Abstract
The atp operon encoding F1Fo ATP synthase in the fermentative obligate anaerobic bacterium Clostridium pasteurianum was sequenced. It consisted of nine genes arranged in the order atpI(i), atpB(a), atpE(c), atpF(b), atpH(δ), atpA(α), atpG(γ), atpD(β), and atpC(ɛ), which was identical to that found in many bacteria. Reverse transcription-PCR confirmed the presence of the transcripts of all nine genes. The amount of ATPase activity in the membranes of C. pasteurianum was low compared to what has been found in many other bacteria. The F1Fo complexes solubilized from membranes of C. pasteurianum and Escherichia coli had similar masses, suggesting similar compositions for the F1Fo complexes from the two bacteria. Western blotting experiments with antibodies raised against the purified subunits of F1Fo detected the presence of eight subunits, α, β, γ, δ, ɛ, a, b, and c, in the F1Fo complex from C. pasteurianum. The F1Fo complex from C. pasteurianum was activated by thiocyanate, cyanate, or sulfhydryl compounds; inhibited by sulfite, bisulfite, or bicarbonate; and had tolerance to inhibition by dicyclohexylcarbodiimide. The target of thiol activation of the F1Fo complex from C. pasteurianum was F1. Thiocyanate and sulfite were noncompetitive with respect to substrate Mg ATP but competitive with respect to each other. The F1 and Fo parts of the F1Fo complexes from C. pasteurianum and E. coli bound to each other, but the hybrid F1Fo complexes were not functionally active.
The F1Fo ATP synthase is a multisubunit enzyme complex found exclusively in cytoplasmic membranes of bacteria, inner membranes of mitochondria, and thylakoid membranes of chloroplasts (4, 14, 54, 59). It synthesizes ATP from ADP and inorganic phosphate (Pi), utilizing the transmembrane chemiosmotic energy of a proton or sodium gradient (4, 14, 18, 59). Reversibly, it hydrolyses ATP to ADP and Pi which, when coupled to proton extrusion, generates chemiosmotic energy (4, 59). Structurally, the enzyme consists of two parts, a membrane-intrinsic Fo and a membrane-extrinsic F1. When detached from the membrane, F1 functions exclusively as ATP hydrolase. The most investigated bacterial F1Fo complex is that from Escherichia coli (F1FoEc) (18, 19, 21). It consists of eight subunits, with a subunit stoichiometry of α3β3γδɛab2c10-12 (21). The α, β, γ, δ, and ɛ subunits constitute the F1 moiety, and the a, b, and c subunits constitute the Fo moiety. All eight subunits are essential for the function of the enzyme complex (14, 58).
The atp operon of the E. coli ATP synthase consists of nine genes arranged in the order atpI(i), atpB(a), atpE(c), atpF(b), atpH(δ), atpA(α), atpG(γ), atpD(β), and atpC(ɛ) (64). A similar composition of the atp operon has been reported for the gram-positive, obligate anaerobic bacteria Moorella thermoacetica (formerly Clostridium thermoaceticum) (11) and Clostridium acetobutylicum (GenBank accession no AAD16419). Some anaerobic bacteria have atp operons consisting of only eight genes. They lack the first gene, atpI, encoding subunit i. Examples are Enterococcus hirae (Streptococcus faecalis) (GenBank accession no M90060), Lactobacillus lactis (GenBank accession no AF059739), and Lactobacillus acidophilus (GenBank accession no AF098522). The atp operon of the anaerobic acetogenic bacterium Acetobacterium woodii consists of 11 genes, including those normally found in bacterial atp operons, plus two additional copies of atpE, encoding the c subunit (55). The A. woodii ATP synthase is sodium rather than proton dependent. It should be noted that the product of the first gene of the atp operon, atpI, has not been found in any purified F1Fo complex (14).
Early work indicated that the F1Fo complex purified from the gram-positive, obligate anaerobic bacterium Clostridium pasteurianum (F1FoCp) could function with only four polypeptides with apparent molecular masses of 65,000, 57,500, 43,000, and 15,000 Da (8, 9). The last polypeptide was not found in the purified F1 ATPase, and it was identified as the dicyclohexylcarbodiimide (DCCD)-binding proteolipid or the c subunit (8, 9). Due to this simple composition, F1FoCp was postulated to be an ancient version of ATP synthase (25, 45). In this study, we show that the composition of the F1FoCp complex, at both the genetic and the protein levels, is very similar to those of the F1Fo complexes from other bacteria (14, 23, 27, 41, 64). Inactivation by sulfite and activation by thiocyanate and sulfhydryl compounds of F1FoCp are also reported.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. pasteurianum strain DSM 525 was obtained from G. Gottschalk, University of Göttingen, Göttingen, Germany. It was grown in a semidefined liquid medium containing 1.0% glucose or glycerol as a carbon source in 3.5-liter batch cultures in 4-liter flasks at 37°C under 100% N2 or CO2 (34). Cells were harvested at the late log phase (after 16 to 18 h of growth; optical density at 600 nm, 1.2 to 1.5) by centrifugation at 6,000 × g, washed in TMG buffer (100 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 10% glycerol), and stored at −80°C until used. Moorella thermoacetica (ATCC 39073) was grown in a semidefined liquid medium with 1% glucose as a carbon source at 58°C under 100% CO2 (12). The following E. coli strains were used: strain DH5α was used as a reference for ATP synthase, strain INVαF′ (Invitrogen, Carlsbad, Calif.) was used as a host for plasmids carrying PCR clones, and ATP synthase-negative (Δunc) mutant DK8 (35) (obtained from M. Futai, Osaka University, Osaka, Japan) was used as a host to screen the genomic library of C. pasteurianum in plasmid pBR322. All E. coli strains were grown and maintained in either Luria-Bertani broth or M9ZB salt medium with glucose (20 mM) as a carbon source (13).
Antibodies used.
All antibodies used in this study were polyclonal. Antibodies against purified M. thermoacetica F1 ATPase (F1Mt) (12) and against a 20-mer synthetic peptide designed from the NH2-terminal end of the δ subunit of F1Mt (11) were raised in rabbits at the Animal Facility of the University of Georgia. Antibodies against purified subunits γ and ɛ of F1 ATPase of E. coli (F1Ec) were obtained from R. Aggeler, University of Oregon. Antibodies against purified subunits a, b, and c of FoEc were obtained from G. Hebestreit-Deckers, University of Osnabrück, Osnabrück, Germany.
DNA source and C. pasteurianum genomic library.
Genomic DNA of C. pasteurianum was isolated by the method of Marmur (46). The C. pasteurianum genomic library was a gift from G. Sawers, John Innes Centre, Norwich, United Kingdom. It was constructed in pBR322 by ligating partial Sau3AI digests of C. pasteurianum genomic DNA into the BamHI site of the plasmid (65).
ATP synthase probes used to screen the C. pasteurianum genomic library.
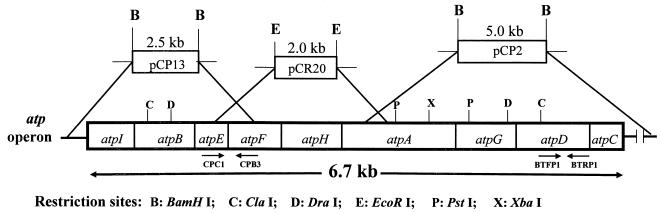
The cloning and sequencing strategy is outlined in Fig. 1. The atp operon of C. pasteurianum was sequenced from three plasmids, pCP2, pCP13, and pCR20. Plasmids pCP2 and pCP13 were isolated from the genomic library after screening with 390- and 490-bp digoxigenin (DIG)-labeled ATPase probes amplified by PCR (see below). Plasmid pCR20 was constructed by cloning a 2.0-kb PCR product containing atp genes into pCR2.1 (Invitrogen). All PCR products were amplified with C. pasteurianum genomic DNA as a template. The following primers were used for amplification of the above PCR products: BTFP1 and BTRP1 for the 390-bp product, CPC1 and CPB3 for the 490-bp product, and CPC1 and CPAL2 for the 2.0-kb PCR product (Table 1). BTFP1 and BTRP1 were degenerate primers, designed from highly conserved amino acid sequences (191ERTREGND198 and 311YVPADDLTD319, respectively) of the F1Ec β subunit (11, 61). CPC1 was designed from a highly conserved sequence (ARQPEA) of the E. coli ATP synthase c subunit, while CPAL2 and CPB3 were designed from known sequences of atpA and atpF encoding the α and b subunits of the F1FoCp complex, respectively. The reaction conditions for PCR were as follows: an initial denaturation at 94°C for 6 min followed by 25 cycles of amplification, with each cycle consisting of 60 s of denaturation at 94°C, 60 s of annealing at 45°C, and 120 s of extension at 72°C. All PCR products were sequenced after being cloned into vector pCR2.1.
