Abstract
Subcellular localization of Ras proteins to the plasma membrane is accomplished in part by covalent attachment of a farnesyl moiety to the conserved CaaX box cysteine. Farnesylation targets Ras to the endoplasmic reticulum (ER), where additional processing steps occur, resulting in translocation of Ras to the plasma membrane. The mechanism(s) by which this occurs is not well understood. In this report, we show that plasma membrane localization of Ras2p in Saccharomyces cerevisiae does not require the classical secretory pathway or a functional Golgi apparatus. However, when the classical secretory pathway is disrupted, plasma membrane localization requires Erf2p, a protein that resides in the ER membrane and is required for efficient palmitoylation of Ras2p. Deletion of ERF2 results in a Ras2p steady-state localization defect that is more severe when combined with sec-ts mutants or brefeldin A treatment. The Erf2p-dependent localization of Ras2p correlates with the palmitoylation of Cys-318. An Erf2p-Erf4p complex has recently been shown to be an ER-associated palmitoyltransferase that can palmitoylate Cys-318 of Ras2p (S. Lobo, W. K. Greentree, M. E. Linder, and R. J. Deschenes, J. Biol. Chem. 277:41268-41273, 2002). Erf2-dependent palmitoylation as well as localization of Ras2p requires a region of the hypervariable domain adjacent to the CaaX box. These results provide evidence for the existence of a palmitoylation-dependent, nonclassical endomembrane trafficking system for the plasma membrane localization of Ras proteins.
Ras proteins are small, plasma membrane-associated guanine nucleotide binding proteins that cycle between GTP- and GDP-bound forms to regulate cell growth and differentiation by interacting with a variety of cellular effectors (9, 15, 31). Mutations that increase the GTP/GDP ratio activate Ras and contribute to cellular transformation in many human cancers (24). Although Ras is initially produced as a cytosolic precursor, Ras must be targeted to the plasma membrane in order to function in signal transduction (12, 49). Membrane association requires a series of posttranslational modifications of a C-terminal motif called the CaaX box (C is Cys, a is generally an aliphatic amino acid, and X is the C-terminal amino acid). These modifications include farnesylation of the CaaX-box Cys, proteolytic removal of the -aaX residues, carboxy methylation, and in most but not all cases, palmitoylation of a second cysteine adjacent to the CaaX box (10, 14, 22, 43).
The sequential modification of the CaaX box is responsible for subcellular targeting of Ras (12). The first step in the modification pathway, farnesylation, has been shown to be sufficient to target Ras to the endoplasmic reticulum (ER), where the CaaX protease and methyltransferase reside (11, 36, 41). The next step, translocation of Ras from the ER to the plasma membrane, requires additional targeting signals. Palmitoylation serves as the second signal for mammalian H-ras, N-ras, and the yeast Ras proteins, whereas a stretch of basic residues (polybasic) provides the signal for K-ras-4B (11, 22). Hancock and colleagues have demonstrated that palmitoylated H-ras protein localizes to the plasma membrane via the classical secretory pathway and is sensitive to brefeldin A, whereas the plasma membrane localization of K-ras-4B protein is resistant to brefeldin A (2). In yeast, palmitoylation is also required for the plasma membrane localization of Ras1p and Ras2p (5). However, the role of palmitoylation has not been clear for at least two reasons. First, despite considerable effort, the protein palmitoyltransferase proposed to modify Ras had not been identified. Second, the subcellular trafficking of yeast Ras from the ER to the plasma membrane has not been defined genetically or biochemically.
In this report we show that the plasma membrane localization of Ras2p in yeast is unaffected by disruption of the classical secretory pathway, suggesting the existence of an alternative or nonclassical pathway for Ras translocation from the ER to the plasma membrane. We find that the proposed alternative pathway requires Erf2p, a component of the recently described palmitoyltransferase for yeast Ras proteins (29). Finally, we show that the C-terminal region of the hypervariable domain of Ras2p is sufficient for palmitoylation in vivo and in vitro, as well as for the ER-to-plasma membrane localization of Ras2p by the nonclassical pathway.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains used in this study are listed in Table 1. An EcoRI/BamHI fragment containing green fluorescent protein (GFP)-Ras2 was cleaved from pGPD-GFP-Ras2 (29) and ligated into YEp55c (37) under the control of the GAL10 promoter. The resulting plasmid, YEp55-GFP-Ras2 (B991), was used as the host to create the C-terminal hypervariable (HV) domain deletion and CCaaX box mutation plasmids. YEp55-GFP-Ras2(Δ286-318) (B912) was created by cutting YEp55-GFP-Ras2 with BspEI to remove the HV domain. The coding region corresponding to the polybasic region of Rho1p was generated by annealing a double-stranded oligonucleotide and ligating to the BspEI site to generate Yep55-GFP-Ras2(polybasic). The same strategy was used to create YEp55-GFP-Ras2(TMD). The C-terminal transmembrane domain (TMD) sequence of Sso1p (amino acids [aa] 266 to 291) was generated by constructing a double-stranded oligonucleotide and ligating to YEp55-GFP-Ras2 digested with BspEI. YEp55-GFP-SCaaX (B993) and YEp55-GFP-CCaaX (B991) were constructed by PCR amplification of the GFP-CCaaX or GFP-SCaaX fragment followed by ligation to YEp55C. To construct YEp55-GFP-Ras2(288-322) (B984), a GFP-Ras2(288-322) fragment was PCR amplified and inserted into vector YEp55C by using EcoRI and BamHI sites.
TABLE 1.
Strains used in this study
| Straina | Genotype |
|---|---|
| LRB937 | MATα sec23-ts his3 leu2 ura3-52 |
| LRB933 | MATα sec14-ts his3 leu2 ura3-52 |
| LRB934 | MATα sec9-ts his3 leu2 ura3-52 |
| LRB938 | MATahis3 leu2 ura3-52 |
| LRB939 | MATα his3 leu2 ura3-52 |
| RJY1438 | MATα erf2::HIS3 his3 leu2 ura3-52 |
| RJY1440 | MATα erf2::HIS3 sec23-ts his3 leu2 ura3-52 |
| RJY1439 | MATaerf2::HIS3 sec14-ts his3 leu2 ura3-52 |
| RJY1441 | MATα erf2::HIS3 sec9-ts his3 leu2 ura3-52 |
| RJY1538 | MATaerg6::KAN his3 leu2 ura3-52 |
| RJY1539 | MATaerg6::KAN erf2::HIS3 his3 leu2 ura3-52 |
| RJY510 | MATα ras1::HIS3 ras2Δ ura3 his3 leu2 (YCp50-Ras1) |
| RJY690 | MATa/α leu2/leu2 ura3/ura3 his3/his3 (pMA210) |
Strains designated LRB were obtained from Lucy Robinson (Louisiana State University Health Sciences Center). RJY1438 to RJY1441 were obtained by single-step gene replacement of LRB939 and LRB937, respectively, with an erf2::HIS3 fragment. RJY1538 was obtained from LRB938 by single-step gene replacement with an erg6::KAN fragment. RJY1539 was obtained from RJY1538 by single-step gene replacement with an erf2::HIS3 fragment. For RJY510, see reference 33. RJY690 is an R. J. Deschenes lab strain transformed with a GAL4 overexpression plasmid (30).
