Abstract
The human pathogen Campylobacter jejuni is one of more than 40 naturally competent bacterial species able to import macromolecular DNA from the environment and incorporate it into their genomes. However, in C. jejuni little is known about the genes involved in this process. We used random transposon mutagenesis to identify genes that are required for the transformation of this organism. We isolated mutants with insertions in 11 different genes; most of the mutants are affected in the DNA uptake stage of transformation, whereas two mutants are affected in steps subsequent to DNA uptake, such as recombination into the chromosome or in DNA transport across the inner membrane. Several of these genes encode proteins homologous to those involved in type II secretion systems, biogenesis of type IV pili, and competence for natural transformation in gram-positive and gram-negative species. Other genes identified in our screen encode proteins unique to C. jejuni or are homologous to proteins that have not been shown to play a role in the transformation in other bacteria.
The gram-negative bacterium Campylobacter jejuni is the most common cause of bacterial gastroenteritis in many industrialized countries (30). In the United States alone, the annual incidence of C. jejuni infection is estimated to be approximately one infection per 100 people (29). Transmission to humans most often occurs via consumption of contaminated poultry, unpasteurized milk, or untreated water. Disease symptoms range from mild, watery diarrhea to a severe, inflammatory diarrhea.
Analysis of numerous C. jejuni isolates has revealed genotype diversity in the species. Based on results from multilocus sequence typing, it is thought that intraspecies recombination plays a large role in generating genetic diversity among C. jejuni strains (11, 56). Recently, horizontal gene transfer was shown to occur in vivo among strains of C. jejuni during experimental infection of chickens (9).
Natural transformation is one potential mechanism for horizontal gene transfer leading to genetic diversity among a population. Natural competence is a physiological state that allows uptake of macromolecular DNA from the environment (13). More than 40 naturally transformable bacterial species have been identified (36). Competent bacteria bind DNA and transport it into the cytoplasm, where it may either recombine into the chromosome or, in the case of plasmid DNA, replicate freely. Like several other human pathogens, including Neisseria gonorrhoeae, Haemophilus influenzae, and Helicobacter pylori (17, 53, 60, 63), C. jejuni is naturally competent for transformation, exhibiting transformation frequencies of ca. 10−4 with chromosomal DNA as the source of DNA (62).
Little is known about the mechanism of transformation of C. jejuni. A plasmid, pVir, from C. jejuni strain 81-176 encodes several proteins homologous to those of conjugation systems in other microbes (3). In the closely related species H. pylori, these type IV secretion system genes are required for natural transformation (25, 26). Two of these pVir-encoded genes, virB11 and comB3, were tested for a role in natural transformation in C. jejuni (3). A virB11 mutant showed no difference in transformation frequency compared to the wild-type strain; a modest defect in transformation efficiency was observed with a comB3 mutant (3).
Since natural transformation may contribute to the genetic diversity seen among different strains of C. jejuni, we sought to characterize the molecular mechanisms of this process. To begin, we identified genes required for natural transformation by taking a predominantly genetic approach. We screened a solo transposon mutant library of C. jejuni 81-176 (22) for mutants that could not be transformed to antibiotic resistance. Eleven genes were identified that, when mutated, reduced the ability of C. jejuni to be transformed by ca. 1,000-fold. Through this genetic approach, we identified chromosomal genes in C. jejuni that encode components of a type II secretion system essential for natural transformation.
MATERIALS AND METHODS
Bacterial strains and media.
C. jejuni 81-176 and its derivatives used in the present study are listed in Table 1. C. jejuni was routinely grown on Mueller-Hinton (MH) agar in microaerophilic conditions at 37°C. When necessary, media were supplemented with antibiotics in the following concentrations: chloramphenicol (15 μg ml−1), kanamycin (50 μg ml−1), trimethoprim (10 μg ml−1), nalidixic acid (30 μg ml−1), streptomycin (100 μg ml−1 or 2 mg ml−1), and cefoperazone (30 μg ml−1). All C. jejuni strains were stored in MH broth with 20% glycerol at −80°C.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 21 |
| JM101 | F′ traD36 proA+B+laclq Δ(lacZ)M15/Δ(lax-proAB) glnV thi | New England Biolabs |
| DH5α/pRK212.1 | contains conjugative plasmid for conjugation of plasmid DNA into Campylobacter | 14 |
| C. jejuni | ||
| 11168 | clinical isolate used for genome sequencing | 43 |
| 81-176 | clinical isolate | 32 |
| DRH153 | 81-176 astA::aphA3; Knr | 22 |
| DRH154 | 81-176 astA::cat; Cmr | This study |
| DRH212 | 81-176 rpsLSm | This study |
| DRH312 | 11168 gyrANA | This study |
| DRH335 | 81-176 gyrANA | This study |
| RSW42 | DRH212 ctsE::cat-rpsL | This study |
| RSW44 | DRH212 ctsX::cat-rpsL | This study |
| RSW46 | RSW44 ΔctsX, in-frame deletion of codons 5 to 190 of CtsX | This study |
| RSW81 | DRH212 ctsP::cat-rpsL | This study |
| RSW82 | DHR212 ctsR::cat-rpsL | This study |
| RSW115 | RSW81 ΔctsP, in-frame deletion of codons 6 to 196 of CtsP | This study |
| RSW136 | RSW42 ΔctsE, in-frame deletion of codons 7 to 514 of CtsE | This study |
| RSW137 | RSW82 ΔctsR, in-frame deletion of codons 4 to 93 of CtsR | This study |
| Plasmids | ||
| pUC19 | Ampr | New England Biolabs |
| pDRH138 | pUC19 with 2.3-kb fragment containing astA cloned into PstI site | This study |
| pDRH142 | pDRH138 with kanamycin cassette cloned into EcoRV site of astA | This study |
| pDRH143 | pDRH138 with chloramphenicol cassette cloned into EcoRV site of astA | This study |
| pDRH265 | pUC19 with 1.4 kb cat-rpsL cloned into SmaI site | 22 |
| pDRH328 | pUC19 with coding sequence of gyrA NAr in BamHI site | This study |
| pRY108 | Campylobacter-E. coli shuttle vector; Knr | 66 |
| pRY109 | pUC19 with Campylobacter cat cassette | 66 |
| pRY112 | Campylobacter-E. coli shuttle vector; Cmr | 66 |
| pILL600 | contains Campylobacter kanamycin cassette | 33 |
| pECO102 | pRY112 derivative with cat promoter in XhoI-BamHI site | This study |
| pRSW100 | pUC19 with 2.8-kb fragment harboring ctsE cloned into XmaI site | This study |
