Abstract
Active genes are insulated from developmentally regulated chromatin condensation in terminally differentiated cells. We mapped the topography of a terminal stage-specific chromatin-condensing protein, MENT, across the active chicken β-globin domain. We observed two sharp transitions of MENT concentration coinciding with the β-globin boundary elements. The MENT distribution profile was opposite to that of acetylated core histones but correlated with that of histone H3 dimethylated at lysine 9 (H3me2K9). Ectopic MENT expression in NIH 3T3 cells caused a large-scale and specific remodeling of chromatin marked by H3me2K9. MENT colocalized with H3me2K9 both in chicken erythrocytes and NIH 3T3 cells. Mutational analysis of MENT and experiments with deacetylase inhibitors revealed the essential role of the reaction center loop domain and an inhibitory affect of histone hyperacetylation on the MENT-induced chromatin remodeling in vivo. In vitro, the elimination of the histone H3 N-terminal peptide containing lysine 9 by trypsin blocked chromatin self-association by MENT, while reconstitution with dimethylated but not acetylated N-terminal domain of histone H3 specifically restored chromatin self-association by MENT. We suggest that histone H3 modification at lysine 9 directly regulates chromatin condensation by recruiting MENT to chromatin in a fashion that is spatially constrained from active genes by gene boundary elements and histone hyperacetylation.
In eukaryotic cells, the DNA is repeatedly wrapped around histone octamers to form nucleosomes. Nucleosome arrays are packed through a hierarchy of folding levels into higher-order chromatin structures with a variable degree of condensation (for reviews, see references 32, 67, 68, and 70). Chromatin condensation is especially tight in heterochromatin, representing the repressed fraction of genetic material (30, 57). Heterochromatin becomes very abundant in the nuclei of terminally differentiated cells, where the majority of formerly active genes are repressed and condensed (20, 21). In this paper we investigate how the unique properties of heterochromatin in terminally differentiated cells integrate the general mechanism of gene regulation by histone modifications.
One of the most intriguing aspects in the process of heterochromatin formation is that chromatin condensation does not spread to the actively transcribed tissue-specific genes, which still maintain an open, nuclease-sensitive conformation (66). For example, in the chicken β-globin domain, a 30-kb region of active DNase-sensitive chromatin is separated from repressed and condensed chromatin by gene boundary elements (2, 11). The repressed chromatin is associated with a special pattern of histone modification, most notably a decreased level of histone H3 and H4 acetylation (28, 43) and an increased level of histone H3 methylated at lysine 9 (H3meK9) (42). The H3meK9 binds to the chromodomain of heterochromatin protein 1 (HP1) (49), and this association has been proposed to determine the repressed state of heterochromatin (35).
In this work we found that HP1 protein levels are reduced (HP1β) or become negligible (HP1α and HP1γ) in differentiated erythrocytes and lymphocytes. Thus, HP1 proteins are not likely to control the developmentally regulated formation of heterochromatin in terminally differentiated blood cells. Therefore, we asked what other factors might direct the expansion of heterochromatin in terminally differentiated and quiescent cells. What is the role of histone modification and chromatin boundaries in spatial regulation of chromatin condensation and protection of active genes from heterochromatin spreading?
Previously we have described MENT (myeloid and erythroid nuclear termination stage-specific protein), an abundant developmentally regulated protein that accumulates in terminally differentiated chicken blood cells, binds to repressed chromatin, and promotes its condensation (22, 24). MENT belongs to the serpin family, and its conserved reactive center loop (RCL) domain is essential for chromatin interactions (34). Since MENT is an obvious candidate for causing heterochromatin spreading in mature blood cells, we asked whether the chromatin boundaries would block MENT spreading to active chromosomal domains. Using in situ cross-linking and chromatin immunoprecipitation (ChIP), we found that the pattern of MENT distribution over the chicken β-globin cluster is opposite to that of acetylated core histones (28, 43) but correlates with increased histone H3 methylation (42). To examine the role of histone modifications in regulating MENT association with chromatin in proliferating and quiescent cells, we constructed a MENT-expressing NIH 3T3 cell line, in which we observed a marked condensation of nuclear chromatin accompanied by a large-scale rearrangement of chromatin marked by methylated histone H3. We also show that histone deacetylase inhibitors increase the level of histone acetylation and inhibit chromatin condensation in vivo and that the histone H3 N-terminal domain dimethylated at Lys 9 cooperates with the RCL domain of MENT to promote chromatin condensation while this domain acetylated at Lys 9 fails to do so. We discuss a molecular model for spatial regulation of chromatin condensation in the genome in which the condensation is promoted by direct interaction of histone H3 N-terminal domains with tissue- and stage-specific chromatin-condensing factors and inhibited by histone hyperacetylation punctuated in the genome by gene boundary elements.
MATERIALS AND METHODS
Materials.
Unless otherwise noted, all fine chemicals were from Sigma. Rabbit polyclonal anti-MENT immunoglobulin G (IgG) (α-MENT) (25), rabbit polyclonal anti-HP1α IgG (α-HP1α) (19), rabbit polyclonal IgG against histone H3 peptide 1-20 in which lysine 9 was trimethylated (α-H3me3K9) (13), and rat monoclonal anti-HP1β (M31) IgG1 (α-HP1β) (69) and anti-HP1γ (M32) IgM (α-HP1γ) (33) were as described previously. Rabbit polyclonal IgGs against histone H3 peptide 6-13 with dimethylated lysine 9 (α-H3me2K9), against histone H3 peptide 1-21 with acetylated lysines 9 and 13 (α-H4acK9,13), against histone H4 peptide 1-10 with acetylated lysine 5 (α-H4acK5), and against histone H4 peptide 7-16 with acetylated lysine 12 (α-H4acK12) were obtained from Upstate Biotechnology (Lake Placid, N.Y.).
Purification of cell nuclei and Western analysis.
Isolation of nuclei from chicken blood cells and cultured cells (HD13 and NIH 3T3), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and detection of MENT protein were conducted as described previously (25). For detection of HP1 proteins and histones, SDS-PAGE was carried out with 15% polyacrylamide and the proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) in 20% methanol. HP1β (69) and HP1α and HP1γ (33) were detected as described previously. Modified histones were visualized with Upstate Biotechnology antibodies (see above) as recommended in the vendor's manual.
Chromatin cross-linking and immunoprecipitation.
We modified the formaldehyde cross-linking ChIP technique (6, 38) for use with chicken blood cell nuclei. Briefly, 1 ml of nuclear suspension in RSB-HEPES buffer (10 mM NaCl, 3 mM MgCl2, 10 mM HEPES [pH 7.5]) at 10 A260 units/ml was cross-linked with 1% formaldehyde at 25°C. For MENT ChIP, the cross-linking continued for 5 or 15 min for erythrocyte and lymphocyte nuclei, respectively. For histone ChIP, erythrocytes were cross-linked for 1 min. The extent of cross-linking was monitored by the disappearance of intact MENT or histone after SDS-PAGE and Western blotting. The cross-linked nuclei were centrifuged at 1,575 × g and lysed in 300 μl of lysis buffer (38) with protease inhibitor cocktail as directed by the supplier (Sigma). After sonication with a Branson Sonifier 250 to shear DNA to lengths of between 200 and 2,000 bp (as estimated by agarose gel electrophoresis), lysates were cleared by centrifugation; the insoluble material was enriched neither with the DNA sequences of interest nor with MENT. An aliquot (10%) of the supernatant was saved to represent unfractionated chromatin. The supernatant was then diluted 10-fold with dilution buffer (0.01% SDS, 1.1% Triton, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.0], 389 mM NaCl) to a final volume of 3 ml.
The cross-linked chromatin suspension was mixed with either 20 μg of α-MENT, 20 μg of preimmune serum, 10 μg of α-H3acK9,13, or 100 μl of α-H3me2K9 and incubated overnight at 4°C. Immune complexes were reacted for 1 h at 25°C with agitation with 30 μl of a 50% suspension of protein A-Sepharose beads (Sigma) equilibrated with wash buffer (lysis buffer diluted 10-fold with dilution buffer). After the reaction, the beads were collected and the supernatant was saved as the unbound chromatin fraction. The beads were washed as described previously (6), and immune complexes were eluted twice with 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at 25°C. Cross-links were reversed by heating, and the released DNA was purified as described previously (38). Denatured samples were applied to Hybond N+ membranes (Amersham) with a slot blot apparatus (100 ng of DNA per slot) together with 1 μg of carrier herring sperm DNA (Gibco-BRL). The membranes were hybridized as described in the Hybond manual with 30 ng of probe DNA 32P labeled with a Random Primer Extension Labeling System (Perkin-Elmer). After hybridization, membranes were washed, exposed to a PhosphorImager (Molecular Dynamics), and quantified with ImageQuant software.
Hybridization probes and data analysis.