The construction of pRS315-Ras2 (B250) and pRS315-Ras2-V19 (B561) has been described previously (4). Glutathione S-transferase (GST)-Ras2p fusions were created by ligating the indicated PCR fragment into the galactose-inducible yeast expression vector pEG(KG) (34). HV domain deletions were constructed by ligating the indicated region of RAS2 to create pEG(KG)-Ras2(288-322) (B1287), pEG(KG)-Ras2(297-322) (B1290), pEG(KG)-Ras2(305-322) (B1289), or pEG(KG)-Ras2(313-322) (B1288). Site-directed mutagenesis was performed to create pEG(KG)-Ras2(K312A) (B1313), pEG(KG)-Ras2(K294A) (B1314), pEG(KG)-Ras2(R297A) (B1319), pEG(KG)-Ras2(K298A) (B1320), and pEG(KG)-Ras2(R297A,K298A) (B1321). The plasmids were confirmed by sequence analysis.
The construction of pRS316-Erf2 (B755) has been described previously (4). For palmitoyltransferase assays, a previously described FLAG-Erf2 and GST-Erf4 operon fusion (pErf2-Erf4) (B1267) was used to express the palmitoyltransferase in XL1 Blue bacteria (Stratagene) (29).
SEC18 was cloned into the galactose-inducible yeast shuttle plasmid pESC-LEU2 (SpeI and BglII sites) after PCR amplification of the wild-type SEC18 open reading frame to create pESC-Sec18. The dominant-negative allele was created by changing Thr-394 to Pro, to create pESC-Sec18(T394P) (B1231). The DNA sequence of the entire SEC18 open reading frame was confirmed prior to use. CDC48, YLL034c, and AFG2 encode proteins related to Sec18p (17, 27). Plasmids expressing dominant-negative alleles of CDC48 (pESC-Cdc48DN) (B1229), YLL034c (pESC-YLL034DN) (B1228), and AFG2 (pESC-Afg2DN) (B1227) were created based on the same amino acid change (T394P) that creates SEC18DN.
Cell growth conditions and fluorescence microscopy.
Cultures were grown in synthetic complete liquid medium lacking leucine (SC-Leu), supplemented with 2% ethanol and 2% glycerol, at 24°C to mid-log phase. Galactose was added (final concentration, 4%), and the culture was shifted to 37°C for 3 h to block the secretory pathway in sec-ts mutants. For brefeldin A treatment, cultures were grown to mid-log phase (30°C); then galactose (4%) and brefeldin A (50 μg/ml) were added, and the cultures were incubated for an additional 3 h. GFP fluorescence was visualized by confocal microscopy (MRC:1024; Bio-Rad). In each experiment for which results are shown, at least 15 fields (approximately 25 cells/field) were observed and five images were taken. Under the conditions used, 80 to 90% of the cells exhibited GFP fluorescence. A representative example is shown in each figure.
Plasmid loss assay.
A previously described plasmid loss assay was used to assess Ras2p function (33). Briefly, RJY510 was transformed with a pRS315 plasmid expressing wild-type or C-terminal mutant RAS2 genes. If the expressed Ras2p protein is able to support Ras-dependent growth, then YCp50(URA3)-Ras1 (B250) can be lost and the strain will survive growth on media containing 5-fluoroorotic acid (5-FOA), a drug that kills uracil prototrophs.
Heat shock assay.
LRB938 harboring pRS315-Ras2(V19) plasmids with the wild-type sequence or the indicated mutant C-terminal sequence was grown for 3 days (at 30°C) to stationary phase and then heat shocked at 55°C for 10 min as described elsewhere (4). Serial dilutions of cells were spotted onto a YEPD (1% yeast extract, 2% peptone, 2% glucose) plate, followed by incubation for 3 days (at 30°C).
In vivo [9,10-3H]palmitic acid labeling.
[3H]palmitic acid labeling was performed essentially as described elsewhere (13). Ras2 proteins were expressed as GST fusion proteins, and glutathione (GSH)-agarose beads were used to isolate the protein fusions (34). Cultures were labeled with 500 μCi of [9,10-3H]palmitic acid (60 Ci/mmol; ICN) in the presence of 3 μg of cerulenin (Sigma)/ml for 4 h. Extracts were prepared, and GST-Ras fusion protein was purified, as described above. The fusion proteins were solubilized in 1/5 volume of protein loading buffer (30 mM Tris-HCl [pH 6.7], 12% [wt/vol] sodium dodecyl sulfate [SDS], 60% [vol/vol] glycerol, and 0.1 μg of bromophenol blue/ml) at 65°C for 5 min, and the proteins were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) (12.5% gel). The gel was dried and subjected to fluorography by using FluoroEnhance according to the manufacturer's instructions.
In vitro Ras palmitoyltransferase assays.
Protein acyltransferase assays were performed as described previously (29). All GST-Ras fusions were constructed from the galactose-inducible vector pEG(KG) (34) and purified as described previously(29). GST-Ras2CCaaX represents the complete Ras2 protein (38 kDa) fused to GST. GST-Ras2(288-322), GST-Ras2(297-322), GST-Ras2(305-322), and GST-Ras2(313-322) consist of GST fused to the indicated amino acids of the Ras2 HV domain (29). The [3H]palmitoyl-coenzyme A (CoA) substrate was synthesized from [3H]palmitic acid (30 to 60 Ci/mmol; NEN) and CoA by using acyl-CoA synthetase (Sigma) and was purified as described previously (44). The protein palmitoylation assay (final volume, 25 μl) was performed by adding the GST-Ras2p (50-pmol) substrate to GST-Erf4p-FLAG-Erf2p GSH beads in 1 mM dithiothreitol-100 mM Tris-HCl (pH 8). The reaction was started by addition of 1 μl of [3H]palmitoyl-CoA (0.5 μM), incubated for 15 min at 30°C, and terminated by addition of 5 μl of a 5× solution of SDS gel loading buffer without dithiothreitol. As a control, the GSH beads containing GST-Erf4p-FLAG-Erf2p were boiled (15 min) prior to the addition of substrates. The assays were analyzed by SDS-PAGE and subjected to fluorography and quantitation as described previously (29).
RESULTS
Posttranslational modification is required for the activated phenotype of Ras2(V19) but not for Ras2-dependent growth in yeast.