| pRSW101 | pUC19 with 2-kb fragment harboring ctsX cloned into XmaI site | This study |
| pRSW102 | pRSW100 with cat-rpsL cloned into NruI site of ctsE | This study |
| pRSW103 | pRSW101 with cat-rpsL cloned into BsgI site of ctsX | This study |
| pRSW105 | pUC19 with ctsE in-frame deletion construct cloned into XmaI site | This study |
| pRSW106 | pUC19 with ctsX in-frame deletion construct cloned into XmaI site | This study |
| pRSW107 | pRSW104 with cat-rpsL cloned into EcoRV site of cj0011c | This study |
| pRSW108 | pUC19 with 1.6-kb fragment with ctsR cloned into XmaI site | This study |
| pRSW110 | pUC19 with 1.6-kb fragment with ctsP cloned into XmaI site | This study |
| pRSW112 | pRSW108 with cat-rpsL cloned into XmaI site of ctsR | This study |
| pRSW113 | pRSW110 with cat-rpsL cloned into XmaI site of ctsP | This study |
| pRSW114 | pUC19 with ctsP in-frame deletion construct cloned into XmaI site | This study |
| pRSW115 | pUC19 with ctsR in-frame deletion construct cloned into XmaI site | This study |
| pRSW119 | pECO102 with ctsF coding sequence (codon 2-stop) cloned into BamHI/XhoI site | This study |
| pRSW120 | pECO102 with ctsE coding sequence (codon 2-stop) cloned into BamHI/XhoI site | This study |
| pRSW121 | pECO102 with ctsX coding sequence (codon 2-stop) cloned into BamHI/XhoI site | This study |
| pRSW122 | pECO102 with ctsP coding sequence (codon 2-stop) cloned into BamHI/XhoI site | This study |
Escherichia coli strains were grown in Luria-Bertani (LB) broth or agar. Antibiotics were used at the following concentrations for E. coli strains when necessary: ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), chloramphenicol (30 μg ml−1), and tetracycline (12.5 μg ml−1). All E. coli strains were stored at −80°C in LB broth with 20% glycerol.
Construction of C. jejuni 81-176 astA::cat and astA::aphA3.
A 2.3-kb fragment containing the gene for arylsulfatase (astA [67]) with 260 bp of upstream and 220 bp of downstream DNA sequence was amplified by PCR from C. jejuni strain 81-176 with primers, each containing a 5′ PstI site. This fragment was digested with PstI and ligated into PstI-digested pUC19 to create pDRH138. To insertionally inactivate astA, pDRH138 was digested with EcoRV and ligated with a PvuII cassette containing cat from pRY109 (66). One plasmid containing astA::cat was obtained and designated pDRH143. pDRH138 was also inactivated by insertion of a SmaI cassette containing aphA3 from pILL600 (33) into the EcoRV site, and the resulting plasmid was designated pDRH142. These suicide plasmids were electroporated into C. jejuni 81-176 and transformants were selected on 20 μg of chloramphenicol ml−1 for pDRH143 or 50 μg of kanamycin ml−1 for pDRH142. A mutant in which wild-type astA had been replaced with astA::cat was obtained, verified by PCR, and designated DRH154. A mutant in which wild-type astA had been replaced with astA::aphA3 was obtained, verified by PCR, and designated DRH153.
Isolation and identification of transformation-deficient mutants.
MH agar plates were spread with 2.5 μg of 81-176 astA::cat chromosomal DNA (DRH154). After a drying step, mutants from a random 81-176 solo library (22) were spread onto MH plates at dilutions sufficient to produce ca. 100 colonies per plate. The plates were incubated for 48 h at 37°C in microaerophilic conditions. Colonies were patched onto MH agar with or without chloramphenicol and again incubated for 48 h. Mutants that did not grow on MH agar with chloramphenicol but did grow on MH alone were saved as potential transformation-deficient mutants.
Putative mutants were tested in a quantitative transformation assay (62) to determine their transformation efficiency. C. jejuni was grown for 48 h and then streaked onto three agar plates, grown overnight for 18 h, and resuspended in MH broth to an optical density at 600 nm of 0.5. Aliquots of 0.5 ml of C. jejuni suspension were added to 1 ml of solidified MH agar in 13-mm test tubes. Cultures were incubated in 5% CO2 at 37°C for 3 h before addition of 1 μg of chromosomal DNA. After incubation for an additional 4 h, dilutions of the bacteria were plated onto MH agar to determine the total number of bacteria and on MH agar containing the appropriate selective antibiotic to determine the number of transformants. Negative controls included strains treated as described above but without the addition of chromosomal DNA. Transformations were conducted in triplicate, and the transformation efficiency reflects the average of the three samples. Experiments were repeated at least three times. The transformation efficiency represents the number of transformants per total number of bacteria per microgram of DNA.
To confirm that transformation-deficient phenotypes were linked to the solo insertion in the mutants, purified chromosomal DNA from each mutant was used to transform 81-176 by natural transformation, and kanamycin-resistant (Knr) colonies were tested for their transformation efficiency. The sequence surrounding each solo insertion in transformation-deficient mutants was determined by inverse PCR (42, 57) and semi-exponential cycle sequencing (51) or by direct sequencing off the chromosome. The location of solo was determined by comparing the resulting sequence to that of the genome sequence of C. jejuni NCTC 11168 (43) and was verified by PCR analysis.
DNA uptake assays.
One to two micrograms of purified chromosomal or plasmid DNA (pRY108 [66]) from both C. jejuni 81-176 and E. coli was labeled by nick translation in a reaction mixture containing 30 μCi of [α-32P]dCTP, using a nick translation kit (Roche). Unincorporated deoxynucleoside triphosphates were removed by gel filtration with G-25 Sephadex Quick-Spin columns (Roche).
For DNA uptake assays, C. jejuni was grown for 48 h and then streaked onto three MH agar plates, grown for 18 h, and resuspended in MH broth to an optical density at 600 nm of 0.5. Aliquots of 500 μl of bacterial suspension were added to Eppendorf tubes and incubated with 0.1 μg of labeled DNA at 37°C for 30 min. Cultures were immediately placed on ice and centrifuged at 4°C. Samples were washed twice with MH broth, resuspended, incubated with 100 μg of DNase I (Roche) ml−1 for 10 min at room temperature, and then washed once in MH broth. Bacterial samples were resuspended in MH broth and transferred to scintillation vials with scintillation fluid for counting in a liquid scintillation counter to determine the amount of radioactivity present. Data are the average of three samples from one experiment, which was repeated at least three times.
Construction of deletion mutants of C. jejuni.
