Abstract
We investigated the formation of X-shaped molecules consisting of joint circular minichromosomes (joint molecules) in Saccharomyces cerevisiae by two-dimensional neutral/neutral gel electrophoresis of psoralen-cross-linked DNA. The appearance of joint molecules was found to be replication dependent. The joint molecules had physical properties reminiscent of Holliday junctions or hemicatenanes, as monitored by strand displacement, branch migration, and nuclease digestion. Physical linkage of the joint molecules was detected along the entire length of the minichromosome and most likely involved newly replicated sister chromatids. Surprisingly, the formation of joint molecules was found to be independent of Rad52p as well as of other factors associated with a function in homologous recombination or in the resolution of stalled replication intermediates. These findings thus imply the existence of a nonrecombinational pathway(s) for the formation of joint molecules during the process of DNA replication or minichromosome segregation.
DNA replication is a highly coordinated process providing an accurate duplication of the chromosomes required for faithful transmission of the genomic information from mother to daughter cells. Studies of mitotic cells have led to a unified model for DNA replication involving three distinct steps; initiation, elongation, and termination (for a review, see reference 57). Several studies seem to corroborate the general features of the bidirectional replication model, which proposes that pairs of divergent replication forks travel away from their origins until they meet the forks coming from opposite directions from the neighboring replicons (22, 42). When two replication forks converge at the end of DNA synthesis, the leading-end polymerases advance toward each other, melting the remaining duplex region, and finish the replication by producing catenated DNA molecules (50, 51). An alternative model for termination proposed that one of the advancing polymerases dislodges from the template by accumulated torsional stress, so that only one fork can advance. In subsequent reaction steps, including strand displacement and branch migration, hemicatenated DNA is formed and resolved by type I topoisomerases (31).
DNA recombination is an integral part of DNA replication and contributes to the structural integrity of the newly replicated genome (for recent reviews, see references 3, 10, and 30). In replicating DNA, X-shaped molecules (xDNA) with properties of Holliday junction intermediates of homologous recombination (21) can be distinguished physically from replicating molecules by two-dimensional gel electrophoresis (5, 43). The formation of such joint molecules was recently studied within the Saccharomyces cerevisiae ribosomal DNA (rDNA) locus (60) and various loci of Physarum polycephalum (6). In both organisms, Holliday junction-like xDNA molecules were more abundant in S phase. Interestingly, mutations in polymerases α and δ stimulated the formation of Holliday junctions in the S. cerevisiae rDNA locus DNA. The elevated levels of Holliday junctions were found to be dependent on RAD52 but not on the yeast homologs of recA.
It has been postulated that these Holliday junctions represent intermediate structures of the repair of DNA lesions at the replication fork (60). DNA samples of naturally synchronous nuclei of Physarum polycephalum were used to demonstrate that replication-dependent xDNA molecules are formed between sister chromatids and that the spontaneous interaction between sister chromatids occurred after the passage of the replication fork. Physical interactions between sister chromatids could result from either DNA repair events induced at sites of DNA lesions or incomplete nascent-strand synthesis (20, 25), or they might be established during replication to maintain a close proximity between the sister chromatids to allow recombinational DNA repair if needed (6).
Joint molecules seem to be a central intermediate of DNA replication and recombination. Therefore, we strove to investigate the molecular structure of joint molecules. For this purpose, we developed a technique that enables the selective enrichment of physically linked, homologous minichromosomes. We found that in vegetatively growing yeast cells, joint molecules are frequently formed during S phase. The joint molecules have the physical properties of Holliday junctions and/or hemicatenates, as deduced by strand displacement, branch migration, and nuclease sensitivity assays. Joints were found all over the circular minichromosome but more often within the zone of replication termination. The formation of joint molecules was not dependent on Rad52p or on various other proteins involved in DNA recombination, replication, and repair. Our results suggest that joint molecules are postreplicative molecules that result from the replication segregation of the newly synthesized minichromosomes.
MATERIALS AND METHODS
Strains, media, and plasmids.
S. cerevisiae A1 (MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δbar1::LYS2) was used for all but one set of experiments, for which S. cerevisiae FF18733 (MATa leu2-3,112 trp1-289 ura3-52 his7-2 lys1-1) and isogenic mutant strains were used. To generate the mutants, the gene of interest was replaced by either the kanamycin resistance gene (MUS81, REV3, SGS1, SRS2, RAD52, and RAD54) or the URA3 gene, which was subsequently inactivated by selection for 5-fluoroorotic acid-resistant and uracil-auxotrophic clones (RAD51 and TOP1). Isogenic strains were obtained either by direct gene replacement in the FF18733 strain (Δrev3 Δsrs2 Δrad51 Δrad52 Δrad54) or by multiple back-crosses of the W303-1B (Δsgs1 Δtop1) and BY-1B (Δmus81) strains into the FF18733 background. All strains were transformed with YRpTRURAP (54) and grown at 30°C in SD minimal medium supplemented with the appropriate amino acids as described previously (47).
α-Factor synchronization.
A1 cells grown in SD to a density of 5 × 106 cells per ml were synchronized with α-factor as previously described (36).
DNA isolation, enrichment of plasmid DNA by polyethylene glycol, psoralen-DNA cross-linking, and preparative gel electrophoresis.
Unless indicated, chemicals were purchased from Sigma. For Fig. 1 and 2, total DNA of early-log-phase cells was isolated and purified on Qiagen G100 columns according to the protocol of the Qiagen genomic DNA handbook. To avoid possible branch migration, total DNA of early-log-phase yeast cells was isolated according to Allers and Lichten (1) with modifications. In brief, 50 ml of cells was harvested by centrifugation, washed with water, and resuspended in 1 ml of spheroplasting buffer (1 M sorbitol, 10 mM EDTA, 5 mM hexamine cobalt trichloride [HCC], 0.1% [vol/vol] β-mercaptoethanol) containing 1,000 U of zymolyase (Seikagaku), and incubated for 45 min at 30°C. Sequentially, 200 μl of water, 25 μl of RNase A (10 mg/ml), 25 μl of proteinase K (20 mg/ml), and solution I (2% cetyltrimethylammonium bromide [CTAB], 1.4 M NaCl, 100 mM Tris [pH 7.6], 25 mM EDTA, 20 mM HCC) were added to the spheroplasts, and the sample was incubated at 37°C for 15 min.