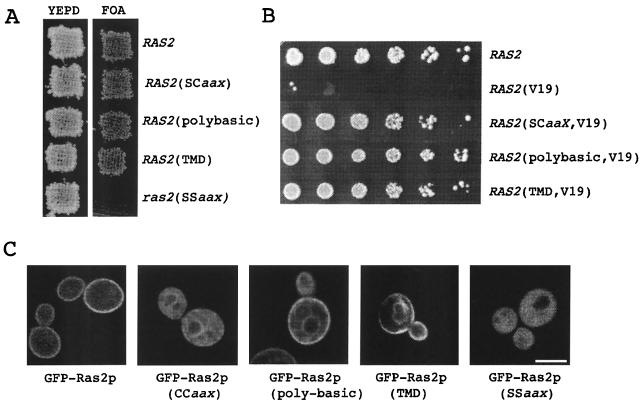
Yeast Ras2p terminates in a canonical CaaX box that directs the farnesylation and palmitoylation of C319 and C318, respectively (Table 2). To evaluate the contributions these modifications make to the function of Ras2p, we examined the abilities of mutants with CaaX box mutations in Ras2p to support Ras-dependent viability. By using a plasmid shuffle assay (33), different Ras2p C-terminal tails were tested for their abilities to confer growth by selecting for the loss of a plasmid-borne copy of wild-type RAS1 [Ycp50(URA3)-Ras1] on a medium supplemented with 5-FOA. As expected, mutating the CaaX box cysteines to serines [Ras2(SSaaX)] created a nonfunctional Ras2 protein (Fig. 1A), indicating that the ability to associate with the plasma membrane is a prerequisite for Ras2p function. Consistent with this idea, we observed that mutants in which the wild-type Ras2p CaaX box had been replaced with different forms of membrane localization signals—farnesylation only [Ras2(SCaaX)], farnesylation combined with a stretch of basic amino acids located immediately upstream of the C terminus of Rho1p [Ras2(polybasic)], and the TMD from plasma membrane-localized protein Sso1p [Ras2(TMD)]—were able to support Ras-dependent growth (Fig. 1A). The morphology and growth rate of these strains were not significantly different from those of strains harboring the wild-type RAS2 allele (data not shown).
TABLE 2.
C-terminal sequences and posttranslational modifications of Ras2 proteins used in this study
| Mutant | C-terminal sequencea | Lipidb |
|---|---|---|
| Ras2(284-322) | -NSKAGQVSNAKQARKQQAAPGGNTSEASKSGSGGCCIIS | Farn, palm |
| Ras2(SCaaX) | -NSKAGQVSNAKQARKQQAAPGGNTSEASKSGSGGSCIIS | Farn |
| Ras2(basic)c | -NS.................GKAKKNTTEKKKKKGSCIIS | Farn |
| Ras2(TMD)d | -NS...........GWLIVFAIIVVVVVVVVVPAVVVKTR | None |
| Ras2(SSaaX) | -NSKAGQVSNAKQARKQQAAPGGNTSEASKSGSGGSSIIS | None |
Underlined sequences are those not normally found in Ras2. Dots indicate a gap in the sequence.
Farn, farnesylation; palm, palmitoylation.
Ras2(basic) contains a C318S mutation and a stretch of basic residues from Rho1 inserted upstream of the CaaX box.
Ras2(TMD) was made by replacing 37 residues from the C terminus of Ras with a C-terminal TMD derived from Ssolp (28).
FIG. 1.
Subcellular localization and the function of wild-type and C-terminal mutated Ras2 proteins. (A) The ability of RAS2 C-terminal mutant proteins to support Ras-dependent growth was assessed using a plasmid loss assay. Ras2 proteins were expressed from a pRS315 plasmid in RJY510, and the cells were plated either onto YEPD plates or onto YEPD plates containing 5-FOA. The ability to grow on YEPD-5-FOA requires expression of a functional Ras2 protein. (B) The effect of the Ras2p C terminus on heat shock sensitivity was analyzed for LRB938 expressing pRS315-Ras2(V19). (C) GFP-Ras fusions were expressed from a galactose-inducible promoter in LRB938 and observed by confocal microscopy. Bar, 5 μm.
Although some of the Ras2p CaaX box mutants we examined supported Ras-dependent growth, they were not able to rescue all Ras2p functions. In mammalian cells, hyperactive RAS alleles cause transformation and tumor formation, whereas in yeast, expression of a hyperactive RAS2 allele results in sensitivity to heat shock and nutrient starvation stresses (42). We examined the heat shock sensitivity of the RAS2 mutants in the context of an activating mutation, a glycine-to-valine change at amino acid position 19, RAS2(V19). Cells expressing Ras2(V19)p-CCaaX were sensitive to heat shock, whereas the C-terminal mutants we examined were resistant and still capable of supporting Ras-dependent growth (Fig. 1B).
Plasma membrane localization of wild-type and CaaX mutant Ras2p.
To evaluate how the lipid modifications contribute to the subcellular localization of Ras2p, we expressed RAS2 wild-type and C-terminal mutant alleles as GFP-Ras2p chimeric proteins in the wild-type strain LRB938 and determined their subcellular localizations. Wild-type GFP-Ras2p, which is able to complement the lethality associated with deleting the RAS1 and RAS2 genes (data not shown), is localized to the plasma membrane, whereas mutating the CaaX box cysteines [GFP-Ras2(SSaaX)] results in a diffused intracellular fluorescence indicative of cytoplasmic localization (Fig. 1C). In contrast, GFP-Ras2(SCaaX), which is farnesylated but not palmitoylated, is localized primarily on endomembranes as well as being detectable on the plasma membrane. The plasma membrane localization defect of GFP-Ras2(SCaaX) can be partially rescued by the insertion of a polybasic domain derived from Rho1p (Fig. 1C). In addition, the fraction of GFP-Ras2(polybasic) protein not at the plasma membrane appears to colocalize with FM4-64, a vacuole-enriched vital dye (data not shown). Finally, unlike the wild-type CaaX box, substitution with a C-terminal TMD of Sso1p directs Ras2p to the plasma membrane (Fig. 1C) while also producing some perinuclear staining that is suggestive of partial ER localization. It would appear that the plasma membrane localization of Ras, like Ras-dependent growth, is supported by a variety of C-terminal membrane localization motifs. Furthermore, intracellular retention of the mutants seems to correlate with their inability to confer heat shock sensitivity.
Plasma membrane localization of Ras2p by classical and nonclassical trafficking pathways.
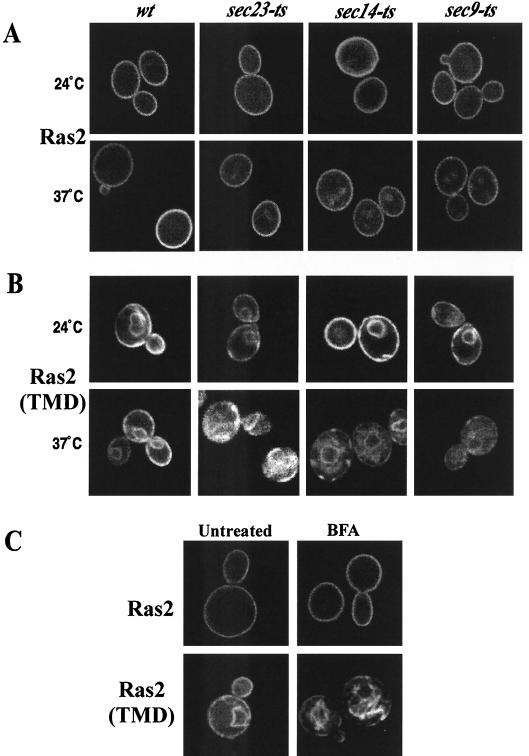
The classical secretory pathway is the best-characterized endomembrane trafficking system for the transport of materials destined for the plasma membrane (40). Recent evidence supports the idea that mammalian H-ras and N-ras require a functional secretory pathway for plasma membrane localization, whereas K-ras-4B does not (2). To examine this question with yeast, we analyzed the subcellular localization of yeast Ras2p in sec23-ts, sec14-ts, and sec9-ts strains. SEC23 encodes a Sar1p GTPase-activating protein required for budding of ER-derived COPII vesicles (51). SEC14 encodes a phospholipid exchange protein that is required for protein export from the Golgi complex (3). SEC9 encodes a SNAP-25 homolog required for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex formation, vesicle docking, and fusion with the plasma membrane (7).