Defined chromosomal deletion mutants were constructed in streptomycin-resistant 81-176 (DRH212) as described by Hendrixson et al. (22). Genes to be deleted were amplified with ca. 500 bp upstream and downstream of the coding sequence by PCR with primers based on the sequence of C. jejuni NCTC 11168 (43). Primers were designed with restriction sites at the 5′ ends to facilitate cloning the PCR products into pUC19. Resulting plasmids were then digested with restriction enzymes that cut only once in the coding sequence of the gene to be deleted. When necessary, 5′ or 3′ overhangs were filled in with T4 DNA polymerase (Invitrogen) to make a blunt end. The genes were then interrupted by ligation of a SmaI cat-rpsL cassette from pDRH265 into the digested site. Constructed suicide plasmids were electroporated (61) into DRH212. Transformants where the insertional mutant replaced the wild-type locus were selected by growth on chloramphenicol, and the interrupted locus was verified by PCR analysis.
Fusions of upstream and downstream DNA fragments surrounding each gene were created by SOEing reactions (23) to create in-frame deletions in the gene of interest. These SOEing products were cloned into pUC19, and the resulting suicide plasmids were electroporated into the appropriate insertionally inactivated mutant. Transformants were selected on 2 mg of streptomycin ml−1 and then screened for loss of the cat-rpsL cassette by screening for chloramphenicol sensitivity on MH agar with 15 μg of chloramphenicol ml−1. Deletion of the appropriate genes was verified by PCR analysis.
Construction of C. jejuni complementing plasmid pECO102.
To construct a plasmid that would allow for complementation of C. jejuni mutants, an 82-bp fragment containing the promoter for the C. jejuni chloramphenicol acetyltransferase (cat) gene from pRY109 (66) was amplified by PCR. To amplify this fragment, two primers were designed. One primer contained a 5′ XbaI site and annealed to the upstream region of the cat promoter. A second primer contained a 5′ BamHI site immediately following and in frame with the start codon of cat. These primers were used to amplify the 82-bp fragment, and the fragment was then digested with XbaI and BamHI. This fragment was then ligated into pRY112 (66) that had been digested with XbaI and BamHI to create pECO102. This plasmid allows for the constitutive expression of various genes by cloning into the BamHI site a coding sequence from codon 2 to the stop codon (codon 2-stop) of the particular sequence. The expressed gene in the plasmid will encode a protein beginning with a methionine residue, followed by a glycine and a serine residue (due to the in-frame BamHI site), and ending with the native protein sequence beginning at the second amino acid through the C terminus of the protein.
Complementation of mutants.
Plasmids for complementation were introduced into E. coli DH5α/pRK212.1 (14), which contains the conjugative machinery to mobilize plasmids into C. jejuni. Conjugations were performed as described by Guerry et al. (19). Briefly, the C. jejuni recipient was grown on MH agar plates in microaerophilic conditions at 37°C for 48 h. Strains were then streaked onto MH plates, grown for 18 h at 37°C in microaerophilic conditions, and then resuspended to an optical density at 600 nm of 1.0. Overnight cultures of LB broth were diluted into fresh broth and grown to an optical density at 600 nm of 0.5. Next, 500 μl of the donor pellet was resuspended in 1 ml of C. jejuni suspension, spotted onto MH agar, and incubated for 5 h. Bacteria were resuspended in MH broth and spread onto MH agar with 10 μg of trimethoprim and 15 μg of chloramphenicol ml−1. After growth for 4 to 5 days, plasmid preparations and PCR were used to confirm that the recipient C. jejuni strain contained the appropriate plasmids.
Construction of nalidixic acid-resistant mutant of C. jejuni strain 81-176.
To isolate a nalidixic acid-resistant (NAr) mutant of C. jejuni 81-176, a lawn of C. jejuni NCTC 11168 (∼2 × 109) was plated onto MH agar containing 20 μg of nalidixic acid ml−1. One 11168 NAr colony (DRH312) was picked and a 2.6-kb fragment containing the coding sequence of gyrANA from this strain was amplified by PCR with primers designed from the genome sequence of 11168. Each primer contained a 5′ BamHI site. This gene was cloned into the BamHI site of pUC19 to create pDRH328. This plasmid was used to electroporate 81-176 to replace wild-type gyrA, and transformants were selected on MH agar containing 20 μg of nalidixic acid ml−1. One NAr mutant was saved and designated DRH335 (81-176 gyrANA).
Nucleotide sequence accession number.
The GenBank nucleotide accession numbers for ctsF from C. jejuni 81-176 and 11168 are AY324656 and AY324657.
RESULTS AND DISCUSSION
Development of a plate DNA transformation assay to screen for mutants.
To identify transposon-generated mutants with reduced ability to undergo natural transformation, we modified the previously described DNA transformation method performed on agar plates (62). Chromosomal DNA from strain DRH154, a derivative of strain 81-176 carrying a cat gene (encoding resistance to chloramphenicol) inserted into the nonessential gene astA (encoding aryl sulfatase) was spread on to agar plates. Dilutions of C. jejuni strain 81-176 were made, and ca. 100 bacteria were spread onto each MH agar plate. After colonies developed, transformed bacteria were identified by replica plating the colonies on MH agar with or without chloramphenicol. Preliminary experiments (results not shown) indicated that 2.5 μg of DNA in this plate assay was sufficient to ensure that virtually 100% of the colonies that arose contained bacteria that had been transformed. To confirm that the resulting chloramphenicol-resistant (Cmr) colonies arose by recombination of the cat gene into astA as opposed to being spontaneous mutants, colonies were screened by PCR to verify the presence of cat. In addition, control experiments were performed to determine the extent of spontaneous chloramphenicol resistance, which was determined to be <10−9. To verify that the plate transformation assay worked with other chromosomal markers and was not simply a consequence of a high rate of recombination at the astA locus, transforming DNA was isolated from strain DRH212, a streptomycin-resistant mutant of C. jejuni 81-176 (22). DNA from this strain was able to transform 81-176 as efficiently as DNA from strain DRH154 (data not shown). Similar transformation efficiencies were also observed with DNA derived from strain DRH153, a derivative of strain 81-176 carrying aphA3 (encoding resistance to kanamycin) inserted into astA and strain DRH335, an NAr mutant of 81-176 (data not shown).
Screening of a solo library of 81-176 for mutants defective in DNA transformation.
