one can also imagine that susceptible alleles play their role by positively selecting a repertoire reactive against a particular self-antigen. The binding of the similar immunodominant epitopes of islet antigens by Ag7 and DQ8 argues in favor of such a view (70, 71). Indeed, the DQ8 molecule can positively select the BDC-2.5 transgenic TCR (72). Here, the 2.5mi+ T cells are selected in high numbers in the thymus, in a manner solely dependent on the susceptible MHC haplotype. Of course, we cannot rule out that H-2b molecules select a parallel, Ab-restricted population that recognizes the same antigen. Yet it is striking that the diabetes-susceptible H-2g7 haplotype can select such an abundant repertoire reactive to pancreatic self. MHC-II molecules were first identified by their capacity, as immune response genes, to present defined foreign antigens and allow the selection of a repertoire that can recognize them (73). Some alleles may selectively bring forth a repertoire targeted to self.
Supplementary Material
Acknowledgments
We thank C. Cantu III, P. Mas, M. McHeyzer-Williams, D. Hohmann, C. Surh, and A. Theofilopoulos for helpful discussions; R. Stefanko, B. Lyles, and A. Guadiz for technical assistance; and E. Hyatt for managing the BDC-2.5 colony. T. Stratmann received a fellowship from the Juvenile Diabetes Foundation International. N. Martin-Orozco received a mentor-based fellowship from the American Diabetes Association. This work was supported by NIH training grant AG00080 (to D. McGavern); grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H. Kikutani); NIH grant AI41439 (to M.B.A. Oldstone); Juvenile Diabetes Research Foundation (JDRF) grant 1-2000-322 and NIH grant DK50561 (to K. Haskins); JDRF Islet Transplantation Center grant “Project 36” and NIH grant 1RO1 DK59658-01 (to C. Benoist and D. Mathis); and NIH grants DK55037 and AG04342 (to L. Teyton).
Footnotes
See the related Commentary beginning on page 826.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: T cell receptor (TCR); glucose-6-phosphate isomerase peptide 282–292 (GPI); phycoerythrin (PE); pancreatic lymph node (PLN); hen-egg lysozyme (HEL); resonance unit (RU); medium fluorescence intensity (MFI).
References
- 1.Tisch R, McDevitt H. Insulin-dependent diabetes mellitus. Cell. 1996;85:291–297. doi: 10.1016/s0092-8674(00)81106-x. [DOI] [PubMed] [Google Scholar]
- 2.Vyse TJ, Todd JA. Genetic analysis of autoimmune diabetes. Cell. 1996;85:311–318. doi: 10.1016/s0092-8674(00)81110-1. [DOI] [PubMed] [Google Scholar]
- 3.Wicker LS, Todd JA, Peterson L. Genetic control of autoimmune diabetes in the NOD mouse. Annu. Rev. Immunol. 1995;13:179–200. doi: 10.1146/annurev.iy.13.040195.001143. [DOI] [PubMed] [Google Scholar]
- 4.Hattori M, et al. The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science. 1986;231:733–735. doi: 10.1126/science.3003909. [DOI] [PubMed] [Google Scholar]
- 5.Acha-Orbea H, McDevitt HO. The first external domain of the nonobese diabetic mouse class II I-A beta chain is unique. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2435–2439. doi: 10.1073/pnas.84.8.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corper AL, et al. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science. 2000;288:505–511. doi: 10.1126/science.288.5465.505. [DOI] [PubMed] [Google Scholar]
- 7.Latek RR, et al. Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice. Immunity. 2000;12:699–710. doi: 10.1016/s1074-7613(00)80220-4. [DOI] [PubMed] [Google Scholar]
- 8.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat. Immunol. 2001;2:501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 9.Owerbach D, Gabbay KH. The search for IDDM susceptibility genes: the next generation. Diabetes. 1996;45:544–551. doi: 10.2337/diab.45.5.544. [DOI] [PubMed] [Google Scholar]
- 10.Todd JA, Wicker LS. Genetic protection from the inflammatory disease type 1 diabetes in humans and animal models. Immunity. 2001;15:387–395. doi: 10.1016/s1074-7613(01)00202-3. [DOI] [PubMed] [Google Scholar]