The plasma membrane localization of GFP-Ras2p was not affected by blocking of the secretory pathway at different points in sec23-ts, sec14-ts, and sec9-ts strains (Fig. 2A). In contrast, the plasma membrane localization of GFP-Ras2(TMD), which is targeted to the plasma membrane by the TMD of Sso1p, was blocked under these conditions (Fig. 2B). The membrane localization of GFP-Yor1p, an ABC transporter that requires the secretory pathway for plasma membrane localization, was also blocked when sec23-ts, sec14-ts, and sec9-ts strains were shifted to the nonpermissive temperature, confirming that the secretory pathway is indeed disabled in the sec-ts mutants (data not shown).
FIG. 2.
The classical secretory pathway is not required for the plasma membrane localization of GFP-Ras2p. (A) GFP-Ras2p was expressed from the galactose-inducible promoter of YEp55 in wild-type (LRB938), sec23-ts (LRB937), sec14-ts (LRB933), and sec9-ts (LRB934) strains. Cultures were grown at 24°C to mid-log phase, galactose (4%) was added, and cells were grown at 24°C (top) or 37°C (bottom) for 3 h. (B) Strains expressing GFP-Ras2(TMD) were treated as described for panel A, and images were captured by use of confocal microscopy 3 h after galactose addition. (C) Effect of brefeldin A on the subcellular localization of GFP-Ras2p and GFP-Ras2(TMD). GFP-Ras2p (top) or GFP-Ras2(TMD) (bottom) was expressed by galactose induction in RJY1538, and cultures were treated with dimethyl sulfoxide (untreated) or 50 μg of brefeldin A/ml in dimethyl sulfoxide (BFA).
GFP-Ras2p localization was also examined in the presence of brefeldin A, a drug that interferes with Golgi function and protein secretion (20, 28). Uptake of brefeldin A by yeast requires depletion of ergosterol, which is accomplished by deleting ERG6 (47). Treatment of strain RJY1538 (erg6Δ) with brefeldin A had no detectable effect on the subcellular distribution of GFP-Ras2p (Fig. 2C). In contrast, GFP-Ras2(TMD), which is dependent on the secretory pathway for plasma membrane localization, was brefeldin A sensitive. In the sec-ts mutants as well as with brefeldin A treatment, GFP-Ras2(TMD) accumulates on endomembrane systems. The apparent plasma membrane localization of GFP-Ras2 in the absence of a functional secretory pathway does not appear to be a product of overexpression. The same result was obtained when GFP-Ras2 was expressed from a MET25-inducible promoter at levels closer to endogenous levels. Although the level of GFP fluorescence was low, the results confirmed the original observation when GFP-Ras2 was expressed from a galactose-inducible promoter. These results demonstrate that the Ras2p C terminus plays a role in the ER-to-plasma membrane trafficking of Ras2p by a mechanism that does not require the classical secretory pathway or a functional Golgi complex.
Ras2p utilizes an Erf2p-dependent mechanism for plasma membrane localization.
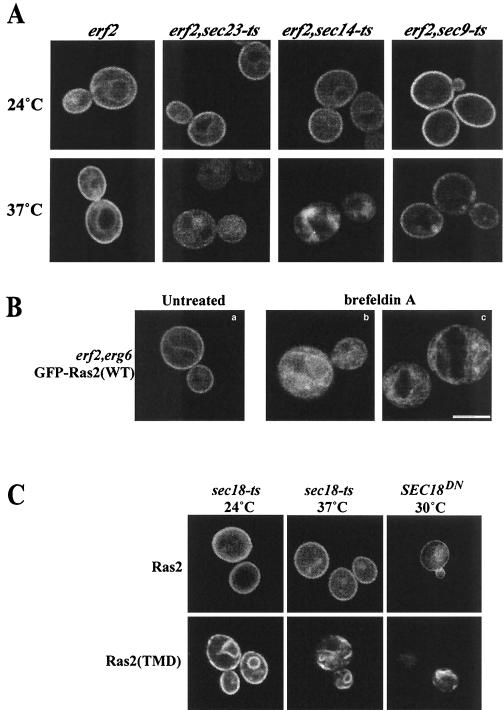
A 41-kDa ER-localized protein, Erf2p,required for efficient palmitoylation of Ras2p, has been described recently (4). Deletion of ERF2 causes partial retention of GFP-Ras2p on internal membranes including the vacuole, decreased palmitoylation of Ras2p, and partial suppression of the heat shock sensitivity of a hyperactive RAS2(V19) allele (4). However, deletion of ERF2 alone does not prevent the plasma membrane localization of Ras or a measurable decrease in cell viability. Erf2p belongs to a family of DHHC (Asp-His-His-Cys) motif C2H2 zinc finger proteins with putative orthologs in yeast and metazoans (4, 6, 35). To test whether Erf2p plays a role in the classical secretory pathway-independent plasma membrane localization of Ras2p, we assayed for Ras2p localization in sec-ts strains lacking ERF2. The plasma membrane localization of GFP-Ras2p was dramatically altered in sec23-ts erf2Δ and sec14-ts erf2Δ strains following a shift to the nonpermissive temperature (Fig. 3A). The GFP fluorescence was shifted from the plasma membrane to intracellular structures, indicating a block of Ras2p trafficking from the ER to the plasma membrane. As expected, introduction of a pRS316-ERF2 plasmid fully rescued the localization defect (data not shown). Intriguingly, the sec9-ts erf2Δ double mutation did not significantly affect the plasma membrane localization of GFP-Ras2p, although an increase in vesicles subtending the plasma membrane was evident, consistent with a reduction in membrane fusion caused by the loss of Sec9p (Fig. 3A). Brefeldin A was also used to block the classical secretory pathway in strains lacking Erf2p to test whether Erf2p is required for the Golgi-independent plasma membrane localization of Ras2p. Addition of brefeldin A to the erf2Δ strain also caused GFP-Ras2p to accumulate within the cell and prevented localization to the cell perimeter (Fig. 3B).
FIG. 3.
