Abstract
A psychrophilic gram-positive isolate was obtained from Antarctic Dry Valley soil. It utilized lactose, had a rod-coccus cycle, and contained lysine as the diamino acid in its cell wall. Consistent with these physiological traits, the 16S ribosomal DNA sequence showed that it was phylogenetically related to other Arthrobacter species. A gene (bgaS) encoding a family 2 β-galactosidase was cloned from this organism into an Escherichia coli host. Preliminary results showed that the enzyme was cold active (optimal activity at 18°C and 50% activity remaining at 0°C) and heat labile (inactivated within 10 min at 37°C). To enable rapid purification, vectors were constructed adding histidine residues to the BgaS enzyme and its E. coli LacZ counterpart, which was purified for comparison. The His tag additions reduced the specific activities of both β-galactosidases but did not alter the other characteristics of the enzymes. Kinetic studies using o-nitrophenyl-β-d-galactopyranoside showed that BgaS with and without a His tag had greater catalytic activity at and below 20°C than the comparable LacZ β-galactosidases. The BgaS heat lability was investigated by ultracentrifugation, where the active enzyme was a homotetramer at 4°C but dissociated into inactive monomers at 25°C. Comparisons of family 2 β-galactosidase amino acid compositions and modeling studies with the LacZ structure did not mimic suggested trends for conferring enzyme flexibility at low temperatures, consistent with the changes affecting thermal adaptation being localized and subtle. Mutation studies of the BgaS enzyme should aid our understanding of such specific, localized changes affecting enzyme thermal properties.
Glycosyl hydrolases (EC 3.2.1 to 3.2.3) cleave the glycosidic bond between two or more carbohydrates or between a carbohydrate and another moiety. Traditionally, glycosyl hydrolases were classified based on functional similarity. More recently, however, Henrissat and his coworkers have organized these enzymes into 90 glycosyl hydrolase families characterized by hydrophobicity plots, amino acid sequence similarities, and reaction mechanisms (10-12). This system also identifies possible common structural domains, thereby defining evolutionary connections and suggesting reaction mechanisms for the glycosyl hydrolases. Based on these criteria, the once-unified group of enzymes that exhibit β-galactosidase activity (EC 3.2.1.23) are now subdivided into four different families: 1, 2, 35, and 42.
Of these, the best studied is family 2, which includes the well-characterized β-galactosidase from Escherichia coli that is encoded by the lacZ gene. Although there is considerable information about the regulation (1), biochemistry (18, 23, 35, 47), reaction mechanism (17, 45), and structure (16) of this LacZ β-galactosidase, few other β-galactosidases within this family have been characterized biochemically (4, 7, 13-15, 26, 27, 43), while most examples exist only as a published sequence. Because of the emphasis on the E. coli LacZ enzyme, opportunities to learn from differences in β-galactosidases from other sources may have been overlooked. Studying these new β-galactosidases can provide insight into the evolution of their genes, suggest structural relationships, yield enzymes with academically and industrially valuable properties, and illuminate the underlying features responsible for thermal adaptation. Further, the characterization of other β-galactosidases offers the advantage of examining enzymes with unique biochemical and structural properties while having a characterized model for comparison.
Because of our interest in studying cold-active enzymes, we have isolated several psychrophilic prokaryotes, studied enzyme properties at low temperatures, and examined the mechanisms proposed for conferring cold activity. As part of our objective of studying cold-active β-galactosidases, we initially isolated psychrophiles from whey-treated fields in central Pennsylvania and cloned genes encoding glycosidases from three different families from a single isolate, B7 (9). In additional work, our researchers showed that this isolate and three others formed a monophyletic clade belonging to a new Arthrobacter species, A. psychrolactophilus, and shared the unique traits of growing at 0 to 5°C and using lactose as a carbon source (20).
Here we report on Arthrobacter strain SB, obtained from an Antarctic Dry Valley sample, the biochemical characterization of its β-galactosidase (BgaS), and its comparison with other enzymes. BgaS appears to be one of the most cold-active enzymes characterized to date, with an optimal activity near 18°C. It maintains at least 50% of its activity at 0°C and loses all activity at 37°C in less than 10 min. Comparisons with purified E. coli β-galactosidase using o-nitrophenyl-β-d-galactopyranoside (ONPG) or lactose as the substrate show that BgaS has a higher catalytic efficiency at 20°C and below. To our knowledge, this is the first study to determine the catalytic efficiency of an enzyme using lactose as the substrate at temperatures below 25°C. The unique cold activity and heat lability of BgaS is of special interest for comparisons and modeling with other family 2 β-galactosidases to discern specific regions and alterations that may confer these traits. The attributes of BgaS also make it a candidate for use in the industrial removal of lactose or as a reporter enzyme for psychrophilic genetic systems.
MATERIALS AND METHODS
Isolation and culture conditions.
Isolate SB was obtained from a soil sample provided by L. E. Casida at Penn State University. The sample had been stored frozen since its collection from the Antarctic Dry Valley region by R. Benoit in the late 1960s. The original enrichment medium was M9 (24) with the addition of 0.2% Casamino Acids, 5 μg of FeSO4/ml, a vitamin solution (Gibco), trace elements (19), and 0.2% cellobiose. Following purification, isolate SB was grown on M9 medium with 0.2% lactose and 0.01% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma, St. Louis, Mo.) to test for the presence of β-galactosidase activity, and in M9 broth medium with either 0.2% lactose or glucose to test for carbon source utilization and additional growth requirements. Trypticase soy agar (TSA) and R2A medium (Difco Laboratories, Detroit, Mich.) with 0.05% glucose or 0.2% peptone and 0.1% lactose were used to determine the temperature range for growth, and phenol red broth (10.0 g of proteose peptone, 5.0 g of NaCl, 0.018 g of phenol red per liter) containing different sugars was used for carbohydrate fermentation studies.
Cell wall analysis.
A cell wall sample was prepared using the short method described by Schleiffer and Kandler (38). The amino acid composition was determined at the Protein and Carbohydrate Structure Facility at the University of Michigan. The total amounts (picomoles) found in two separate experiments were averaged to give 49,000 picomoles of amino acid.
16S rRNA gene amplification and cloning of β-galactosidase genes.
Genomic DNA was obtained from isolate SB using the Puregene isolation kit (Gentra, Minneapolis, Minn.) with a modification of the gram-negative protocol of heating the sample at 80°C for 15 min. The 16S rRNA gene was amplified from chromosomal DNA by PCR with Ready-To-Go beads (Amersham, Piscataway, N.J.) and universal primers 8F and 1492R (31, 44). The product was sequenced at The Penn State Nucleic Acid Sequencing Facility with an ABI model 370 sequencer.
Fragments from 2 to 6 kb of partially digested isolate SB genomic DNA were purified (37), ligated into vector pΔα18 (42), and transformed into E. coli JM109 cells. The transformation mixture was plated onto Luria-Bertani medium (37) containing 100 μg of ampicillin/ml, 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) (Fisher, Pittsburgh, Pa.), and 0.01% of the chromogen X-Gal. Two colonies hydrolyzed X-Gal at 37°C, while four others became blue due to X-Gal hydrolysis within 5 h after a shift to 18°C. Restriction analysis showed that the inserts from the transformants that hydrolyzed X-Gal at 18°C appeared to be the same. Plasmid DNA from one of these transformants was purified, and the insert was sequenced at The Penn State Nucleic Acid Facility on an ABI 370 automated sequencer. The complete double-stranded sequence was obtained and used in the comparisons.