FIG. 1.
Cloning and sequencing strategy. The atp operon of C. pasteurianum was sequenced from three plasmids, pCp2, pCp13, and pCR20. Plasmids pCp2 and pCp13 were obtained from the genomic library after screening with DIG-labeled PCR products, and pCR20 was obtained by cloning into pCR2.1 of a 2.0-kb PCR product containing atp genes amplified from C. pasteurianum genomic DNA. The primers used for PCR are indicated below the target genes.
TABLE 1.
Primers for PCR and RT-PCR experiments
| Primer | Gene | Sequence (5′→3′) |
|---|---|---|
| BTFP1 | atpD | GA(AG)CAG(TGA)AC(TAG)(AC)G(TGA)GA(AG)GG(TAA)AA(TC)G |
| BTRP1 | atpD | GTI(AG)(AG)TC(AG)TC(ATC)GC(TAC)G(AT)AC(AG)TA |
| CPAL2 | atpA | CTCTTGCTGGTGGTCTTC |
| CPC1 | atpE | GC(AT)CG(AT)CA(AT)CC(AT)GAAGC |
| CPB3 | atpF | TAAGTTCTGCCTCATTGG |
| CPBTP1 | atpD | ATAGAGCAAGAAGGATTC |
| CPBTP2 | atpD | CATACAAAATCACTCCTT |
| CPIP1 | atpI | AGTTATAGCAGGATTGAT |
| CPIP2 | atpI | TTAATCACTTCCTTTCTT |
| CPAP1 | atpB | TACTTGAAAAATTTACGC |
| CPAP2 | atpB | TAGTTATTCTTCCTCTGC |
| CPBP1 | atpF | GGAAGAGCTGAGAAATTA |
| CPBP2 | atpF | GCTACAGCATATCTCCTA |
| CPEP1 | atpC | ATCTACAACTTCTGGTGG |
| APEP2 | atpC | AATCCTTTCTTCTGCTCT |
RNA isolation and RT-PCR.
Total RNA was isolated from whole cells of C. pasteurianum with a Qiagen RNeasy midi-kit. RNA was treated with RNase-free DNase I (Roche Molecular Biochemicals, Indianapolis, Ind.) prior to use in hybridization or reverse transcription (RT)-PCR experiments. For RT-PCR experiments, total RNA (2 μg) was treated with DNase I and reverse transcribed into cDNA with either Superscript (Bethesda Research Laboratories, Rockville, Md.) or Omniscript (Qiagen) reverse transcriptase according to the manufacturer's directions. The cDNA synthesized in these reactions was used as a template in PCRs to amplify different atp genes.
Hybridization experiments.
Colony and Northern hybridization experiments were carried out with the nonradioactive Genius System (Roche Applied Science, Indianapolis, Ind.) and DIG-labeled PCR products as probes (11, 13). RNA samples were denatured with glyoxal (5) prior to use in Northern hybridization experiments.
Membrane preparation and ATPase assays.
Membranes of M. thermoacetica and E. coli DH5α were prepared by breaking cells in a French press (12). Membranes of C. pasteurianum were prepared by a modified lysozyme method (8). Briefly, cell paste (30 to 40 g [wet weight]) was suspended in 150 ml of lysis buffer (100 mM Tris-HCl [pH 8.0], 0.5 M sucrose) containing lysozyme (2.0 mg/ml) and incubated at 37°C for 1 h. Highly viscous cell lysate obtained at this step was centrifuged at 10,000 × g for 10 min. The supernatant was discarded, and the pellet containing protoplasts was suspended in TMG buffer containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF), DNase I, and RNase I (0.2 mg/ml each) and passed through a French press. Intact cells and cell debris were removed by centrifugation at 6,000 × g for 10 min, and the supernatant was centrifuged at 100,000 × g for 1 h. The pellet containing membranes was washed twice and then suspended in TMG buffer containing 1 mM Na ATP. The ATPase (ATP hydrolysis) activity was assayed at 37°C with a reaction mixture containing100 mM Tris-HCl (pH 8.0) and 5 mM MgCl2 and with 2 mM Na ATP or Mg ATP as a substrate. The reaction was started by adding ATP and stopped by adding sodium dodecyl sulfate (SDS) to a final concentration of 1%. Pi released by ATP hydrolysis was measured colorimetrically as described previously (12). Unless otherwise stated, membranes used in all ATPase assays were prepared from glucose-grown cells. For inhibition or activation studies, reaction mixtures containing the enzyme were incubated with the activator or inhibitor at 37°C for 10 min prior to ATPase assays. Each experiment was carried out in triplicate, and the reported results are the average of three independent experiments with a standard deviation of ±0% to 10%.
Solubilization of F1Fo and F1 from membranes of C. pasteurianum and E. coli.
Washed membranes (4 to 5 mg/ml) of C. pasteurianum and E. coli were suspended in TMG buffer containing 1 mM Na ATP and 0.5 mM PMSF. For the solubilization of F1Fo, n-dodecyl-β-maltoside (DM) (Anatrace, Maumee, Ohio) was added to the membrane suspension to a final concentration of 1% (wt/vol). The suspension was incubated on ice for 30 min and then centrifuged at 100,000 × g for 1 h. The pellet was discarded, and the supernatant containing F1Fo was saved. To concentrate F1Fo, the enzyme was precipitated with 60% ammonium sulfate in the presence of 1% Na cholate as described by Hicks and Krulwich (27). The precipitate containing mostly F1Fo was dissolved in TMG buffer containing 0.02% DM and 1 mM Mg ATP, and the solution was dialyzed against three changes of the same buffer. Solubilization of F1 from washed membranes of E. coli (10, 60) and C. pasteurianum (8) was carried out by extraction with buffer containing EDTA. Soluble F1 ATPase of C. pasteurianum (F1Cp) and F1Ec were concentrated on YM100 membranes with an Amicon concentrator and partially purified by gel filtration on TSK gel G3000SW (TosoHaas, Montgomeryville, Pa.) by using a fast protein liquid chromatography system (Amersham Pharmacia, Piscataway, N.J.). Fractions with the highest ATPase activity were pooled, dialyzed against TMG buffer containing 1 mM Na ATP, and concentrated as described above.
In vitro reconstitution of F1-depleted membranes with exogenously added F1.
Washed membranes of C. pasteurianum and E. coli were stripped of F1 by repeated extractions with EDTA buffer (8, 10, 60). Stripped membranes were washed and suspended (2 mg of protein per ml) in reconstitution buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol [vol/vol], 0.1 mM PMSF). Partially purified F1Cp or F1Ec was added to F1-depleted membranes, and the mixture was incubated at 37°C for 30 min and centrifuged at 100,000 × g for 1 h. The pellet containing reconstituted membranes was washed twice in reconstitution buffer prior to ATPase assays. Maximum binding of F1 to F1-depleted membranes occurred at a ratio of 1 U of F1 per 1 mg of F1-depleted membranes, irrespective of their origins. This ratio was maintained in all reconstitution reactions.
Other methods.
Protein was estimated by using the Lowry method as described previously (12). Polyacrylamide gel electrophoresis (PAGE) in the presence (denatured) or absence (native) of SDS was carried out as described by Laemmli (40). Western blotting experiments were carried out according to the instructions of Bio-Rad (Hercules, Calif.) (12, 13). The ATP-driven proton-pumping activities of intact and reconstituted membranes were determined by measuring the quenching of fluorescent dye acridine orange as described by Moriyama et al. (48). The reaction mixture (3.0 ml) contained 10 mM Tricine-choline (pH 8.0), 140 mM KCl, 5 mM MgCl2, 1 μg of valinomycin/ml, 1 μM acridine orange, and membranes (50 to 300 μg of protein). The reactions started by adding Na ATP (1 mM) and the changes in fluorescence (emission, 530 nm; excitation, 490 nm) were recorded with a Shimadzu fluorescence spectrometer (model RF2150). Standard techniques were used for all DNA manipulation experiments (57). Oligonucleotide synthesis and DNA sequencing reactions were carried out at the Molecular Genetics Instrumentation Facility of the University of Georgia.