Effect of deletion of ERF2 on the subcellular localization of GFP-Ras2p in wild-type, sec-ts mutant, and brefeldin A-treated cells. (A) YEp55-GFP-Ras2p was expressed in erf2::HIS3 (RJY1438), erf2::HIS3 sec23-ts (RJY1440), erf2::HIS3 sec14-ts (RJY1439), or erf2::HIS3 sec9-ts (RJY1441) strains. Cultures were grown and induced as described in the legend to Fig. 2, and images were captured 3 h after galactose induction. (B) Effect of brefeldin A on the localization of GFP-Ras2p in strains lacking Erf2p. YEp55-GFP-Ras2p was expressed in an erg6::Kanr erf2::HIS3 (RJY1539) strain that was either left untreated (a) or treated with brefeldin A (50 μg/ml) (b and c). Bar, 5 μm. (C) Sec18p is not required for the plasma membrane localization of wild-type GFP-Ras2. GFP-Ras2p (top) or GFP-Ras2(TMD) (bottom) was expressed by galactose induction in RJY1538 harboring a chromosomal sec18-ts mutation or in LRB938 carrying a plasmid expressing a SEC18DN allele. GFP-Ras2p or GFP-Ras2(TMD) was expressed as described in the legend to Fig. 2, and images were captured 3 h after galactose induction.
Erf2-dependent plasma membrane localization of Ras2p does not require Sec18p, the yeast homolog of NSF.
Following the addition of the farnesyl and palmitoyl moieties, Ras2p remains membrane associated, suggesting that the nonclassical pathway from the ER to the plasma membrane may involve a vesicle-mediated pathway. We examined whether plasma membrane localization of GFP-Ras2p requires Sec18p, a homolog of N-ethylmaleimide-sensitive factor (NSF) that is believed to be required for all SNARE-dependent vesicle fusion events in yeast (17, 19, 50). As shown in Fig. 3C, switching a sec18-ts strain to the nonpermissive temperature or expressing the dominant-negative form, SEC18DN, had no detectable effect on the plasma membrane localization of GFP-Ras2p but, as expected, did block the localization of GFP-Ras2(TMD). This result led us to test whether Sec18p-related ATPases play a role in Ras2p trafficking. CDC48, YLL034c, and AFG2 encode proteins that are components of the ER and are reported to play a role in ER membrane fusion events (17, 27). Dominant-negative alleles of CDC48, YLL034c, and AFG2 were constructed by engineering a proline substitution at the position of the conserved residue Thr-394. The result was the same as that observed with SEC18DN: GFP-Ras2p localization was not affected by expression of dominant-negative alleles of CDC48, YLL034c, or AFG2 (data not shown). The simplest conclusion from these studies is that a vesicle-mediated mechanism is not involved in the Erf2-dependent trafficking of Ras2p. However, we cannot rule out Sec18-independent processes.
The HV domain is sufficient for Erf2p-dependent plasma membrane localization of Ras2p.
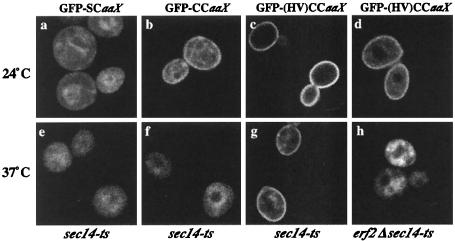
We examined the role of the last 35 residues of the HV domain (aa 288 to 322) in Ras2p localization via the nonclassical pathway. The subcellular localization of GFP-SCaaX, GFP-CCaaX, and GFP-(HV)CCaaX was examined in sec14-ts and sec14-ts erf2 mutant strains (Fig. 4). GFP-SCaaX, which is farnesylated but not palmitoylated, localizes primarily on endomembranes. GFP-CCaaX, which we presume to be inefficiently palmitoylated like GST-CCaaX (see below), is primarily localized on endomembranes. Addition of the HV domain residues 288 to 322 [GFP-(HV)CCaaX] results in plasma membrane localization comparable to that of full-length GFP-Ras2p. Plasma membrane localization of GFP-(HV)CCaaX is unaffected by the loss of SEC14. However, GFP-(HV)CCaaX localization is partially altered in the erf2Δ mutant, and is abolished in the sec14-ts erf2Δ double mutant, when the cells are shifted to the nonpermissive temperature (Fig. 4), mimicking the patterns seen with the full-length GFP-Ras2p construct. This suggests that the CaaX box and the C-terminal region of the HV domain are sufficient for plasma membrane localization of Ras2p through an Erf2p-dependent mechanism.
FIG. 4.
The HV domain of Ras2p is sufficient for Erf2p-dependent plasma membrane localization. The indicated GFP fusion proteins were expressed in sec14-ts (LRB933) (a through c and e through g) or erf2::HIS3 sec14-ts (RJY1439) (d and h) cells by galactose induction, and the cultures were then incubated at 24°C (a through d) or 37°C (e through h) for 3 h.
The HV domain is important for Erf2p-dependent palmitoylation of Ras2p.
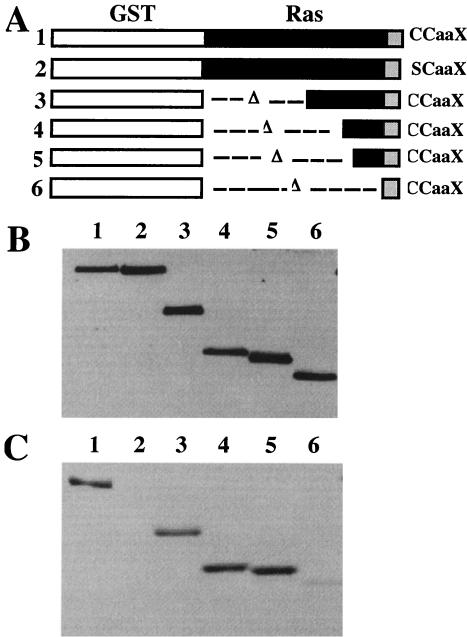
In vivo [3H]palmitate labeling studies were performed to determine if the HV region influenced the palmitoylation of Ras2p. A series of GST-Ras2p fusion constructs was created to define the sequence requirement for palmitoylation (Fig. 5A). All of the GST-Ras2p proteins were expressed at comparable levels (Fig. 5B). GST-Ras2p was efficiently labeled with [3H]palmitate, whereas the Cys-318-to-Ser-318 mutant [pEGRas2(SCaaX)] was not (Fig. 5C, lane 2). Deleting the GTP binding domain and most of the HV domain to create GST-Ras2(288-322) had no significant effect on [3H]palmitate incorporation (Fig. 5C, lanes 1 and 3 to 5). However, removal of 30 residues proximal to the CCaaX box significantly reduced [3H]palmitate incorporation (Fig. 5C, lane 6). The reduction was specific for palmitoylation based on the observations that GST-Ras2(316-322) undergoes farnesylation and CaaX proteolysis (data not shown).
FIG. 5.