The chicken β-globin genomic probes 5 (1,340 bp MspI-MspI fragment from plasmid pCBG4) and 7 (1,364-bp MspI-MspI fragment from pCAβG1) and the ovalbumin genomic probe (910-bp PvuII fragment of the chicken ovalbumin gene coding region from plasmid pOv12) were described previously (24). The β-globin probes 1 (590-bp BglI-BamHI fragment from pCBG D), 2 (1,300-bp KpnI-KpnI fragment from pCBG C), 3 (1,200-bp SspI-BamHI fragment from pCBG C), 4 (900-bp BamHI-BamHI fragment from pCBG B), 8 (1,300-bp HindIII-PstI fragment from pCBG23 [p556]), 9 (900-bp KpnI-PstI fragment from pCBG23 [p556]), 11 (500-bp BamHI-EcoRI fragment from pCBG28 [p520]), and 12 (100-bp NcoI-NcoI fragment from pCBG26 [p520]) were kindly provided by Mark Reitman (56). Probe 6 (517 bp) was generated by PCR amplification from chicken genomic DNA with oligonucleotides 5′GGAGAACTACCGTGGGCAACAA and 5′GGGAGTTGAGCTGCAGAAGGAA. Probe 10 (891 bp) was generated by PCR amplification from chicken genomic DNA with oligonucleotides 5′CTGAAAATCCAGCACGGGAAGC and 5′GCAGATGATCCCCAGTCCAATC.
In all experiments we compared hybridizations of DNA retained after immunoprecipitation with anti-MENT and control preimmune serum as well as DNA from the unbound material. Hybridization data were analyzed and plotted by using Microsoft Excel and KaleidaGraph (Synergy Software). Relative enrichment (RE) was calculated for each series of experiments (one antibody and one cell type) according to the formula RE = [(HN − HNcont)/HNtot]/[(Hmax − Hmaxcont)/Hmaxtot], where HN is the hybridization signal after immunoprecipitation with one type of antibody probed with DNA probe N, HNcont is the hybridization signal after immunoprecipitation of the same material with control preimmune serum probed with N, HNtot is the hybridization signal from total cross-linked DNA of the same material hybridized with probe N, Hmax is the hybridization signal obtained with the same antibody probed with DNA that provides the maximal hybridization signal in the given experimental series (max), Hmaxcont is the hybridization signal after immunoprecipitation with control preimmune serum probed with max, and Hmaxtot is the hybridization signal from total cross-linked DNA probed with max.
Chromatin isolation and reconstitution and agarose gel electrophoresis.
Soluble chromatin was isolated from chicken erythrocyte nuclei treated with micrococcal nuclease as described previously (25). To remove the histone H3 N-terminal domains, soluble chromatin solution (A260 = 10) was treated with 33 μl of agarose-conjugated trypsin (Sigma T-1763) per ml for 90 min at 37°C, and the reaction was stopped by spinning down the immobilized trypsin for 60 s at 16,000 × g and adding 1 mM phenylmethylsulfonyl fluoride. The trypsin-digested chromatin was loaded onto a 5 to 25% sucrose gradient containing 10 mM Tris (pH 7.5), 1 mM EDTA, and 0.5 M NaCl and centrifuged for 22 h at 60,000 × g in an SW-27 rotor (Beckman). The gradient fraction containing chains of 6 (± 1) nucleosomes (trypsin-digested oligonucleosome arrays) was collected and dialyzed against 10 mM HEPES (pH 7.5)-2 mM EDTA.
Recombinant wild-type MENT (MENTwt) and the MENTov mutant were expressed in Escherichia coli and isolated as described previously (34). For binding studies with oligonucleosomes, 4 μl of 10 mM HEPES (pH 7.5)-40 mM NaCl containing various concentrations of MENT (0.05 to 0.2 mg/ml) was mixed with 4 μl of native oligonucleosome preparation (0.1 mg of DNA per ml) in 10 mM HEPES (pH 7.5)-40 mM NaCl and 2 μl of 10 mM HEPES (pH 7.5)-40 mM NaCl containing various concentrations (1 to 10 μM) of peptides containing 21 N-terminal amino acids of histone H3 and either two methyl groups (N-H3me2) (catalog no. 12-430; Upstate), one acetyl group (N-H3ac) (catalog no. 12-431; Upstate), or no modification (N-H3ac) (catalog no. 12-403, Upstate) at lysine 9. The reaction mixtures were incubated on ice for 30 min and centrifuged for 10 min at 18,000 × g and 4°C. The DNA concentrations in the supernatants and pellets were determined by UV spectrophotometry (A260 = 1 for 50 μg/ml of DNA). Supernatants were mixed with 5% glycerol and electrophoresed in 1% agarose (Sigma type IV) in a buffer containing 20 mM HEPES (pH 7.5) and 0.2 mM EDTA as described previously (25).
Construction and propagation of stable MENT-expressing cell lines.
NIH 3T3-lacI cells were obtained by stable transfection of NIH 3T3 cells (ATCC CRL-658), first with a Lac repressor-expressing vector pCMVLacI (Stratagene) as described in the vendor's manual and then, after establishment of stable LacI-positive clones, with either pOPRSVI/MCS vector (Stratagene) containing a MENTwt cDNA insert or pRC/CMV vector (Invitrogen) containing the MENTov mutation as described previously (34). Cells were grown in Dulbecco's modified Eagle's medium with 4.5 g of glucose (Mediatech) per liter, 1.5 g of sodium bicarbonate per liter, 1× antibiotic-antimycotic solution, and 10% (vol/vol) calf serum in a 5% CO2 incubator at 37.5°C. At 24 h after transfection, cells were replated into 100-mm-diameter dishes and grown in the presence of 500 μg of Geneticin (Gibco-BRL) per ml. Single colonies were isolated by using sterile cloning disks (Fisher) and were transferred to individual wells of 24-well plates. After reaching confluence, cell clones were replated and examined by immunofluorescence microscopy and Western blotting with α-MENT to select the clones with high levels of MENT expression.
Immunofluorescence microscopy and image analysis.
For fluorescence microscopy, NIH 3T3 cells expressing MENT were grown, fixed, and stained with primary antibodies (α-MENT [1:1,000] and α-H3me2K9 [1:500]) and red fluorescent secondary antibodies (1:600) (Alexa Fluor 594 goat anti-rabbit IgG; Molecular Probes) as described previously (34). To block the stained cells for labeling with another set of rabbit antibodies, the specimens were washed as before, incubated for 30 min with phosphate-buffered saline (PBS) containing 3.5% bovine serum albumin (BSA) and 1% whole rabbit serum (Gibco-BRL), washed with PBS as before, and incubated for 30 min with PBS containing 3.5% BSA and a 1:100 dilution of goat anti-rabbit Fab fragments (Jackson Immunoresearch Labs). The specimens were then washed and treated with new primary antibodies (α-H3me3K9; 1:1,000) and green fluorescent secondary antibodies (dilution 1:600) (Alexa Fluor 488 goat anti-rabbit IgG; Molecular Probes) as described above.
Nuclei from embryonic erythrocytes, adult erythrocytes, and lymphocytes resuspended in 40 mM NaCl-10 mM Tris (pH 7.5)-1 mM EDTA were attached to poly-l-lysine-coated microscope slides (Sigma) over 10 min, fixed with 4% paraformaldehyde (EM Sciences), washed with PBS for 5 min, and permeabilized in KCM (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.1% [vol/vol] Triton X-100) for 15 min. Samples were then blocked in KCM containing 2% (wt/vol) BSA and 10% nonfat dry milk for 1 h. The specimens were overlaid with KCM containing BSA, dry milk, and α-MENT (dilution 1:500) and incubated for 1 h at room temperature. Specimens were washed two times in KCM and overlaid with KCM containing BSA and dry milk for 5 min, and then fluorescent secondary antibodies (Alexa Fluor 594 goat anti-rabbit IgG) were added and incubated for 1 h at room temperature. The samples were finally stained with 0.1 μg of Hoechst 33258 per ml in PBS, mounted, and analyzed under a fluorescence microscope, followed with image deconvolution as described previously (34).
RESULTS
MENT, but not constitutive heterochromatin proteins, accumulates during avian blood cell differentiation.
Methylation of histone H3 at lysine 9 (H3meK9) has been suggested to be a primary signal in establishing the heterochromatin/euchromatin segregation in concert with histone methyltransferase and the ubiquitous H3meK9-binding protein HP1 (35, 42). Since the amount of cytologically detectable heterochromatin is dramatically increased during terminal blood cell differentiation (14, 40), we compared the levels of H3meK9 and HP1 in proliferating and terminally differentiated avian hemopoietic cells. In parallel, we also monitored the level of MENT, a heterochromatin protein expressed during terminal avian blood cell differentiation (25).
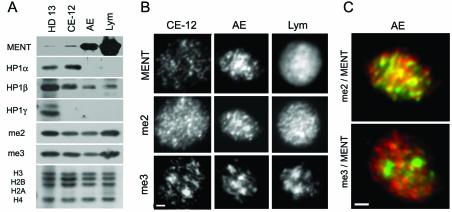
When we probed Western blots containing proteins from the nuclei of proliferating chicken promyelocytic cell line HD13 with antibodies against the three known HP1 variants, HP1α, HP1β, and HP1γ (33, 69), we observed prominent bands with all three probes (Fig. 1A). In equal amounts of nuclear material from mature terminally differentiated cells (adult erythrocytes and lymphocytes), the HP1 levels were either lower (HP1β) or undetectable (HP1α and HP1γ), suggesting that these proteins are not involved in the developmentally regulated chromatin condensation seen in mature terminally differentiated cells. In contrast, the level of MENT was low in 12-day erythrocytes and HD13 cells but was high in adult erythrocytes and especially high in chicken lymphocytes (Fig. 1A). Thus, the level of MENT, but not of HP1, is consistent with the global silencing and chromatin condensation seen in mature erythrocytes and lymphocytes.