The lacZ gene was obtained from E. coli strain ATCC 23848 genomic DNA obtained using the Puregene isolation kit with a modification of the gram-negative protocol of heating the sample at 80°C for 15 min. The lacZ and lacY genes were amplified from chromosomal DNA using the enzyme Pfu DNA polymerase and the following primers: 5′-ATGATTACGGATTCACTGGCC-3′ and 5′-TTAAGCGACTTCATTCACCTG-3′. Amplified product was blunt-end cloned into pΔα18, and the lacZ gene was amplified using the enzyme Pfu DNA polymerase and the following primers: 5′-ATGATTACGGATTCACTGGCC-3′ and 5′-TTATTATTATTTTTGACACCA-3′. The amplified lacZ gene was then blunt-end ligated into pΔα18. Constructs were subjected to restriction digests to demonstrate that they yielded the patterns expected for the E. coli lacZ gene.
Phylogenetic analysis of 16S rRNA and β-galactosidase genes.
The SB isolate double-stranded 16S rRNA gene sequence was compared with those from the Ribosomal Database Project (http://rdp.cme.msu.edu/html) and the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) (21, 22) and aligned using the Clustal W program found in the BioEdit platform (version 5.0.6; Department of Microbiology, North Carolina State University [http://www.mbio.ncsu.edu/BioEdit/bioedit.html]). The alignment was used in maximum parsimony, maximum likelihood, and distance analyses utilizing the PAUP package (version 4.0b10; School of Computational Sciences and Informational Technology, Florida State University [http://paup.csit.fsu.edu]). The sequence data were analyzed using the maximum parsimony method (heuristic search), the maximum likelihood method (with a transition/transversion ratio determined from the data matrix), and the distance method (neighbor-joining algorithm and Jukes-Cantor model), with 10,000 bootstrap replicates being performed for this method. The initial distance analysis used the neighbor-joining algorithm and an uncorrected p distance measure using the PAUP program. The results with distance trees were compared using the Jukes-Cantor, F81, F84, Kimura two-parameter, Kimura three-parameter, Tamura-Nei, Tajima-Nei, HKY85, and the general time-reversible models with equal and unequal rates (gamma parameters with shapes of 0.5, 1.0, 2.0, 3.0, 4.0, and 5.0) for variable sites. Trees generated by all three methods were congruent. A distance matrix was generated using the same alignment by the PAUP program, using the Jukes-Cantor, F81, F84, Kimura two-parameter, Kimura three-parameter, Tamura-Nei, Tajima-Nei, HKY85, and the general time-reversible models with equal rates for variable sites, and the matrices were similar.
The BioEdit alignment of the bgaS gene sequence was analyzed utilizing the PAUP package and methods described for the 16S rRNA genes. Trees generated by all methods were congruent, with minor variations in branch lengths when unequal rates for variable sites were allowed, and the distance matrix was generated by the PAUP program.
Enzyme purification.
The enzyme used for determining the N-terminal amino acid sequence and oligomeric state was purified from E. coli DH5α cells carrying a pET22b(+) construct of bgaS (method I). Cells were grown at 37°C for 5 h in Luria-Bertani containing ampicillin and then transferred to 18°C, induced with 1 mM IPTG (final concentration), and incubated an additional 16 h. The cells were harvested by centrifugation, resuspended in Z buffer without β-mercaptoethanol (designated as modified Z buffer and containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) (24), and disrupted by passage through a French pressure cell (18,000 lb/in2). Ammonium sulfate was added to 25% saturation, the precipitate was removed by centrifugation, ammonium sulfate was added to 60%, and the precipitate was collected. The resuspended precipitate was applied to an affinity column containing p-aminophenyl β-d-thiogalactopyranoside (Sigma). BgaS was eluted from the column with modified Z buffer containing 300 mM NaCl, and fractions with β-galactosidase activity were collected. Enzyme preparations were dialyzed and stored at 5°C in modified Z buffer.
The enzyme used for other biochemical characterizations was purified from E. coli MC1061 (DE3) cells containing a pET28a(+) vector (Stratagene Cloning Systems, La Jolla, Calif.) construct containing either the bgaS or lacZ gene inserted to create an N-terminal six-histidine fusion (H-BgaS or H-LacZ) with their protein (method II). An E. coli transformant containing either construct was grown and disrupted as described above. Following centrifugation, the supernatant was mixed with an equal volume of ice-cold column wash buffer (modified Z buffer; 300 mM NaCl, 5 mM imidazole; pH 7.0), and loaded onto a TALON SuperFlow column (ClonTech, Palo Alto, Calif.). The column was washed with eight column volumes of buffer, and protein was eluted with elution buffer (modified Z buffer; 300 mM NaCl, 150 mM imidazole). The eluted enzyme fractions containing either H-BgaS or H-LacZ were dialyzed against 3 liters of modified Z buffer and stored at 5°C.
Thrombin treatment.
To prevent heat inactivation while removing the His tag from H-BgaS, the enzyme was incubated with thrombin (Novagen, Madison, Wis.) at 4°C, rather than the recommended 20 or 37°C, for 48 h. The reaction was monitored by measuring the hydrolysis of the 48-kDa test protein into cleaved polypeptides by polyacrylamide gel electrophoresis. The His tag and thrombin were separated from the BgaS enzyme by passage through a G-20 Sephadex column. The resulting BgaS enzyme was used for kinetic studies.
N-terminal and oligomeric state determinations.
Fractions of the BgaS enzyme purified by method I were combined and subjected to electrophoresis on a sodium dodecyl sulfate-7.5% polyacrylamide gel. The single Coomassie blue-stained band was transferred to a polyvinylidene difluoride membrane, excised, and placed in a sterile microcentrifuge tube. The N-terminal sequence was determined at the Protein Sequencing Facility of the Hershey Medical Center of The Pennsylvania State University.
Sedimentation coefficients were obtained during ultracentrifugation for 1.5 h in an Optima XL-A analytic ultracentrifuge (Beckman), using an absorbance optical system consisting of a xenon flash lamp scanning at 280 nm. Each centrifugation was performed using a 1-mg/ml sample concentration of purified BgaS in a double-sector cell at 208,000 × g at either 4 or 25°C, scanning every 5 min at 25°C and every 2 min at 4°C. Scanning at each temperature resulted in a pattern that was indicative of a single species.
Biochemical characterization.
All protein concentrations were measured using the bicinchoninic acid method (46) with bovine serum albumin (Promega) as a standard. The thermodependency of enzyme activity was determined by incubating the enzyme in modified Z buffer containing 2.2 mM ONPG for 5 min at temperatures ranging from 0 to 40°C for BgaS and 0 to 65°C for E. coli LacZ. Reactions were stopped by the addition of 1 M Na2CO3, and hydrolysis of the nitrophenyl group was detected spectroscopically at 420 nm. Since the assay conditions can affect the ratio of active to heat-inactivated enzyme, especially at high temperatures, all conditions were carefully standardized and enzyme addition was used to initiate the reactions. The thermodependency of activity results were highly reproducible using these methods. The thermostability of BgaS was determined by incubating the enzyme at 15, 30, and 37°C, removing aliquots for up to 120 min. The enzyme was then immediately assayed for ONPG activity at 15°C in the same manner as the thermodependency of activity assays. The optimal pH values were determined by assaying with 2.2 mM ONPG for 5 min at 15°C in buffers ranging in pH from 4.0 to 10.0 in increments of 0.5 pH units. The buffers used were 0.1 M sodium acetate-acetic acid for pH 4.0 to 6.0, 0.1 M phosphate for pH 6.0 to 8.0, and 0.1 M potassium chloride-boric acid for pH 8.0 to 10.0.