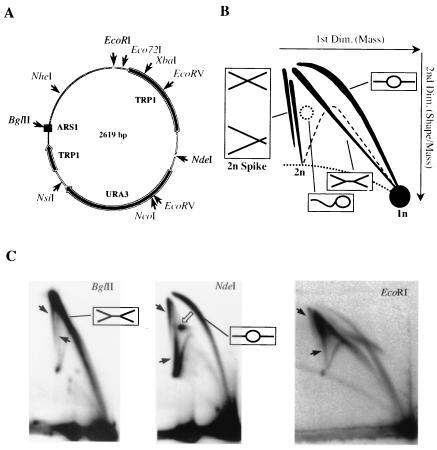
FIG. 1.
Two-dimensional gel analysis of YRpTRURAP. (A) Schematic representation of YRpTRURAP. Shown are relevant restriction sites (thin arrows), the origin of replication (ARS1, black box), and the TRP1 and URA3 genes (thick arrows) (for details, see reference 54). (B) Diagram of the migration patterns of replication and recombination intermediates of singly-cut minichromosome after two-dimensional gel electrophoresis. The direction of first- and second-dimension electrophoresis (arrows), migration pattern bubble- and double-Y-shaped molecules (black lines; interpretation in boxes), as well as simple Y-shaped molecules (dashed lines) are indicated. The double spike represents the migration pattern of two populations of 2n-shaped molecules. See text for details. (C) Two-dimensional gel electrophoretic analysis of YRpTRURAP from an exponential-phase culture. Prior to analysis, Qiagen-purified DNA was linearized with the restriction enzyme indicated on top. Depicted are the resulting replicating molecules (interpretation in boxes), different populations of molecules in the 2n spike (black arrows), and δ-like structures (open arrow). To achieve sufficient resolution of the 2n spikes of the EcoRI-digested DNA, the second-dimension electrophoresis was prolonged (17 h).
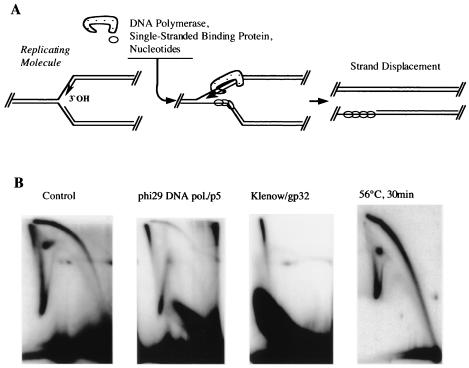
FIG. 2.
2n spike contains strand displacement-resistant but branch migration-sensitive population of molecules. (A) Scheme depicting the effect of DNA polymerase and single-stranded-DNA binding protein on replicating molecules. Initiation of strand displacement at the leading strand (arrow, 3′OH) is indicated. (B) Two-dimensional gel electrophoretic analysis of YRpTRURAP replicating and recombining intermediates linearized with NdeI (see Fig. 1A) following the treatment indicated above the autoradiograms. Note that different gel running conditions (1.5% agarose in the second dimension) were used in the three first panels and that the DNA was purified on Qiagen columns.
The proteins were extracted with 300 μl of chloroform-isoamyl alcohol (24:1), and the supernatant was transferred to a new tube. DNA was precipitated by adding 2 volumes of solution II (1% CTAB, 50 mM Tris [pH 7.6], 10 mM EDTA, 5 mM HCC) and centrifugation for 5 min at full speed in a microcentrifuge. The DNA was resuspended in 0.5 ml of solution III (1.4 M NaCl, 10 mM Tris [pH 7.6], 1 mM EDTA, 1 mM HCC), precipitated with 1 volume of isopropanol, and centrifuged at full speed for 10 min in a microcentrifuge. The pellet was briefly rinsed with 70% ethanol, air dried, resuspended in 100 μl of 1× restriction buffer containing MgCl2, and stored at 4°C. To enrich for plasmid DNA, 27 μl of 2.5 M NaCl-20% polyethylene glycol 8000 was added, and the solution was incubated at 4°C for 2 h. The chromosomal DNA was precipitated by centrifugation in a microcentrifuge at 13,000 rpm at 4°C. The supernatant was applied on a G50 column equilibrated with 1 mM MgCl2-1 mM Tris (pH 7.6), and the flowthrough was precipitated with 250 mM NaCl and 1 volume of isopropanol. The pellet was rinsed with 70% ethanol, air dried, resuspended in 20 μl of 1 mM MgCl2-10 mM Tris (pH 7.6), and stored at 4°C.
Preparative gel electrophoresis and DNA isolation were basically done as described earlier (17). Enriched minichromosomal DNA was digested with Eco72I (Fermentas) and psoralen cross-linked under the conditions stated below. Per slot, about 500 ng of minichromosomal DNA was electrophoresed in a 0.5% low-gelling-temperature agarose gel (SeaPlaque; FMC) at 1 V/cm for 20 h. A gel slice at the position corresponding to the 1n and 2n-sized minichromosomal DNA was cut out, and the DNA was recovered by digesting the agarose with AgarACE enzyme (Promega). In a 1.5-ml tube, a 250-mg gel slice was heated at 72°C for 5 min and cooled to 42°C and then 1 U of agarose was added. The sample was further incubated at 42°C for 2 h, and the DNA was recovered by subsequent isopropanol precipitation with HindIII-digested λ phage DNA added as a carrier.
Restriction enzyme, T4 endonuclease VII, and mung bean and S1 nuclease digestion.
Restriction enzyme, mung bean, and S1 nuclease digests were carried out in the conditions recommended by the supplier (Roche). T4 endonuclease VII digests were done as described (17).
Branch migration, strand displacement assay, and psoralen cross-linking.
To induce branch migration, restriction enzyme-digested DNA was applied on a G50 column equilibrated in 1 mM Tris (pH 7.6), precipitated with isopropanol, resuspended in 20 μl of branch migration buffer (10 mM EDTA, 1 mM Tris, pH 8.0), and incubated at 56°C for 30 min. For the strand displacement reaction, either Klenow or φ29 polymerase was used. Stepwise, 1 μl of nucleotide mix (5 mM each dATP, dCTP, dGTP, and dTTP; Pharmacia) and 0.5 μl of gp32 protein (4 μg/μl; USB), 0.5 μl of Klenow (2 U/μl; Roche), or 0.5 μl of p5 single-strand binding protein (4 μg), and 1 μl of φ29 DNA polymerase (20 ng) were added to 18 μl of restriction enzyme-digested DNA in 1× restriction buffer. The sample was incubated for 30 min at 37°C. Possible branch migration and strand displacement were inhibited by addition of 1 μl of 4,5′,8-trimethylpsoralen and cross-linking with a 366-nm UV lamp (model B-100 A; Ultra Violet Products, Inc., San Gabriel, Calif.) at a distance of 6 cm for 5 min as previously described (11).