- 11.Carrasco-Marin E, Shimizu J, Kanagawa O, Unanue ER. The class II MHC I-Ag7 molecules from non-obese diabetic mice are poor peptide binders. J. Immunol. 1996;156:450–458. [PubMed] [Google Scholar]
- 12.Ridgeway WM, Fasso M, Fathman CG. A new look at MHC and autoimmune disease. Science. 1999;284:749–751. doi: 10.1126/science.284.5415.749. [DOI] [PubMed] [Google Scholar]
- 13.Stratmann T, et al. The I-Ag7 MHC class II molecule linked to murine diabetes is a promiscuous peptide binder. J. Immunol. 2000;165:3214–3225. doi: 10.4049/jimmunol.165.6.3214. [DOI] [PubMed] [Google Scholar]
- 14.Markees TG, et al. NOD mice have a generalized defect in their response to transplantation tolerance induction. Diabetes. 1999;48:967–974. doi: 10.2337/diabetes.48.5.967. [DOI] [PubMed] [Google Scholar]
- 15.Kishimoto H, Sprent J. A defect in central tolerance in NOD mice. Nat. Immunol. 2001;2:1025–1031. doi: 10.1038/ni726. [DOI] [PubMed] [Google Scholar]
- 16.André I, et al. Checkpoints in the progression of autoimmune disease: lessons from diabetes models. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2260–2263. doi: 10.1073/pnas.93.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delovitch TL, Singh B. The nonobese diabetic mouse as a model of autoimmune diabetes: immune dysregulation gets the NOD. Immunity. 1997;7:727–738. doi: 10.1016/s1074-7613(00)80392-1. [DOI] [PubMed] [Google Scholar]
- 18.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 19.Lafaille JJ, Nagashima K, Katsuki M, Tonegawa S. High incidence of spontaneous autoimmune encephalomyelitis in immunodeficient anti-myelin basic protein T cell receptor transgenic mice. Cell. 1994;78:399–408. doi: 10.1016/0092-8674(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 20.Goverman J, et al. Transgenic mice that express a myelin basic protein-specific cell receptor develop spontaneous autoimmunity. Cell. 1993;72:551–560. doi: 10.1016/0092-8674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 21.Verdaguer J, et al. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J. Exp. Med. 1997;186:1663–1676. doi: 10.1084/jem.186.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 23.Kouskoff V, et al. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 24.Klein TC, Doffinger R, Pepys MB, Ruther U, Kyewski B. Tolerance and immunity to the inducible self antigen C-reactive protein in transgenic mice. Eur. J. Immunol. 1995;25:3489–3495. doi: 10.1002/eji.1830251242. [DOI] [PubMed] [Google Scholar]
- 25.Zal T, Volkmann A, Stockinger B. Mechanisms of tolerance induction in major histocompatibility complex class II-restricted T cells specific for a blood-borne self-antigen. J. Exp. Med. 1994;180:2089–2099. doi: 10.1084/jem.180.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 27.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 28.Pankewycz O, Strom TB, Rubin-Kelley VE. Islet-infiltrating T cell clones from non-obese diabetic mice that promote or prevent accelerated onset diabetes. Eur. J. Immunol. 1991;21:873–879. doi: 10.1002/eji.1830210403. [DOI] [PubMed] [Google Scholar]
- 29.Daniel D, Gill RG, Schloot N, Wegmann D. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur. J. Immunol. 1995;25:1056–1062. doi: 10.1002/eji.1830250430. [DOI] [PubMed] [Google Scholar]
- 30.Haskins K, Portas M, Bradley B, Wegmann D, Lafferty K. T-lymphocyte clone specific for pancreatic islet antigen. Diabetes. 1988;37:1444–1448. doi: 10.2337/diab.37.10.1444. [DOI] [PubMed] [Google Scholar]
- 31.Haskins K, Portas M, Bergman B, Lafferty K, Bradley B. Pancreatic islet-specific T-cell clones from nonobese diabetic mice. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8000–8004. doi: 10.1073/pnas.86.20.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haskins K, McDuffie M. Acceleration of diabetes in young NOD mice with a CD4+ islet-specific T cell clone. Science. 1990;249:1433–1436. doi: 10.1126/science.2205920. [DOI] [PubMed] [Google Scholar]