FIG. 1.
Heterochromatin proteins in chicken blood cell nuclei. (A) Proteins from the nuclei of chicken HD13 cells, 12-day embryo chicken erythrocytes (CE-12), adult erythrocytes (AE), and chicken lymphocytes (Lym) were analyzed by SDS-PAGE and Western blotting with antibodies against MENT, HP1α, HP1β, HP1γ, H3me2K9 (me2), and H3me3K9 (me3) as indicated. The bottom panel shows electrophoresis of histones from the same material stained with Coomassie blue R-250. (B) Immunofluorescence microscopy of nuclei of 12-day embryo chicken erythrocytes, adult erythrocytes, and chicken lymphocytes stained with antibodies against MENT, H3me2K9, and H3me3K9 as indicated. Bar, 1 μm. (C) Overlays of immunofluorescence micrographs of adult chicken erythrocyte nuclei. Top panel, double staining with antibodies against MENT (red) and H3me2K9 (green). Bottom panel, double staining with antibodies against MENT (red) and H3me3K9 (green). Bar, 1 μm.
To detect H3meK9 in Western blots and nuclear immunostaining, we used antibodies against an H3 peptide containing dimethylated lysine 9 (α-H3me2K9) (Upstate Biotechnology) and an H3 peptide containing trimethylated lysine 9 (α-H3me3K9 (13). α-H3me3K9 but not α-H3me2K9 stains pericentromeric constitutive heterochromatin (13). The α-H3me3K9 antibody has no cross-reaction with the α-H3me2K9, nor does it recognize methylated K27 of histone H3 (63). Western blotting (Fig. 1A) showed that the levels of methylated histone H3 recognized by both antibodies do not change significantly between the proliferating and the mature blood cells. This result is consistent with metabolic stability of histone H3 methylation at K9 (1).
To determine if MENT is associated with any specific structure in the nuclear chromatin, we stained the nuclei of chicken 12-day embryo erythrocytes, adult erythrocytes, and lymphocytes with α-MENT, α-H3me2K9, and α-H3me3K9 (Fig. 1B). In agreement with the results of Western analysis, MENT had a very low level in embryo erythrocytes compared to adult erythrocytes and lymphocytes. In adult erythrocytes, α-MENT and α-H3me2K9 had similar focal patterns. In contrast, α-MENT and α-H3me2K9 in lymphocytes showed a broad distribution, suggesting that these markers spread to most of the nuclear chromatin in lymphocytes. In all cells, α-H3me3K9 had a focal pattern consistent with its association with constitutive heterochromatin (13). Figure 1C shows a sharp contrast between mostly overlapping MENT and H3me2K9 (yellow) and nonoverlapping MENT (red) and H3me3K9 (green) in adult erythrocyte nuclei. Thus, in adult chicken erythrocytes MENT is colocalized with the dimethylated but not with the trimethylated histone H3(K9).
Sharp transitions in the MENT level at the boundaries of the active chicken β-globin in chicken erythrocytes.
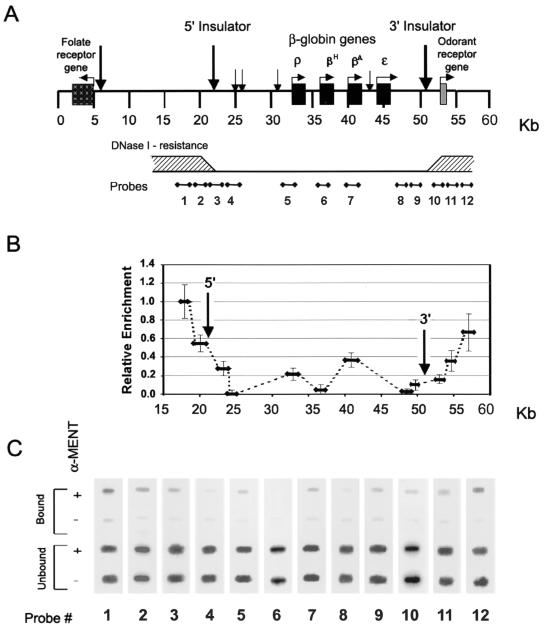
Previous studies of chromatin organization at the chicken β-globin gene cluster in embryonic and adult erythrocytes (3, 28, 64) have revealed a 30-kb domain of active DNase I-sensitive chromatin conformation. The DNase-sensitive domain, which includes all four β-globin genes and their transcriptional regulatory elements, is surrounded by DNase I-resistant condensed heterochromatin (Fig. 2A). Two boundary elements located at the borders of the DNase I-sensitive domain have been suggested to protect active chromatin from spreading of chromatin-condensing factors (2, 11). Since MENT is an abundant chromatin-condensing protein expressed in adult erythrocytes, we asked whether the active DNase-sensitive β-globin domain is protected from MENT. To map its localization on chicken erythrocyte chromatin in situ, we used formaldehyde ChIP analysis followed by slot hybridization probed with DNA derived from the chicken β-globin locus (56). We sequenced the DNAs of all of our probes to confirm the absence of repeated elements. The positions of the probes in relation to the important structural elements and the region of DNase sensitivity at the β-globin domain are shown in Fig. 2A.
FIG. 2.
Mapping of MENT on the β-globin domain in chicken erythrocytes. (A) A scheme of the β-globin cluster shows the positions of β-globin genes and the map numbering system from reference 43. Vertical arrows indicate the positions of DNase I-hypersensitive sites, including those corresponding to the 5′ and 3′ insulators (58). The lower horizontal bar shows a cumulative scheme of DNase I resistance (shaded areas indicate resistance) derived from several experiments (3, 28, 58, 64). The scheme also shows the positions of hybridization probes 1 to 12 used throughout this work. (B) MENT levels on the chicken β-globin cluster. The graph shows the relative enrichment of MENT at the regions corresponding to the β-globin probes 1 to 12. Error bars indicate standard deviations. (C) Slot hybridization analysis of immunoprecipitated MENT-DNA complexes from the chicken β-globin cluster. After formaldehyde cross-linking of isolated nuclei, DNA-MENT complexes were immunoprecipitated either with α-MENT (+) or with preimmune serum (−). DNA was purified and hybridized with probes 1 to 12 derived from the chicken β-globin cluster.
ChIP mapping shows a dramatic difference in MENT levels upstream and downstream of the β-globin domain boundaries (Fig. 2B and C). Specifically, the MENT concentration varies ca. 50-fold between probe 1 and probe 4 (Fig. 2C and 3B). The maximal level of MENT binding (probe 1) corresponds to the area where DNase I-resistant heterochromatin separates the active β-globin cluster from the folate receptor gene (54). The midtransition point in the MENT level corresponds to the 5′ boundary region described by Chung et al. (11). Two local minimums of MENT are located at the globin-proximal sites downstream of the 5′ boundary (probe 4) and upstream of the 3′ boundary (probe 8). Remarkably, both points of minimal MENT level are located within the DNase-sensitive domain at similar distances from the boundary elements. The level of MENT increases less abruptly over the 3′ boundary region, reaching about 60% of the maximal level at the position downstream of the silent odorant receptor gene (probe 12). The concentration of MENT upstream and downstream of the β-globin cluster is considerably higher than that at the constitutively silent ovalbumin gene (Fig. 3B), suggesting that the regions of MENT accumulation do not merely reflect the state of gene repression but are associated with repressed chromatin flanking active genes.
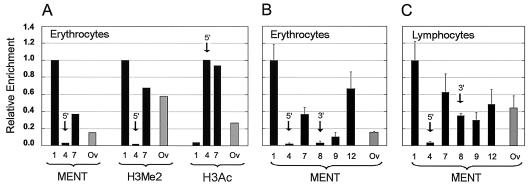
FIG. 3.
MENT levels at selected DNA sequences from chicken blood cells. (A) The histograms show relative enrichment of MENT, H3me2K9 (H3me2), and H3acK9,13 (H3ac) detected by ChIP in chicken erythrocytes. (B and C) The histograms show the relative enrichment of MENT (as related to the probe 1) detected by ChIP with selected genome probes in chicken erythrocytes (A) and lymphocytes (B). DNA probes 1, 4, 5, 7, 8, and 12 (black bars) from the β-globin domain (see Fig. 2A) and Ov (gray bars) from the chicken ovalbumin-coding region, positions 2375 to 3285 (17), were used. Arrows show the probes corresponding to the MENT minimums at the 5′ boundary and the 3′ boundary of the chicken β-globin gene (Fig. 2B).