Agarose gel electrophoresis, Southern transfer, and hybridization.
The procedures described by Brewer and Fangman (8) for two-dimensional gels were used with the following minor modifications. The first dimension was 0.5% agarose (Life Technologies) and was run at 1 V/cm for 20 h. The second dimension was run at 4 V/cm for 12 h in 1.0% or 1.2% agarose. Alkaline Southern blotting and hybridization were done as previously described (35) with a gel-purified, randomly 32P-labeled fragment containing YRpTRURAP as a probe.
Quantification of replicative intermediates and joint molecules.
All measurements were done with the Molecular Dynamics ImageQuant software. In brief, after running the first dimension, equal loading of monomer DNA was confirmed by visual inspection. Monomer DNA was cut off, and the remaining DNA was separated in a second-dimension gel. As depicted in Fig. 4A, the signals obtained for the bubble arc, δ-molecules, and the two 2n spikes were quantified with the indicated areas. To exclude nonspecific background signal, signal obtained from the same areas at a membrane region distant from the replicative intermediates was subtracted.
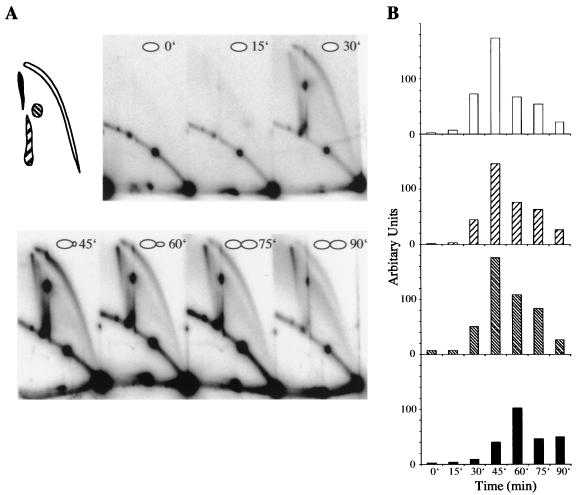
FIG. 4.
Two-dimensional gel electrophoretic analysis of YRpTRURAP from a synchronized culture. (A) Yeast cells were synchronized by α-factor arrest. After release from arrest, samples were withdrawn from the culture, and CTAB-purified DNA was digested with NdeI (see Fig. 1A). The time points taken are indicated on the autoradiogram. The regions used for quantification are indicated in the left diagram. (B) The quantitative data from A were plotted as a function of time. Replicating molecules include the bubble arc (white bars), lasso-type molecules (right dashed bars), and 2n-sized replicative intermediates (left hatched bars). While replicating molecules were most abundant after 45 min, the majority of SDR molecules were apparent 60 min after α-factor release (black bars).
RESULTS
Discrimination of 2n molecules by two-dimensional gel electrophoresis.
We made use of YRpTRURAP, a small multicopy circular minichromosome that contains an origin of replication (ARS1) and the URA3 gene as a selection marker (Fig. 1A) (54). DNA replicative intermediates were separated according to their mass and shape by two-dimensional agarose gel electrophoresis (Fig. 1B) (8). Depending on the restriction enzyme used to linearize the minichromosomal DNA, we mainly observed the appearance of double-Y (BglII), bubble-shaped (NdeI), or a mixture of asymmetric bubble and asymmetric double-Y intermediates (EcoRI) (Fig. 1C). This result is in agreement with previous studies (8, 23, 42) which showed that bidirectional replication of a yeast minichromosome terminates in a region opposite the origin of replication (ARS1).
In the NdeI-digested DNA, a prominent spot (Fig. 1C, open arrow) appeared that consisted of molecules with a migration behavior different from that of the bubble-shaped molecules. Replicating simian virus 40 (SV40) molecules with similar migration behavior were previously isolated from preparative two-dimensional gels. Analysis of these molecules by electron microscopy showed that they have a δ-like structure (J. Wu and J. M. Sogo, unpublished data). Remarkably, two distinct populations of molecules were resolved into two 2n spikes, which presumably correspond to either nearly fully replicated molecules or recombining molecules (black arrows). The faster migration of the nearly fully replicated molecules corresponds to the expected mobility of structures close to but smaller than 2n. To avoid possible loss of 2n molecules, in some experiments minichromosome replication and recombination intermediates were stabilized during DNA isolation by the presence of cetyltrimethylammonium bromide (CTAB) (1) and after various enzymatic treatments by psoralen-DNA cross-linking (36, 43).
2n-sized molecules resistant to strand displacement.
We reasoned that the specific removal of replicating DNA molecules would allow the identification of recombining molecules among the 2n-sized species and vice versa. Resolution of replicating molecules was achieved by the addition of a DNA polymerase, single-stranded-DNA binding protein, and nucleotides which together induce a strand displacement reaction as described for the replication of the linear phage φ29 (Fig. 2A) (7). Strand displacement by the φ29 P2/P5 complex led to a strong reduction of bubble- and double-Y-shaped replication intermediates, and the 2n spike representing the faster-migrating molecules (Fig. 2B). Other DNA polymerases in combination with a single-stranded-DNA binding protein were also shown to have strand displacement activity (58). After Klenow and gp32 treatment, nearly all the replicating molecules disappeared except the fraction of strand displacement-resistant (SDR) molecules, which were already observed after P2/P5 treatment. Recombining molecules show properties similar to those of very late replicating molecules. However, they are easily destroyed by branch migration. To confirm that the SDR molecules show the properties of recombination intermediates, we induced a branch migration reaction by depletion of multivalent cations and heat treatment. After 30 min of incubation under branch migration-promoting conditions, the replication intermediates were still present, while most of the SDR molecules were resolved and disappeared.
SDR molecules are nuclease sensitive.
Because the SDR molecules were sensitive to branch migration, we assumed that they represented recombination intermediates. The classical intermediate during homologous recombination is a Holliday junction (21). Previous studies showed that T4 endonuclease VII cleaves synthetic Holliday junctions efficiently but also recognizes and processes other related DNA structures (24, 26, 27), including replication intermediates (17). While T4 endonuclease VII converts replicating molecules into double-stranded fragments of size n and smaller, the cleavage of Holliday junction-like structure should generate only n-sized minichromosomes (Fig. 3A). T4 endonuclease VII treatment led to the disappearance of most of the branched molecules (Fig. 3B). We noted that some of these molecules were only partially cleaved by T4 endonuclease VII (only one arm of the 2n molecules was cleaved), which can be seen by the appearance of a faint simple Y arc (right panel). This reduction in T4 endonuclease VII cleavage efficiency most likely resulted from psoralen cross-linking of the DNA, because partially digested molecules were virtually absent in non-cross-linked DNA (data not shown).