- 33.Nakano N, Kikutani H, Nishimoto H, Kishimoto T. T cell receptor V gene usage of islet beta cell-reactive T cells is not restricted in non-obese diabetic mice. J. Exp. Med. 1991;173:1091–1097. doi: 10.1084/jem.173.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candeias S, Katz J, Benoist C, Mathis D, Haskins K. Islet-specific T-cell clones from nonobese diabetic mice express heterogeneous T-cell receptors. Proc. Natl. Acad. Sci. U. S. A. 1991;88:6167–6170. doi: 10.1073/pnas.88.14.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergman B, Haskins K. Islet-specific T-cell clones from the NOD mouse respond to beta-granule antigen. Diabetes. 1994;43:197–203. doi: 10.2337/diab.43.2.197. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez A, Andre-Schmutz I, Carnaud C, Mathis D, Benoist C. Damage control, rather than unresponsiveness, effected by protective DX5+ T cells in autoimmune diabetes. Nat. Immunol. 2001;2:1117–1125. doi: 10.1038/ni738. [DOI] [PubMed] [Google Scholar]
- 37.Lühder F, Höglund P, Allison JP, Benoist C, Mathis D. Cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) regulates the unfolding of autoimmune diabetes. J. Exp. Med. 1998;187:427–432. doi: 10.1084/jem.187.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horwitz MS, et al. Diabetes induced by Coxsackie virus: initiation by bystander damage and not molecular mimicry. Nat. Med. 1998;4:781–785. doi: 10.1038/nm0798-781. [DOI] [PubMed] [Google Scholar]
- 39.Balasa B, Van Gunst K, Sarvetnick N. The microbial product lipopolysaccharide confers diabetogenic potential on the T cell repertoire of BDC-2.5/NOD mice: implications for the etiology of autoimmune diabetes. Clin. Immunol. 2000;95:93–98. doi: 10.1006/clim.2000.4855. [DOI] [PubMed] [Google Scholar]
- 40.Judkowski V, et al. Identification of MHC class II-restricted peptide ligands, including a glutamic acid decarboxylase 65 sequence, that stimulate diabetogenic T cells from transgenic BDC-2.5 nonobese diabetic mice. J. Immunol. 2001;166:908–917. doi: 10.4049/jimmunol.166.2.908. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida K, et al. Evidence for shared recognition of a peptide ligand by a diverse panel of NOD-derived, islet-specific, diabetogenic T cell clones. Int. Immunol. 2002;14:1439–1447. doi: 10.1093/intimm/dxf106. [DOI] [PubMed] [Google Scholar]
- 42.Peccoud J, Dellabona P, Allen P, Benoist C, Mathis D. Delineation of antigen contact residues on an MHC class II molecule. EMBO J. 1990;9:4215–4223. doi: 10.1002/j.1460-2075.1990.tb07869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez A, et al. Genetic control of diabetes progression. Immunity. 1997;7:873–883. doi: 10.1016/s1074-7613(00)80405-7. [DOI] [PubMed] [Google Scholar]
- 44.Basu D, Horvath S, Matsumoto I, Fremont DH, Allen PM. Molecular basis for recognition of an arthritic peptide and a foreign epitope on distinct MHC molecules by a single TCR. J. Immunol. 2000;164:5788–5796. doi: 10.4049/jimmunol.164.11.5788. [DOI] [PubMed] [Google Scholar]
- 45.Matsumura M, Saito Y, Jackson MR, Song ES, Peterson PA. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules isolated from transfected Drosophila melanogaster cells. J. Biol. Chem. 1992;267:23589–23595. [PubMed] [Google Scholar]
- 46.Scott CA, Garcia KC, Carbone FR, Wilson IA, Teyton L. Role of chain pairing for the production of functional soluble IA major histocompatibility complex class II molecules. J. Exp. Med. 1996;183:2087–2095. doi: 10.1084/jem.183.5.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia KC, et al. Alphabeta T cell receptor interactions with syngeneic and allogeneic ligands: affinity measurements and crystallization. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13838–13843. doi: 10.1073/pnas.94.25.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N. Y.). 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 49.Morton TA, Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- 50.McGavern DB, Christen U, Oldstone MBA. Molecular anatomy of antigen-specific CD8+ T cell engagement and synapse formation in vivo. Nat. Immunol. 2002;3:918–925. doi: 10.1038/ni843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto I, Staub A, Benoist C, Mathis D. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 52.Davis MM, et al. Ligand recognition by alpha beta T cell receptors. Annu. Rev. Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 53.Skinner PJ, Daniels MA, Schmidt CS, Jameson SC, Haase AT. Cutting edge: in situ tetramer staining of antigen-specific T cells in tissues. J. Immunol. 2000;165:613–617. doi: 10.4049/jimmunol.165.2.613. [DOI] [PubMed] [Google Scholar]