ChIP experiments by two other groups showed that repressed chromatin upstream of the 5′ boundary of the chicken β-globin domain is associated with a decreased level of histone H3 and H4 acetylation (28, 43) and an increased level of histone H3 methylation at lysine 9 (42). In this work we used a different protocol for immunoprecipitation of MENT that involves formaldehyde cross-linking. To confirm that the MENT-enriched region in our experiments corresponds to the area of high histone methylation and low acetylation detected earlier, we immunoprecipitated the cross-linked chromatin with antibodies against methylated and acetylated histone H3. Probing this material with DNAs from selected gene probes (Fig. 3A) showed a substantial enrichment of H3me2K9 and depletion of H3ac2K9 at the repressed chromatin upstream of the β-globin boundary (probe 1). In contrast, the area containing the minimal level of MENT (probe 4) had a very low level of H3me2K9 and a high level of H3ac2K9. This result is in good agreement with the earlier experiments (28, 42, 43). In the middle of the β-globin domain the concentration of MENT reaches a local peak close to the position of the transcriptionally active βA-globin gene and its upstream intergenic region (probe 7). Interestingly, the level of MENT at site 7 correlates with increased histone H3 dimethylation (Fig. 3A). An earlier ChIP mapping of the entire β-globin domain has also documented a significant peak of H3 dimethylation in this area (42). In contrast, our ChIP with trimethylated histone H3 has shown almost no variation between the different probes and no correlation with MENT (not shown), consistent with clearly distinct localization of MENT and H3me3K9 in chicken erythrocytes (Fig. 1C). Taken together with the immunofluorescence (Fig. 1C) showing a general colocalization of MENT with H3me2K9, our results demonstrate that in chicken erythrocytes the MENT level is correlated positively with methylation and negatively with acetylation of lysine 9 in histone H3.
MENT occupies the inactive β-globin domain but spares its 5′ boundary in lymphocytes.
Erythrocytes and lymphocytes originate from the same hemopoietic precursor cells, but their differentiation pathways separate before the onset of β-globin gene transcription so that β-globin is not expressed in the lymphoid cells. The DNase I-hypersensitive sites marking the active β-globin gene regulatory elements are present in the precursor hemopoietic cells and are gradually decreased during terminal lymphoid differentiation (36). Chicken lymphocytes have a ca. 5-fold higher total level of MENT than erythrocytes (25) (Fig. 1A). From our immunofluorescence studies (Fig. 1B) we anticipated that MENT might become much more evenly distributed in the lymphocyte chromatin. Indeed, using α-MENT ChIP we observed much less variation in MENT between different loci in lymphocytes than in erythrocytes (compare Fig. 3B and C). For example, the level of MENT at the internal β-globin probe 7 was only 1.6 times lower than that at the 5′ flank of the β-globin domain (probe 1) in lymphocytes compared, with a threefold difference in erythrocytes. Most remarkably, the MENT level at the 3′ boundary of the β-globin domain in lymphocytes (probe 8 in Fig. 3C) was dramatically higher than that at the same region in erythrocytes (probe 8 in Fig. 3B). These results show that the β-globin domain becomes occupied by MENT during lymphoid differentiation and that the transitions in the MENT level at the active β-globin gene in erythrocytes are not due to a DNA sequence specificity of the protein.
Although MENT occupied the 3′ flank of the inactive β-globin domain in lymphocytes, at the 5′ end (probe 4) we still observed a prominent minimum in the MENT level similar to that in erythrocytes. This is consistent with persistence of the 5′ DNase-hypersensitive site in non-globin-expressing cells, the ability of the 5′ but not the 3′ boundary to protect from position effect variegation (58), and the presence of a very high peak of histone acetylation at the 5′ β-globin boundary in the lymphoid DT40 cells (43). Thus, a constitutive hypersensitive site remains insulated from MENT even when the surrounding chromosomal domain succumbs to the transcriptional silencing and chromatin condensation.
Modifications at lysine 9 in the histone H3 N-terminal domain directly affect MENT interaction with chromatin in vitro.
Our colocalization (Fig. 1C) and ChIP (Fig. 2 and 3) experiments suggested to us that MENT association with chromatin might be regulated by the histone modification pattern, in particular promoted by dimethylation and inhibited by acetylation of lysine 9 in histone H3. Lysine 9 is located in the N-terminal domain of histone H3, which may be selectively removed by trypsin without unfolding the nucleosome structure (23) but causing a partial unfolding of chromatin higher-order structure (41). Limited digestion of histone N termini with trypsin has been used before to demonstrate an essential and specific role of N-terminal peptides of histone H2B in metaphase chromosome condensation (15) and of the N-terminal domain of histone H3 in chromatin higher-order folding (41). Here we decided to use limited trypsin digestion to examine whether the histone H3 N-terminal domain is involved in MENT interaction with chromatin in vitro.
Previously we used an agarose gel shift assay to observe that reconstitution of soluble chromatin with increasing concentrations of native chicken MENT caused reversible chromatin self-association and precipitation (25). Now we expressed MENT in E. coli and isolated MENTwt and a mutant in which the MENT RCL domain is replaced with an inactive RCL domain from ovalbumin (MENTov). In transiently transfected cells, this mutation dramatically changes MENT association with chromatin and blocks the MENT-induced chromatin condensation (34), thus providing an efficient control for the specificity of MENT interaction with chromatin.
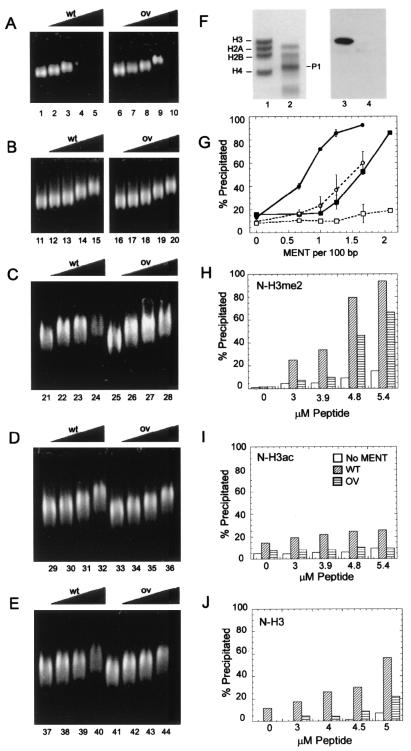
After we reconstituted soluble chromatin with either wild-type or mutant MENT, we observed that MENTwt interacts with soluble nucleosome arrays and causes a strong band shift (Fig. 4A, lanes 1 to 5) and an increasing precipitation (Fig. 4G) at MENT/DNA ratios of 1:200 to 1:50 bp. MENTov has a similarly strong interaction with chromatin as detected by band shift (Fig. 4A, lanes 6 to 10) but a significantly reduced ability to precipitate chromatin (Fig. 4G). We concluded that the RCL mutation inhibited the chromatin-condensing effect of MENT in vitro.
FIG. 4.
Cooperative interaction of MENTwt with the dimethylated N-terminal domain of histone H3. (A) Agarose gel electrophoresis of native chicken chromatin reconstituted with MENTwt (lanes 1 to 5) and MENTov (lanes 6 to 10); (B) agarose gel electrophoresis of trypsin-digested chromatin reconstituted with MENTwt (lanes 11 to 15) and MENTov (lanes 16 to 20); (C) agarose gel electrophoresis of trypsin-digested chromatin reconstituted with 5 μM dimethylated N-H3me2 and increasing concentrations of MENTwt (lanes 21 to 24) and MENTov (lanes 25 to 28); (D) agarose gel electrophoresis of trypsin-digested chromatin reconstituted with 5 μM acetylated N-H3ac and increasing concentrations of MENTwt (lanes 29 to 32) and MENTov (lanes 33 to 36); (E) agarose gel electrophoresis of trypsin-digested chromatin reconstituted with 5 μM unmodified N-H3 and increasing concentration MENTwt (lanes 37 to 40) and MENTov (lanes 41 to 44). MENT/DNA ratios (moles per 100 bp) were 0 (lanes 1, 6, 11, 16, 21, 25, 29, 33, 37, and 41), 0.5 (lanes 2, 7, 12, 17, 22, 26, 34, 38, and 42), 1 (lanes 3, 8, 13, 18, 23, 27, 35, 39, and 43), 2 (lanes 4, 9, 14, 19, 24, 28, 36, 40, and 44), and 4 (lanes 5, 10, 15, and 20). (F) SDS-PAGE (lanes 1 and 2) and Western blotting with α-H3me2K9 (lanes 3 and 4) of native chicken erythrocyte chromatin before (lanes 1 and 3) and after (lanes 2 and 4) digestion with trypsin. Note the absence of intact histone H3 and the disappearance of H3me2K9 in the trypsindigested sample. P1 shows the position of the main histone H3 trypsin digestion product (23). (G) Graph showing the percentage of precipitated material in native chromatin reconstituted with MENTwt (•) and MENTov (▪) and in trypsin-digested chromatin reconstituted with MENTwt (○) and MENTov (□). Ratios of MENT/DNA were as indicated. Error bars indicate standard deviations. (H to J) Histograms showing the percentage of precipitated material in trypsin-digested chromatin reconstituted with no MENT (open bars), 1 mol of MENTwt/60 bp (diagonally hatched bars), and 1 mol of MENTov/60 bp (horizontally hatched bars) plus increasing concentrations of N-H3me2 (H), N-H3ac (I), and N-H3 (J). Histone peptide concentrations were as indicated.
We removed the histone H3 N termini by treatment of soluble chromatin with immobilized trypsin (41) and purification on sucrose gradients containing 0.5 M NaCl. SDS-PAGE (Fig. 4F) shows that limited trypsinization resulted in complete disappearance of intact histone H3 (lane 2). Coomassie blue staining (lane 2) shows an accumulation of the histone band P1, which has been previously identified as histone H3 lacking 26 amino acids from its N terminus (23). A Western blot, showing no reaction with α-H3me2K9 in the trypsin-digested chromatin (lane 4), is consistent with elimination of the N-terminal domain of H3 containing K9. The trypsin-digested chromatin continues to bind both MENTwt and MENTov as detected by the gel shift assay (Fig. 4B), thus demonstrating that the cleaved H3 N-terminal domains were not necessary for MENT binding. However, in the trypsin-digested chromatin, we observed a dramatic reduction in chromatin precipitation by MENT (Fig. 4G), suggesting that the intact histone H3 N domains were important for chromatin self-association and precipitation by MENT.