Requirements for metal ions were tested by incubating the enzyme in 20 mM phosphate buffer containing 100 mM EDTA for 1.5 h at 0°C. The enzyme was then loaded onto a Sephadex G-25 (Sigma) column and eluted with 20 mM phosphate buffer. Fractions containing protein were pooled and assayed for activity in 20 mM phosphate buffer containing various concentrations of MgCl2, MnCl2, CaCl2, CoCl2, CuCl2, NaCl, and KCl.
Substrate specificity was tested by incubating the enzyme at 15°C for 5 min in modified Z buffer containing a 2.2 mM final concentration of various nitrophenyl substrates. Substrates tested were ONPG, p-nitrophenyl-β-d-galactoside (PNPG), o-nitrophenyl-β-d-fucopyranoside, p-nitrophenyl-β-d-mannoside, o-nitrophenyl-β-d-glucoside, p-nitrophenyl-β-d-xyloside, p-nitrophenyl-β-d-cellobioside, p-nitrophenyl-β-d-arabinoside, p-nitrophenyl-β-d-lactoside, p-nitrophenyl-β-d-galacturonide, p-nitrophenyl-β-d-glucuronide, and p-nitrophenyl-α-d-galactoside (Sigma).
Kinetic assays for BgaS were performed at 5, 10, 18, and 20°C in modified Z buffer with various concentrations of ONPG and at 5 and 18°C in modified Z buffer (plus hexokinase, glucose-6-phosphate dehydrogenase, and NADP) with various concentrations of lactose. Kinetic assays for LacZ were performed at 20°C with ONPG as the substrate. The Ki values were determined using various concentrations of the inhibitors d-galactose and d-galactal with ONPG as the substrate. For kinetics with ONPG, hydrolysis of the nitrophenyl group was detected at 420 nm. For kinetics with lactose, production of NADPH, created through the coupled assay of hexokinase and glucose-6-phosphate dehydrogenase (Sigma), was detected at 412 nm using a Genesys2 spectrophotometer (Spectronic Instruments, Inc., Rochester, N.Y.). Kinetic and Ki values were calculated by using the analysis program EnzymeKinetics (version 1.5; Trinity Software, Fort Pierce, Fla.) and verified using the Windows Non-Lin program (M. Johnson and D. Yphantis, University of Virginia, Charlottesville).
Cysteine titrations were performed by incubating native BgaS enzyme in titration buffer (150 mM phosphate buffer containing 2 mM dithiobisnitrobenzoic acid [DTNB]). Reactions were scanned in a Genesys2 spectrophotometer at 412 nm at 5-min intervals. The total number of cysteines was determined by unfolding the protein in titration buffer containing 8 M urea (final concentration). Reactions were then assayed in the same manner as the folded protein.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the 16S rRNA gene from the Arthrobacter isolate SB and bgaS are AY327445 and AY327444, respectively.
RESULTS
Isolate characterization.
Isolate SB was selected for further study because cells grew at low temperatures and hydrolyzed the chromogen X-Gal as an indicator of β-galactosidase activity. The isolate was a gram-positive, nonmotile, strict aerobe that was unable to ferment glucose, sucrose, or lactose. It grew at temperatures from 0 to 25°C but did not form colonies on TSA or R2A medium at 30°C. It was non-beta-hemolytic oxidase negative, and growth was inhibited by the addition of 5% NaCl to the medium. The SB cells had no vitamin requirements and were able to use lactose as a sole carbon source.
Microscopic examination indicated that the cells were rod shaped during exponential growth and coccoidal during stationary phase, a property associated with members of the Arthrobacter genus. Another feature found in Arthrobacter species is the presence of lysine, rather than meso-diaminopimelic acid, as the diamino acid found in the peptidoglycan. An analysis of the amino acids in isolate SB cell walls demonstrated that lysine was present at 10% of the total amino acids. The percentages for other important amino acids were as follows: alanine, 35.2; threonine, 12.0; serine, 2.9; glycine, 5.6; glutamic acid, 11.8. Neither diaminopimelic acid nor ornithine was detected.
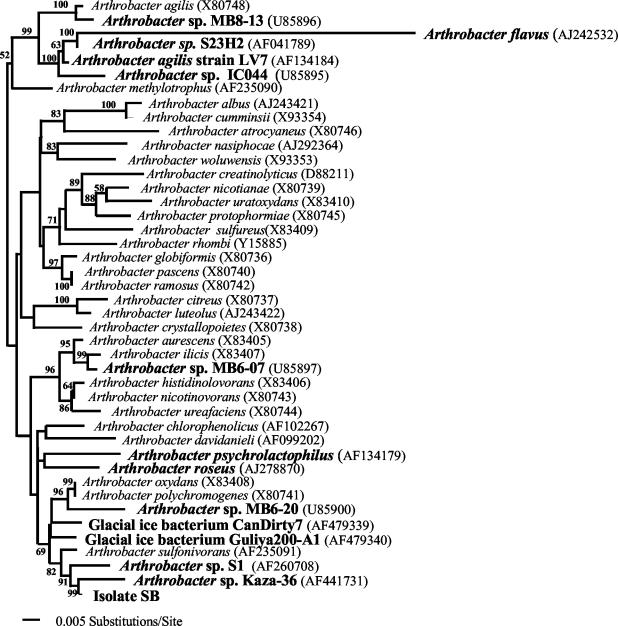
To examine the phylogenetic position of our isolate, we amplified and sequenced the 16S rRNA gene (Fig. 1). Analysis of the sequence showed that it clustered with other organisms isolated from cold environments: Siberian soil (Arthrobacter sp. strain S1) and a cold desert in the Spiti Valley (Arthrobacter sp. strain Kaza-36). These three isolates formed a well-supported cluster with Arthrobacter sulfonivorans, which was recently isolated from root soil (3). A. sulfonivorans grows at 5°C and has optimal growth at 20 to 25°C, so it would also be considered a psychrophile according to the definition of Neidhardt et al. (28). The Jukes-Cantor evolutionary distance matrix indicated that isolate SB could be a strain of A. sulfonivorans (distance of 1.13%). The evolutionary distances were greater between isolate SB and Arthrobacter nicotinovorans, Arthrobacter oxydans, and Arthrobacter polychromogenes (1.99, 1.40, and 1.48%, respectively). All other distances from characterized strains were found to be larger than 2%.
FIG. 1.
Rectangular phylogram derived from a maximum parsimony, maximum likelihood, and distance methods search of 16S rRNA gene sequence data with 10,000 bootstrap replicates for 43 taxa, each comprising 1,613 nucleotide positions. The tree was rooted using the sequences of Arthrobacter agilis, Arthrobacter flavus, Arthrobacter methylotrophus, Arthrobacter sp. IC044, Arthrobacter sp. MB8-13, Arthrobacter sp. S23H2, and A. agilis strain LV7 as the outgroup. Bolded organism names are known psychrophiles. Accession numbers for the 16S rRNA gene sequences of all organisms are in parentheses.
Cloning of β-galactosidase genes.