Identification of sequences required for efficient palmitoylation of Ras2p. (A) Schematic diagram of a series of GST-Ras2p fusion proteins with the indicated deletions of Ras2p sequences. (B) Immunoblot analysis of steady-state levels of Ras2 protein. Total-cell extracts were prepared from strain RJY690 expressing GST-Ras2(CCaaX) (lane 1), GST-Ras2(SCaaX) (lane 2), GST-Ras2(190-322) (lane 3), GST-Ras2(273-322) (lane 4), GST-Ras2(288-322) (lane 5), or GST-Ras2(316-322) (lane 6). Ras2p was visualized by using the anti-Ras antibody Y13-259, and immune complexes were visualized by chemiluminescence (ECL; Amersham). (C) Fluorography of a duplicate of the gel in panel B detects [9,10-3H]palmitic acid-labeled GST-Ras2p.
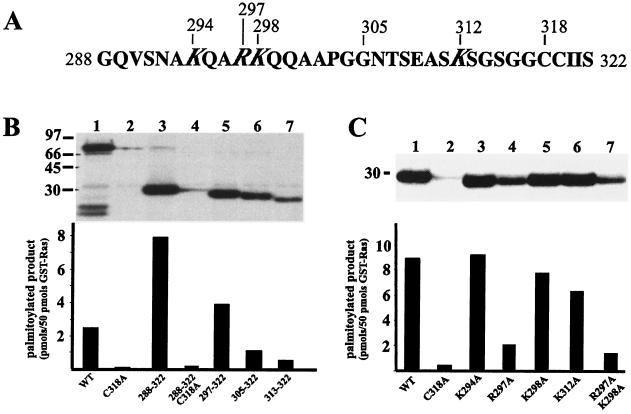
Recently, it was reported that an ER-localized complex, Erf2p-Erf4p, carries out the palmitoylation of Ras2p (29), implicating palmitoylation as one of the driving forces in the ER-to-plasma membrane translocation of Ras proteins. The results presented above (Fig. 4) indicate that the C-terminal region of the Ras2 HV domain is sufficient for the Erf2-dependent plasma membrane localization of Ras2 via a nonclassical pathway. We therefore examined the role of this region of the HV domain in Ras2 palmitoylation by the Erf2p-Erf4p palmitoyltransferase (29). The sequence of this region is shown in Fig. 6A. GST-Ras2(288-322) was purified from yeast, quantified by immunoblot analysis (data not shown), and tested in the palmitoylation assay. As seen in Fig. 6B, the C terminus of the Ras HV domain (aa 297 to 322) is sufficient for efficient palmitoylation of Ras2 in vitro. Removal of sequences between aa 297 and the CCaaX box results in a decrease in palmitoylation, similar to what is seen in vivo (Fig. 5).
FIG. 6.
In vitro palmitoylation of wild-type and mutant Ras2 proteins. (A) Sequence of the C-terminal region of the Ras2p HV domain (aa 288 to 322). Amino acid positions corresponding to mutations in basic amino acid residues are indicated. (B) Equal amounts (50 pmol of fusion protein) of GST-Ras2p (lane 1), GST-Ras2(C318A) (lane 2), GST-Ras2p(288-322) (lane 3), GST-Ras2p(288-322) (C318A) (lane 4), GST-Ras2p(297-322) (lane 5), GST-Ras2p(305-322) (lane 6), and GST-Ras2p(313-322) (lane 7) were incubated with Erf2p-Erf4p and [3H]palmitate. Products were analyzed by SDS-PAGE and fluorography (top), or [3H]palmitate incorporation was determined by excising the band, adding scintillation fluid, and counting (bottom). (C) The requirement for basic amino acid residues within the HV domain was investigated by mutating Arg and Lys residues to Ala. Equal amounts (50 pmol of fusion protein) of GST-Ras2 (lane 1), GST-Ras2(C318A) (lane 2), GST-Ras2(K294A) (lane 3), GST-Ras2(R297A) (lane 4), GST-Ras2(K298A) (lane 5), GST-Ras2(K312A) (lane 6), and GST-Ras2(R297A,K298A) (lane 7) were incubated with Erf2p-Erf4p and[3H]palmitate. Products were analyzed by SDS-PAGE and fluorography (top), or [3H]palmitate incorporation was determined by excising the band, adding scintillation fluid, and counting (bottom).
To further define the sequence requirement for the Erf2p-Erf4p-dependent palmitoylation in Ras2p, residues within the HV domain were mutated. It has been suggested that basic residues may be important for Ras2p palmitoylation (33). Based on this notion, we engineered several Ras2p derivatives that substitute alanine for either lysine or arginine. As seen in Fig. 6C, there was a differential effect on palmitoylation depending on the basic residue mutated. Mutating Lys-294 to Ala had no measurable effect on palmitoylation, whereas the Arg-297-to-Ala mutation caused the most dramatic decrease in Erf2p-Erf4p-dependent palmitoylation. There was a small effect of mutating Lys-298 or Lys-312. We have also performed a quantitative plasmid loss assay on these mutants and found that the in vivo function of the Ras mutants correlates very well with the in vitro palmitoylation results (X. Dong and R. J. Deschenes, unpublished data) The positions of the basic residues appear to dictate their importance in the specificity of the palmitoylation reaction.
DISCUSSION
A general theme is emerging where farnesylation directs CaaX box proteins to the ER for further processing by a CaaX protease and prenylcysteine-dependent carboxyl methyltransferase (11, 36, 41). However, the mechanism by which Ras translocates from the ER to the plasma membrane is not known. In the case of mammalian H-ras and N-ras, and yeast Ras1p and Ras2p, palmitoylation is also required for translocation from the ER to the plasma membrane. K-ras-4B, on the other hand, utilizes a polybasic region rather than palmitoylation as a signal for plasma membrane localization. This difference has two known functional consequences. First, K-ras-4B and H-ras localize to distinct plasma membrane microdomains (39). Second, plasma membrane localization of H-ras requires a functional secretory pathway, whereas K-ras-4B is transported to the plasma membrane even when the classical secretory pathway is blocked (2).
In this report we show that blocking the secretory pathway by brefeldin A or by mutations in early, middle, and late secretory pathway components has little or no effect on Ras2p localization in yeast. These results do not rule out a role for the classical secretory pathway but clearly show that it is not required for ER-to-plasma membrane translocation of Ras2p. Deletion of ERF2 in combination with sec23-ts (ER-to-Golgi transport), sec14-ts (intra-Golgi transport), and sec18-ts (vesicle transport) prevents Ras2p localization with the plasma membrane. On the other hand, deletion of ERF2 alone results in partial mislocalization of GFP-Ras2p to the vacuole membrane but overall does not preclude Ras2p from reaching the plasma membrane. Based on these observations, we propose that prenylated Ras2p translocates from the ER to the plasma membrane by either the classical secretory pathway or the Erf2p-dependent pathway. Post-Golgi components of the secretory pathway are not required even in the absence of Erf2p, since Ras2p localization appears normal in sec9-ts erf2Δ (Fig. 3) and sec4-ts erf2Δ (data not shown) double mutants. Parallel pathways from the post-Golgi vesicles to the plasma membrane have been described that could account for the ability of Ras to reach the plasma membrane in the absence of Sec9 and Sec4 proteins (21, 23).
