Abstract
Hereditary hemochromatosis (HH) is a common autosomal recessive disease characterized by increased iron absorption and progressive iron storage that results in damage to major organs in the body. Recently, a candidate gene for HH called HFE encoding a major histocompatibility complex class I-like protein was identified by positional cloning. Nearly 90% of Caucasian HH patients have been found to be homozygous for the same mutation (C282Y) in the HFE gene. To test the hypothesis that the HFE gene is involved in regulation of iron homeostasis, we studied the effects of a targeted disruption of the murine homologue of the HFE gene. The HFE-deficient mice showed profound differences in parameters of iron homeostasis. Even on a standard diet, by 10 weeks of age, fasting transferrin saturation was significantly elevated compared with normal littermates (96 ± 5% vs. 77 ± 3%, P < 0.007), and hepatic iron concentration was 8-fold higher than that of wild-type littermates (2,071 ± 450 vs. 255 ± 23 μg/g dry wt, P < 0.002). Stainable hepatic iron in the HFE mutant mice was predominantly in hepatocytes in a periportal distribution. Iron concentrations in spleen, heart, and kidney were not significantly different. Erythroid parameters were normal, indicating that the anemia did not contribute to the increased iron storage. This study shows that the HFE protein is involved in the regulation of iron homeostasis and that mutations in this gene are responsible for HH. The knockout mouse model of HH will facilitate investigation into the pathogenesis of increased iron accumulation in HH and provide opportunities to evaluate therapeutic strategies for prevention or correction of iron overload.
Keywords: major histocompatibility complex class I protein, iron, liver, gene targeting
Hereditary hemochromatosis (HH) is a common autosomal recessive disease of iron metabolism leading to increased iron storage. Its frequency is estimated to be 1 in 300–400 and the carrier frequency to be 1 in 8–10 individuals of Northern European descent (1–5). In HH, loss of the normally tight regulation of intestinal iron absorption leads to an increase in transferrin saturation and progressive iron deposition in the cytoplasm of parenchymal cells of various organs and tissues, including the liver, pancreas, heart, joints, and endocrine glands. The clinical consequences of iron overload include cirrhosis of the liver and hepatocellular cancer, diabetes, heart failure, arthritis, and hypogonadism. Recently, Feder et al. (6) reported a candidate gene for HH that was identified by positional cloning and that encodes a novel major histocompatibility complex class I-like protein called HFE. They found 83% of HH patients are homozygous for the same missense mutation (C282Y). A few were compound heterozygotes for C282Y and a second mutation (H63D). These findings were confirmed by several other studies (7–11).
The 343-aa human HFE protein contains an extracellular peptide-binding region (α1 and α2), an Ig-like domain (α3), a transmembrane region, and a short cytoplasmic tail. The C282Y mutation disrupts a critical disulfide bond in the α3 domain of the HFE protein, which prevents its binding to β2-microglobulin (β2M) and its presentation on the cell surface (12, 13). Parkkila et al. (14) showed that the HFE protein is expressed in many segments of the gut and has a unique subcellular localization in the crypts of the small intestine that suggests a role for the HFE protein in regulating iron absorption in the small intestine.
The findings that β2M-deficient mice developed progressive iron overload similar to that seen in HH patients initially suggested the involvement of a major histocompatibility complex class I gene in HH (15–17). To test the hypothesis that deficiency or functional derangement of the HFE gene product is the molecular basis of HH, we generated mice with a targeted HFE mutation. HFE-deficient mice exhibit profound abnormalities in iron homeostasis. The abnormally high transferrin saturations and excessive accumulation of iron in liver, even on a standard diet, demonstrate that the HFE gene is involved in the regulation of homeostasis. Until now, no animal model has been found that completely recapitulates the biochemical abnormalities and histopathology of human HH (18). The mouse homozygous for the HFE gene knockout provides such a model for human HH and the observations reported here demonstrate that the mechanisms regulating iron absorption are conserved between mouse and human.