β-Galactosidase assays of Arthrobacter strain SB cell extracts at different temperatures showed a bimodal curve with one peak near 15°C and another around 35°C, suggesting the presence of more than one enzyme (data not shown). To examine this further, a chromosomal library was prepared, and ampicillin-resistant transformants were selected and screened for the ability to hydrolyze X-Gal. Among the approximately 10,000 transformants, two hydrolyzed X-Gal during incubation at 37°C. After plates were transferred to 18°C, an additional four colonies began hydrolyzing X-Gal. Two transformants with each phenotype were purified for initial characterization. The results showed the presence of two different activities, one with an optimum at 15°C and the other with an optimum above 35°C. We focused on the ones with the lower temperature optima because we are interested in cold-active enzymes. Restriction digests of these inserts showed that they were derived from the same fragment. Sequence data from the shortest construct contained an open reading frame encoding a full-length β-galactosidase gene, bgaS.
Gene and N-terminal amino acid sequences.
The sequence of bgaS was analyzed and found to encode a 1,053-amino-acid protein with a predicted mass around 114 kDa. The fragment had a high G+C content (67 mol%), which is typical for Arthrobacter species. In order to examine the BgaS enzyme, it was purified using method I in Materials and Methods. A single protein band migrating near 116 kDa was observed following sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie blue. This band was excised and used to determine the N-terminal amino acid sequence of AQFTASPPA. This corresponded to the predicted nucleotide sequence except for the absence of a methionine at the beginning, which was apparently cleaved in the E. coli host strain.
Phylogenetic relationship among the family 2 enzymes.
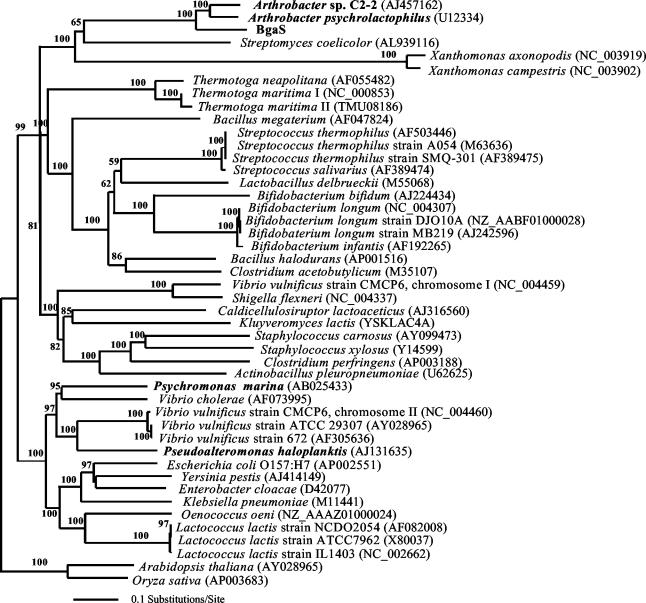
A comparison of the bgaS sequence with those from the NCBI database showed that it was most closely related to two lacZ-like genes, one from an Antarctic Arthrobacter sp. C2-2 (71% similar) and the other from Arthrobacter psychrolactophilus (66% similar) (Fig. 2). Although biochemical data are not available for the β-galactosidase from Arthrobacter sp. C2-2, it is interesting that the closely related enzyme from A. psychrolactophilus has a temperature optimum around 40°C and is quite stable at 37°C (42).
FIG. 2.
Rectangular phylogram derived from maximum parsimony, maximum likelihood, and distance methods search of family 2 β-galactosidase gene sequences, with 10,000 bootstrap replicates for 46 taxa each containing 4,449 nucleotide positions. The tree was rooted using the sequences of Arabidopsis thaliana and Oryza sativa as the outgroup. Gene sequences from organisms shown in bold are known or hypothesized to encode cold-active β-galactosidases. Accession numbers for all the β-galactosidase gene sequences are in parentheses.
Alignment with protein sequences from the family 2 β-galactosidases (data not shown) showed that the two glutamic acid residues (461 and 537) involved in catalysis in the E. coli LacZ enzyme were conserved in BgaS. The alignments also showed that the conserved region 60 residues toward the N terminus from the general acid-base catalyst, typical of members of the family 2 glycosyl hydrolases, was also conserved in BgaS. The combination of the phylogenetic analysis and conservation of important residues established that BgaS is a member of the family 2 glycosyl hydrolases.
Determination of pH range and substrate specificity.
The initial assays followed procedures used for the E. coli LacZ enzyme, so we examined these conditions to determine if they were optimal for the BgaS enzyme. The activity was measured in a variety of buffers with pH values from 4.0 to 10.0, and activity was found from pH 6.0 to 9.5 with the greatest activity at pH 7.0 and 50% activity around pH 6.7 and 8.5 (data not shown). This optimum was similar to the pH 7.2 optimum found for the E. coli LacZ enzyme (41). The substrate specificity was determined by assaying with several chromogenic substrates. The enzyme had about 84% of the ONPG activity when PNPG was the substrate, but it showed less than 2% of the ONPG activity with any of the other substrates tested (Table 1).
TABLE 1.
Percent activity of purified H-BgaS with various nitrophenyl-derived chromogenic substrates
| Substrate | Activitya (%) |
|---|---|
| o-Nitrophenyl-β-d-galactoside | 100 |
| p-Nitrophenyl-β-d-galactoside | 84.1 |
| o-Nitrophenyl-β-d-fucopyranoside | <2 |
| p-Nitrophenyl-β-d-cellobioside | <2 |
| o-Nitrophenyl-β-d-glucoside | <2 |
| p-Nitrophenyl-α-d-galactoside | <2 |
| p-Nitrophenyl-β-d-mannoside | <2 |
| p-Nitrophenyl-β-d-xyloside | <2 |
| p-Nitrophenyl-β-d-arabinoside | <2 |
| p-Nitrophenyl-β-d-lactoside | <2 |
| p-Nitrophenyl-β-d-galacturonide | <2 |
| p-Nitrophenyl-β-d-glucuronide | <2 |
The values are relative to the 100% value observed with ONPG (27.0 U/mg).
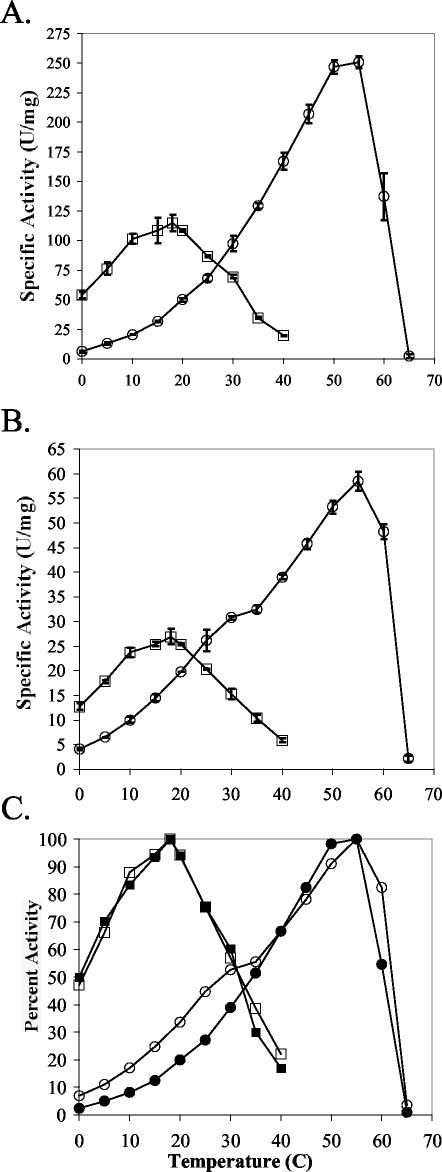
Initial characterization.