We then took the complexes of MENTwt and MENTov with trypsin-digested nucleosome arrays (1:100 bp) and reconstituted them with increasing concentrations of the 21-amino-acid N-terminal peptide of histone H3 containing either two methyl groups (N-H3me2), one acetyl group (N-H3ac), or no modification at K9 (N-H3). Figure 4H shows that association with the dimethylated histone H3 N-terminal peptide restored the selective precipitation of chromatin by MENTwt. In a reciprocal experiment, we took the complexes of chromatin with 5 μM N-H3me2 and added increasing concentrations of MENTwt and MENTov. Again the precipitation was much stronger with MENTwt than with MENTov (Fig. 4C), showing that the intact RCL domain was required for cooperation with dimethylated histone H3 in chromatin condensation.
Strikingly, reconstitution with increasing concentrations of the similar 21-amino-acid N-terminal peptide of histone H3 containing an acetyl group at K9 (N-H3ac) had no apparent effect on chromatin interaction with MENT. Neither the electrophoretic mobility (Fig. 4D) nor precipitation of chromatin complexes with MENTwt and MENTov (Fig. 4I) was significantly affected by the acetylated peptide. Moreover, reconstitution with unmodified peptide (N-H3) also showed significantly reduced precipitation (Fig. 4E and J) compared with dimethylated peptide. We thus concluded that MENT specifically and cooperatively interacts with the methylated N-terminal domain of histone H3 to promote chromatin self-association. Further, this interaction requires an intact RCL and is inhibited by histone H3 acetylation at K9.
Histone deacetylase inhibitors and RCL mutations have a similar effect on MENT interaction with nuclear chromatin in vivo.
The correlation of MENT distribution with the histone modification pattern over the β-globin domain (Fig. 2) and our in vitro experiments (Fig. 4) suggested that the histone modifications could directly regulate chromatin condensation by directing MENT binding to certain chromosomal loci and/or inhibiting MENT binding to active chromatin. Because MENT is expressed in terminally differentiated quiescent blood cells that do not proliferate and do not support genetic manipulations, we decided to test this possibility by experiments with a model cellular system. We chose the cell line NIH 3T3, in which a transition from a proliferating to a quiescent stage can be easily induced by contact inhibition or serum deprivation (37, 39). Previously, we found that transient expression of MENT in NIH 3T3 cells caused significant chromatin condensation and inhibited cell proliferation. We have also identified mutations in the RCL domain of MENT that eliminated the MENT-induced phenotype in transiently transfected cells (34), thus providing efficient controls for the specificity of the effect of ectopic MENT on chromatin.
We established a stable NIH 3T3 cell line expressing MENTwt from a cDNA construct driven by an IPTG (isopropyl-β-D-thiogalactopyranoside)-inducible promoter (see Materials and Methods). This cell line (3T3/MENT-63) was used to express MENTwt in all of our experiments. Immunofluorescence of the MENT-expressing cells showed a strong nuclear signal detected with α-MENT antibodies (Fig. 5A, panel 1). In previous work, we established the specificity of these antibodies for MENT-expressing cells (26, 34). Here we confirmed the absence of antibody cross-reactivity by the lack of signal in MENT-expressing 3T3/MENT-63 cells stained with control preimmune serum and in control cells (NIH 3T3-lacI) transfected with vector only (not shown).
FIG. 5.
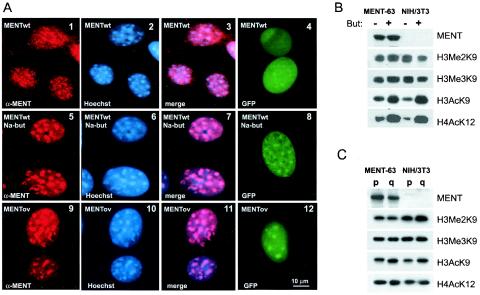
A histone deacetylase inhibitor and a mutation in the RCL domain change the MENT location in nuclear chromatin. (A) Immunofluorescence microscopy of proliferating 3T3/MENT-63 cells (panels 1 to 3), the same cells incubated in the presence of 10 mM sodium butyrate for 48 h (panels 5 to 7), and 3T3/MENTov-16 cells stably expressing a MENT-ovalbumin swap mutant (panels 9 to 11) fixed in acetone-methanol and stained with α-MENT (panels 1, 5, and 9) and Hoechst 33258 (panels 2, 6, and 10). Panels 3, 7, and 11, overlays of images stained with α-MENT and Hoechst. Panels 4, 8, and 12, immunofluorescence microscopy of unfixed, unstained proliferating NIH 3T3 cells transiently expressing GFP fusions with MENTwt (panels 4 and 8) and MENT-ovalbumin swap (panel 12). The cells in panel 8 were incubated for 48 h with 10 mM sodium butyrate. Bar, 10 μm. (B) Protein samples from the nuclei of 3T3/MENT-63 cells stably expressing MENTwt (MENT-63) and control NIH 3T3-lacI cell clones stably transfected with nonexpressing vector (NIH 3T3) incubated in the absence (−) or presence (+) of 10 mM sodium butyrate (But) for 48 h were analyzed by SDS-PAGE and Western blotting with α-MENT, α-H3me2K9, α-H3me3K9, α-H3acK9, and α-H4acK12 as indicated. (C) Protein samples from the nuclei of proliferating (p) and quiescent (q) 3T3/MENT-63 cells and control NIH 3T3-lacI cells were analyzed by SDS-PAGE and Western blotting with α-MENT, α-H3me2K9, α-H3me3K9, α-H3acK9, and α-H4acK12 as indicated.
As detected by semiquantitative analysis of the Western blotting shown in Fig. 5B, the 3T3/MENT-63 cell line grown in the presence of IPTG for 48 h had accumulated 0.07 molecule of MENT per 200 bp of DNA, i.e., a level about 3.5-fold higher than that in erythrocytes. The 3T3/MENT-63 cells had approximately twofold-slower proliferation than control cells transfected with vector only (NIH 3T3-lacI) or cells (3T3/MENTov-16) expressing the RCL mutant, MENTov. Additionally, their nuclei became notably smaller than those in the MENTov cells (compare panel 2 with panel 10 in Fig. 5A) and the control cells (Fig. 6, panel 6).
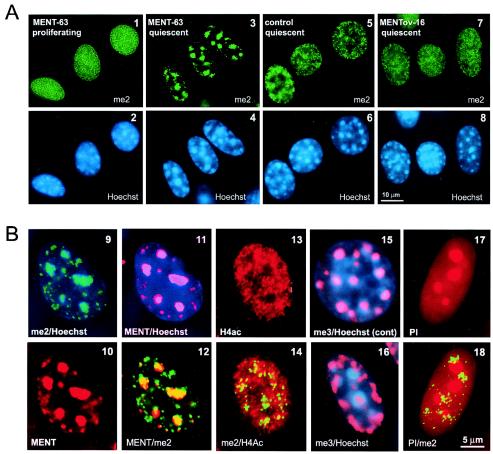
FIG. 6.
Rearrangement of chromatin marked by histone H3 methylation in quiescent MENT-expressing cells. (A) Immunofluorescence microscopy of 3T3/MENT-63 cells (panels 1 to 4), control NIH 3T3-lacI cells (panels 5 and 6), and 3T3/MENTov-16 cells (panels 7 and 8). Cells were fixed in the proliferating state (panels 1 and 2) and the quiescent state (panels 3 to 8) and then stained with α-H3me2K9 (me2) (panels 1, 3, 5, and 7) and Hoechst 33258 (panels 2, 4, 6, and 8). Bar, 10 μm. (B) Immunofluorescence microscopy of 3T3/MENT-63 cells (panels 9 to 14 and 16 to 18) and control NIH 3T3-lacI cells (panel 15). Cells were fixed in the quiescent state and then stained with α-H3me2K9 (green) (panels 9, 12, 14, and 18), α-MENT (red) (panels 10 to 12), α-H4acK12 (red) (panels 13 and 14), α-H3me3K9 (red) (panels 15 and 16), Hoechst 33258 (blue) (panels 9, 11, 15, and 16), and propidium iodine (red) (panels 17 and 18). Bar, 10 μm.
NIH 3T3 cells, like other mouse cell lines, have a well-defined focal distribution of nuclear heterochromatin that can be visualized with Hoechst 33258, a fluorescent dye that preferentially stains AT-rich DNA in pericentromeric heterochromatin (31). Figure 5A (panels 1 to 3) shows that in proliferating 3T3/MENT-63 cells, MENT is widely distributed in the nuclei, with the protein found both within the Hoechst-positive heterochromatic blocks and over the euchromatic areas.