A library of random C. jejuni 81-176 mutants created by solo transposition (22) was screened for mutants defective for DNA transformation. Approximately 4,500 mutants were screened by the plate transformation method with 2.5 μg of astA::cat chromosomal DNA from DRH154 as the DNA source. When colonies arose they were patched onto MH agar with or without chloramphenicol. Fifty-seven mutants did not grow on MH agar containing chloramphenicol, indicating that they may harbor a solo insertion in a gene required for transformation. A quantitative transformation assay (19, 62) was used to determine the transformation efficiency of each individual mutant. Wild-type 81-176 was transformed with an efficiency ranging from ca. 10−3 to 10−4; 34 of the 57 putative transformation mutants we isolated had transformation efficiencies of approximately 1 to 100% of that of wild-type 81-176. These mutants were not characterized further (data not shown).
The other 23 mutants we isolated had more severe transformation defects, with efficiencies ca. 1,000 times lower than the wild-type strain (Table 2). Several of these solo insertion sites were identical, indicating that the strains are siblings, and are not indicated on Table 2. As a preliminary confirmation that the transformation defect was due to the solo insertion and not to an unlinked spontaneous mutation, we performed the equivalent of a genetic backcross by transforming wild-type 81-176 with chromosomal DNA from each mutant to kanamycin resistance (encoded in the solo transposon). When tested with the quantitative transformation assay, the resulting Knr strains had transformation defects similar to the original solo mutant strains. We conclude from these experiments that the transformation-deficient phenotypes of the original strains were likely due to the solo insertion.
TABLE 2.
Location of solo in C. jejuni transformation mutants
| Transposon locationa | Coding sequence descriptionb | Transformation efficiencyc |
|---|---|---|
| NAd-81-176 | NA | 2.3 × 10−3 |
| cj1352 (ceuB) | Enterochelin uptake permease | 6.70 × 10−8 |
| cj1343c (ctsG) | Putative periplasmic protein | 8.00 × 10−8 |
| cj1076 (proC) | Putative PCA reductase | 2.5 × 10−7 |
| cj1077 (ctsT) | Putative periplasmic protein | 3.32 × 10−7 |
| 3′ of ansA (C23) | Not in a coding sequence | 5.47 × 10−8 |
| cj1028c (ctsW) | Purine/pyrimidine phosphoribosyltransferase | <10−9 |
| cj1473c (ctsP) | Putative ATP/GTP binding protein | 2.48 × 10−7 |
| cj1475c (ctsR) (2) | Hypothetical protein | 1.78 × 10−7-1.60 × 10−8 |
| cj1471c (ctsE) (4) | Putative type II secretion system E protein | 1.51 × 10−7-6.00 × 10−7 |
| cj1470c (ctsF) | Pseudogene type II secretion system F protein | 2.94 × 10−7 |
| cj1474c (ctsD) (4) | Putative type II secretion system D protein | 1.37 × 10−7-9.04 × 10−8 |
The number in parentheses indicates the number of independent solo insertions identified in each gene
The gene designation and proposed function are based on the annotated genome sequence of C. jejuni strain 11168 (43).
The transformation efficiency is calculated as the number of transformants per total number of CFU per microgram of DNA.
NA, not applicable.
A partial type II secretion system required for competence.
Sequence analysis of the mutants revealed the locations of the solo insertions. Figure 1 shows chromosomal regions containing each gene identified in our screen. Genes identified by our screen fall into three broad groups: (i) those with homology to genes required for diverse macromolecular transport processes in a range of bacterial species, (ii) those with homology to genes from other bacteria whose role in competence has not been shown before, and (iii) those with no known homologies. Because of the genetic screen used to identify these genes, we propose to designate them as cts for Campylobacter transformation system. Genes in the first category include cj1343c, cj1470c, cj1471c, and cj1474c (Table 2 and Fig. 1), which are most similar to several genes from type II secretion systems responsible for transport of macromolecules such as pilus subunits, toxins, and other exoenzymes (49, 50). Type II systems are also associated with competence in some bacteria, including N. gonorrhoeae (31) and Bacillus subtilis (13). Type II secretion systems have components in each compartment of the cell and orthologous genes from different species often share letter designations across systems, although in some cases presented below the orthologues were named prior to the adoption of this general convention. Genes identified in our screen that are likely to encode protein orthologues of type II proteins in other bacteria were given the letter designation of that protein family; and those lacking homology were given other letter designations (Table 2). The type II secretion apparatus is typically comprised of a minimum of 12 proteins; of those, our analysis identified perhaps five, as detailed below.
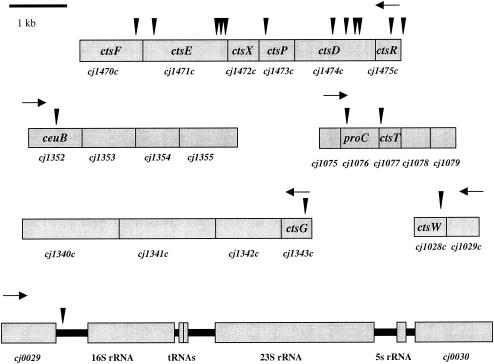
FIG. 1.
Regions encompassing genes identified as playing a role in transformation and location of solo in these genes. Triangles represent solo insertion sites. Black arrows indicate the direction of transcription.
The type II cts homologues we identified through transposon mutagenesis are termed CtsD (Cj1474c), CtsE (Cj1471c), CtsF (Cj1470c), and CtsG (Cj1343c) (Table 2). CtsD has weak homology to the PilQ protein (e value = 0.038), which is thought to be an outer membrane pore involved in natural transformation and type IV pilus biogenesis in N. gonorrhoeae (12). CtsE is homologous to ComGA of B. subtilis (e value = 2.6 × 10−52) and PilT of N. gonorrhoeae (e value = 2.2 × 10−30), both of which are required for transformation (7, 64). CtsF shares similarity to PilG of N. gonorrhoeae (e value = 4 × 10−13) and is also required for transformation and pilus biogenesis in that species (59).
CtsG (Cj1343c) has limited similarity at its N terminus to that of other type II system G orthologues and is annotated as such in the C. jejuni sequence (43). Proteins of this class in type II secretion systems are similar to subunits of type IV pili and other molecules termed pseudopilins, which may be important for the development of the secretion apparatus per se (24). These molecules are produced as precursor proteins that are subsequently processed at their N termini by a prepilin peptidase (49, 50). The N-terminal leader that is cleaved by prepilin peptidases is usually positively charged and is followed by a region of ca. 20 primarily hydrophobic amino acids (34). The peptidase cleaves at a conserved site [G*F(M)XXXE, where X represents hydrophobic amino acids, and “*” represents the cleavage site] (40, 55).