The thermodependency of activity and the thermal lability of BgaS activity were examined using enzyme purified by method I. The enzyme had a temperature optimum between 15 and 20°C (Fig. 3A) and lost 90% of its activity within 10 min at 35°C. These results showed that BgaS was extremely cold active and heat labile and was of substantial interest for further biochemical characterization. We found, however, that about 90% of the enzyme produced from the pET22b(+) construct formed inclusion bodies and that the purification procedure yielded enzyme preparations with varying specific activities. Attempts to resolubilize the inclusion bodies were unsuccessful, as were experiments using different hosts, the arabinose-inducible vector (pBAD), and varying IPTG concentrations. Thus, a vector carrying the bgaS gene and encoding an N-terminal His tag was constructed which allowed a more rapid purification and reproducible yield of enzyme activity. However, we noted that the overall specific activity was reduced (25 U/mg versus 100 U/mg at 18°C). Because we wanted to compare the BgaS characteristics with those found for the E. coli enzyme, we prepared a similar construct with the lacZ gene as described in Materials and Methods. Interestingly, the LacZ specific activities also decreased by fourfold (60 U/mg versus 250 U/mg at 55°C).
FIG. 3.
Influence of temperature on the β-galactosidase activities of BgaS (squares) and LacZ (circles) (A), H-BgaS (squares) and H-LacZ (circles) (B), and the relative activities of BgaS (filled squares), H-BgaS (open squares), LacZ (filled circles), and H-LacZ (open circles) (C). The specific activity corresponding to 100% activity was 115, 26.9, 250.8, and 58.5 U/mg for BgaS, H-BgaS, LacZ, and H-LacZ, respectively. One unit is defined as the amount of enzyme needed to release 1 μmol of o-nitrophenyl/min.
Comparison of BgaS and LacZ properties.
The properties of the BgaS and LacZ enzymes were compared to their His-tagged BgaS (H-BgaS) and LacZ (H-LacZ) counterparts to determine whether other characteristics were altered. The H-BgaS enzyme retained the pH optimum and substrate specificity found for the enzyme purified by method I (data not shown). The thermodependency of enzyme activity was also measured for both the H-BgaS and the E. coli H-LacZ enzymes (Fig. 3A and B). In order to determine whether the His-tagged enzymes had the same thermal dependency of activity profile as their nontagged counterparts, the relative activities of all four enzymes were compared (Fig. 3C). These results showed that although the His-tagged versions had reduced activity, the enzymes maintained their original thermal properties. The comparison of the BgaS and LacZ enzymes also demonstrated that the BgaS enzymes were more active at temperatures at or below 20°C than their LacZ counterparts (Fig. 3A and B).
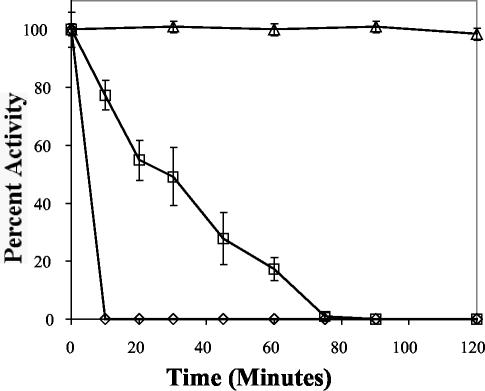
Our initial results with purified enzyme and those from the thermodependency of activity studies suggested that the BgaS enzyme was extremely heat labile. Incubation of the H-BgaS enzyme at different temperatures substantiated these results, as both forms of the enzyme were inactivated within 10 min at 37°C and within 1 h at 30°C but remained stable at 15°C for at least 2 h (Fig. 4). This loss of activity was not reversed by continued incubation at 15 or 4°C for 24 h. Thus, unlike the report of a phosphoglycerate kinase where the catalytic activity was reduced while the thermostability was increased by the introduction of a His tag (2), our results showed that the thermodependency of activity and thermostability remained the same with the His-tagged enzymes even though the activity decreased fourfold (Fig. 3 and 4).
FIG. 4.
Thermostability of purified H-BgaS at 15°C (triangles), 30°C (squares), and 37°C (diamonds). Specific activity corresponding to 100% activity was 26.9 U/mg.
Metal requirements.
To examine possible metal requirements for H-BgaS, it was treated with various concentrations of EDTA. Unlike the Pseudoalteromonas haloplanktis β-galactosidase, which was inhibited by 5 mM EDTA (13), no activity loss was detected until the EDTA concentration reached 100 mM, at which the enzyme was inactivated after a 1.5-h treatment. Activity of H-BgaS was relatively unchanged by the addition of Co2+, Cu2+, Ca2+, or Na+ (2, 2, 1.5, or 5% increase, respectively). However, activity was stimulated by the addition of Mg2+ (75%), Mn2+ (13%), or K+ (12%). The metal requirements were also examined by dialyzing the enzyme against morpholinepropanesulfonic acid (MOPS) buffer containing various metals. H-BgaS lost all activity after overnight dialysis against 100 mM MOPS but retained activity when Mg2+ and K+ (78%), Mg2+(60%), Mn2+ (20%), or K+ (10%) was added to the dialysis buffer. These experiments confirmed the requirements for Mg2+, Mn2+, or K+.
Kinetic and inhibitor studies.
The results of the thermodependency-of-activity studies showed that the specific activities of the BgaS enzyme were greater than those of the LacZ enzyme at or below 20°C (Fig. 3A and B). To further examine the activity of H-BgaS, the Km and Vmax values were determined at four temperatures with ONPG as the substrate (Table 2). The H-BgaS enzyme had a greater catalytic activity at 20°C than the H-LacZ enzyme (184 versus 166 s−1 mM−1) and had substantial activity at 5°C, where the activity of the H-LacZ enzyme was severely reduced (Fig. 3B). In order to determine if the His tag had the same effect on kinetic values that it had on specific activity, we cleaved the His tag from the BgaS and LacZ enzymes by using thrombin and determined the kinetic constants at 18 and 20°C. The fivefold kcat increase and small decrease in Km values led to a 10-fold increase in catalytic efficiency at 20°C for BgaS, and it retained a higher catalytic activity than the thrombin-treated LacZ enzyme at 20°C. The values for the H-BgaS enzyme at different temperatures were used to construct an Arrhenius plot, and an energy of activation of 20.3 kJ/mol was calculated (data not shown).
TABLE 2.
Kinetic parameters for purified BgaS, H-BgaS, LacZ, and H-LacZ at various temperatures, using ONPG as the substrate
| Enzyme | Temp (°C) | Vmax (U/mg) | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| H-BgaS | 5 | 22.7 ± 0.4 | 0.7 ± 0.03 | 43.0 ± 0.8 | 61.4 |
| H-BgaS | 10 | 30.2 ± 5.2 | 0.7 ± 0.05 | 57.2 ± 9.5 | 81.7 |
| H-BgaS | 18 | 52.7 ± 4.3 | 0.8 ± 0.05 | 100 ± 8.0 | 116.3 |
| H-BgaS | 20 | 58.1 ± 5.6 | 0.6 ± 0.06 | 1,10.1 ± 10.6 | 183.6 |
| H-LacZ | 20 | 16.3 ± 0.1 | 0.2 ± 0.04 | 31.7 ± 0.3 | 166.0 |
| BgaSa | 20 | 76.9 | 0.3 | 583.5 | 1,823 |
| LacZ | 20 | 52.2 | 0.2 | 219.6 | 1,098 |
Incubated with thrombin at 4°C to remove the N-terminal His tag.
The kinetic constants for H-BgaS were also determined at two temperatures (5 and 18°C) with lactose as the substrate (Table 3). H-BgaS had a catalytic efficiency of 0.27 and 0.53 at 5 and 18°C, respectively. Both of these values are higher than those reported for a LacZ enzyme (0.15) without a His tag at 20°C (13).