To examine, whether MENT can be spatially regulated by histone acetylation, we incubated the 3T3/MENT-63 and control cells in presence of 10 mM sodium butyrate, a histone deacetylase inhibitor. With both MENT-expressing and control cells, we observed a notable increase in H4acK12 and H3acK9, both markers of active chromatin (Fig. 5B). Remarkably, 3T3/MENT-63 incubated in the presence of sodium butyrate shows a major change in MENT distribution, with the protein becoming predominantly associated with heterochromatin (Fig. 5A, panels 5 to 7). In parallel to the change in MENT localization, the nuclei in butyrate-treated cells return to normal size (Fig. 5A, panel 6), suggesting that histone hyperacetylation reversed chromatin condensation by MENT. The Western blotting in Fig. 5B shows no decrease in MENT levels in the butyrate-treated cells, indicating that the loss of the protein from euchromatic sites is not the result of protein degradation or decreased expression. Incubation of the 3T3/MENT-63 cells with another deacetylase inhibitor, 100 nM trichostatin, produced an effect very similar to that observed with sodium butyrate (not shown).
To test the effect of the deacetylase inhibitors on MENT distribution in living cells, we constructed an N-terminal protein fusion of MENTwt with green fluorescent protein (GFP-MENTwt). After transfecting GFP-MENTwt in NIH 3T3 cells, we observed an even distribution of GFP-MENT in the nuclei. When cells expressing GFP-MENTwt were treated with 10 mM sodium butyrate, we observed a notable relocation of MENT to heterochromatic foci (compare panels 4 and 8 in Fig. 5A), demonstrating that the effect of the deacetylase inhibitors on MENT compartmentalization is not an artifact of cell fixation and the antigen-antibody exposure.
Our previous experiments with transiently expressed MENT (34) and the in vitro experiments described above (Fig. 4) suggested that the RCL domain is important for MENT interaction with chromatin. To further examine the function of the RCL domain in vivo, we established the stably transfected 3T3/MENTov-16 cell line expressing MENTov. Figure 5A (panels 9 to 11) shows that the 3T3/MENTov-16 cells have a phenotype strikingly resembling that of the butyrate-treated cells expressing MENTwt (panels 5 to 7), with the focal heterochromatin pattern of MENT and decondensed nuclei. The GFP-MENTov fusion in living NIH 3T3 cells also showed a focal localization (panel 12). The effects of histone deacetylase inhibitors and the RCL mutation on MENT localization in vivo (Fig. 5) strongly support the results of the in vitro experiments (Fig. 4) showing that both the RCL and unacetylated histone N-terminal domain(s) mediate MENT interactions with chromatin.
Remodeling of chromatin associated with methylated histone H3(K9) in MENT-expressing quiescent cells.
The colocalization of MENT and α-H3me2K9 in chicken erythrocytes (Fig. 1B) and the specific interaction of MENT with the dimethylated N-terminal domain of histone H3 in vitro (Fig. 4) prompted us to ask whether MENT can target dimethylated histone H3 in the 3T3/MENT-63 cells. In proliferating 3T3/MENT-63 cells, MENT is broadly distributed in nuclear chromatin (Fig. 5A, panel 1). When we stained these cells with α-H3me2K9, we observed a similarly uniform distribution of the label unrelated to heterochromatic foci (compare panels 1 and 2 in Fig. 6). The H3me2K9 staining pattern was not significantly different from that in control NIH 3T3 cells (not shown), consistent with previously reported H3me2K9 localization in nuclear chromatin (45, 52).
Since MENT normally acts in quiescent vertebrate cells, we compared the chromatin organizations in proliferating and quiescent NIH 3T3 cells. Remarkably, after we drove MENT-expressing NIH 3T3 cells into quiescence by contact growth inhibition, we observed a dramatic change in the distribution of histone H3me2K9 in 3T3/MENT-63 compared with control cells. While in the quiescent MENT-expressing cells the H3me2K9 became notably concentrated at the Hoechst-positive chromatin blocks (Fig. 6, panels 3 and 4 and the image overlay in panel 9), in the control quiescent cells it remained broadly spread throughout the nucleus (Fig. 6, panels 5 and 6). Western blotting (Fig. 5C) showed no significant change in the level of H3me2K9 between the proliferating and quiescent 3T3/MENT-63 cells, indicating that the dimethylated H3 relocates to pericentromeric heterochromatin rather than being lost in the noncentromeric areas. In a striking parallel to dimethylated histone H3, MENT also became focal and colocalized with H3me2K9 and pericentromeric heterochromatin in quiescent cells (see MENT staining and overlays with me2 and Hoechst staining in Fig. 6, panels 10 to 12). Thus, MENT and dimethylated histone H3(K9) undergo coordinated spatial transitions during quiescence of MENT-expressing NIH 3T3 cells.
In comparison to the 3T3/MENT-63 cells, cells expressing the RCL mutant (3T3/MENTov-16), showed no pericentromeric concentration of H3me2K9 (Fig. 6, panels 7 and 8). Incubation of quiescent 3T3/MENT-63 with histone deacetylase inhibitors similarly prevented the association of H3me2K9 with chromocenters (data not shown). Note that in both 3T3/MENTov-16 and sodium butyrate-treated 3T3/MENT-63 cells (Fig. 5A), MENT is associated with pericentromeric heterochromatin and thus is dissociated from the noncentromeric H3me2K9. These results show that both the RCL mutation and histone hyperacetylation inhibit MENT association with chromatin marked by dimethylated histone H3.
We asked whether the MENT-induced redistribution of modified histones to heterochromatin is specific for dimethylated histone H3. From our ChIP experiments we predicted that chromatin remodeling by MENT should not involve acetylated core histones. Indeed, the quiescent 3T3/MENT-63 cells stained with antibodies against H4acK12 showed a broad distribution of the modified histone that did not correlate with either Hoechst (not shown) or H3me2K9 staining (compare panels 13 and 14 in Fig. 6).
In contrast to the broadly distributed H3me2K9 (45), histone H3 trimethylated at lysine 9 (H3me3K9) is normally associated with pericentromeric heterochromatin (13). Indeed, in control cells the α-H3me3K9 antibodies showed strong focal staining precisely colocalized with Hoechst-positive chromatin blocks (Fig. 6, panel 15). Interestingly, in the quiescent MENT-expressing cells the chromatin marked by H3me3K9 simultaneously reduced its preferential association with chromocenters and became more abundant at the nuclear periphery (Fig. 6, panel 16), in the zones free of H3me2K9 and MENT. Together with the data obtained with primary chicken erythrocytes (Fig. 1C), this result confirms that MENTwt preferentially targets chromatin modified by histone H3(K9) dimethylation rather than trimethylation. Chromatin methylation at the pericentromeric heterochromatin is thought to be controlled by a positive feedback regulation imposed by interacting methylated histone H3, histone methyltransferase, and HP1 (35). It appears likely that MENT association with methylated histone H3 at the chromocenters interferes with this regulatory pathway, leading to a partial exclusion of the enzyme responsible for histone trimethylation from constitutive heterochromatin.
Finally, we questioned whether the chromatin remodeling observed in quiescent MENT-expressing cells was associated with any other major rearrangements or defects in nuclear morphology. In the quiescent 3T3/MENT-63 cells we observed no disassembly of the Hoechst-stained constitutive heterochromatin (Fig. 6, panel 4) such as has been reported for cells treated for several days with histone deacetylase inhibitors (62). To test whether the expression of MENT could cause a collapse of nuclear chromatin and relocation of the total chromatin mass to centromeres (as observed, for example, during apoptosis [29]), we counterstained MENT-expressing cells with propidium iodine, a general nucleic acid stain. The propidium iodine-stained cells showed bright spots corresponding to the nucleoli but no lesions or otherwise damaged chromatin, in particular at sites counterstained with H3me2K9 (compare panels 17 and 18 in Fig. 6). Moreover, the quiescent MENT-expressing cells with pericentromeric H3me2K9 were completely viable and, after trypsinization and replating, resumed proliferation, nevertheless showing a strong delay in exiting the quiescent state. The reactivated proliferating cells had a phenotype indistinguishable from that of the original 3T3/MENT-63 cells (Fig. 5A, panels 1 and 2). We thus concluded that the MENT-induced chromatin transitions were reversible and compatible with cell viability.
DISCUSSION
The chicken β-globin locus has been widely employed as a model genetic system, leading to the description of several important structural features of active chromatin. These include long-range nuclease sensitivity (66), developmentally regulated hypersensitive sites associated with transcriptional regulatory elements (61), and structural insulation of active chromatin from silencing (12). We have now mapped the distribution of a developmentally regulated chromatin-condensing protein, MENT, across the β-globin gene cluster. Our results clearly establish MENT as the first chromatin-condensing protein whose access to the active chromatin is blocked at the boundary elements of an active vertebrate gene.
Genetic studies with yeast revealed a general mechanism for heterochromatin spreading that is mediated by nonhistone proteins interacting with histones and regulated by histone modifications (9, 18, 27). In budding yeast, a histone acetyltransferase tethered to a chromatin locus can act as a boundary element that inhibits heterochromatin spreading by histone hyperacetylation (16). In fission yeast, histone H3 methylation at lysine 9 that is confined to heterochromatin mediates the chromatin association with Swi6, a homologue of the vertebrate heterochromatin protein HP1 (47). In vertebrates, studies of the β-globin domain boundaries have also shown a principal role for histone acetylation in insulating genes from position-dependent silencing (46, 53). Since histone acetylation and methylation at one lysine side chain are mutually exclusive (55), acetylation has been proposed to insulate active genes by blocking interactions of chromatin-condensing proteins with methylated histone H3(K9) (8, 42).