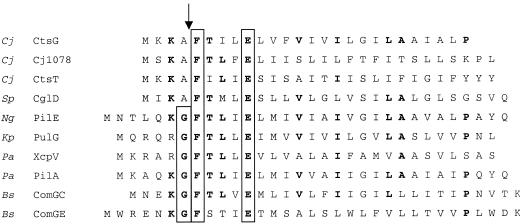
The predicted amino terminus of CtsG, has a variant of the above site, AFXXXE (Fig. 2). In other systems, experimentally replacing the glycine in the first position with alanine does not inhibit cleavage of the signal sequence (55). Furthermore, prepilin-like proteins naturally containing this nonconsensus cleavage site play a role in transformation in both Streptococcus pneumoniae and Streptococcus gordonii (37, 45). The translated amino acid sequence of another gene isolated in our screen, ctsT (cj1077), also has a putative peptidase cleavage site (Fig. 2). In addition, cj1078, which is immediately downstream of ctsT, would encode a protein with a peptidase cleavage site (Fig. 2). Although cj1078 was not identified in our screen, its proximity to ctsT and proC, another gene identified in our analysis (see below), suggests that it is involved in the transformation process as well. To identify a candidate prepilin peptidase that may play a role in competence, we performed a basic local alignment search tool (BLAST) search of the C. jejuni genome sequence with a B. subtilis processing peptidase that is involved in natural transformation (ComC) (6) and found one protein, Cj0825, with homology to ComC. The two proteins share 29% identity and 46% similarity over 109 amino acids. In B. subtilis, ComC is required for processing of ComGC, ComGD, ComGE, and ComGG, and comC mutants are deficient for DNA binding and uptake (8). The identification of this gene among transformation mutants identified by another group and reported recently in a meeting abstract suggests that this gene could encode the peptidase required for cleaving the prepilin molecules discussed above (D. L. Wilson et al., Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003, abstr. D-049, p. 211, 2003).
FIG. 2.
Alignment of N-terminal regions of CtsG, CtsT, and Cj1078 to pre-pilins and pre-pilin-like proteins from S. pneumoniae (45), N. gonorrhoeae (65), K. pneumoniae (47), P. aeruginosa (41, 44), and B. subtilis (1, 6). The processing site is indicated by an arrow. The conserved G, F, and E residues are boxed, and residues conserved among a number of the prepilins are indicated in boldface.
Type II secretion systems such as Cts of C. jejuni share similarity to systems used in the assembly of type IV pili (24). Such a pilus plays a role in transformation in several species, including N. gonorrhoeae, Legionella pneumophila, Pseudomonas stutzeri, and others (15, 18, 54). Since C. jejuni has several proteins similar to pre-pilin-like molecules, we used electron microscopy to determine whether C. jejuni cells formed a pilus-like structure under conditions in which the bacterium is competent. We did not observe any structures that resembled a pilus (data not shown), a finding consistent with the findings of Gaynor et al. (16).
In our sequence analysis of these insertions, we discovered that cj1470c (ctsF) is annotated in the C. jejuni 11168 genome as a probable pseudogene. We identified a solo insertion in cj1470c in strain 81-176 among our transformation mutants and, because there are no downstream genes whose expression could be affected by an upstream transposon insertion, we speculated that it might indeed encode an intact protein. We sequenced a 1.4-kb region of the genome spanning cj1470c in strains 81-176 and 11168 and found 21 base changes between the two sequences, 17 of which were in the coding region of cj1470c. Most of these are silent mutations, although a change in the sequence at amino acid 190 results in a conservative change from an alanine to a valine.
A key difference in the two sequences is at position 1404347 of the 11168 sequence, where we identified an insertion of an A/T base pair in both 81-176 and 11168 that is not in the published sequence of strain 11168. Thus, instead of a run of five A/T pairs, there are actually six, which shifts the reading frame so that in strain 11168 it encodes a protein of 393 amino acids and not 279 amino acids as predicted by the genome sequence (43). The coding sequence from the cj1470c homologue in strain 81-176 encodes a protein of 392 amino acids, one less than in strain 11168, due to a frameshift at the 3′ end of the gene. Both of these sequences are deposited in GenBank.
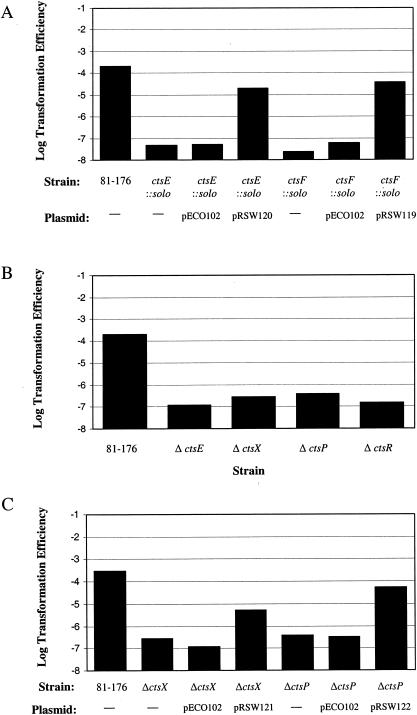
Several solo insertions in our screen mapped to genes arranged in a likely operon that includes ctsR, ctsD, ctsP, ctsE, and ctsF (Fig. 1). For several of these genes, we performed experiments to rule out polarity effects of the solo insertions. We expressed ctsE in the ctsE::solo strain by using an expression plasmid, pECO102, constructed for this purpose. The cloned gene complemented the ctsE transposon insertion by over 2 orders of magnitude (Fig. 3A), although it did not restore transformation efficiency to full wild-type levels, which is a general observation for all of mutants in this putative operon (Fig. 3A and C; see below). We speculate that this may be due to a requirement for precise coordination of expression levels to achieve optimal (wild type) levels of transformation, which may not occur when one of the gene products is expressed from pECO102.
FIG. 3.
Transformation efficiency of in-frame deletion mutants and complemented constructs. The data represent the average of three samples per strain from one experiment. Experiments were repeated at least three times. (A) Transformation efficiency of C. jejuni solo mutants complemented with pECO102 or derivative containing the coding sequence for the appropriate gene; (B) transformation efficiency of C. jejuni deletion mutants; (C) transformation efficiency of C. jejuni deletion mutants complemented with pECO102 or derivative containing the coding sequence from deleted gene.