Ras proteins exhibit a high degree of sequence conservation throughout the amino-terminal GTP binding domain. In mammals, the first 85 aa of H-ras, N-ras, K-ras-4A, and K-ras-4B, which include the effector binding regions, are identical, and the next 80 aa exhibit 85% homology. The isoforms of Ras carry out distinct functions based on differences in subcellular localization, which are directed by the Ras C-terminal HV domain (48). We have shown that 35 aa of the HV domain which include the CaaX box represent the minimal region required for palmitoylation in vivo and in vitro. In addition, the localization pattern of GFP(HV)CCaaX and its translocation via the Erf2p-dependent pathway are indistinguishable from those of full-length Ras2p. Thus, palmitoylation and plasma membrane localization through the Erf2-dependent pathway appear to require the same part of the Ras2p HV region. To date the sequence requirement for palmitoylation has been elusive. This could be because there are multiple palmitoyltransferases (for example, the Erf2 orthologs). The availability of a Ras palmitoyltransferase in yeast will allow a detailed analysis of the sequence requirements for protein palmitoylation for the first time. It appears that one important mechanism of recognition is via electrostatic interactions with basic amino acid side chains, such as those of the HV domain of Ras.
The ERF2 gene was identified in a genetic screen for mutants incapable of utilizing palmitoylation-dependent Ras alleles (4). Two distinguishing features of the Erf2p family of proteins, which are found in yeast and metazoans (35), are the presence of four putative transmembrane spanning regions and a DHHC (Asp-His-His-Cys) cysteine-rich domain (CRD). Saccharomyces cerevisiae encodes three DHHC CRD genes related to ERF2: PSL10, YNL326, and YOL003. In addition, there are three related DHYC CRD genes: YDR459, AKR1, and AKR2. Deletion of the ERF2-related genes alone or together with erf2Δ did not produce any new phenotypes or additional effects on the phenotype of erf2Δ mutants (data not shown). Akr1p has recently been shown to exhibit palmitoyltransferase activity using the yeast casein kinase II as a substrate (38). Mammalian orthologs of ERF2 can be readily identified based on their size, the presence of four putative transmembrane sequences, and the DHHC CRD motif (35). At present, it is not known if orthologs of Erf2p and Akr1p play a role in the palmitoylation of other proteins.
If the classical secretory pathway and the Golgi complex are not required, how does translocation of Ras from the ER to the plasma membrane occur? One possibility is that Ras transiently dissociates from the ER membrane and regains plasma membrane localization as a result of diffusion. Since prenylation is an irreversible step, this would presumably require a mechanism to shield or bury the lipid when Ras is not membrane bound, as is the case for the escort proteins that allow prenylated Rab proteins to shuttle between soluble and membrane-bound forms (1). The prenylated Rab acceptor protein PRA1 has been proposed to be a receptor and escort protein for Ras and other prenylated proteins in mammalian cells (16). The yeast homolog of PRA1, Yip3p, interacts with Rab GTPases and VAMP2-interacting protein, but an interaction with Ras proteins was not examined (8). The inability to detect a cytosolic pool of lipid-modified Ras protein in yeast would argue against a mechanism involving Ras escort protein such as Rab GDI proteins. If Ras trafficking is restricted to a vesicle-mediated process, then vesicle budding and fusion should be involved. However, if this is the case, then it must occur in the absence of Sec18p, the yeast homolog of NSF that is involved in most, if not all, vesicle-membrane fusions in yeast (17, 19, 50). The elucidation of palmitoylation-dependent mechanisms of Ras translocation from the ER to the plasma membrane will require further work.
Other studies have implicated nonclassical pathways for targeting lipid-modified peptides and proteins to the plasma membrane. One of the earliest examples involved the prenylated yeast mating pheromone a-factor, which is exported from cells even when the secretory pathway is blocked (25, 32). Later, two groups reported that in mammalian cells lines such as COS-1, CHO, MDCK, and BHK, K-ras-4B shows little endomembrane or vesicular staining (2, 11). Furthermore, blocking the secretory pathway by low temperatures or brefeldin A treatment has little effect on K-ras-4B. Instead, K-ras-4B presumably translocates by a different mechanism, as inferred from its interaction with microtubules in NIH 3T3 cells (45). Intriguingly, other lipid modifications, such as the N-terminal acylation of the Src family member Fyn, also allow plasma membrane localization in the presence of brefeldin A (46). However, there are cases in which the classical secretory pathway does play a role in the trafficking of lipid-modified proteins. H-ras and N-ras are temporarily localized to the ER and Golgi complex before they reach the plasma membrane, and blocking the secretory pathway prevents the plasma membrane localization of N-ras and H-ras. SNAP-25, GAP-43 (18), and Rho2p (Dong and Deschenes, unpublished) also appear to require an intact secretory pathway for plasma membrane targeting. In general, the subcellular trafficking and ultimate localization of Ras and other lipid-modified proteins depend on multiple factors including the type of lipid modification, sequences within the protein, and the interaction with specific proteins such as Erf2p.
The functional implications of utilizing either the sec-dependent or the sec-independent (Erf2p-dependent) pathway for the translocation of Ras to the plasma membrane are not fully understood at this time. However, examination of the activated RAS2(V19) alleles may provide a clue. Our results demonstrate that a fully processed C terminus is not required to support Ras-dependent growth but is required for the heat shock-sensitive phenotype of strains expressing the activated RAS2(V19) allele (Fig. 1). There are several possible explanations for this observation. First, the interaction between Ras and adenylyl cyclase might involve complete C-terminal processing, including palmitoylation. Kataoka and colleagues have demonstrated lipid modification-dependent activation of adenylyl cyclase in vitro (26). A second possibility is that prenylation and palmitoylation are required for targeting of Ras to specific microdomains where localization would play a role in regulating the engagement of effectors. Evidence that palmitoylation plays this role in mammalian cells has been presented (48). To date we have been unable to isolate lipid rafts containing wild-type and mutant yeast Ras proteins in yeast. Future work includes defining the precise roles of palmitoylation and membrane localization in the function of wild-type and activated mutants of Ras.
Acknowledgments
We thank Lucy Robinson (Louisiana State University Health Sciences Center) for generously providing the sec-ts mutant strains used in this study and Jan Fassler, Lois Weisman, and members of the Deschenes laboratory for their comments on the manuscript.
This work was supported by Public Health Service grant CA-50211 from the National Cancer Institute to R.J.D.