Here we show that these present models for heterochromatin spreading are in remarkable agreement with the spatial regulation of MENT in nuclei. First, MENT colocalizes with methylated K9 of histone H3 in chicken erythrocytes (Fig. 1B and C) and in 3T3/MENT-63 cells (Fig. 6), and it comaps (Fig. 2 and 3) with this modification at the β-globin domain (42). Second, MENT's ectopic expression in NIH 3T3 cells causes a significant redistribution of chromatin marked by methylated histone H3 (Fig. 6). Third, the histone H3 N domain dimethylated at lysine 9 cooperates with the RCL domain of MENT to promote chromatin condensation (Fig. 4). Fourth, its spreading at the β-globin locus (Fig. 2) is strikingly opposite to the level of histone acetylation (43). Finally, the MENT location in nuclear chromatin is regulated by deacetylase inhibitors (Fig. 5). By the mode of spatial regulation, MENT closely resembles HP1, a prototype heterochromatin protein. In contrast to HP1, MENT is accumulated, not depleted, in condensed chromatin of terminally differentiated cells (Fig. 1) and effectively condenses chromatin at its natural protein/DNA ratio (25). Our studies thus establish MENT as the first vertebrate chromatin-condensing protein spatially regulated by histone modifications. The experimental system employing stably expressed MENT provides a novel approach to study the mechanism of chromatin condensation even in cells that do not normally express MENT.
MENT does not contain any homology to the chromodomain, the domain in HP1 that binds methylated histone H3 lysine 9. However, our experiments revealed a key role of the RCL domain as a principal structural element mediating MENT interactions with chromatin. The RCL domain is conserved among the members of the serpin (for serine protease inhibitor) protein family, to which MENT belongs (22). In other serpins, RCL mediates strong protein-protein interactions that involve a large-scale conformational transition of the RCL domain from the stressed to the relaxed (S→R) conformation (60). Remarkably, the mutations of the conserved RCL amino acids that are essential for the S→R transition disrupt chromatin condensation (34) and the remodeling of H3me2K9 (Fig. 6) by MENT. Our other findings, in particular the importance of RCL for chromatin condensation (Fig. 4) but not for chromatin and DNA binding in vitro (data not shown), strongly support the idea that the RCL domain is directly involved in histone binding. In addition to MENT, several other closely related serpins have been recently found in mammalian nuclei (4, 10). One such nuclear serpin, LEI (serpin B1), accumulates in terminally differentiated mammalian granulocytes and interacts with histones, and its nuclear localization is affected by histone deacetylase inhibitors and RCL mutations in a manner very similar to that for MENT (E. Popova and S. A. Grigoryev, unpublished data). Our work thus uncovers a direct role for the conserved RCL domain in chromatin condensation that may illuminate the nuclear function(s) of MENT-related serpins.
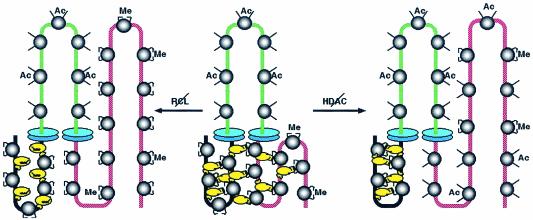
How does MENT interact with histone H3 N-terminal domains to promote chromatin condensation? Unlike HP1, MENT does not need the N-terminal domain of H3 to bind chromatin (Fig. 4), thus indicating a quite different mechanism for heterochromatin interaction. Indeed, in addition to the RCL, MENT contains another important domain, the M loop, which is located on the opposite pole of the MENT molecule (22) and is essential for centromeric heterochromatin targeting (34). Previously we have observed that chromatin condensation caused by MENT is associated with the formation of protein bridges that hold nucleosome arrays together even if their DNA is cleaved by nucleases (25). Here we propose that the M loop and the RCL domains together make such a bridge that mediates the chromatin condensation. As modeled in Fig. 7, while the M loop binds to the AT-rich centromeric heterochromatin DNA (the M loop contains an AT-rich DNA binding motif, the AT hook), the RCL domain interacts with dimethylated and deacetylated N-terminal domain of histone H3 in noncentromeric chromatin. Simultaneous binding via the AT hook to centromeric constitutive heterochromatin and, via RCL, to noncentromeric chromatin would allow MENT to connect distant chromatin fibers by protein bridges, causing the redistribution of noncentromeric chromatin marked by a high level of dimethylated histone H3 into close proximity to centromeric heterochromatin. A chromosomal domain containing active hyperacetylated chromatin is insulated from MENT spreading by boundary elements that create zones of especially strong hyperacetylation, blocking MENT spreading from the domain flanks (Fig. 7, center structure). After such rearrangement, the inactive chromatin marked by H3me2K9 would form zones of local concentration consistent with the H3me2K9 foci colocalized with MENT in mature erythrocytes (Fig. 1C) and quiescent 3T3/MENT-63 cells (Fig. 6) and with the high level of MENT at the boundaries of the β-globin gene (Fig. 2). The protein bridges formed by MENT may facilitate pairing of boundary elements that has been proposed to be important for their barrier function (5). The RCL mutation disrupts MENT-histone bridges (Fig. 7, left structure), prevents chromatin remodeling (Fig. 6), and sends MENT to constitutive heterochromatin (Fig. 5). Histone hyperacetylation interferes with MENT binding with histone H3 (Fig. 7, right structure) and also sends MENT to constitutive heterochromatin (Fig. 5). Although we cannot exclude the possibility that MENT causes demethylation of noncentromeric histone H3 rather than the relocation of total chromatin as we propose in Fig. 7, in view of the metabolic stability of histone H3 methylation at K9 (1) and the lack of changes in histone H3 methylation levels associated with MENT expression, deacetylase inhibition, and quiescence (Fig. 5B and C), we consider histone demethylation a less likely possibility.
FIG. 7.
A model for chromatin condensation regulated by MENT, histone modifications, and gene boundary elements. (Central structure) MENTwt (yellow) binds simultaneously to constitutive heterochromatin (black) and silenced euchromatin (red) marked by H3 dimethylated at lysine 9 (Me). MENT binds to chromatin cooperatively and forms bridges between chromatin fibers that bring euchromatin containing dimethylated histone H3 close to constitutive heterochromatin, thus causing chromatin condensation. A chromosomal domain containing active hyperacetylated chromatin (green) is insulated from MENT spreading by boundary elements (blue disks) that create zones of especially strong hyperacetylation blocking MENT spreading from the domain flanks. (Right structure) Histone deacetylase (HDAC) inhibitors increase the acetylation level of noncentromeric chromatin, interfere with MENT-histone interactions, and promote MENT binding to constitutive heterochromatin. (Left structure) RCL mutations disrupt MENT interaction with chromatin marked by dimethylated histone H3 and also promote MENT binding to constitutive heterochromatin.
In actively proliferating and differentiating cells, genes are inactivated through modification of chromatin higher-order structure that involves a spatial separation of active and repressed chromatin and association of repressed genes with pericentromeric heterochromatin (44, 59). Remarkably, during terminal differentiation, when large-scale silencing inactivates many genes, the levels of key repressor factors such as Ikaros (7) and HP1 isotypes (this work) decline, in some cases to nondetectable levels. With the apparent simplification of the repressive apparatus, the large-scale chromatin condensation caused by developmentally regulated factors such as MENT should ensure a complete inactivation of unused housekeeping genes that might otherwise trigger illegitimate cell proliferation and oncogenic transformation (65). Several recent studies have revealed a role of noncentromeric histone H3 methylation in silencing of euchromatic genes associated with cell cycle-dependant repression, G (0), and quiescence (48, 50, 51). Our results, especially the focalization of chromatin marked with H3me2K9 in mature erythrocytes (Fig. 1C) and the chromatin remodeling in quiescent MENT-expressing NIH 3T3 cells (Fig. 6), indicate that noncentromeric histone H3 methylation may mark a class of chromosomal regions for coordinated condensation and silencing at the terminal differentiation stage. Work aimed at identification of other factors involved in regulating chromatin condensation and the genes undergoing coordinated structural rearrangement during terminal differentiation is now in progress.
Acknowledgments
We thank M. Reitman (Bethesda, Md.) for sending us DNA of the β-globin cluster, T. Graf (New York, N.Y.) for the HD-13 cell line, and E. Popova, I. Boulynko, and L. Carrel (Hershey, Pa.) for valuable comments.
This work was supported in part by NIH grants GM-59118 to S.A.G. and GM-36477 to K.S.Z. and by a grant from the Russian Foundation for Basic Research (02-04-48571) to V.L.K.
Natalia E. Istomina, Sain S. Shushanov, and Evelyn M. Springhetti contributed equally to this work.