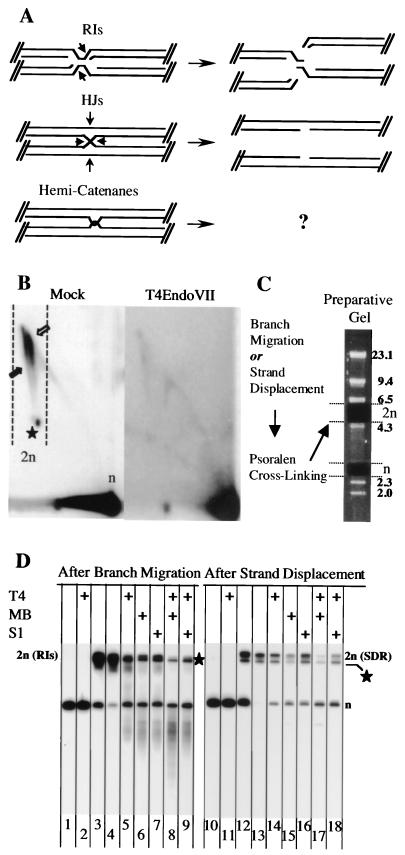
FIG. 3.
Enzymatic characterization of 2n molecules. (A) Putative shapes of 2n molecules. Indicated are the nuclease cleavage sites necessary for structure resolution (arrows). RIs, replicative intermediates; HJs, Holliday junctions. (B) Two-dimensional gel analysis of mock-treated (left) and T4 endonuclease VII-treated (right) minichromosomes. CTAB-purified DNA from cells 90 min after release from α-factor was linearized with Eco72I (see Fig. 1). The locations of the n- and 2n-sized molecules (arrows and star, respectively, within the dashed lines) are indicated. Note that the experimental conditions led to clear detection of both 2n spikes. In contrast, two-dimensional gel analysis of Eco72I-digested DNA isolated from synchronized cells 45 min after α-factor release (data not shown) gave a result similar to that obtained with EcoRI-digested DNA (see Fig. 1C). (C) Schematic illustration for the isolation of n- and 2n-sized molecules from a preparative gel. After branch migration or strand displacement, molecules were further stabilized by psoralen-DNA cross-linking. With this treat-ment, structural changes are prevented during the high-temperature DNA extraction procedure from low-melting-point agarose. To remove the correct agarose gel plugs, TRURAP DNA was mixed with HindIII-digested lambda marker DNA (fragment sizes are indicated in kilobases to the right). (D) Southern blot analysis of 2n molecules after branch migration (lanes 1 to 9) or strand displacement followed by psoralen-DNA cross-linking (lanes 10 to18). The n-sized molecules (lanes 1, 2, 10, and 11) and 2n-sized molecules were isolated once (lanes 3 and 12) or twice (lanes 4 to 9 and 13 to 18) from preparative gels. The DNA was either mock treated (lanes 1, 3, 4, 10, 12, and 13) or treated with T4 endonuclease VII (T4), mung bean nuclease (MB), or S1 nuclease (S1), as indicated on top. The migration of the n-sized molecules is indicated to the right. The star indicates putatively linear molecules of size 2n (see text for details). The smear-like signal between the n and 2n molecules (lanes 5 to 7) most likely resulted from partially cleaved replication intermediates. Note that whereas T4 endonuclease VII specifically cleaves the leading arm of the replication forks (17), the mung bean and S1 nucleases can also cut the lagging arm of the replication forks.
Next, we enriched for very late replication intermediates (after branch migration), SDR molecules (after strand displacement), and monomers by monodirectional preparative gel electrophoresis (outlined in Fig. 3C). As illustrated in Fig. 3D, 2n molecules were strongly enriched after twofold purification by preparative gel electrophoresis (lanes 4 and 13). However, we repeatedly isolated a fragment that migrated slightly faster than the SDR and replication intermediate molecules (Fig. 3D, star). Although we cannot explain the origin of this fragment, it most likely corresponds to 2n molecules with migration properties similar to those of linear molecule (see also Fig. 3B, star). These linear 2n molecules probably arose from partial digestion of circular minichromosome multimers. Linear 2n molecules are expected to migrate slightly faster than branched molecules of nearly the same size.
T4 endonuclease VII did not alter the structure of isolated plasmid monomers (Fig. 3D, lanes 2 and 11). However, replication intermediates (Fig. 3D, lane 5) and SDR molecules (Fig. 3D, lane 14) were converted into smaller molecules. While cut-replicating molecules were size n and smaller, SDR molecules were converted solely into n-sized products, as predicted (Fig. 3A). Four-way junctions and replication intermediates (3, 19, 48) are expected to contain a short distorted region or short single-stranded DNA stretches, respectively. We reasoned that these structures might be sensitive to nucleases, which act on gapped (mung bean nuclease) and/or nicked (S1 nuclease) DNAs (33). Both replicating and SDR molecules were affected by mung bean (Fig. 3D, lanes 6 and 15) and S1 nuclease treatment (Fig. 3D, lanes 7 and 16). Like T4 endonuclease VII, these nucleases cut replication intermediates into molecules of size n and smaller, while SDR molecules were converted solely into n-sized molecules. Notably, double digestion of the DNAs with T4 endonuclease VII and mung bean (Fig. 3D, lanes 8 and 17) or S1 (Fig. 3D, lanes 9 and 18) nuclease led to an additive digestion of replicating and SDR molecules. This additive effect might be best explained by a complementary mode of cleavage and sequence specificity of the nucleases tested.
SDR molecules are S-phase dependent.