REFERENCES
- 1.Andres, D. A., M. C. Seabra, M. S. Brown, S. A. Armstrong, T. E. Smeland, F. P. M. Cremers, and J. L. Goldstein. 1993. cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73:1091-1099. [DOI] [PubMed] [Google Scholar]
- 2.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bankaitis, V. A., J. R. Aitken, A. E. Cleves, and W. Dowhan. 1990. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347:561-562. [DOI] [PubMed] [Google Scholar]
- 4.Bartels, D. J., D. A. Mitchell, X. Dong, and R. J. Deschenes. 1999. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6775-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya, S., L. Chen, J. R. Broach, and S. Powers. 1995. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc. Natl. Acad. Sci. USA 92:2984-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohm, S., D. Frishman, and H. W. Mewes. 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennwald, P., B. Kearns, K. Champion, S. Keränen, V. Bankaitis, and P. Novick. 1994. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 79:245-258. [DOI] [PubMed] [Google Scholar]
- 8.Calero, M., and R. N. Collins. 2002. Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem. Biophys. Res. Commun. 290:676-681. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 10.Casey, P. J., P. A. Solski, C. J. Der, and J. E. Buss. 1989. p21ras is modified by a farnesyl isoprenoid. Proc. Natl. Acad. Sci. USA 86:8323-8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 12.Der, C. J., and A. D. Cox. 1991. Isoprenoid modification and plasma membrane association: critical factors for ras oncogenicity. Cancer Cells 3:331-340. [PubMed] [Google Scholar]
- 13.Deschenes, R. J., and J. R. Broach. 1987. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol. Cell. Biol. 7:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deschenes, R. J., J. B. Stimmel, S. Clarke, J. Stock, and J. R. Broach. 1989. RAS2 protein of Saccharomyces cerevisiae is methyl-esterified at its carboxyl terminus. J. Biol. Chem. 264:11865-11873. [PubMed] [Google Scholar]
- 15.Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa, C., J. Taylor, and A. B. Vojtek. 2001. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J. Biol. Chem. 276:28219-28225. [DOI] [PubMed] [Google Scholar]
- 17.Frohlich, K. U. 2001. An AAA family tree. J. Cell Sci. 114:1601-1602. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalo, S., and M. E. Linder. 1998. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol. Biol. Cell 9:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, T. R., and S. D. Emr. 1991. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J. Cell Biol. 114:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, T. R., P. A. Scott, and S. D. Emr. 1993. Brefeldin A reversibly blocks early but not late protein transport steps in the yeast secretory pathway. EMBO J. 12:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurunathan, S., D. David, and J. E. Gerst. 2002. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 21:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 23.Harsay, E., and A. Bretscher. 1995. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 131:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joneson, T., and D. Bar-Sagi. 1997. Ras effectors and their role in mitogenesis and oncogenesis. J. Mol. Med. 75:587-593. [DOI] [PubMed] [Google Scholar]
- 25.Kuchler, K., R. E. Sterne, and J. Thorner. 1989. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 8:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda, Y., N. Suzuki, and T. Kataoka. 1993. The effect of posttranslational modifications on the interaction of Ras2 with adenylyl cyclase. Science 259:683-686. [DOI] [PubMed] [Google Scholar]
- 27.Latterich, M., K. U. Frohlich, and R. Schekman. 1995. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82:885-893. [DOI] [PubMed] [Google Scholar]
- 28.Lippincott-Schwartz, J., L. C. Yuan, J. S. Bonifacino, and R. D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobo, S., W. K. Greentree, M. E. Linder, and R. J. Deschenes. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268-41273. [DOI] [PubMed] [Google Scholar]
- 30.Ma, J., and M. Ptashne. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847-853. [DOI] [PubMed] [Google Scholar]
- 31.Macara, I. G., K. M. Lounsbury, S. A. Richards, C. McKiernan, and D. Bar-Sagi. 1996. The Ras superfamily of GTPases. FASEB J. 10:625-630. [DOI] [PubMed] [Google Scholar]
- 32.McGrath, J. P., and A. Varshavsky. 1989. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature 340:400-404. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, D. A., L. Farh, T. K. Marshall, and R. J. Deschenes. 1994. A polybasic domain allows nonprenylated Ras proteins to function in Saccharomyces cerevisiae. J. Biol. Chem. 269:21540-21546. [PubMed] [Google Scholar]
- 34.Mitchell, D. A., T. K. Marshall, and R. J. Deschenes. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715-722. [DOI] [PubMed] [Google Scholar]
- 35.Putilina, T., P. Wong, and S. Gentleman. 1999. The DHHC domain: a new highly conserved cysteine-rich motif. Mol. Cell. Biochem. 195:219-226. [DOI] [PubMed] [Google Scholar]
- 36.Romano, J. D., W. K. Schmidt, and S. Michaelis. 1998. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell 9:2231-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose, M. D., and J. R. Broach. 1990. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-229. [DOI] [PubMed] [Google Scholar]
- 38.Roth, A. F., Y. Feng, L. Chen, and N. G. Davis. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1:98-105. [DOI] [PubMed] [Google Scholar]
- 40.Schekman, R. 1992. Genetic and biochemical analysis of vesicular traffic in yeast. Curr. Opin. Cell Biol. 4:587-592. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, W. K., A. Tam, K. Fujimura-Kamada, and S. Michaelis. 1998. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 95:11175-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigal, I. S., J. B. Gibbs, J. S. D'Alonzo, and E. M. Scolnick. 1986. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc. Natl. Acad. Sci. USA 83:4725-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stimmel, J. B., R. J. Deschenes, C. Volker, J. Stock, and S. Clarke. 1990. Evidence for an S-farnesylcysteine methyl ester at the carboxyl terminus of the Saccharomyces cerevisiae RAS2 protein. Biochemistry 29:9651-9659. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, D. C., N. Weber, L. R. Hogge, and E. W. Underhill. 1990. A simple enzymatic method for the preparation of radiolabeled erucoyl-CoA and other long-chain fatty acyl-CoAs and their characterization by mass spectrometry. Anal. Biochem. 184:311-316. [DOI] [PubMed] [Google Scholar]
- 45.Thissen, J. A., J. M. Gross, K. Subramanian, T. Meyer, and P. J. Casey. 1997. Prenylation-dependent association of Ki-Ras with microtubules: evidence for a role in subcellular trafficking. J. Biol. Chem. 272:30362-30370. [DOI] [PubMed] [Google Scholar]
- 46.van't Hof, W., and M. D. Resh. 1997. Rapid plasma membrane anchoring of newly synthesized p59fyn: selective requirement for NH2-terminal myristoylation and palmitoylation at cysteine-3. J. Cell Biol. 136:1023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel, J. P., J. N. Lee, D. R. Kirsch, M. D. Rose, and E. S. Sztul. 1993. Brefeldin A causes a defect in secretion in Saccharomyces cerevisiae. J. Biol. Chem. 268:3040-3043. [PubMed] [Google Scholar]
- 48.Walsh, A. B., and D. Bar-Sagi. 2001. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J. Biol. Chem. 276:15609-15615. [DOI] [PubMed] [Google Scholar]
- 49.Willumsen, B. M., A. D. Cox, P. A. Solski, C. J. Der, and J. E. Buss. 1996. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene 13:1901-1909. [PubMed] [Google Scholar]
- 50.Wilson, D. W., C. A. Wilcox, G. C. Flynn, E. Chen, W. J. Kuang, W. J. Henzel, M. R. Block, A. Ullrich, and J. E. Rothman. 1989. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature 339:355-359. [DOI] [PubMed] [Google Scholar]
- 51.Yoshihisa, T., C. Barlowe, and R. Schekman. 1993. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science 259:1466-1468. [DOI] [PubMed] [Google Scholar]