REFERENCES
- 1.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation. Dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. C., A. G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291:447-450. [DOI] [PubMed] [Google Scholar]
- 3.Bellard, M., M. T. Kuo, G. Dretzen, and P. Chambon. 1980. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 8:2737-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, C. H., E. J. Blink, C. E. Hirst, M. S. Buzza, P. M. Steele, J. Sun, D. A. Jans, and P. I. Bird. 2001. Nucleocytoplasmic distribution of the ovalbumin serpin pi-9 requires a nonconventional nuclear import pathway and the export factor crm1. Mol. Cell. Biol. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17:664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K. E., J. Baxter, D. Graf, M. Merkenschlager, and A. Fisher. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3:207-218. [DOI] [PubMed] [Google Scholar]
- 8.Burgess-Beusse, B., C. Farrell, M. Gaszner, M. Litt, V. Mutskov, F. Recillas-Targa, M. Simpson, A. West, and G. Felsenfeld. 2002. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA 99:16433-16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277:4778-4781. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, T. L., and R. R. Schleef. 1999. Identification of a nuclear targeting domain in the insertion between helices C and D in protease inhibitor-10. J. Biol. Chem. 274:11194-11198. [DOI] [PubMed] [Google Scholar]
- 11.Chung, J. H., A. C. Bell, and G. Felsenfeld. 1997. Characterization of the chicken β-globin insulator. Proc. Natl. Acad. Sci. USA 94:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, J. H., M. Whiteley, and G. Felsenfeld. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505-514. [DOI] [PubMed] [Google Scholar]
- 13.Cowell, I. G., R. Aucott, S. K. Mahadevaiah, P. S. Burgoyne, N. Huskisson, S. Bongiorni, G. Pantera, L. Fanti, S. Pimpinelli, R. Wu, D. M. Gilbert, W. Shi, R. Fundele, H. Morrisin, P. Jeppesen, and P. B. Singh. 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111:22-36. [DOI] [PubMed] [Google Scholar]
- 14.Dardick, I., and G. Setterfield. 1976. Volume of condensed chromatin in developing primitive-line erythrocytes of chick. Exp. Cell Res. 100:159-171. [DOI] [PubMed] [Google Scholar]
- 15.de la Barre, A. E., D. Angelov, A. Molla, and S. Dimitrov. 2001. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 20:6383-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugaiczyk, A., S. L. Woo, D. A. Colbert, E. C. Lai, M. L. Mace, Jr., and B. W. O'Malley. 1979. The ovalbumin gene: cloning and molecular organization of the entire natural gene. Proc. Natl. Acad. Sci. USA 76:2253-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 19.Filesi, I., A. Cardinale, S. van Der Sar, I. G. Cowell, P. B. Singh, and S. Biocca. 2002. Loss of heterochromatin protein 1 (HP1) chromodomain function in mammalian cells by intracellular antibodies causes cell death. J. Cell Sci. 115:1803-1813. [DOI] [PubMed] [Google Scholar]
- 20.Francastel, C., D. Schubeler, D. I. Martin, and M. Groudine. 2000. Nuclear compartmentalization and gene activity. Nat Rev. Mol. Cell. Biol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 21.Grigoryev, S. A. 2001. Higher-order folding of heterochromatin: protein bridges span the nucleosome arrays. Biochem. Cell. Biol. 79:227-241. [PubMed] [Google Scholar]
- 22.Grigoryev, S. A., J. Bednar, and C. L. Woodcock. 1999. MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J. Biol. Chem. 274:5626-5636. [DOI] [PubMed] [Google Scholar]
- 23.Grigoryev, S. A., and I. A. Krasheninnikov. 1982. Transient unfolding of trypsin-digested chromatin core particles. Eur. J. Biochem. 129:119-125. [DOI] [PubMed] [Google Scholar]
- 24.Grigoryev, S. A., V. O. Solovieva, K. S. Spirin, and I. A. Krasheninnikov. 1992. A novel nonhistone protein (MENT) promotes nuclear collapse at the terminal stage of avian erythropoiesis. Exp. Cell Res. 198:268-275. [DOI] [PubMed] [Google Scholar]
- 25.Grigoryev, S. A., and C. L. Woodcock. 1998. Chromatin structure in granulocytes. A link between tight compaction and accumulation of a heterochromatin-associated protein (MENT). J. Biol. Chem. 273:3082-3089. [DOI] [PubMed] [Google Scholar]
- 26.Grigoryev, S. A., and C. L. Woodcock. 1993. Stage-specific expression and localization of MENT, a nuclear protein associated with chromatin condensation in terminally differentiating avian erythroid cells. Exp. Cell Res. 206:335-343. [DOI] [PubMed] [Google Scholar]
- 27.Grunstein, M. 1998. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93:325-328. [DOI] [PubMed] [Google Scholar]
- 28.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 13:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendzel, M., W. Nishioka, Y. Raymond, C. Allis, D. Bazett-Jones, and J. Th'ng. 1998. Chromatin condensation is not associated with apoptosis. J. Biol. Chem. 273:24470-24478. [DOI] [PubMed] [Google Scholar]
- 30.Hennig, W. 1999. Heterochromatin. Chromosoma 108:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Hilwig, I., and A. Gropp. 1973. Decondensation of constitutive heterochromatin in L cell chromosomes by a benzimidazole compound (“33258 Hoechst”). Exp. Cell Res. 81:474-477. [DOI] [PubMed] [Google Scholar]
- 32.Horn, P. J., and C. L. Peterson. 2002. Chromatin Higher Order Folding-Wrapping up Transcription. Science 297:1824-1827. [DOI] [PubMed] [Google Scholar]
- 33.Horsley, D., A. Hutchings, G. W. Butcher, and P. B. Singh. 1996. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Cell Genet. 73:308-311. [DOI] [PubMed] [Google Scholar]
- 34.Irving, J. A., S. S. Shushanov, R. N. Pike, E. Y. Popova, D. Bromme, T. H. Coetzer, S. P. Bottomley, I. A. Boulynko, S. A. Grigoryev, and J. C. Whisstock. 2002. Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J. Biol. Chem. 277:13192-13201. [DOI] [PubMed] [Google Scholar]
- 35.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 36.Jimenez, G., S. D. Griffiths, A. M. Ford, M. F. Greaves, and T. Enver. 1992. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc. Natl. Acad. Sci. USA 89:10618-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knosp, O., H. Talasz, and B. Puschendorf. 1991. Histone acetylation and histone synthesis in mouse fibroblasts during quiescence and restimulation into S-phase. Mol. Cell. Biochem. 101:51-58. [DOI] [PubMed] [Google Scholar]
- 38.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 39.Ladha, M. H., K. Y. Lee, T. M. Upton, M. F. Reed, and M. E. Ewen. 1998. Regulation of exit from quiescence by p27 and cyclin D1-CDK4. Mol. Cell. Biol. 18:6605-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leitch, A. R. 2000. Higher levels of organization in the interphase nucleus of cycling and differentiated cells. Microbiol. Mol. Biol. Rev. 64:138-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leuba, S. H., C. Bustamante, J. Zlatanova, and K. van Holde. 1998. Contributions of linker histones and histone H3 to chromatin structure: scanning force microscopy studies on trypsinized fibers. Biophys. J. 74:2823-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 43.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundgren, M., C.-M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 45.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 46.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 48.Nicolas, E., C. Roumillac, and D. Trouche. 2003. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol. Cell. Biol. 23:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 51.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 52.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 53.Pikaart, M., F. Recillas-Targa, and G. Felsenfeld. 1998. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12:2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prioleau, M. N., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 56.Reitman, M., and G. Felsenfeld. 1990. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol. Cell. Biol. 10:2774-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 58.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman, G. A., P. I. Bird, R. W. Carrell, F. C. Church, P. B. Coughlin, P. G. Gettins, J. A. Irving, D. A. Lomas, C. J. Luke, R. W. Moyer, P. A. Pemberton, E. Remold-O'Donnell, G. S. Salvesen, J. Travis, and J. C. Whisstock. 2001. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276:33293-33296. [DOI] [PubMed] [Google Scholar]
- 61.Stalder, J., A. Larsen, J. D. Engel, M. Dolan, M. Groudine, and H. Weintraub. 1980. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20:451-460. [DOI] [PubMed] [Google Scholar]
- 62.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 63.Tamaru, H., X. Zhang, D. McMillen, P. B. Singh, J.-I. Nakayama, S. Grewal, C. D. Allis, X. Cheng, and E. U. Selker. Trimethylation of histone H3 lysine-9 associated with methylated DNA in Neurospora. Nat. Genet. 34:75-79. [DOI] [PubMed]
- 64.Verreault, A., and J. O. Thomas. 1993. Chromatin structure of the beta-globin chromosomal domain in adult chicken erythrocytes. Cold Spring Harbor Symp. Quant. Biol. 58:15-24. [DOI] [PubMed] [Google Scholar]
- 65.Weintraub, H. 1985. Assembly and propagation of repressed and depressed chromosomal states. Cell 42:705-711. [DOI] [PubMed] [Google Scholar]
- 66.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848-856. [DOI] [PubMed] [Google Scholar]
- 67.Widom, J. 1998. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27:285-327. [DOI] [PubMed] [Google Scholar]
- 68.Woodcock, C. L., and S. Dimitrov. 2001. Higher order structure of chromatin and chromosomes. Curr. Opin. Gen. Dev. 11:130-135. [DOI] [PubMed] [Google Scholar]
- 69.Wreggett, K. A., F. Hill, P. S. James, A. Hutchings, G. W. Butcher, and P. B. Singh. 1994. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet. 66:99-103. [DOI] [PubMed] [Google Scholar]
- 70.Zlatanova, J., S. H. Leuba, and K. van Holde. 1999. Chromatin structure revisited. Crit. Rev. Eukaryot. Gene Expr. 9:245-255. [DOI] [PubMed] [Google Scholar]