In addition to complementing the transposon insertions, we constructed strains with nonpolar mutations in the form of in-frame deletions in ctsE, ctsR, and ctsP (RSW136, RSW137, and RSW115, respectively) (Table 1). Each strain we constructed this way was ca. 1,000-fold reduced in transformation efficiency compared to the wild-type strain, a finding similar to what we observed with the original solo-induced mutants (Fig. 3B and Table 2). Based on these results, we conclude that the phenotypes of most of the transposon mutants are not due to polarity. We have yet to rule out the possibility of a polarity effect in the ctsD::solo insertion mutant. However, given its homology to genes required for transformation in other systems and the fact that this gene is surrounded by genes required for the transformation of C. jejuni, we consider it likely that ctsD is required for this process as well and that the insertion does not affect transformation because of polarity.
Also within the putative ctsR operon is cj1472c, in which we did not recover insertions from our transposon screen. Based on its chromosomal location, we nevertheless hypothesized that the encoded protein, which is annotated as a putative membrane protein, may play a role in transformation. We therefore constructed a strain deleted of cj1472c, which we termed strain RSW46. In our standard transformation assay, RSW46 is transformed 1,000 times less efficiently than wild-type strain 81-176 (Fig. 3B). Given the transformation defect of strain RSW46, which lacks complete cj1472c, we gave this gene a cts designation and have termed it ctsX, as noted on Fig. 3B.
Other cts genes.
Two other classes of cts genes not obviously associated with the type II secretion system discussed above were identified during our screen for competence-deficient mutants. One class consists of the C. jejuni homologues of ceuB (cj1352), proC (cj1076), and cj1028c (which we termed ctsW)—three genes whose putative products have well-described functions in other bacteria (Table 2). CeuB is an enterochelin-specific permease, thought to be an integral membrane protein involved in transport of enterochelin for iron acquisition in Campylobacter coli (48). Its potential role in transformation has not been reported. Pyrroline 5-carboxylate reductase (PCA reductase), encoded by proC, catalyzes the reduction of pyrroline-5-carboxylate to proline as the third step in the conversion of glutamate to proline (10). ProC is homologous to ComER of B. subtilis (e value = 8.1e-11), which, despite its name, was found not to function in transformation (27). CtsW is annotated in the 11168 genome as a purine/pyrimidine phosphoribosyltransferase (43). A homologue of indeterminate function has been identified in H. pylori and Thermotoga maritima. Whether T. maritima is naturally competent has yet to be demonstrated (39). CtsW also has very weak homology to ComFC of B. subtilis (e value = 0.1), and B. subtilis strains carrying comFC mutations have a 10-fold decrease in transformation efficiency relative to the wild type (35). The ComF operon is required for DNA transport but not binding (13).
The third class of mutant includes cj1475c and cj1473c (Table 2). BLAST searches with these open reading frames (ORFs) revealed no significant homology to any proteins in the database. We have therefore termed these genes ctsR and ctsP to denote them as components of the Campylobacter transformation system. In one strain, mutant C23, solo is in a region between cj0029 and the coding region for 16S rRNA.
DNA uptake ability of cts mutants.
The steps in transformation can be separated into two discrete, experimentally distinguishable steps: DNA binding and DNA uptake. In assays for DNA binding, radioactively labeled DNA is incubated with cells, after which they are collected and washed by centrifugation; binding is measured as the total cell-associated radioactivity after washing. DNA uptake can be examined by measurement of the amount of labeled DNA in a DNase I-resistant form. In order to determine the stage at which the transformation defect lies with our isolated solo mutants, we performed DNA binding and uptake assays.
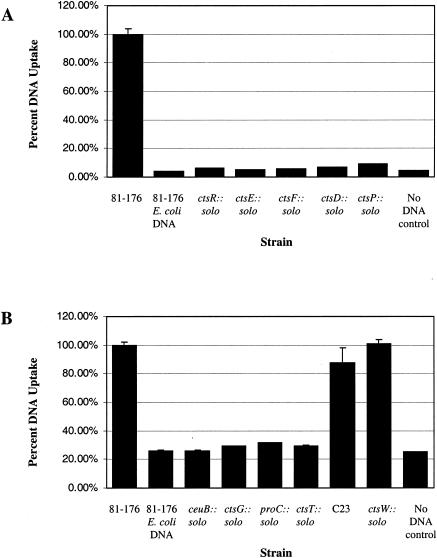
We first examined DNA-binding and DNA uptake abilities of strains carrying insertions within the type II secretion homologues ctsF, ctsE, and ctsD, along with ctsP and ctsR, which are linked in a putative operon. Although the DNA-binding activities of the mutant strains were consistently lower than the wild-type levels, the variability from assay to assay prevented a definitive conclusion regarding the DNA-binding ability of these mutants relative to the wild-type (data not shown). In contrast, the results of DNA uptake assays were very consistent; each mutant takes up DNA at levels no greater than the background level, which is ca. 10% of the wild-type level (Fig. 4A).
FIG. 4.
DNA uptake assays. The data are the average of three samples from one experiment, which was repeated at least three times. Values are normalized to the wild-type levels. Wild-type 81-176 samples with no added radiolabeled DNA serve as a control. Since C. jejuni does not take up E. coli DNA (62), we performed DNA uptake experiments with radiolabeled DH5α DNA which served as a negative control. (A) Uptake of radiolabeled 81-176 chromosomal DNA by C. jejuni 81-176 and mutants isolated in the operon spanning from ctsR to ctsF; (B) uptake of radiolabeled 81-176 chromosomal DNA by 81-176 and solo mutants from other locations in the genome.
DNA uptake levels were examined in the other six solo mutants unlinked to the putative ctsR operon (Fig. 4B). Several of these mutants (ceuB::solo, ctsG::solo, proC::solo, and ctsT::solo) take up DNA at background levels, much like the strains with solo insertions in the operon ranging from ctsR to ctsF. In these mutants and in the type II secretion system mutants, we were unable to distinguish whether the transformation defect lies in transport of DNA across the outer membrane or if the defect is in the binding step of transformation, since the results from our DNA-binding assays were not consistent. However, the defect clearly lies in one of the earlier steps of transformation. In contrast, mutant C23 and ctsW::solo take up DNA at levels approaching that of the wild type (Fig. 4B). This suggests that the transformation defect in these two mutants (i.e., C23 and ctsW::solo) lies in a later stage of transformation, such as transport to the cytoplasm or recombination with the chromosome.
Identification of ORFs with homology to known competence proteins.