TABLE 3.
Kinetic parameters for purified H-BgaS, using lactose as the substrate
| Temp (°C) | Vmax (U/mg) | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| 5 | 2.5 ± 1.1 | 19.4 ± 2.0 | 4.7 | 0.27 |
| 18 | 28 ± 1.5 | 11.5 ± 1.6 | 5.2 | 0.53 |
The Ki values for d-galactose (substrate analogue) and d-galactal (transition-state analogue) were determined using ONPG as the substrate. Values calculated for d-galactose (12 mM) were similar to those reported for LacZ (10 to 20 mM) (34, 35), whereas values calculated for d-galactal (0.218 mM) were significantly higher than those reported for LacZ (0.016 mM) (23). Dixon plots generated for both inhibitors showed a pattern of competitive inhibition (data not shown).
Oligomeric state and heat lability.
We used analytic ultracentrifugation to examine the oligomeric state of the BgaS enzyme to determine whether the active form was a multimer. The results gave a corrected sedimentation coefficient of 20 × 10−13 s at 4°C, which corresponded to a molecular mass of 463,000 g/mol, consistent with the active enzyme being a homotetramer. We also investigated the oligomeric state of enzyme above its thermal optimum to determine whether the loss of activity at moderate temperatures (Fig. 3 and 4) could be correlated with a change in subunit association. The sedimentation coefficient at 25°C was 4 × 10−13 s, which corresponded to a molecular mass of 112,000 g/mol. The ratio (MW4°C/MW25°C) of the two calculated masses was 4:1, which is consistent with the BgaS homotetramer found at 4°C dissociating into inactive monomers at 25°C. Although monomers of the LacZ enzyme have also been reported to be inactive, the tetramer is stable at 25°C and its monomers can reassociate to form active enzyme (29). In contrast, the BgaS enzyme did not regain activity after dissociating into monomers, even when cooled and incubated at 4°C for up to 24 h. These findings suggest that an inherent structural feature of the BgaS enzyme, not found in LacZ, causes its irreversible dissociation into inactive monomers at low temperatures.
Comparison with other enzymes.
Enzyme analyses often include amino acid composition comparisons, because some hypotheses regarding the thermal adaptation of proteins involve the frequency of particular bonds and amino acid side chains. Common suggestions for cold-active enzymes include fewer prolines or arginines, a lower Arg/(Arg plus Lys) ratio, a decrease in hydrophobic residues coupled with an increase in polar residues, and a decrease in the number of disulfide bonds (6, 30, 32, 36). These differences are often observed during pairwise comparisons of enzymes with different thermal properties. In an initial comparison between the cold-active BgaS and the representative mesophilic LacZ enzyme (Table 4), several amino acids did show differences; however, with the exception of the decrease in cysteine composition, these trends did not fit patterns mentioned for cold-active proteins. To further explore the difference in cysteine residues, we constructed a model of BgaS by using the known coordinates for the LacZ enzyme. This examination showed that the cysteines were dispersed throughout all five domains in the LacZ structure, whereas the cysteines clustered in and near the active site in our BgaS model but were not close enough to form disulfide bonds (data not shown). Furthermore, the reduction in cysteine residues may not be significant because it has been reported that the LacZ enzyme does not contain disulfide bonds (29), and titration experiments with DTNB with the folded BgaS enzyme suggest that at least three of the four cysteines are not reduced (data not shown).
TABLE 4.
Amino acid compositions from family 2 glycosyl hydrolases
| Residue(s) | No. of residues
|
||
|---|---|---|---|
| E. coli K-12 LacZ | Arthrobacter SB isolate | A. psychrolactophilus | |
| Alanine | 77 | 119 | 107 |
| Arginine | 66 | 81 | 74 |
| Asparagine | 47 | 31 | 31 |
| Aspartic acid | 64 | 76 | 77 |
| Cysteine | 16 | 4 | 4 |
| Glutamine | 58 | 23 | 23 |
| Glutamic acid | 62 | 63 | 61 |
| Glycine | 71 | 109 | 88 |
| Histidine | 34 | 27 | 28 |
| Isoleucine | 39 | 32 | 33 |
| Leucine | 96 | 76 | 95 |
| Lysine | 20 | 9 | 13 |
| Methionine | 23 | 15 | 9 |
| Phenylalanine | 38 | 35 | 31 |
| Proline | 62 | 70 | 74 |
| Serine | 60 | 78 | 75 |
| Threonine | 55 | 59 | 54 |
| Tryptophan | 39 | 32 | 33 |
| Tyrosine | 31 | 25 | 25 |
| Valine | 64 | 89 | 81 |
| Total residues | 1,025 | 1,053 | 1,017 |
| Arg/(Arg+Lys) ratio | 0.8 | 0.9 | 0.8 |
| Hydrophobic residues (AGILMFPWV) | 509 | 577 | 551 |
| Polar residues (RNDCEQHKSTY) | 513 | 476 | 465 |
Based on our previous examination of possible structural features contributing to the thermostability of glycosyl hydrolases (32), we proposed that pairwise comparisons of enzymes could lead to misleading conclusions because the differences may be due to evolutionary changes that are independent of any inherent thermal properties (40). In this study, we had the advantage of a β-galactosidase with a temperature optimum around 40°C from a related psychrophilic Arthrobacter species. Thus, comparison of the more cold-active BgaS enzyme with that from A. psychrolactophilus would be expected to show composition changes associated with a transition from a cold-active to a more mesophilic enzyme (Table 4). However, the results were not consistent with the suggested trends, except for a decrease in the number of prolines. In addition, we averaged the amino acid compositions reported for the bacterial family 2 glycosyl hydrolases from psychrophiles, mesophiles, and thermophiles used in determining the phylogenetic relationships (Fig. 2). Although the total number of glycines increased for the psychrophiles, other trends were not observed (data not shown), suggesting that changes conferring thermal adaptation are more subtle and are not reflected in an overall amino acid composition.
DISCUSSION
We have isolated a psychrophilic strain, SB, and characterized its β-galactosidase to enhance the understanding of cold-active enzymes. The phylogenetic analysis of the 16S ribosomal DNA sequence from this isolate placed it in the genus Arthrobacter, which is consistent with its growth and morphological characteristics. The sequence of a gene encoding a cold-active β-galactosidase cloned from the isolate predicts that it produces a 1,053-amino-acid protein that is slightly larger than E. coli LacZ (47). The sequence of this bgaS gene shows it is most closely related to β-galactosidases from two other Arthrobacter species, one of which was also from Antarctica, but the other was from Pennsylvania farmland.
The initial characterization of purified BgaS showed that it had an exceptionally low temperature optimum near 18°C and was heat labile above 20°C. These results raised interesting questions regarding its kinetic properties, which required additional purified enzyme. In order to obtain preparations with reproducible specific activities, we created an N-terminal His tag version of BgaS and, subsequently, the LacZ enzyme for comparison. This allowed rapid and reproducible purification of both proteins; however, the N-terminal His tag caused a fourfold decrease in the specific activities (Fig. 3). This effect was surprising for the LacZ protein, because its gene has been used routinely as a reporter fusion protein and the N-terminal alpha region of the enzyme can be deleted and added separately in alpha complementation to produce an active, though less stable, enzyme (8, 47). Thus, it would seem that the N-terminal region of LacZ would be relatively immune to the addition of the His tag sequence.