Nucleotide sequence accession number.
The nucleotide sequence of the atp operon of C. pasteurianum has been assigned accession number AF283808 in the GenBank, EMBL, and DDBJ libraries.
RESULTS
Cloning, sequencing, and analysis of the primary structure of ATP synthase subunits encoded by the C. pasteurianum atp operon.
Fig. 1 outlines the strategy used to clone and sequence the C. pasteurianum atp operon. This operon contains nine open reading frames (ORFs) (ORF1 through ORF9) within a 7.5-kb DNA fragment. The deduced amino acid sequences encoded by ORF2 to ORF9 shared between 22 and 72% identical residues with the corresponding sequences for the eight subunits of ATP synthases from several sources. The deduced amino acid sequence encoded by ORF1 had 19% residues identical to those encoded by the first gene, atpI, of the E. coli atp operon, indicating that it corresponded to atpI. The nine genes of the C. pasteurianum atp operon are arranged in the order atpI(i), atpB(a), atpE(c), atpF(b), atpH(δ), atpA(α), atpG(γ), atpD(β), and atpC(ɛ), an organization identical to those of the atp operons of many bacteria (11, 23, 55, 64).
All atp genes have ATG start codons. The reading frames of all atp genes were AT rich, with an average GC content of 32%. A putative promoter structure with the sequence 5′-GTTGAAA-N17-TCTAAT-3′, resembling the E. coli consensus σ70 promoter sequence 5′-cTTGACA-N17-21-TATAaT-3′, was found 48 bp upstream of atpI. Long intergenic regions were found between atpB and atpE (88 bp) and between atpE and atpF (39 bp). No secondary promoter structure was apparent in these intergenic regions. The deduced molar masses (Da) of the subunits were as follows: i, 13,818; a, 25,634; c, 8,374; b, 18,466; δ, 20,702; α, 54,943; γ, 31,300; β, 50,766; and ɛ, 11,866. With the exception of that for the ɛ subunit, these masses are comparable to those of the corresponding ATP synthase subunits from other sources (11, 27, 55, 64). The F1Ec ɛ subunit has 139 residues, while the F1Cp ɛ subunit has 108 residues, lacking 31 residues at the carboxyl-terminal end. Deletion mutation of the E. coli ɛ subunit has shown that this segment is not necessary for the function of the enzyme (37). The natural absence of this segment at the COOH-terminal end of the C. pasteurianum ATP synthase ɛ subunit supports this finding.
The deduced amino acid sequences for the α, β, and c subunits of the C. pasteurianum ATP synthase are more conserved (between 40 and 72% identical residues) than those for the a, b, γ, δ, and ɛ subunits (between 22 and 39% identical residues) relative to sequences for the corresponding ATP synthases from other origins. Several motifs and residues of the highly conserved α, β, and c subunits are apparent. They include the nucleotide-binding domain or Walker motifs (169GDRQTGKT176 in the α subunit and 152GGAGVGKT159 in the β subunit), the so-called γ-subunit-interacting (383DELSEED389) region of the β subunit, the 43ARQP46 sequence in the hydrophilic loop of the c subunit, and the DCCD-binding acidic residue E64 of the c subunit (1, 19, 64).
The atp operon of C. pasteurianum is transcribed into a single polycistronic mRNA.
Initially, total C. pasteurianum RNA was subjected to Northern hybridization experiments with different ATP synthase probes, including those used in the screening of the genomic library. The hybridization signals were very weak and almost undetectable. Increasing the amount of RNA or varying the stringency of hybridization conditions had little effect on the hybridization signals. Therefore, we used more sensitive RT-PCR experiments to verify the transcripts of the atp operon of C. pasteurianum. Total RNA (2 μg) was treated with RNase-free DNase and reverse transcribed with primer CPEP2 (Table 1), designed on the basis of the 3′ end of atpC, the last gene of the atp operon (Fig. 1). The cDNA synthesized from this reaction was used as a template in PCR to amplify atpI, atpB, atpF, and atpC. These genes were targeted because their products were reported to be absent from the purified ATP synthase from C. pasteurianum (8, 9). The gene-specific primers used in PCR were CPIP1 and CPIP2 for atpI, CPAP1 and CPAP2 for atpB, CPBP1 and CPBP2 for atpF, and CPEP1 and CPEP2 for atpC. A control PCR was carried out for each set of primers with DNase-treated RNA as a template. PCR products of the expected length (∼200 bp) were amplified in reactions with cDNA as a template (data not shown) but not in those with DNase-treated RNA as a template. These results suggest that the PCR products were amplified exclusively from cDNA, which was apparently synthesized from full- to partial-length polycistronic transcripts of the atp operon of C. pasteurianum, as reported for other bacteria (11, 55).
Solubilization of F1FoCp and F1Cp from membranes by extraction with detergent and EDTA.
Washed membranes (9.4 mg of protein) of C. pasteurianum were extracted with DM and EDTA to solubilize F1Fo and F1, respectively. The total ATPase activities solubilized from membranes by DM (1.198 μmol of Pi · min−1) and EDTA (0.942 μmol of Pi · min−1) were found to be higher than that of the same amount of untreated membranes (0.080 μmol of Pi · min−1) (Table 2). A similar increase in the activity of soluble F1FoCp was observed with other detergents, e.g., Triton X-100 (1% [vol/vol]) and octylglucopyranoside (1% [wt/vol]). The increase in the ATPase activity of F1Fo after solubilization from membranes by DM has been reported for other bacteria (27). The increase in the activity of F1Cp after solubilization from membranes by EDTA is unusual, but a similar finding has been reported for ATPase in E. coli mutants with abnormalities in the functions of the Fo moiety of the enzyme complex (39). DCCD and Na azide inhibited both membrane-bound (MB) and soluble F1FoCp or F1Cp (Table 2). The levels of inhibition by DCCD (100 μM) were 53% for MB F1FoCp and 55% for soluble F1FoCp. The levels of inhibition by Na azide (1 mM) were 72% for MB F1Cp and 68% for soluble F1Cp. In contrast, the levels of inhibition of MB F1Ec (or F1FoEc) at the above concentrations of DCCD and Na azide were between 92 and 98%. A higher tolerance of MB and soluble F1FoCp or F1Cp to inhibition by DCCD and Na azide is not unusual, as similar results were reported for several gram-positive bacteria (3, 26, 28, 32, 49). A comparison of the ATPase activities of DM- and EDTA-extracted membranes with that of untreated membranes indicated that about 87 and 70% of ATPase activities were solubilized from membranes by DM and EDTA, respectively (Table 2). The residual ATPase activities of DM- and EDTA-extracted membranes were more tolerant to inhibition by DCCD or Na azide than that of untreated membranes; therefore, these activities could be attributed to other types of ATPases.
TABLE 2.
ATPase activities of MB and soluble F1Cp or F1FoCp after extraction with EDTA and detergenta
| Treatment | Protein (mg) | Activityb
|
Activity (U · mg−1) in the presence of c:
|
||
|---|---|---|---|---|---|
| U | U · mg−1 | DCCD (100 μM) | Na azide (1 mM) | ||
| None | 9.40 | 0.800 | 0.085 | 0.045 | 0.024 |
| Membrane + DM | |||||
| Residual membrane | 4.39 | 0.108 | 0.024 | 0.020 | 0.019 |
| Extracts (F1Fo) | 5.24 | 1.198 | 0.228 | 0.125 | 0.073 |
| Membrane + EDTA | |||||
| Residual membrane | 7.32 | 0.240 | 0.033 | 0.022 | 0.020 |
| Extracts (F1) | 2.22 | 0.942 | 0.424 | 0.330 | 0.135 |
For solubilization of F1Fo, washed membranes (2 mg/ml) were suspended in TMG buffer containing Na ATP (1 mM), DM (1% [wt/vol]), and PMSF (0.1 mM); the suspension was incubated on ice for 1 h and centrifuged at 100,000 × g for 1 h. The supernatant containing soluble F1Fo was collected, and the pellet containing residual membranes was washed and resuspended in TMG buffer. For solubilization of F1, washed membranes (1 mg/ml) were subjected to similar treatments, except that the membranes were suspended in 100 mM Tris-HCl (pH 8.0)-15 mM EDTA-2 mM LiCl-1 mM ATP. EDTA was removed from the extracts by dialysis against TMG buffer containing 1 mM Na ATP. The pellet containing residual membranes was washed and resuspended in TMG buffer. ATPase assays were done with Na ATP (2 mM) as a substrate.