Prior work has identified several genes with homology to those involved in transformation in H. pylori. Bacon et al. identified the type IV secretion system on pVir, which is homologous to the type IV system required for transformation in H. pylori (3). In addition, C. jejuni was previously reported to have a DprA homologue with the strongest homology to HP0333 of H. pylori (2). DprA has been shown to be important in transformation in both H. influenzae and H. pylori (2, 28, 58). We performed BLAST searches against the C. jejuni genome to identify other potential transformation gene homologues that may have been missed in our screen of the solo transposon library. For these we used the translated amino acid sequence from genes required for natural transformation in other bacteria. This search identified Cj0011c, which is homologous to ComEA of B. subtilis, with 56% identity and 82% similarity between the translated amino acid sequence of the two genes over 62 amino acids (out of a total of 79 for the predicted ORF). S. pneumoniae also requires a ComEA ortholog for transformation (45). ComEA of B. subtilis (and ComE of N. gonorrhoeae) binds DNA and is required for DNA uptake (5, 27, 46). Further work is needed to confirm the role of this ComEA homologue in transformation: at present, our attempts to construct a nonpolar mutation in cj0011c have not been successful.
Analysis of the role of pVir encoded genes in transformation.
As noted above, Bacon et al. have shown that C. jejuni strain 81-176 carries a plasmid, pVir, that contains seven ORFs whose products are homologous to type IV secretion system proteins (3, 4). In H. pylori several of these homologues, including ComB4 and ComB7 through ComB10, are important for transformation (25, 26). In H. pylori the virD4 homologue (HP1006) is not required for transformation (52).
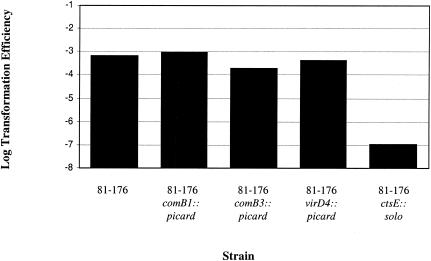
We tested the role of several of these type IV secretion system homologues for their role in natural transformation in C. jejuni (Fig. 5). Mutants contained the transposon picard, carrying cat (encoding chloramphenicol resistance) inserted in the indicated gene (D. R. Hendrixson and V. J. DiRita, unpublished observations). Transformation assays were conducted with 1 μg of DRH335 DNA (a spontaneous NAr mutant of 81-176) as the donor source. Our results confirm that strains with mutations in comB3 have a slightly reduced level of transformation relative to the wild-type (3), whereas mutants with transposon insertions in comB1 and virD4 transformed at levels near wild type (Fig. 5). The transformation efficiency of ctsE::solo is shown for comparison in Fig. 5. Clearly, mutations in the pVir genes have a less profound effect on transformation.
FIG. 5.
Transformation efficiency of type IV secretion system mutants of pVir. The results are from one experiment conducted in triplicate, and the experiment was repeated at least three times.
Conclusions.
We used transposon mutagenesis to begin identifying genes whose products are required for natural transformation in C. jejuni. Bacon et al. identified plasmid-encoded genes in strain 81-176 that are similar to those of type IV secretion systems, which play a role in natural transformation in the closely related organism H. pylori (3, 25, 26). Unlike the situation in H. pylori, where mutations in several type IV secretion system genes eliminated transformation, a comB3 mutant in C. jejuni had a transformation efficiency that was approximately five times lower than the wild-type, while a virB11 mutant displayed a wild-type transformation efficiency (3). In addition, we tested two other type IV secretion system homologues, virD4 and comB1, and concluded that they are not important for natural transformation in C. jejuni.
In contrast, the 11 mutants we isolated by solo transposon mutagenesis in C. jejuni are considerably more defective in their ability to undergo transformation; the efficiency with which the cts mutants are transformed is ca. 1,000-fold less than that of the wild-type strain 81-176. These data indicate that transformation of C. jejuni does not appear to proceed solely via the type IV system similar to that required for transformation in H. pylori. The genes identified in our screen indicate that a putative type II secretion system in C. jejuni is responsible for promoting natural transformation in C. jejuni. The respective roles of the type IV plasmid-encoded system characterized to some extent by Bacon et al. and the type II system identified here obviously await future study.
Many naturally competent organisms have a group of core proteins similar to several of the Cts proteins that are required for DNA binding and uptake. These are encoded by the comG operon and comC in B. subtilis, where they were first characterized (1, 20, 38). In general, these proteins comprise a large family of related proteins that have been found to function in a variety of systems, including type II secretion systems, biogenesis of type 4 fimbriae, competence for natural transformation, and filamentous phage morphogenesis (24). In B. subtilis the comG operon and comC are required for the binding of DNA to the cell surface and in N. gonorrhoeae they are required for DNA uptake (7, 64).
Due to their functions, this family of proteins has been designated PSTC proteins (for pilus, secretion, twitching motility, and competence) (13). Several of the proteins encoded by the genes identified in our study fall into this family. The PSTC proteins can be divided into five classes, and C. jejuni appears to have a member from each class, four of which were identified by our transposon screen and one of which was identified by BLAST searches against the C. jejuni genome. Table 3 summarizes key features of each class of PSTC proteins and indicates the classes that the Cts proteins fall into.
TABLE 3.
C. jejuni homologues of PSTC proteins
| Class | C. jejuni protein | Class characteristics | B. subtilis protein |
|---|---|---|---|
| I | CtsE | Walker A box | ComGA |
| II | CtsF | Membrane protein | ComGB |
| III | CtsG CtsT Cj107a | Conserved N-terminal sequence resembling cleavage sites of type IV prepilin proteins | ComGC ComGD ComGe ComGG |
| IV | Cj0825b | Prepilin peptidase | ComC |
| V | CtsD | Secretin | Not present |
Cj1078 has not yet been demonstrated to play a role in transformation in C. jejuni. It is downstream of CtsT, which also has a putative prepilin peptidase cleavage site.
Cj0825 has been described as having a role in transformation by Wilson et al. (Abstr. 103rd Gen. Meet. Am. Soc. Microbiol.)
Mutants of C. jejuni lacking the PSTC proteins CtsD, CtsE, CtsF, and CtsG decrease DNA uptake to background levels (Table 2). In B. subtilis, ComG proteins are thought to enable DNA binding by allowing access of DNA to ComEA, the receptor for DNA binding, at the cell surface (46). In N. gonorrhoeae, ComG homologues are essential for the DNA uptake step of transformation (64). These C. jejuni PTSC proteins may play a role in DNA binding, but this was difficult to determine definitively because DNA-binding assays with the mutant strains gave widely variable results, although the binding levels were consistently lower than the wild-type level. Perhaps there are redundant systems for DNA binding on the cell surface of C. jejuni that are not affected by these mutations, and this possibility is under investigation.
It appears that C. jejuni uses components of other transformation systems and some novel components to bring in DNA from the environment. Further study will focus on the roles of identified proteins in the process of transformation, as well as on dissection of the transformation apparatus.