Zou and Rothstein described the appearance of recombinant DNA molecules in the rDNA locus of S. cerevisiae during the S phase of the cell cycle (60), while work done in Physarum polycephalum showed that the formation of joint DNA molecules was tightly linked to DNA replication and occurred in late S phase (6). These findings prompted us to analyze the formation of SDR molecules during the cell cycle in more detail. Cells were synchronized in G1 with α-factor, and following release from the arrest, samples were withdrawn at various time points to isolate the plasmid DNA. NdeI-digested and psoralen cross-linked molecules were analyzed by two-dimensional gel electrophoresis and Southern blotting (Fig. 4A). The areas depicted in Fig. 4A were used to quantify the levels of SDR molecules and replication intermediates (Fig. 4B). As shown in Fig. 4A, SDR molecules were not present at the G1/S boundary, but their formation was stimulated during S phase. Separate quantification of the various types of replication intermediates revealed that replication was most strongly stimulated approximately 45 min after α-factor release (Fig. 4B). However, relative to the replication intermediates, the peak level of SDR molecules was shifted to 60 min after the release, and half of them were still present after 90 min. Thus, the formation and resolution of SDR molecules appeared to be retarded relative to the replication intermediates. This might be interpreted as meaning that SDR molecule formation is a postreplicative process (6) or that SDR molecules are resolved later than replicating DNA.
Formation of SDR molecules is Rad52 independent.
Next, we wanted to examine the genetic requirements for the formation of SDR molecules. Since the SDR molecules are only present during S phase, we mainly focused on genes involved in the control of replication-associated recombination. These included members of the RAD52 epistasis group (for reviews, see references 40 and 52) and the genes encoding the Holliday junction resolution endonuclease Mus81p (for a review, see reference 18), the Rev3p component of the DNA polymerase ζ involved in translesion synthesis (for a review, see reference 53), the Sgs1p and Srs2p helicases (for a review, see reference 28), and the Top1p topoisomerase (for a review, see reference 59). To get comparable levels of replicating cells and thus recombination intermediates, cultures were grown to the mid-exponential phase, and cells were harvested at optical densities at 600 nm of 0.5 to 0.6.
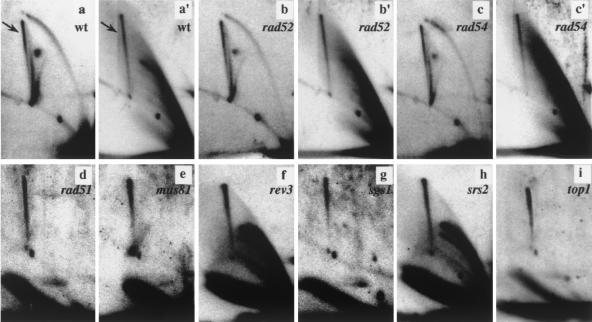
Prior to psoralen cross-linking and two-dimensional gel analysis (Fig. 5), NdeI-digested DNA was either mock treated (panels a, b, and c) or incubated with Klenow polymerase, gp32, and nucleotides to induce strand displacement (panels a′, b′, c′, and d to i). Previous studies showed that virtually all mitotic homologous recombination in S. cerevisiae depends on a functional Rad52 protein (25). Strikingly, the absence of Rad52p did not affect the level of SDR molecule formation (compare panels a′ and b′) and, in accordance with previous observations that Holliday junctions accumulate in DNA polymerase α- and δ-deficient cells via a RecA homolog-independent mechanism (60), the RAD51 and RAD54 genes were also dispensable (Fig. 5, panels c′ and d). While mutations of genes in the RAD52 epistasis group were likely to reduce the level of SDRs, the lack of enzymes possibly involved in Holliday junction resolution might be expected to increase the level of Holliday junction. We can exclude that the Mus81p-containing Holliday junction resolvase complex is required for SDR molecule resolution because joint molecule accumulation was not affected by deletion of MUS81 (Fig. 5, panel e).
FIG. 5.
SDR molecule formation does not depend on proteins involved in recombination. Two-dimensional gel electrophoretic analysis of CTAB-purified and NdeI-digested minichromosomes before (panels a to c) and after (panels a′ to c′ and d to i) strand displacement. DNAs from the wild type (wt, panels a and a′) and the isogenic rad52 (panels b and b′), rad54 (panels c and c′), rad51 (panel d), mus81 (panel e), rev3 (panel f), sgs1 (panel g), srs2 (panel h), and top1 (panel i) strains are shown. The SDR molecules are indicated (arrows). We noticed a variation in the efficiency of the strand displacement reaction that was apparently related to the sample preparation. No apparent effects on replication intermediate distribution were detected on the mutant strains analyzed with respect to the wild type (compare panel a with panels b and c, and data not shown).
Although we analyzed DNA from cells which were not exposed to DNA-damaging conditions, we could not rule out the possibility that SDR molecules might originate from DNA repair or translesion bypass events at sites of stalled replication forks. For this purpose, we tried to channel the resolution of aberrant replication structures into the homologous recombination pathway by mutation of either the translesion DNA polymerase Rev3p (Fig. 5, panel f) or the Sgs1p and Srs2p helicases (Fig. 5, panels g and h) (15). Remarkably, deletion of these genes did not increase the level of SDR molecules. Taken together, these results appear to be in conflict with the notion that the SDR molecules are intermediates of homologous recombination, e.g., Holliday junction-like molecules, and support the idea that they consist of hemicatenanes. Type I-like topoisomerase are able to catalyze the formation and resolution of hemicatenanes (34). Interestingly, Top1p was also dispensable for the formation and resolution of SDR molecules (Fig. 5, panel i). However, we cannot strictly discount the possibility that Top1 is involved in SDR processing but other topoisomerases substitute for its activity in the top1 mutant background (41). Similarly, the SDR spike was clearly detectable in top3 mutant cells (data not shown).
Joining of SDR molecules is detected throughout the entire minichromosome.
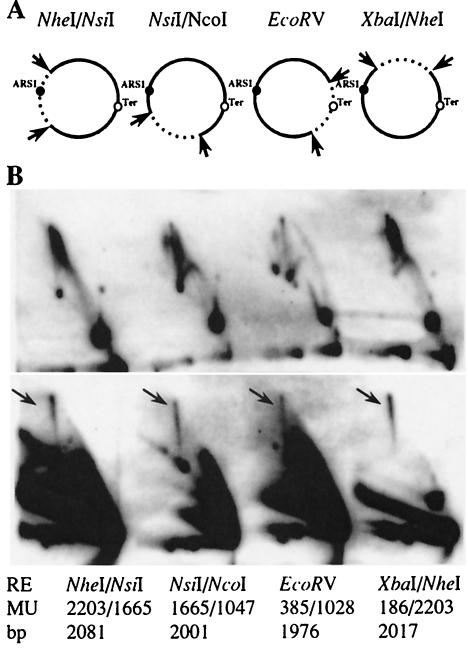
The formation and resolution of SDR molecules during S phase were apparently retarded compared to the replication intermediates. Thus, the separation process of the newly replicated minichromosomes could lead to the appearance of SDR molecules (31, 48). We therefore analyzed the frequency with which joints occur at different sites along the minichromosome, including a region opposite the origin of replication, where the majority of replication termination takes place (see Fig. 1C). Prior to two-dimensional gel analysis, the minichromosomal DNA was digested with selected restriction enzymes to generate various deletions of about 500 bp (Fig. 6A). With the radioactively labeled full-length minichromosome as the probe, we highlighted the joint molecules corresponding to the 2-kb fragments of the minichromosome DNA before (Fig. 6B, top panel) and after (Fig. 6B, bottom panel) strand displacement. Note that the 500-bp fragment migrated out of the gel and could therefore not be detected. The presence of the 2n spikes after strand displacement was clearly detected in all restriction fragments analyzed (Fig. 6B, arrows). This result indicates that, besides replication termination, other parameters, e.g., replication elongation, determine the physical linking of SDR molecules. Interestingly, only cleavage of the DNA with EcoRV resulted in a decrease in the signal intensity of SDR molecules (data not shown, but compare spike signals in Fig. 6B).