Because the overall activity of the H-BgaS enzyme was reduced, we reexamined the pH optimum, substrate specificity, thermostability, and thermodependency properties to make certain they were the same as those of the nontagged enzyme before determining the kinetic values. Biochemical testing confirmed that these properties remained the same for the two enzymes. The thermal optimum of H-BgaS was 18°C, one of the lowest on record, and it retained a high level (50%) of its activity at 0°C. To date, most reported β-galactosidases from other psychrophilic microorganisms have higher thermal optima that are generally over 40°C (5, 7, 9, 13, 39). One β-galactosidase from P. haloplanktis had an optimum of about 45°C (13), whereas one from a closely related Pseudoalteromonas species had a reported optimum of 26°C, with about 28% of its maximal activity remaining at 5°C (7).
Of particular interest was the high activity of the BgaS enzyme below 25°C. When adding equal amounts of purified protein, BgaS had 2.1 and 5.0 times more activity than the LacZ enzyme at 20 and 10°C, respectively (Fig. 3A). We found kcat values for the BgaS and LacZ enzymes to be 584 and 220 s−1, respectively, at 20°C with ONPG as a substrate (Table 2). The report for the P. haloplanktis enzyme showed a kcat value of 203 s−1 at 20°C, and it was noted that the Km values sharply increased above 15°C with ONPG (13). Both the H-BgaS and the BgaS enzymes had higher catalytic efficiencies than their respective LacZ counterpart at 20°C (Table 2). In addition, we have reported on kinetic studies with lactose as the substrate at temperatures below 25°C which demonstrate a higher catalytic efficiency for the BgaS enzyme at 5°C than values reported for E. coli LacZ at 25°C (13). Thus, the low temperature optimum, activity remaining at 0°C, and kcat values make BgaS a unique cold-active β-galactosidase among those studied and an ideal candidate for the commercial removal of lactose from milk where activity at refrigerated temperatures is critical.
Another notable feature of the BgaS enzyme was its heat lability at temperatures over 20°C. Reduced thermostability is a property often associated with cold-active enzymes (6, 36, 49); however, the reasons for the loss of activity are often unknown. To examine the possibility that the BgaS enzyme could be dissociating into inactive monomers at higher temperatures, we examined its oligomeric state by ultracentrifugation. The results showed that the BgaS enzyme is indeed a tetramer at 4°C, where it is active, and has a sedimentation coefficient consistent with it becoming a monomer during ultracentrifugation at 25°C. Future structural studies of the BgaS enzyme are planned that could provide important insights into the contacts and forces influencing the stability and reassociation of β-galactosidase tetramers.
Researchers are interested in determining the structural features that dictate the thermal properties of enzymes so that proteins can be engineered with desired properties. In attempts to discern key differences, investigators have compared the structures of enzymes with different temperature optima, generally mesophilic and thermophilic proteins. Unfortunately, no rules have emerged predicting precise changes leading to specific thermal traits. Our results published here and in previous findings (32) show that many proposed trends, including overall amino acid composition changes, disappear when using averages for several proteins or comparing evolutionarily related enzymes. Evidence from random mutagenesis experiments (25, 33, 48) suggests that small regional changes not only can alter an enzyme's thermostability but also broaden its temperature range of activity. In addition, our investigators have found that two amino acid changes in a family 42 β-galactosidase (BgaB) expand its temperature range by 20°C (N. Panasik, unpublished data). These data all point to the notion that amino acid sequence and gross structural comparisons will not lead to answers about enzyme adaptation to different thermal pressures (2, 25, 48-50). Instead, there is growing evidence (50) that these answers will be found in subtle, synergistic, and cooperative intramolecular interactions. In order to examine this for the especially cold-active β-galactosidase described here, we are currently using random mutagenesis of the bgaS gene to obtain enzymes with altered thermal properties. Results from these mutation studies will facilitate more direct tests regarding the need for enzyme flexibility and specific amino acids to sustain activity at low temperatures.
Acknowledgments
We thank members of our research group for helpful discussions and suggestions and L. E. Casida and R. Benoit for collection and storage of the Antarctic soil samples. We also thank Frank Ruch at Protein Scientific, Inc., for helpful discussions concerning the commercial applications of cold-active β-galactosidases and Robert Simpson for his help with the analytic ultracentrifugation experiments.
This work was supported by Department of Energy grant DE-FG02-93ER20117. James Coker was partially supported by NSF Research Training Grant DBI-9602232. Peter Sheridan received partial funding from an Alfred P. Sloan Foundation Fellowship in Molecular Evolution from the National Science Foundation, The Penn State Astrobiology Center NASA-Ames cooperative agreement NCC2-1057, and grant NSF/IGERT DGE-9972759 for the Biogeochemical Research Initiative for Education.
REFERENCES
- 1.Beckwith, J. R., and D. Zipser. 1970. The lactose operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bentahir, M., G. Feller, M. Aittaleb, J. Lamotte-Brasseur, T. Himri, J.-P. Chessa, and C. Gerday. 2000. Structural, kinetic, and calorimetric characterization of the cold-active phosphoglycerate kinase from the Antarctic Pseudomonas sp. TACII18. J. Biol. Chem. 275:11147-11153. [DOI] [PubMed] [Google Scholar]
- 3.Borodina, E., D. P. Kelly, P. Schumann, F. A. Rainey, N. L. Ward-Rainey, and A. P. Wood. 2002. Enzymes of dimethylsulfone metabolism and the phylogenetic characterization of the facultative methylotrophs Arthrobacter sulfonivorans sp. nov., Arthrobacter methylotrophus sp. nov., and Hyphomicrobium sulfonivorans sp. nov. Arch. Microbiol. 177:173-183. [DOI] [PubMed] [Google Scholar]
- 4.Burchhardt, G., and H. Bahl. 1991. Cloning and analysis of the β-galactosidase-encoding gene from Clostridium thermosulfurogenes EM1. Gene 106:13-19. [DOI] [PubMed] [Google Scholar]
- 5.Coombs, J. M., and J. E. Brenchley. 1999. Biochemical and phylogenetic analyses of a cold-active β-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl. Environ. Microbiol. 65:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Amico, S., P. Claverie, T. Collins, D. Georlette, E. Gratia, A. Hoyoux, M.-A. Meuwis, G. Feller, and C. Gerday. 2002. Molecular basis of cold adaptation. Phil. Trans. R. Soc. London 357:917-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes, S., B. Geueke, O. Delgado, J. Coleman, and R. Hatti-Kaul. 2002. β-Galactosidase from a cold-adapted bacterium: purification, characterization and application for lactose hydrolysis. Appl. Microbiol. Biotechnol. 58:313-321. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher, C. N., and R. E. Huber. 1999. Stabilities of uncomplemented and complemented M15 beta-galactosidase (Escherichia coli) and the relationship to alpha-complementation. Biochem. Cell Biol. 77:109-118. [DOI] [PubMed] [Google Scholar]