Acknowledgments
We thank Hank Seifert, Eric Skaar, and Dorothy Sorenson for help with the transmission electron microscopy. We also thank Erin O'Rourke for the construction of pECO102.
This work was supported in part by USDA grant 2001-2991 to V.J.D. R.S.W. is a trainee of the Genetics Training Program at the University of Michigan.
REFERENCES
- 1.Albano, M., R. Breitling, and D. Dubnau. 1989. Nucleotide sequence and genetic organization of the Bacillus subtilis ComG operon. J. Bacteriol. 171:5386-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., D. A. Israel, K. Kugugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analysis of the pVir plasmid of Campylobacter jejuni. J. Bacteriol. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, I., and E. C. Gotschlich. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J. Bacteriol. 183:3160-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, Y. S., and D. Dubnau. 1995. ComC is required for the processing and translocation of ComGC, a pilin-like competence protein of Bacillus subtilis. Mol. Microbiol. 15:543-551. [DOI] [PubMed] [Google Scholar]
- 7.Chung, Y. S., and D. Dubnau. 1998. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J. Bacteriol. 180:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, Y. S., F. Breidt, and D. Dubnau. 1998. Cell surface localization and processing of the ComG proteins required for DNA binding during transformation in Bacillus subtilis. Mol. Microbiol. 29:905-913. [DOI] [PubMed] [Google Scholar]
- 9.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. M. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 10.Deutch, A. H., C. J. Smith, K. E. Rushlow, and P. J. Kretschmer. 1982. Escherichia coli Δ1-pyrroline-5-carboxylate reductase: gene sequence, protein overproduction, and purification. Nucleic Acids Res. 10:7701-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake, S. L., and M. Koomey. 1995. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol. 18:975-986. [DOI] [PubMed] [Google Scholar]
- 13.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 14.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae: a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 16.Gaynor, E. C., N. Ghori, and S. Falkow. 2001. Bile-induced “pili” in Campylobacter jejuni are bacteria-independent artifacts of the culture medium. Mol. Microbiol. 39:1546-1549. [DOI] [PubMed] [Google Scholar]
- 17.Goodgal, S. H., and R. M. Herriott. 1961. Studies on transformations of Haemophilus influenzae. I. Competence. J. Gen. Physiol. 44:1201-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graupner, S., V. Frey, R. Hashemi, M. G. Lorenz, G. Brandes, and W. Wackernagel. 2000. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 182:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, J., M. Albano, and D. Dubnau. 1987. Isolation and characterization of Tn917 lac-generated competence mutants of Bacillus subtilis. J. Bacteriol. 1987:3104-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Ltd., London, United Kingdom.
- 24.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 25.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 26.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 27.Inamine, G. S., and D. Dubnau. 1995. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J. Bacteriol. 169:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karudapuram, S. X. Zhao, and G. J. Barcak. 1995. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J. Bacteriol. 177:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kendall, E. J. C., and E. I. Tanner. 1982. Campylobacter enteritis in general practice. J. Hyg. 88:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 31.Koomey, M. 1998. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS 106(Suppl. 84):56-61. [DOI] [PubMed] [Google Scholar]
- 32.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 33.Labigne-Roussel, A., P. Courcoux, and L. Tompkins. 1988. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaPointe, C. F., and R. K. Taylor. 2000. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275:1502-1510. [DOI] [PubMed] [Google Scholar]
- 35.Londoño-Vallejo, J. A., and D. Dubnau. 1993. comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol. Microbiol. 9:119-131. [DOI] [PubMed] [Google Scholar]
- 36.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lunsford, R. D., and A. G. Roble. 1997. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J. Bacteriol. 179:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohan, S., J. Aghion, N. Guillen, and D. Dubnau. 1989. Molecular cloning and characterization of comC, a late competence gene of Bacillus subtilis. J. Bacteriol. 171:6043-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. G. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 40.Nunn, D., and S. Lory. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 88:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunn, D., and S. Lory. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175:4375-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic application of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 44.Pasloske, B. L., B. B. Finlay, and W. Paranchych. 1985. Cloning and sequencing of the Pseudomonas aeruginosa PAK pilin gene. FEBS Lett. 183:408-412. [DOI] [PubMed] [Google Scholar]
- 45.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Provvedi, R., and D. Dubnau. 1999. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 31:271-280. [DOI] [PubMed] [Google Scholar]
- 47.Reyss, I., and A. P. Pugsley. 1990. Five additional genes in the pulC-O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023 which are required for pullulanase secretion. Mol. Gen. Genet. 222:176-184. [DOI] [PubMed] [Google Scholar]
- 48.Richardson, P. T., and S. F. Park. 1995. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 141:3181-3191. [DOI] [PubMed] [Google Scholar]
- 49.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 51.Sarkar, G., and M. E. Bolander. 1997. Direct sequencing of unpurified PCR-amplified DNA by semiexponential cycle sequencing (SECS). Mol. Biotechnol. 8:269-277. [DOI] [PubMed] [Google Scholar]
- 52.Smeets, L. C., J. J. E. Bijlsma, E. J. Kuipers, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2000. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Microbiol. Lett. 27:99-102. [DOI] [PubMed] [Google Scholar]
- 53.Sparling, P. F. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strom, M. S., and S. Lory. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa: effect on leader peptide cleavage, N-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656-1664. [PubMed] [Google Scholar]
- 56.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tchetina, E., and E. B. Newman. 1995. Identification of Lrp-related genes by inverse PCR and sequencing: regulation of two mal operons of Escherichia coli by leucine-responsive regulatory protein. J. Bacteriol. 10:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomb, J. F., G. J. Barcak, M. S. Chandler, R. J. Redfield, and H. O. Smith. 1989. Transposon mutagenesis, characterization, and cloning of transformation genes of Haemophilus influenzae. J. Bacteriol. 171:3769-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tønjum, T., N. E. Freitag, E. Namork, and M. Koomey. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16:451-464. [DOI] [PubMed] [Google Scholar]
- 60.Tsuda, M., M. Karita, and T. Nakazawa. 1993. Genetic transformation in Helicobacter pylori. Microbiol. Immunol. 37:85-89. [DOI] [PubMed] [Google Scholar]
- 61.van Vliet, A. H. M., A. C. Wood, J. Henderson, K. Wooldridge, and J. M. Ketley. 1997. Genetic manipulation of enteric Campylobacter species. Methods Microbiol. 27:407-419. [Google Scholar]
- 62.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]
- 64.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]
- 65.Wolfgang, M., J. P. M. van Putten, S. F. Hayes, and M. Koomey. 1999. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31:1345-1357. [DOI] [PubMed] [Google Scholar]
- 66.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 67.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]