FIG. 6.
Frequency of SDR molecules in various regions of YRpTRURAP. (A) CTAB-purified plasmid DNA was digested with different restriction enzymes (arrow) to create two fragments of about 500 bp and 2 kb in length. The relative location of ARS1 (black dot) and the zone of replication termination (Ter, empty dot) are indicated. (B) Two-dimensional gel electrophoresis of the fragments before (top panel) and after (bottom panel) strand displacement. The SDR molecules are marked (black arrow). Cleavage sites of the restriction enzymes (RE) relative to EcoRI in map units (MU; EcoRI = map unit 0; see Fig. 1) and the expected size of the 2-kb fragment (in base pairs) are indicated at the bottom.
DISCUSSION
In this work, we analyzed the molecular structure and formation of xDNA molecules, which occur during replication of vegetatively growing yeast cells. Our results confirm that minichromosome replication in S. cerevisiae initiates at the ARS1 origin and proceeds in a bidirectional manner (8, 23, 42). Termination of replication did not occur at a specific site diametrically opposed to the origin but most frequently along the hemisphere opposite the ARS, as evidenced by a vertical spike in two-dimensional gel analyses. This vertical spike has been observed in replicating DNA of a variety of organisms (6, 9, 16, 44, 60). Yet, in contrast to other studies, our experimental conditions revealed the existence of a second so-called 2n spike. This led us to propose that one of the 2n spikes consisted of recombinant structures, while the other consisted of nearly fully replicated minichromosomes.
The structural similarities of late replicating and joint DNA molecules have always been an impediment in the analysis of xDNA. We reasoned that the free 3′ hydroxyl group present in the elongating strand of a replicating molecule could serve as a primer for subsequent DNA synthesis by a DNA polymerase (see Fig. 2A). Indeed, the replication intermediates were a substrate for strand displacement by φ29 or Klenow polymerase, in combination with single-stranded-DNA binding protein. Since joint DNA was sensitive to branch migration, we were able to subdivide the 2n spikes into two physically distinct categories of DNA molecules. One 2n spike contained (slightly slower migrating) molecules resistant to strand displacement and sensitive to branch migration (SDR molecules), while molecules in the second 2n spike (termed replication intermediates) showed the opposite properties.
SDR molecules with intact phosphodiester backbones consist of either Holliday junctions or hemicatenated DNA (see also Fig. 3A). To discriminate between these possibilities, we subjected the SDR molecules to nuclease digestion. Several nucleases have been shown to cleave synthetic Holliday junctions (46). Among these, T4 endonuclease VII has the poorest DNA sequence specificity and also exhibits general activity on branched DNA (26). T4 endonuclease VII cleaved SDR molecules into linear duplex products of size n. Resolution of the SDR molecules into linear duplex products of size n supports the idea that they consist of Holliday junctions or T4 endonuclease VII-sensitive hemicatenanes. Remarkably, digestion of SDR molecules with nucleases that have a single-strand specificity led to the same result. The crystal structures of synthetic Holliday junctions showed minor distortions from B-DNA at the junction (14), but it remains to be elucidated to what extent such distortions are recognized by the nucleases. Thus, nuclease digestions also allowed us to distinguish between replication intermediates and SDR molecules, but they did not discriminate between Holliday junctions and hemicatenanes
Since the appearance of joint molecules in the S. cerevisiae rDNA array (60) as well as in a single-copy DNA fragment of P. polyceaphalum (6) and Schizosaccharomyces pombe (44) was shown to be tightly restricted to S phase, we examined the cell cycle dependency of SDR molecule formation. SDR molecules were essentially absent at the G1/S boundary (after α-factor arrest), although in haploid cells the minichromosomes exist in multiple copies and therefore could serve as a substrate for joint molecule formation by homologous recombination. The appearance of SDR molecules was dependent on entry of the cells into S phase and appeared to be retarded relative to the formation of replication intermediates. Thus, either the SDR molecules are formed postreplicative, or they are the result of a process that is active only in late S phase. In accordance with other reports (6), we noted that hydroxyurea further retarded the formation of joint molecules (90 min after release in hydroxyurea; data not shown). ARS1 is a replication origin that is activated during early S phase, and the presence of hydroxyurea still led to replication activation. However, the slowing down of DNA synthesis by hydroxyurea (49) delays the termination of replication and the appearance of the joint molecules, suggesting that these events are connected.
We envisaged that SDR molecules are intermediates of the repair or the bypass of DNA lesions during replication or replication termination. We monitored SDR molecule levels after the disruption of genes that control the DNA strand exchange reaction during homologous recombination (RAD52, RAD51, and RAD54) (40) or the formation, prevention, and resolution of reversed forks, including a lesion bypass polymerase, DNA helicases, and a Holliday junction resolvase (REV3, SGS1, SRS2, and MUS81) (29). Strikingly, disruption of all of these genes did not alter the level of SDR molecule formation. It is particularly surprising that Rad52p is dispensable, because this protein is vital for mitotic homologous recombination and Holliday junction formation in rDNA (60) and cannot be substituted for by other proteins, including its structural homologue, Rad59 (2). Thus, either a yet-to-be-identified protein(s) is able to form SDR recombinant molecules by a process independent of Rad51p, Rad52p, and Rad54p or the SDR molecules do not represent intermediates of homologous recombination.