One unit of activity is defined as 1 μmol of P1 released per min.
DCCD or Na azide was added to assay mixtures, which were incubated at 37°C for 10 min prior to the start of ATPase assays by the addition of ATP.
Low level of the F1Fo complex in C. pasteurianum membranes.
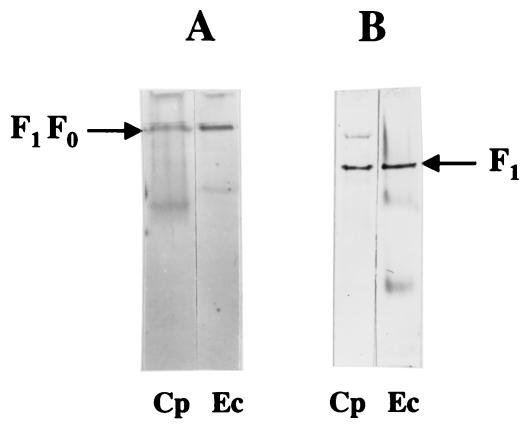
Washed membranes of C. pasteurianum had a low level of ATPase activity, 0.082 μmol · min−1 · mg−1, about one-fourth that of E. coli membranes, 0.328 μmol · min−1 · mg−1. To test whether the enzyme was latent or not, washed membranes of C. pasteurianum were subjected to treatments that are known to activate MB F1Fo. Limited proteolysis with trypsin had no effect, while treatment with detergents or anions had a marginal effect on the activity of MB F1FoCp (Table 3), suggesting that the level of MB F1FoCp could be low; this notion is further supported by the poor transcription of the atp genes of C. pasteurianum, as the atp transcripts were almost undetectable on Northern blots. For further verification, whole-cell extracts of C. pasteurianum, E. coli, and M. thermoacetica were subjected to Western blotting experiments with antibodies raised against purified F1Mt. The antibodies reacted with the α and β subunits of F1Cp and F1Mt but only with the β subunit of F1Ec (Fig. 2), indicating the antigenic similarity of the latter subunit in the three bacteria. Strong antigenic similarity of the β subunit of F1 has also been reported for other bacteria (42, 53, 60). The β subunit of F1Mt was more like the β subunit of F1Cp (73% identical residues) than the β subunit of F1Ec (68% identical residues). The intensity of the immunosignal corresponding to the β subunit was much stronger in E. coli extracts than in C. pasteurianum extracts (Fig. 2). Since the level of the β subunit reflects the amount of the F1Fo complex, these results support a low level of F1Fo in C. pasteurianum.
TABLE 3.
Influence of different effectors of ATPase on activities of MB F1FoCp and F1FoEca
| Detergent or anion | Results for membranes from:
|
|||
|---|---|---|---|---|
|
C. pasteurianum
|
E. coli
|
|||
| U · mg−1 | % | U · mg−1 | % | |
| None | 0.080 | 100 | 0.328 | 100 |
| NaHCO3 (20 mM) | 0.057 | 71 | 0.406 | 125 |
| Na2SO3 (20 mM) | 0.009 | 12 | 0.436 | 133 |
| Na2S2O5 (20 mM) | 0.016 | 26 | 0.360 | 110 |
| NaSCN (20 mM) | 0.156 | 195 | 0.138 | 42 |
| KOCN (20 mM) | 0.102 | 128 | 0.065 | 20 |
| Methanol (10%) | 0.114 | 143 | 0.456 | 139 |
ATPase assays were done with Na ATP (2 mM) as a substrate as described in Materials and Methods. Effectors were incubated with membranes in assay mixtures at 37°C for 10 min prior to the start of ATPase assays.
FIG. 2.
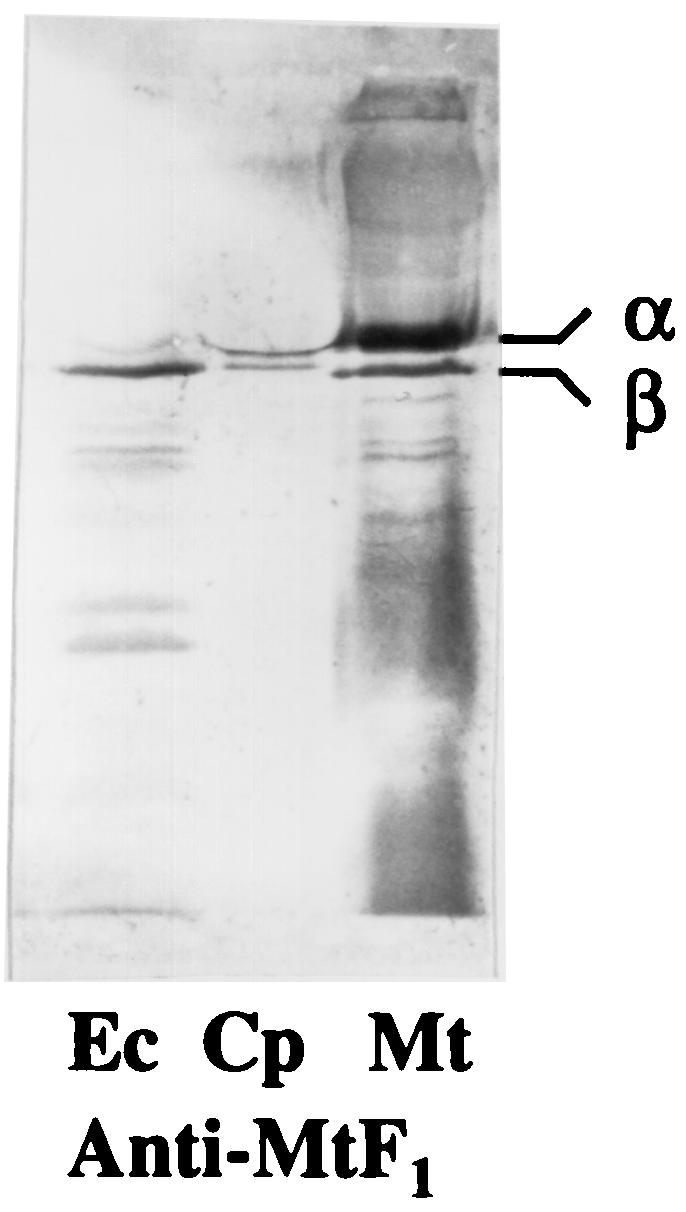
Immunoblots of whole-cell extracts (50 μg of protein) from E. coli (Ec), C. pasteurianum (Cp), and M. thermoacetica (Mt) after reaction with antibodies against purified F1Mt. Proteins were separated by SDS-PAGE (10% acrylamide) and transblotted onto polyvinylidene difluoride membranes prior to the immunoreactions.
F1FoCp has a more complex subunit composition than previously reported.