MATERIALS AND METHODS
HFE Targeting Vector.
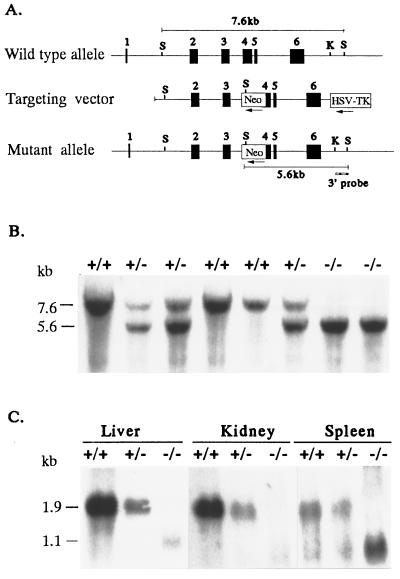
We used a mouse cDNA probe to isolate a single BAC clone from a 129/SvJ mouse BAC library (Genome Systems). Genomic sequencing of 3- to 5-kb plasmid subclones (6) identified four overlapping clones covering ≈15 kb, which contain the entire mouse HFE gene. The 3.5-kb Xhol–EcoRV fragment containing part of intron 1, exon 2 and exon 3, and part of intron 3 was inserted upstream of the Neo gene of plasmid PGKNeobpATK (Stratagene) digested with the compatible XhoI and EcoRV. The 2.6-kb HincII–XhoI fragment containing approximately half of exon 4 and exon 5 and 6 was subcloned between the Neo gene and thymidine kinase (Fig. 1A). Thus, the 1.7-kb PGKneo cassette replaced a 360-bp EcoRV–HincII fragment encompassing a portion of exon 4 and intron 3. The location of the genomic probe used to screen for homologous recombination is shown in Fig. 1A.
Figure 1.
Targeted disruption of the HFE gene. (A) Structure of the HFE gene (Top), the targeting construct (Middle), and the predicted structure of the disrupted HFE gene after homologous recombination (Bottom). Only the relevant restriction sites are shown: S, SacI site; K, KpnI site. The numbered solid boxes represent exons. Neo and HSV-TK (herpes simplex virus thymidine kinase) refer to the positive and negative selective markers, respectively. The position of the 3′ external probe is indicated. The horizontal lines show the positions of the 7.6-kb and 5.6-kb restriction fragments diagnostic for wild-type and properly targeted alleles, respectively. (B) Southern blot analysis of SacI-digested genomic DNA of eight mice from one litter resulting from a cross between two HFE+/− mice. The blot was hybridized with the 3′ KpnI–SacI probe. The wild-type and mutant alleles are indicated by 7.6- and 5.6-kb SacI fragments, respectively. (C) Northern blot analysis of liver, kidney, and spleen HFE mRNA from HFE+/+, HFE+/−, and HFE−/− mice. Twenty micrograms of total RNA from liver, kidney, or spleen RNA were analyzed by Northern blotting with mouse HFE 32P-labeled riboprobe. The 1.9-kb mRNA transcript present in +/+ mice was reduced in amounts in HFE+/− tissues, and absent in tissues from HFE−/− mutant mice.
Gene Targeting in Embryonic Stem (ES) Cells and Generation of Mutant Mice.