Reversed replication forks have been proposed to arise at sites of stalled DNA replication (20, 49). A reversed fork would allow the bypass of DNA lesions on the leading-strand template (20), or could be cleaved by Holliday junction resolvases, generating a broken replication fork (45). Obviously, the processing of a reversed replication fork might be involved in the generation of SDR molecules but disruption of either REV3, SGS1, SRS2, or MUS81 had no noticeable effect on the level of SDR molecules. This result is in line with a delayed appearance of SDR molecules relative to the progression of replication forks. If reversed forks were the source of SDR molecules, one might have expected contemporaneous formation of replication intermediates and X-shaped DNA.
Finally, we envisioned the possibility that SDRs might be an intermediate in the late steps of replication termination. Topoisomerases are needed to unlink DNA during and after replication (56). The effects of topoisomerase inactivation in S. cerevisiae include minichromosome multimerization (55) and aberrant replication termination and recombination (32). Type I topoisomerases carry out reactions that involve the breaking of only one strand of DNA (4). Therefore, strains carrying mutations in TOP1 should display increased levels of SDR molecules if this protein is needed to resolve hemicatenanes. However, the disruption of TOP1 did not affect either the formation or the resolution of SDR molecules. It is conceivable that Top1p is dispensable for SDR molecule resolution because it might be substituted for by another type I or even a type II enzyme (41).
What is the origin of SDR molecules? All functions tested were dispensable for the formation and/or the resolution of replication-dependent SDR yeast minichromosomes. This finding is a priori surprising and implies that the source of SDR molecule formation is not a homologous recombination event between sister chromatids (25). Our observation that the crossing of minichromosomes is more frequent in a zone of replication termination (EcoRV cleavage led to a decrease in the signal intensity of the SDR molecules; see Fig. 6B) suggests that the formation of a substantial amount of SDR molecules is associated with termination of replication. Electron microscopy should provide an elegant method for visualization of SDR molecules and for mapping the branch point of joint molecules.
SV40 molecules have the structural features of joint circular minichromosomes. In electron micrographs of SV40 molecules, we frequently observed replicated DNA molecules with point connections at the level of single-stranded DNA (48). Individual micrographs of psoralen cross-linked molecules spread under denaturing condition (31, 48) have not determined whether the two circular molecules were joined by strand exchange or by hemicatenation. However, most of the molecules were observed to lack the open Holliday junction configuration, which should be seen more frequently than crossed configurations if a strand exchange process indeed took place between the two rings (12). Statistically, a homologous recombination event between identical DNA sequences should occur with equal frequency along the entire molecule. However, in psoralen cross-linked SV40 minichromosomes, the joins also mainly coincided with the region of replication termination (48).
Our experimental data support the idea that SDR molecules are mainly made up of hemicatenated minichromosomes. The hemicatenated molecules are formed postreplicatively and mark an intermediate in replication segregation. If hemicatenanes exist as structures, as proposed by Laurie et al. (31), the parental strands should remain linked to one another, while the two newly synthesized daughter strands should remain unlinked. One might predict that pulling apart the crossing of the parental strands could structurally resemble a double Holliday junction structure. Thereby, a hemicatenane could be spontaneously converted into a recombination-like intermediate that would not lead to recombinant molecules and that would be independent of strand invasion-promoting activities but dependent on resolving activities. It will be challenging to see whether our results can be reproduced in linear chromosomes or if the SDR molecules are a particularity of small, circular minichromosomes.
In chromosomes, the hemicatenanes could be formed after the passage of the replication fork (6), giving rise to replicating bubbles carrying a hemicatenane (39). This would be consistent with roughly half of the SDR molecules' being located outside the replication termination zone. Furthermore, the accumulation of SDR molecules at the termination zone of replication could be a consequence of the relatively long half-life of molecules in the late stage of replication relative to molecules with proceeding replication forks. This fit well with the data obtained from EcoRV-digested DNA, where, after removal of the termination zone, both strand displacement-sensitive and SDR molecules were less abundant. Further experimentation in different recombination-deficient backgrounds might help to untangle the genetic and enzymatic requirements for SDR molecule formation and resolution. Moreover, selective strand displacement and psoralen-DNA cross-linking will offer a new tool to study SDR molecule and Holliday junction formation stimulated by defective proteins (37, 60) or DNA damage (38).
Acknowledgments
We thank D. Fitzgerald, H. Gaillard, A. Aguilera, and M. Lopes for stimulating discussion and careful reading of the manuscript, U. Suter for continuous encouragement, and Margaret Fäsi for excellent technical assistance. We are grateful to B. Kemper and M. Salas for generously providing the T4 endonuclease VII and φ29 DNA polymerase and the p5 enzymes, respectively.
This work was supported by grants from the Swiss National Science Foundation (31-63418) and the Bonizzi-Theler Stiftung (to P.S.).
REFERENCES
- 1.Allers, T., and M. Lichten. 2000. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 28:e6. [DOI] [PMC free article] [PubMed]
- 2.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 3.Barre, F. X., B. Soballe, B. Michel, M. Aroyo, M. Robertson, and D. Sherratt. 2001. Circles: the replication-recombination-chromosome segregation connection. Proc. Natl. Acad. Sci. USA 98:8189-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, A. D., and A. Maxwell. 1993. DNA topology. Oxford University Press, Oxford, UK.