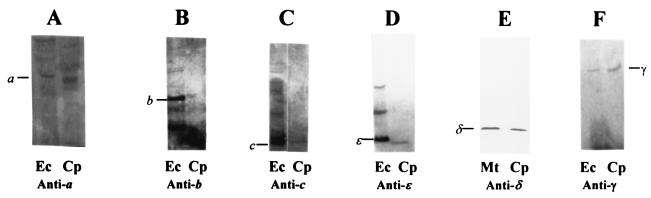
The F1Cp, F1FoCp, F1Ec, and F1FoEc complexes solubilized from membranes were subjected to native PAGE and Western blotting experiments with antibodies raised against purified F1Mt (12). Immunosignals of F1Cp and F1FoCp corresponded to the signals of F1Ec, and F1FoEc, respectively (Fig. 3), suggesting that these complexes have similar masses and perhaps similar compositions. The antibodies against purified F1Mt demonstrated the presence of the α and β subunits of F1FoCp (Fig. 2). To identify other subunits of F1FoCp, antibodies against purified subunits γ, ɛ, a, b, and c of F1FoEc and those against a synthetic peptide (1MSENQNVARRYARALFNIARE20) designed on the basis of the NH2-terminal end of the δ subunit of F1Mt were used (11). Nine residues of this peptide (8RRYAXALXNIA19) were identical to the corresponding sequence at the NH2-terminal end of the F1Cp δ subunit. Figure 4A to F show that the antibodies against each of the purified ATP synthase subunits reacted with the corresponding subunits of the F1FoCp complex, demonstrating the presence of all eight subunits typically found in bacterial ATP synthases. The interactions between the antigen and the corresponding antibodies were much weaker for the subunits of FoCp than the subunits of F1Cp, a result which is not unusual, as reported for other bacteria (60). A comparison of the molar masses of the F1FoCp, F1FoEc, and M. thermoacetica F1Fo subunits, as shown on Western blots, indicated the expected similarities in the masses for subunits α, β, γ, δ, and c from the three bacteria and the expected differences in the masses for subunits a, b, and ɛ.
FIG. 3.
Immunoblots of soluble F1Fo (A) and F1 (B) from C. pasteurianum (Cp) and E. coli DH5α (Ec) after reactions with antibodies against purified F1Mt. (A) Soluble F1FoCp after precipitation with ammonium sulfate from DM extracts (1.0% [wt/vol]) of C. pasteurianum membranes (30 μg) and soluble F1FoEc in DM extracts (1.0% [wt/vol]) of E. coli membranes (30 μg). (B) Partially purified F1Cp (30 μg) and F1Ec (30 μg). Proteins were separated by native PAGE (7.5% acrylamide) and transblotted onto polyvinylidene difluoride membranes prior to immunoreactions.
FIG. 4.
Immunoblots of concentrated DM extracts of C. pasteurianum membranes (Cp, 60 μg of protein) and whole-cell extracts of E. coli (Ec, 30 μg of protein) and M. thermoacetica (Mt, 30 μg of protein) after reactions with antibodies against ATP synthase subunits a (A), b (B), c (C), and ɛ (D) of F1FoEc, subunit δ of F1Mt (E), and purified γ of F1Ec (F). DM extracts of C. pasteurianum membranes were concentrated by precipitation with ammonium sulfate as described in Materials and Methods. Proteins were separated by SDS-PAGE (10% acrylamide) and transblotted onto polyvinylidene difluoride membranes prior to immunoreactions.
F1FoCp is activated by thiocyanate and inhibited by sulfite.
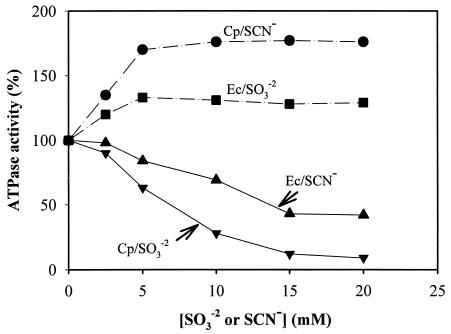
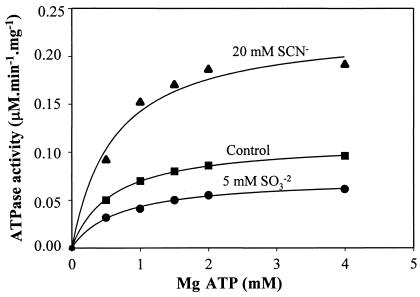
MB F1FoCp responded differently to anions than did MB F1FoEc. Sulfite (SO32−) and bisulfite (S2O52−) activated MB F1FoEc but inhibited MB F1FoCp (Table 3). On the other hand, thiocyanate and cyanate activated F1FoCp but inhibited F1FoEc (Table 3 and Fig. 5). Thiocyanate treatment did not release F1 from C. pasteurianum membranes, suggesting that the activation was not due to the solubilization of the enzyme. The activation of F1FoCp by thiocyanate or cyanate and its inhibition by sulfite or bisulfite were reproducible with different preparations of membranes from cells grown on either glucose or glycerol as a carbon source. Soluble F1FoCp and soluble F1Cp also exhibited activation or inhibition by oxyanions similar to that of MB F1FoCp (Table 4). The thiocyanate activation or sulfite inhibition of F1FoCp at variable concentrations of the substrate (Mg ATP) followed Michaelis-Menten enzyme kinetics (Fig. 6). Double-reciprocal plots of the inhibition of ATPase activity versus fixed substrate concentrations ([SO3−]) were found to be noncompetitive with respect to Mg ATP (data not shown). The Km for Mg ATP was 0.62 ± 0.03 mM (mean and standard deviation). A similar Km in the presence of SCN− or SO32− suggested that the affinity of the enzyme for the substrate (Mg ATP) was not affected by either of these anions, a finding previously reported by others (16, 30, 50, 62). The Kis for sulfite inhibition, calculated from plots of the inhibition of ATPase activity versus [SO3−] (Dixon plots; data not shown) were 3.1 and 2 mM in the presence and absence of 5 mM NaSCN, respectively. The increase in the Ki for the inhibitory anion in the presence of the activating anion is a normal response of F1Fo ATPase also reported by others (16). It has been suggested that both activating and inhibiting anions compete for the same regulatory site(s) of F1Fo complexes (30, 31, 44, 62). Accordingly, thiocyanate was found to antagonize the effect of sulfite on F1FoCp or vice versa, and sulfite was found to antagonize the effect of thiocyanate on F1FoEc or vice versa (data not shown).
FIG. 5.
Effect of NaSCN and Na2SO3 on ATPase activities of MB F1FoCp and F1FoEc. Washed membranes were incubated with NaSCN or Na2SO3 at the indicated concentrations in ATPase assay reaction mixtures at 37°C for 10 min prior to ATPase assays. The ATPase activities of untreated membranes of C. pasteurianum (0.080 μmol of Pi · min−1 · mg−1) and E. coli (0.333 μmol of Pi · min−1 · mg−1) were taken as 100% for calculation of the percent activation or inhibition of ATPase activity by NaSCN or Na2SO3. Symbols: •, activation of MB F1FoCp by NaSCN; ▪, activation of MB F1FoEc by Na2SO3; ▴, inhibition of MB F1FoCp by Na2SO3; ▾, inhibition of MB F1FoCp by Na2SO3.
TABLE 4.
Effect of anions on ATPase activities of MB and soluble F1Cp or F1FoCpa
| Anion (mM) | Results for:
|
|||||
|---|---|---|---|---|---|---|
| Membranes
|
Soluble F1Cpb
|
Soluble F1FoCpc
|
||||
| U · mg−1 | % | U · mg−1 | % | U · mg−1 | % | |
| None | 0.092 | 100 | 0.428 | 100 | 0.162 | 100 |
| KSCN (10) | 0.185 | 202 | 0.831 | 190 | 0.458 | 283 |
| KOCN (10) | 0.120 | 131 | 0.599 | 140 | 0.210 | 130 |
| Na2SO3 (10) | 0.015 | 16 | 0.128 | 30 | 0.040 | 25 |
Anions were incubated in assay mixtures at 37°C for 10 min prior to the start of ATPase assays.
Soluble F1Cp was partially purified from EDTA extracts by gel filtration as described in Materials and Methods.
Soluble F1FoCp was obtained from DM extracts of membranes after precipitation with ammonium sulfate as described in Materials and Methods.
FIG. 6.
ATPase activity of C. pasteurianum membranes at various concentrations of substrate (Mg ATP) in the absence (▪) or presence of 20 mM NaSCN (▴) or 5 mM Na2SO3 (•).
Activation of F1FoCp by sulfhydryl reagents.