- 54.Haanen JB, et al. In situ detection of virus- and tumor-specific T-cell immunity. Nat. Med. 2000;6:1056–1060. doi: 10.1038/79573. [DOI] [PubMed] [Google Scholar]
- 55.Chen HD, et al. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 56.You S, et al. Detection and characterization of T cells specific for BDC2.5 T cell-stimulating peptides. J. Immunol. 2003;170:4011–4020. doi: 10.4049/jimmunol.170.8.4011. [DOI] [PubMed] [Google Scholar]
- 57.Höglund P, et al. Initiation of autoimmune diabetes by developmentally regulated presentation of islet cell antigens in the pancreatic lymph nodes. J. Exp. Med. 1999;189:331–339. doi: 10.1084/jem.189.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhardwaj V, Kumar V, Geysen HM, Sercarz EE. Degenerate recognition of a dissimilar antigenic peptide by myelin basic protein-reactive T cells. Implications for thymic education and autoimmunity. J Immunol. 1993;151:5000–5010. [PubMed] [Google Scholar]
- 59.Zhang Y, et al. In situ beta cell death promotes priming of diabetogenic CD8 T lymphocytes. J. Immunol. 2002;168:1466–1472. doi: 10.4049/jimmunol.168.3.1466. [DOI] [PubMed] [Google Scholar]
- 60.Jingwu Z, et al. Myelin basic protein-specific T lymphocytes in multiple sclerosis and controls: precursor frequency, fine specificity, and cytotoxicity. Ann. Neurol. 1992;32:330–338. doi: 10.1002/ana.410320305. [DOI] [PubMed] [Google Scholar]
- 61.Casrouge A, et al. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. J. Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 62.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 63.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murali-Krishna K, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 65.Zimmerman C, Brduscha-Riem K, Blaser C, Zinkernagel RM, Pircher H. Visualization, characterization, and turnover of CD8+ memory T cells in virus-infected hosts. J. Exp. Med. 1996;183:1367–1375. doi: 10.1084/jem.183.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blattman JN, et al. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casanova JL, Maryanski JL. Antigen-selected T-cell receptor diversity and self-nonself homology. Immunol. Today. 1993;14:391–394. doi: 10.1016/0167-5699(93)90140-G. [DOI] [PubMed] [Google Scholar]
- 68.Kanagawa O, Martin SM, Vaupel BA, Carrasco-Marin E, Unanue ER. Autoreactivity of T cells from nonobese diabetic mice: an I-Ag7-dependent reaction. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1721–1724. doi: 10.1073/pnas.95.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nepom GT, Kwok WW. Molecular basis for HLA-DQ associations with IDDM. Diabetes. 1998;47:1177–1184. doi: 10.2337/diab.47.8.1177. [DOI] [PubMed] [Google Scholar]
- 70.Chao C-C, Sytwu H-K, Chen EL, Toma J, McDevitt HO. The role of MHC class II molecules in susceptibility to type I diabetes: identification of peptide epitopes and characterization of the T cell repertoire. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9299–9304. doi: 10.1073/pnas.96.16.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu B, Gauthier L, Hausmann DHF, Wucherpfennig KW. Binding of conserved islet peptides by human and murine MHC class II molecules associated with susceptibility to type I diabetes. Eur. J. Immunol. 2000;30:2497–2506. doi: 10.1002/1521-4141(200009)30:9<2497::AID-IMMU2497>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 72.Wen L, Wong FS, Sherwin R, Mora C. Human DQ8 can substitute for murine I-A(g7) in the selection of diabetogenic T cells restricted to I-A(g7) J. Immunol. 2002;168:3635–3640. doi: 10.4049/jimmunol.168.7.3635. [DOI] [PubMed] [Google Scholar]
- 73.McDevitt HO, Benacerraf B. Genetic control of specific immune responses. Adv. Immunol. 1969;11:31–74. doi: 10.1016/s0065-2776(08)60477-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.