- 5.Bell, L., and B. Byers. 1983. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal. Biochem. 130:527-535. [DOI] [PubMed] [Google Scholar]
- 6.Benard, M., C. Maric, and G. Pierron. 2001. DNA replication-dependent formation of joint DNA molecules in Physarum polycephalum. Mol. Cell 7:971-980. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, L., A. Bernad, J. M. Lazaro, G. Martin, C. Garmendia, and M. Salas. 1989. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 264:8935-8940. [PubMed] [Google Scholar]
- 8.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 9.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 10.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 11.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dressler, D., and H. Potter. 1982. Molecular mechanisms in genetic recombination. Annu. Rev. Biochem. 51:727-761. [DOI] [PubMed] [Google Scholar]
- 13.Duckett, D. R., A. I. Murchie, A. Bhattacharyya, R. M. Clegg, S. Diekmann, E. von Kitzing, and D. M. Lilley. 1993. The structure of DNA junctions and their interaction with enzymes. Eur. J. Biochem. 211:285-295. [DOI] [PubMed] [Google Scholar]
- 14.Eichman, B. F., M. Ortiz-Lombardia, J. Aymami, M. Coll, and P. Shing Ho. 2002. The inherent properties of DNA four-way junctions: Comparing the crystal structures of Holliday junctions. J. Mol. Biol. 320:1037-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 16.Gomez, M., and F. Antequera. 1999. Organization of DNA replication origins in the fission yeast genome. EMBO J. 18:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber, M., R. E. Wellinger, and J. M. Sogo. 2000. Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell. Biol. 20:5777-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haber, J. E., and W. D. Heyer. 2001. The fuss about Mus81. Cell 107:551-554. [DOI] [PubMed] [Google Scholar]
- 19.Hargreaves, D., D. W. Rice, S. E. Sedelnikova, P. J. Artymiuk, R. G. Lloyd, and J. B. Rafferty. 1998. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 A resolution. Nat. Struct. Biol. 5:441-446. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 21.Holliday, R. 1964. A mechanism for gene conversion in fungi. Genet. Res. 5:282-304. [DOI] [PubMed] [Google Scholar]
- 22.Huberman, J. A., and A. D. Riggs. 1968. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 32:327-341. [DOI] [PubMed] [Google Scholar]
- 23.Huberman, J. A., L. D. Spotila, K. A. Nawotka, S. M. El-Assouli, and L. R. Davis. 1987. The in vivo replication origin of the yeast two micron plasmid. Cell 51:473-481. [DOI] [PubMed] [Google Scholar]
- 24.Jensch, F., and B. Kemper. 1986. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 5:181-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadyk, L. C., and L. H. Hartwell. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132:387-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemper, B., F. Jensch, M. von Depka-Prondzynski, H. J. Fritz, U. Borgmeyer, and K. Mizuuchi. 1984. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb. Symp. Quant. Biol. 49:815-825. [DOI] [PubMed] [Google Scholar]
- 27.Kleff, S., and B. Kemper. 1988. Initiation of heteroduplex-loop repair by T4-encoded endonuclease VII in vitro. EMBO J. 7:1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, H. L. 2000. A radical solution to death. Nat. Genet. 25:132-134. [DOI] [PubMed] [Google Scholar]
- 29.Klein, H. L., and K. N. Kreuzer. 2002. Replication, recombination, and repair. Going for the gold. Mol. Cell 9:471-480. [DOI] [PubMed] [Google Scholar]
- 30.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurie, B., V. Katritch, J. Sogo, T. Koller, J. Dubochet, and A. Stasiak. 1998. Geometry and physics of catenanes applied to the study of DNA replication. Biophys. J. 74:2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levac, P., and T. Moss. 1996. Inactivation of topoisomerase I or II may lead to recombination or to aberrant replication termination on both SV40 and yeast 2 micron DNA. Chromosoma 105:250-260. [DOI] [PubMed] [Google Scholar]
- 33.Lilley, D. M., and L. R. Hallam. 1984. Thermodynamics of the ColE1 cruciform. Comparisons between probing and topological experiments with single topoisomers. J. Mol. Biol. 180:179-200. [DOI] [PubMed] [Google Scholar]
- 34.Lucas, I., and O. Hyrien. 2000. Hemicatenanes form upon inhibition of DNA replication. Nucleic Acids Res. 28:2187.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucchini, R., and J. M. Sogo. 1992. Different chromatin structures along the spacers flanking active and inactive Xenopus rRNA genes. Mol. Cell. Biol. 12:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucchini, R., and J. M. Sogo. 1994. Chromatin structure and transcriptional activity around the replication forks arrested at the 3′ end of the yeast rRNA genes. Mol. Cell. Biol. 14:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 1998. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. USA 95:6739-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neecke, H., G. Lucchini, and M. P. Longhese. 1999. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 18:4485-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olavarrieta, L., M. L. Martinez-Robles, P. Hernandez, D. B. Krimer, and J. B. Schvarztman. 2002. Knotting dynamics during DNA replication. Mol. Microbiol. 46:699-707. [DOI] [PubMed] [Google Scholar]
- 40.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saavedra, P. A., and J. A. Huberman. 1986. Both DNA topoisomerases I and II relax 2μm plasmid in living yeast cells. Cell 45:65-70. [DOI] [PubMed] [Google Scholar]
- 42.Santamaria, D., E. Viguera, M. L. Martinez-Robles, O. Hyrien, P. Hernandez, D. B. Krimer, and J. B. Schvartzman. 2000. Bi-directional replication and random termination. Nucleic Acids Res. 28:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwacha, A., and N. Kleckner. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83:783-791. [DOI] [PubMed] [Google Scholar]
- 44.Segurado, M., M. Gómez, and F. Antequera. 2002. Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol. Cell 10:907-916. [DOI] [PubMed] [Google Scholar]
- 45.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 46.Sharples, G. J. 2001. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 39:823-834. [DOI] [PubMed] [Google Scholar]
- 47.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Media, p. 61-64. In J. B. Hicks (ed.), Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sogo, J. M., H. Stahl, T. Koller, and R. Knippers. 1986. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J. Mol. Biol. 189:189-204. [DOI] [PubMed] [Google Scholar]
- 49.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 50.Sundin, O., and A. Varshavsky. 1980. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell 21:103-114. [DOI] [PubMed] [Google Scholar]
- 51.Sundin, O., and A. Varshavsky. 1981. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell 25:659-669. [DOI] [PubMed] [Google Scholar]
- 52.Sung, P., K. M. Trujillo, and S. Van Komen. 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451:257-275. [DOI] [PubMed] [Google Scholar]
- 53.Sutton, M. D., and G. C. Walker. 2001. Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination. Proc. Natl. Acad. Sci. USA 98:8342-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thoma, F. 1986. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J. Mol. Biol. 190:177-190. [DOI] [PubMed] [Google Scholar]
- 55.Trigueros, S., and J. Roca. 2001. Circular minichromosomes become highly recombinogenic in topoisomerase-deficient yeast cells. J. Biol. Chem. 276:2243-2248. [DOI] [PubMed] [Google Scholar]
- 56.Ullsperger, C. J., A. V. Vologodskii, and N. R. Cozarelli. 1995. Nucleic acids and molecular biology, p. 115-142. In D. M. J. Lilley and F. Eckstein (ed.), Unlinking of DNA by topoisomerases during DNA replication. Springer-Verlag, Berlin, Germany.
- 57.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 58.Walker, G. T., M. C. Little, J. G. Nadeau, and D. D. Shank. 1992. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc. Natl. Acad. Sci. USA 89:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, L., J. K. Karow, and I. D. Hickson. 1999. Genetic recombination: Helicases and topoisomerases link up. Curr. Biol. 9:R518-R520. [DOI] [PubMed] [Google Scholar]
- 60.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]