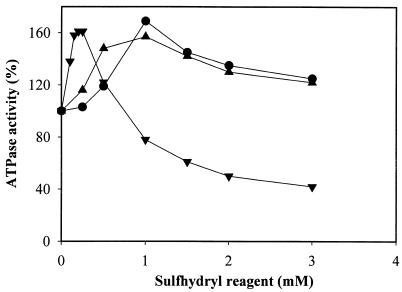
Sulfhydryl compounds are known to activate F1Fo from plants and photosynthetic bacteria (29, 51, 52). Here we show that the sulfhydryl compounds dithiothreitol (DTT), β-mercaptoethanol (β-ME), and cysteine activated both MB and soluble F1FoCp or F1Cp (Table 5 and Fig. 7). None of these compounds had any effect on F1FoEc (data not shown). The activation of F1FoCp by sulfhydryl reagents increased in the following order: cysteine < ME < DTT. Maximum activation of the enzyme occurred in the presence of 0.2 mM β-ME and 1 mM DTT or cysteine. At higher concentrations, all sulfhydryl compounds were inhibitory, and the inhibition was significantly higher with β-ME than with DTT or cysteine (Fig. 7). The activation of soluble F1Cp by sulfhydryl compounds was comparable to that of MB or soluble F1FoCp (Table 5), suggesting that F1 is the target of thiol activation. Thiol activation of F1 was previously reported only for enzymes from chloroplasts and cyanobacteria (29, 51, 52).
TABLE 5.
Effect of sulfhydryl reagents on ATPase activities of MB and soluble F1Cp and F1FoCpa
| Sulfhydryl reagent (mM) | Results for:
|
|||||
|---|---|---|---|---|---|---|
| Membranes
|
F1Cp
|
F1FoCp
|
||||
| U · mg−1 | % | U · mg−1 | % | U · mg−1 | % | |
| None | 0.078 | 100 | 0.436 | 100 | 0.156 | 100 |
| DTT (2) | 0.104 | 143 | 0.628 | 144 | 0.217 | 139 |
| Cysteine (2) | 0.105 | 141 | 0.610 | 140 | 0.212 | 136 |
| β-ME (0.5) | 0.096 | 124 | 0.545 | 125 | 0.193 | 124 |
Sulfhydryl reagents were added to assay mixtures containing membranes, soluble F1Cp, or soluble F1FoCp, and mixtures were incubated at 37°C for 10 min prior to the start of ATPase assays. Solutions of sulfhydryl reagents were prepared fresh before each experiment. Soluble F1FoCp and soluble F1Cp were obtained as described in Materials and Methods.
FIG. 7.
Activation of MB F1FoCp by sulfhydryl reagents. Symbols: •, DTT; ▾, cysteine; ▴, β-ME.
The F1 and Fo portions of the F1FoCp and F1FoEc complexes are not functionally compatible.
Inhibition studies indicated that both MB F1FoCp and soluble F1FoCp are relatively tolerant to inhibition by DCCD compared to Na azide (Table 2). Washed membranes of C. pasteurianum failed to generate a proton gradient from ATP hydrolysis (data not shown). A general interpretation of these results is that ATP hydrolysis by F1 could be uncoupled from proton translocation through Fo. To further investigate this notion, F1-depleted membranes of C. pasteurianum (FoCp) and E. coli (FoEc) were reconstituted with F1 from the above bacteria. DCCD-sensitive ATPase activity and ATP-driven proton pumping were observed only with reconstituted membranes consisting of F1Ec and FoEc (data not shown) and not with other combinations when F1 and Fo were from different bacteria. Previously, we reported similar results for F1 and Fo of the ATP synthases from E. coli and the gram-positive anaerobic bacterium M. thermoacetica (10). These results suggest that the F1 and Fo portions of the ATP synthases from E. coli and gram-positive anaerobic bacteria are not functionally compatible, as previously reported (10).
DISCUSSION
This report is a correction of an earlier report that the functionally active F1FoCp complex consists of only four subunits instead of the eight commonly found in most bacteria. Here we have shown that F1FoCp had a composition similar to that of E. coli ATP synthase. The atp operon of C. pasteurianum consists of nine genes, like the atp operon of E. coli. The RT-PCR experiments confirmed the presence of all atp genes, including those encoding a, b, δ, and ɛ subunits, which were previously not found in purified F1FoCp (8, 9). Native PAGE analyses (Fig. 3) of intact F1 and F1Fo complexes suggested similar molar masses for F1Cp and F1Ec or F1FoCp and F1FoEc. Western blotting experiments with antibodies against the corresponding subunits of the F1FoEc and M. thermoacetica F1Fo complexes confirmed the presence of all eight ATP synthase subunits in F1FoCp. Apparently, the missing subunits were lost during the purification of the F1FoCp complex reported by Clarke and coworkers (8, 9), a finding which is not unusual for anaerobic bacteria (11, 12, 56). One may speculate that the conditions used in the purification steps were too harsh for the integrity of the enzyme complex, e.g., excessive use of chromatograpic steps, absence of protease inhibitors in purification buffers, and use of sonication for the solubilization of F1Fo from membranes. In most instances, F1Fo complexes have been purified under milder conditions with fewer chromatographic steps (12, 21, 27, 41, 56).
The low level of activity of MB F1Cp could have been influenced by Fo, as reported for E. coli mutants with a Gly213 → Cys substitution in the highly conserved fourth transmembrane helix (TMH4) of the a subunit of ATP synthase (39). TMH4 of the ATP synthase a subunit is the crucial segment of the subunit that interacts with the c subunit during proton translocation through membrane-integral Fo of the F1Fo complex (14, 18, 33, 63). Analysis of the primary structure of the a subunit of F1FoCp revealed unusual features in its TMH4. It contains two Cys residues, Cys160 and Cys165, that are complementary to Gly208 and Gly213 in the corresponding segment of the ATP synthase a subunit from E. coli and other sources. Recently, by site-directed mutagenesis it was shown that the replacement of Gly213 with Cys in the a subunit inhibited the growth of E. coli on succinate, increased the tolerance to DCCD inhibition, and blocked the ATP-driven proton-pumping activity of the membranes (33, 39). These properties resemble those of the MB F1FoCp complex.
F1FoCp exhibits unusual responses to oxyanions. Sulfite has been shown to be a strong activator (2, 6, 15-17, 27, 30-32, 44) and thiocyanate has been shown to be a strong inhibitor (7, 30, 31) of F1Fo ATP synthase, findings that were also demonstrated in this study for F1FoEc. In contrast, sulfite inhibited and thiocyanate activated F1FoCp, and they were competitive with respect to each other but noncompetitive with respect to ATP (Fig. 6). It is not clear whether the two anions compete for the same or a different regulatory site(s) in the enzyme complex.
Thiol activation of F1Fo was previously reported only for the enzymes in plants and photosynthetic bacteria (29, 51, 52). The activation was shown to occur because of the reduction of an intrapeptide disulfide bond between two cysteine residues within the γ subunit of the enzyme complex (47). Analysis of the primary structure of the subunits of the F1FoCp complex revealed the presence of six Cys residues, four in the α subunit and one each in the β and γ subunits. The possibility of the formation of intra- or interpeptide disulfide bonds between these Cys residues was investigated by homology modeling based on the structure of the bovine F1 ATPase (1). Measurement of the relative distances between the Cys residues indicated a close proximity between Cys47 and Cys73 (within 6 Å) of the F1Cp α subunit, while the distances between other Cys residues, including those of the β and γ subunits, were much longer (data not shown). Therefore, the possibility of disulfide bond formation between Cys47 and Cys73 of the F1Cp α subunit is reasonable. In a database search, Cys residues complementary to Cys47, Cys73, and Cys193 were found to be present in the α subunit of F1 ATPase from two other clostridial species, C. acetobutylicum (GenBank accession no. G972520) and Clostridium perfringens (GenBank accession no BAB81895). It is not known whether F1Fo complexes from these bacteria are subject to thiol modulation.
DCCD inhibits the activity of F1Fo by reacting with a key carboxylate residue (Glu or Asp) in the second transmembrane helix of the c subunit (19, 20, 24). It has been shown that the binding of DCCD to the carboxylate residue is strongly influenced by the regions of the two helices of the c subunit (20, 24) as well as by the regions of the a subunit (TMH4) that interact with this residue (19, 63). The above factors might contribute to the weaker binding of DCCD to the c subunit of F1FoCp, resulting in increased tolerance to the inhibitor.