- 9.Gutshall, K. R., D. E. Trimbur, J. J. Kasmir, and J. E. Brenchley. 1995. Analysis of a novel gene and β-galactosidase isozyme from a psychrotrophic Arthrobacter isolate. J. Bacteriol. 177:1981-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrissat, B., and G. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637-644. [DOI] [PubMed] [Google Scholar]
- 13.Hoyoux, A., I. Jennes, P. Dubois, S. Genicot, F. Dubail, J. M. Francois, E. Baise, G. Feller, and C. Gerday. 2001. Cold-adapted β-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microbiol. 67:1529-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, R. E., N. J. Roth, and H. Bahl. 1996. Quaternary structure, Mg2+ interactions, and some kinetic properties of the β-galactosidase from Thermoanerobacterium thermosulfurigenes EM1. J. Protein Chem. 15:621-629. [DOI] [PubMed] [Google Scholar]
- 15.Hung, M.-N., Z. Xia, N.-T. Hu, and B. H. Lee. 2001. Molecular and biochemical analysis of two β-galactosidases from Bifidobacterium infantis HL96. Appl. Environ. Microbiol. 67:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson, R. H., X.-J. Zhang, R. F. DuBose, and B. W. Matthews. 1994. Three-dimensional structure of β-galactosidase from E. coli. Nature 369:761-766. [DOI] [PubMed] [Google Scholar]
- 17.Juers, D. H., T. D. Heightman, A. Vasella, J. D. McCarter, L. Mackenzie, S. G. Withers, and B. W. Matthews. 2001. A structural view of the action of Escherichia coli (lacZ) β-galactosidase. Biochemistry 40:14781-14794. [DOI] [PubMed] [Google Scholar]
- 18.Juers, D. H., R. H. Jacobson, D. Wigley, X.-J. Zhang, R. E. Huber, D. E. Tronrun, and B. W. Matthew. 2000. High resolution refinement of β-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for α-complementation. Protein Sci. 9:1685-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krieg, N. R. 1981. Enrichment and isolation, p. 112-142. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 20.Loveland-Curtze, J., P. P. Sheridan, K. R. Gutshall, and J. E. Brenchley. 1999. Biochemical and phylogenetic analyses of psychrophilic isolates belonging to the Arthrobacter subgroup and description of Arthrobacter psychrolactophilus sp. nov. Arch. Microbiol. 171:355-363. [DOI] [PubMed] [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Liburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosome Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak, B. L., N. Larsen, M. L. McCaughey, R. Overbeek, G. J. Olsen, K. Fogel, J. Blandy, and C. R. Woese. 1994. The ribosomal database project. Nucleic Acids Res. 22:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Bilbao, M., R. E. Holdsworth, L. A. Edwards, and R. E. Huber. 1991. A highly reactive β-galactosidase (Escherichia coli) resulting from a substitution of an aspartic acid for Gly-794. J. Biol. Chem. 266:4979-4986. [PubMed] [Google Scholar]
- 24.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Miyazaki, K., and F. H. Arnold. 1999. Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J. Mol. Evol. 49:716-720. [DOI] [PubMed] [Google Scholar]
- 26.Moller, P. L., F. Jorgensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular β-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakao, M., M. Harada, Y. Kodama, T. Nakayama, Y. Shibano, and T. Amachi. 1994. Purification and characterization of a thermostable β-galactosidase with high transgalactosylation activity from Saccharopolyspora rectivirgula. Appl. Microbiol. Biotechnol. 40:657-663. [Google Scholar]
- 28.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell, p. 506. Sinauer Associates Inc., Sunderland, Mass.
- 29.Nichtl, A., J. Buchner, R. Jaenicke, R. Rudolph, and T. Scheibel. 1998. Folding and association of β-galactosidase. J. Mol. Biol. 282:1083-1091. [DOI] [PubMed] [Google Scholar]
- 30.Oikawa, T., K. Yamanaka, T. Kazuoka, N. Kazuika, and K. Soda. 2001. Psychrophilic valine dehydrogenase of the Antarctic psychrophile, Cytophaga sp. KUC-1—purification, molecular characterization and expression. Eur. J. Biochem. 268:4375-4383. [DOI] [PubMed] [Google Scholar]
- 31.Pace, N. R., D. L. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 32.Panasik, N., Jr., J. E. Brenchley, and G. K. Farber. 2000. Distribution of the structural features contributing to thermostability in mesophilic (α/β)8 barrel glycosyl hydrolases. Biochim. Biophys. Acta 1543:189-201. [DOI] [PubMed] [Google Scholar]
- 33.Perl, D., U. Mueller, U. Heinemann, and F. X. Schmid. 2000. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 39:1251-1255. [DOI] [PubMed] [Google Scholar]
- 34.Roth, N. J., and R. E. Huber. 1996. The β-galactosidase (Escherichia coli) reaction is partly facilitated by interactions of His540 with the C6 hydroxyl of galactose. J. Biol. Chem. 271:14296-14301. [DOI] [PubMed] [Google Scholar]
- 35.Roth, N. J., B. Rob, and R. E. Huber. 1998. His-357 of β-galactosidase (Escherichia coli) interacts with the C3 hydroxyl in the transition state and helps to mediate catalysis. Biochemistry 37:10099-10107. [DOI] [PubMed] [Google Scholar]
- 36.Russell, N. J. 2000. Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extremophiles 4:83-90. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan, P. P., and J. E. Brenchley. 2000. Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic antarctic Planococcus isolate. Appl. Environ. Microbiol. 66:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheridan, P. P., N. Panasik, Jr., J. M. Coombs, and J. E. Brenchley. 2000. Approaches for deciphering the structural basis of low temperature enzyme activity. Biochim. Biophys. Acta 1543:417-433. [DOI] [PubMed] [Google Scholar]
- 41.Tenu, J. P., O. M. Viratelle, J. Garnier, and J. Yon. 1971. pH dependence of the activity of beta-galactosidase from Escherichia coli. Eur. J. Biochem. 20:363-370. [DOI] [PubMed] [Google Scholar]
- 42.Trimbur, D. E., K. R. Gutshall, P. Prema, and J. E. Brenchley. 1994. Characterization of a psychrotrophic Arthrobacter gene and its cold-active β-galactosidase. Appl. Environ. Microbiol. 60:4544-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Laere, K. M., T. Abee, H. A. Schols, G. Beldman, and A. G. Voragen. 2000. Characterization of a novel beta-galactosidase from Bifidobacterium adolescentis DSM 20083 active towards transgalactooligosaccharides. Appl. Environ. Microbiol. 66:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, A., and D. R. Rose. 1997. Mechanism of catalysis by retaining β-glycosyl hydrolyases. Curr. Opin. Struct. Biol. 7:645-651. [DOI] [PubMed] [Google Scholar]
- 46.Wiehelman, K., R. Braun, and J. Fitzpatrick. 1988. Investigation of the bicinchoninic acid protein assay: identification of the groups responsible for color formation. Anal. Biochem. 175:231-237. [DOI] [PubMed] [Google Scholar]
- 47.Zabin, I., and A. V. Fowler. 1970. β-Galactosidase, the lactose permease protein, and thiogalactoside transacetylase, p. 89-119. In J. H. Miller and J. R. Beckwith (ed.), The lactose operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Zartler, E. R., F. E. Jenney, M. Terrell, M. K. Eidsness, M. W. W. Adams, and J. H. Prestegard. 2001. Structural basis for thermostability in aporubredoxins from Pyrococcus furiosus and Clostridium pasteurianum. Biochemistry 40:7279-7290. [DOI] [PubMed] [Google Scholar]
- 49.Zecchinon, L., P. Claverie, T. Collins, S. D'Amico, D. Delille, G. Feller, D. Georlette, E. Gratia, A. Hoyoux, M.-A. Meuwis, G. Sonan, and C. Gerday. 2001. Did psychrophilic enzymes really win the challenge? Extremophiles 5:313-321. [DOI] [PubMed] [Google Scholar]
- 50.Zierenberg, R. A., M. W. W. Adams, and A. J. Arp. 2000. Life in extreme environments: hydrothermal vents. Proc. Natl. Acad. Sci. USA 97:12961-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]