Unlike most aerobic bacteria and some anaerobic bacteria, e.g., M. thermoacetica, Moorella thermoautotrophica, or A. woodii, C. pasteurianum lacks the ability to synthesize ATP from ATP synthase via chemiosmosis (25, 43). C. pasteurianum is a fermentative anaerobic bacterium. Depending on growth conditions, several fermentative bacteria, e.g., Salmonella enterica serovar Typhimurium (22), Enterococcus hirae (36), and Lactobacillus acidophilus (38), utilize ATP synthase exclusively in the direction of ATP hydrolysis to regulate cytoplasmic pH. A similar function might be attributed to the F1FoCp ATP synthase, as suggested by Harris (25). The unusual domain structure (TMH4) of the ATP synthase a subunit might have some regulatory role in the ATP synthase function of the enzyme complex, similar to that observed in E. coli mutants (33, 39). However, the physiological relevance of this possibility could be questioned without mutational changes in the clostridial enzyme.
Acknowledgments
We thank Bijoy Mohanty for helpful suggestions for the RNA work.
This work was funded by grant DE-FG02-93ER20127 from the Department of Energy.
REFERENCES
- 1.Abrahams J. P., A. G. Leslie, R. Lutter, and J. E. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370:521-628. [DOI] [PubMed] [Google Scholar]
- 2.Bakels, R. H., H. S. Van Walraven, J. E. Van Wielink, Z.-D. I. G. Van Der Zwet-De, B. E. Krenn, K. Krab, J. A. Berden, and R. Kraayenhof. 1994. The effect of sulfite on the ATP hydrolysis and synthesis activity of membrane-bound H+-ATP synthase from various species. Biochem. Biophys. Res. Commun. 201:487-492. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya, S., P. C. Banerjee, and P. K. Das. 1990. A plasma-membrane associated ATPase from the acidophilic bacterium Acidiphilium cryptum. Biochem. Cell Biol. 68:1222-1225. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, P. D. 1997. The ATP synthase-a splendid molecular machine. Annu. Rev. Biochem. 66:717-749. [DOI] [PubMed] [Google Scholar]
- 5.Burnett, W. V. 1997. Northern blotting of RNA denatured in glyoxal without buffer re-circulation. BioTechniques 22:668-671. [DOI] [PubMed] [Google Scholar]
- 6.Cappellini, P., P. Turina, V. Fregni, and B. A. Melandri. 1997. Sulfite stimulates the ATP hydrolysis activity of but not proton translocation by the ATP synthase of Rhodobacter capsulatus interferes with the activation by delta muH+. Eur. J. Biochem. 248:496-506. [DOI] [PubMed] [Google Scholar]
- 7.Cerdan, E., M. L. Campo, E. Santiago, and N. Lopez-Moratalla. 1987. Effect of nucleotides and inhibitory anions on mitochondrial ATPase. Its pH dependence. Rev. Esp. Fisiol. 43:281-285. [PubMed] [Google Scholar]
- 8.Clarke, D. J., and J. G. Morris. 1979. The proton-translocating adenosine triphosphatase of the obligately anaerobic bacterium Clostridium pasteurianum. 2. ATP synthetase activity. Eur. J. Biochem. 98:613-620. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, D. J., F. M. Fuller, and J. G. Morris. 1979. The proton-translocating adenosine triphosphatase of the obligately anaerobic bacterium Clostridium pasteurianum. 1. ATP phosphohydrolase activity. Eur. J. Biochem. 98:597-612. [DOI] [PubMed] [Google Scholar]
- 10.Das, A., and L. G. Ljungdahl. 1993. F0 and F1 parts of ATP synthases from Clostridium thermoautotrophicum and Escherichia coli are not functionally compatible. FEBS Lett. 317:17-21. [DOI] [PubMed] [Google Scholar]
- 11.Das, A., and L. G. Ljungdahl. 1997. Composition and primary structure of the F1Fo ATP synthase from the obligately anaerobic bacterium Clostridium thermoaceticum. J. Bacteriol. 179:3746-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, A., D. M. Ivey, and L. G. Ljungdahl. 1997. Purification and reconstitution into proteoliposomes of the F1Fo ATP synthase from the obligately anaerobic gram-positive bacterium Clostridium thermoautotrophicum. J. Bacteriol. 179:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, A., E. D. Coulter, D. M. Kurtz, Jr., and L. G. Ljungdahl. 2001. Five-gene cluster in Clostridium thermoaceticum consisting of two divergent operons encoding rubredoxin oxidoreductase-rubredoxin and rubrerythrin— type A flavoprotein— high-molecular-weight rubredoxin. J. Bacteriol. 183:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckers-Hebestreit, G., and K. Altendorf. 1996. The F0F1-type ATP synthases of bacteria: structure and function of the F0 complex. Annu. Rev. Microbiol. 50:791-824. [DOI] [PubMed] [Google Scholar]
- 15.Du, Z. Y., and P. D. Boyer. 1990. On the mechanism of sulfite activation of chloroplast thylakoid ATPases and the relation of ADP tightly bound at a catalytic site to the binding change mechanism. Biochemistry 29:402-407. [DOI] [PubMed] [Google Scholar]
- 16.Ebel, R. E., and H. A. Lardy. 1975. Stimulation of rat liver mitochondrial adenosine triphosphatse by anions. J. Biol. Chem. 250:191-196. [PubMed] [Google Scholar]
- 17.Fermin Pacheco-Moses, Jose J. Garcia, J. S. Rodriguez-Zavala, and R. Moreno-Scanchez. 2000. Sulfite and membrane energization induce two different active states of the Paracoccus denitrificans F0F1-ATPase. Eur. J. Biochem. 267:993-1000. [DOI] [PubMed] [Google Scholar]
- 18.Fillingame, R. H. 1997. Coupling H+ transport and ATP synthesis in F1F0-ATP synthases: glimpses of interacting parts in a dynamic molecular machine. J. Exp. Biol. 200:217-224. [DOI] [PubMed] [Google Scholar]
- 19.Fillingame, R. H., W. Jiang, O. Y. Dmitriev, and P. C. Jones. 2000. Structural interpretations of F0 rotary function in the Escherichia coli F1F0 ATP synthase. Biochim. Biophys. Acta 1458:387-403. [DOI] [PubMed] [Google Scholar]
- 20.Fillingame, R. H., M. Oldenburg, and D. Fraga. 1991. Mutation of alanine 24 to serine in subunit c of the Escherichia coli F1F0 ATP synthase reduces reactivity of aspartyl 61 with dicyclohexylcarbodiimide. J. Biol. Chem. 266:20934-20939. [PubMed] [Google Scholar]
- 21.Foster, D. L., and R. H. Fillingame. 1982. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J. Biol. Chem. 257:2009-2015. [PubMed] [Google Scholar]
- 22.Foster, J. W., and H. K. Hall. 1991. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J. Bacteriol. 173:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Futai, M., T. Noumi, and M. Maeda. 1989. ATP synthase (H+-ATPase): results by combined biochemical and molecular biological approaches. Annu. Rev. Biochem. 58:111-136. [DOI] [PubMed] [Google Scholar]
- 24.Girvin, M. E., and R. H. Fillingame. 1994. Hairpin folding of subunit c of F1F0 ATP synthase: 1H distance measurements to nitroxide-derivatized aspartyl-61. Biochemistry 33:665-674. [DOI] [PubMed] [Google Scholar]
- 25.Harris, D. A. 1981. The coupling ATPase complex: an evolutionary view. BioSystems 14:113-121. [DOI] [PubMed] [Google Scholar]
- 26.Hensel, M., G. Deckers-Hebestreit, and K. Altendorf. 1991. Purification and characterization of the F1 portion of the ATP synthase (F1F0) of Streptomyces lividans. Eur. J. Biochem. 202:1313-1319. [DOI] [PubMed] [Google Scholar]
- 27.Hicks, B., and T. A. Krulwich. 1990. Purification and reconstitution of the F1F0-ATP synthase from alkaliphilic Bacillus firmus OF4. Evidence that the enzyme translocates H+ but not Na+. J. Biol. Chem. 265:20547-20554. [PubMed] [Google Scholar]
- 28.Hicks, D. B., and T. A. Krulwich. 1986. The membrane ATPase of alkalophilic Bacillus firmus RAB is an F-type ATPase. J. Biol. Chem. 261:12896-12902. [PubMed] [Google Scholar]
- 29.Hicks, D. B., and C. F. Yocum. 1986. Properties of the cyanobacterial coupling factor ATPase from Spirulina platensis. II. Activity of the purified and membrane-bound enzymes. Arch. Biochem. Biophys. 245:230-237. [DOI] [PubMed] [Google Scholar]
- 30.Iraburu, M. J., M. L. Lopez-Zabalza, and E. Santiago. 1994. Catalytic and regulatory sites in CF1. Rev. Esp. Fisiol. 50:55-62. [PubMed] [Google Scholar]
- 31.Ivaschenko, A. T., and K. R. Uteulin. 1983. Effect of anions on the ATPase activity of submitochondrial particles. Biokhimia 48:11-16. [PubMed] [Google Scholar]
- 32.Ivey, D. M., and L. G. Ljungdahl. 1986. Purification and characterization of the F1 ATPase from Clostridium thermoaceticum. J. Bacteriol. 165:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, W., and R. H. Fillingame. 1998. Interacting helical faces of subunits a and c in the F1F0 ATP synthase of Escherichia coli defined by disulfide cross-linking. Proc. Natl. Acad. Sci. USA 95:6607-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kell, D. B., M. W. Peck, G. Rodger, and J. G. Morris. 1981. On the permeability to weak acids and bases of the cytoplasmic membrane of Clostridium pasteurianum. Biochem. Biophys. Res. Commun. 99:81-88. [DOI] [PubMed] [Google Scholar]
- 35.Klionsky, D. J., W. S. A. Brusilow, and R. D. Simoni. 1984. In vivo evidence for the role of the epsilon subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J. Bacteriol. 160:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi, H. 1987. Regulation of cytoplasmic pH in streptococci, p. 255-269. In J. Reijer and A. Peterkofsky (ed.), Sugar transport and metabolism in Gram-positive bacteria. Ellis Horwood Ltd., Chichester, United Kingdom.
- 37.Kuki, M., T. Noumi, M. Maeda, A. Amemura, and M. Futai. 1988. Functional domains of epsilon subunit of Escherichia coli H+-ATPase (F0F1). J. Biol. Chem. 263:17437-17442. [PubMed] [Google Scholar]
- 38.Kullen, M. J., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33:152-161. [DOI] [PubMed] [Google Scholar]
- 39.Kuo, P. H., and R. K. Nakamoto. 2000. Intragenic and intergenic suppression of the Escherichia coli ATP synthase subunit a mutation of Gly-213 to Asn: functional interactions between residues in the proton transport site. Biochem. J. 347:797-805. [PMC free article] [PubMed] [Google Scholar]
- 40.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 41.Laubinger, W., and P. Dimroth. 1988. Characterization of the ATP synthase of Propionigenium modestum as a primary sodium pump. Biochemistry 27:7531-7537. [DOI] [PubMed] [Google Scholar]
- 42.Laubinger, W., G.-D. Hebestreit, K. Altendorf, and P. Dimroth. 1990. A hybrid adenosinetriphosphatase composed of F1 of Escherichia coli and F0 of Propionigenium modestum is a functional sodium pump. Biochemistry 29:5458-5463. [DOI] [PubMed] [Google Scholar]
- 43.Ljungdahl, L. G. 1994. The acetyl-CoA pathway and the chemiosmotic generation of ATP during acetogenesis, p. 63-87. In H. L. Drake (ed.), acetogenesis. Chapman & Hall, New York, N.Y.
- 44.Lopez-Zabalza, M. J., A. J. Iriarte, J. Huaman, N. Lopez-Moratalla, and E. Santiago. 1980. Effect of bicarbonate and other anions on the oxidized and reduced forms of F1-ATPase. Rev. Esp. Fisiol. 36:421-426. [PubMed] [Google Scholar]
- 45.Maloney, P. C., and T. H. Wilson. 1985. The evolution of ion pumps. BioScience 35:143-148. [Google Scholar]
- 46.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 47.Miki, J., M. Maeda, Y. Mukohata, and M. Futai. 1988. The gamma-subunit of ATP synthase from spinach chloroplasts, primary structure deduced from the cloned cDNA sequence. FEBS Lett. 232:221-226. [DOI] [PubMed] [Google Scholar]
- 48.Moriyama, Y., A. Iwamoto, H. Hanada, M. Maeda, and M. Futai. 1991. One-step purification of Escherichia coli H+-ATPase (F0F1) and its reconstitution into liposomes with neurotransmitter transporters. J. Biol. Chem. 266:22141-22148. [PubMed] [Google Scholar]
- 49.Muntyan, M. S., I. V. Mesyanzhinova, Y. M. Milgrom, and V. P. Skulachev. 1990. The F1-type ATPase in anaerobic Lactobacillus casei. Biochim. Biophys. Acta 1016:371-377. [DOI] [PubMed] [Google Scholar]
- 50.Murataliev, M. B., and P. D. Boyer. 1992. The mechanism of stimulation of Mg ATPase activity of chloroplast F1-ATPase by non-catalytic adenine-nucleotide binding. Acceleration of the ATP-dependent release of inhibitory ADP from a catalytic site. Eur. J. Biochem. 209:681-687. [DOI] [PubMed] [Google Scholar]
- 51.Nalin, C. M., and R. E. McCarty. 1984. Role of disulfide bond in the gamma subunit in activation of ATPase of chloroplast coupling factor 1. J. Biol. Chem. 259:7275-7280. [PubMed] [Google Scholar]
- 52.Norling, B., G. Kelemen, and L. Ernster. 1986. Agents inducing high Mg2+-ATPase activity of isolated coupling factor 1 from spinach chloroplasts. Biochem. Biophys. Res. Commun. 141:636-642. [DOI] [PubMed] [Google Scholar]
- 53.Noumi, T., N. Oka, H. Kanazawa, and M. Futai. 1986. Mutational replacements of conserved amino acid residues in the beta subunit resulted in defective assembly of H+-translocating ATPase (F0F1) in Escherichia coli. J. Biol. Chem. 261:7070-7075. [PubMed] [Google Scholar]
- 54.Pedersen, P. L. 1994. ATP synthase. The machine that makes ATP. Curr. Biol. 4:1138-1141. [DOI] [PubMed] [Google Scholar]
- 55.Rahlfs, S., S. Aufurth, and V. Müller. 1999. The Na+-F1F0-ATPase operon from Acetobacterium woodii. Operon structure and presence of multiple copies of atpE which encode proteolipids of 8- and 18-kda. J. Biol. Chem. 274:33999-34004. [DOI] [PubMed] [Google Scholar]
- 56.Redlinger, J., and V. Müller. 1994. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1F0 type enzyme. Eur. J. Biochem. 223:275-283. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 58.Schneider, E., and K. Altendorf. 1985. All three subunits are required for the reconstitution of an active proton channel (F0) of Escherichia coli ATP synthase (F1F0). EMBO J. 4:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Senior, A. E. 1988. ATP synthesis by oxidative phosphorylation. Physiol. Rev. 68:177-231. [DOI] [PubMed] [Google Scholar]
- 60.Steffens, K., A. Di Gioia, G. Deckers-Hebestreit, and K. Altendorf. 1987. Structural and functional relationship of ATP synthases (F1F0) from Escherichia coli and the thermophilic bacterium PS3. J. Biol. Chem. 262:8334-8338. [PubMed] [Google Scholar]
- 61.Sumi, M., M. H. Sato, K. Denda, T. Date, and M. Yoshida. 1992. DNA fragment homologous to F1-ATPase β subunit was amplified from genomic DNA of Methanosarcina barkeri: indication of an archaebacterial F1-type ATPase. FEBS Lett. 314:207-210. [DOI] [PubMed] [Google Scholar]
- 62.Uteulin, K. R., and A. T. Ivashchenko. 1982. Peculiarities of anion actions on mitochondrial ATPase. Biokhimia 47:1641-1644. [PubMed] [Google Scholar]
- 63.Vik, S. B., J. C. Long, L. Wada, and D. Zhang. 2000. A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim. Biophys. Acta 1458:457-466. [DOI] [PubMed] [Google Scholar]
- 64.Walker, J. E., M. Saraste, and N. J. Gay. 1982. The unc operon: nucleotide sequence, regulation and structure of ATP synthase. Biochim. Biophys. Acta 768:164-200. [DOI] [PubMed] [Google Scholar]
- 65.Weidner, G., and G. Sawers. 1996. Molecular characterization of the genes encoding pyruvate formate-lyase and its activating enzyme of Clostridium pasteurianum. J. Bacteriol. 178:2440-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]