The targeting vector (25 μg) was linearized with BamHI and introduced into the 129/Sv-derived ES cell line RW4 (Genome Systems, St. Louis) (1 × 107 cells) by electroporation (230 V and 500 μF) in a Bio-Rad gene pulser. After 24 hr, the cells were placed under selection with 400 μg/ml G418 (GIBCO/BRL) and 2 μM gangciclovir (Syntex) for 6 days. Genomic DNA of resistant clones was digested with SacI and hybridized with the 3′ 0.9-kb KpnI–SacI external probe (Fig. 1A). Three independent targeted ES clones, HFE 17, 23, and 124, were used for injection into the blastocysts of C57BL/6J mice and transferred into pseudopregnant female mice as described (19). Chimeric male offspring were bred to C57BL/6J females and the agouti F1 offspring were tested for transmission of the disrupted allele by Southern blot analysis of SacI-digested genomic DNA by using the 3′ external probe (Fig. 1 A and B). Heterozygous matings of the F1 mice were carried out to produce homozygous F2 mutant mice. Mice were fed with a standard diet (LM-485 Teklad sterilized mouse diet 7012, Harlan, which contains 0.02% wt/wt iron) or, in one 14-day iron loading experiment, with a Purina 5001 Plus butylated hydroxytoluene control diet containing 0.02% wt/wt iron with or without supplemental 2% (wt/wt) carbonyl iron (Harlan). Food was removed 14 hr before blood collections and isolation of tissues for measurement of iron content and morphological examination.
Northern Blot Analysis.
Total cellular RNA was isolated from tissues of HFE+/+, HFE+/−, and HFE−/− mice using a guanidinium/phenol solution (RNA-Stat60, Tel-Test, Friendswood, TX). Twenty micrograms of RNA from each source was denatured in formaldehyde-containing buffer and electrophoresed in 1% agarose, 2.2 M formaldehyde gels. Equivalent loading of intact RNA was assured by visualization of ethidium bromide stained 28S and 18S ribosomal RNA bands. Transcript sizes were estimated by using RNA standards (Promega). The RNA was transferred to Nytran membranes (Schleicher & Schuell), immobilized by UV crosslinking, and prehybridized at 65°C in 50% formamide, 5× SSPE (standard saline phosphate/EDTA), 5× Denhardt’s, 50 mM NaPO4 (pH 6.5), 200 μg/ml salmon sperm DNA, 1 mM EDTA, 0.1%SDS. Blots were hybridized overnight at 65°C with 32P-labeled riboprobe, washed in 2× SSPE at reverse transcriptase (RT) for 20 min, 0.2× SSPE at RT for 20 min, twice in 0.2× SSPE/0.1% SDS at 65°C for 20 min, and autoradiographed.
Transferrin Saturation.
Mice were bled retroorbitally. Serum iron and total iron binding capacity (TIBC) were measured by using the protocol of Fielding (20). For the results shown in Fig. 2, 200 μl of serum from each animal was used for analysis of iron and TIBC by using a kit from Sigma, and the assays were performed by Genox, using a Cobas Fara II chemical analyzer. Transferrin saturation was calculated as (serum iron ÷ TIBC) × 100%.
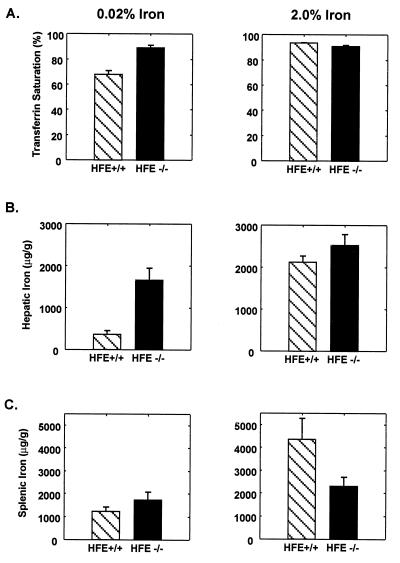
Figure 2.
Increased transferrin saturation and hepatic iron content in HFE−/− mice. Values represent means ± SE. (Left) The transferrin saturation (A), hepatic iron content (B), and splenic iron content (C) in HFE+/+ and HFE−/− mice fed 0.02% iron diets from weaning until sacrifice at 10 weeks of age. Both the transferrin saturation and hepatic iron content were increased in the HFE−/− mice (P < 0.001 and P < 0.01, respectively). (Right) The transferrin saturation (A), hepatic iron content (B), and splenic iron content (C) in HFE+/+ and HFE−/− mice fed control diets supplemented with 2% wt/wt carbonyl iron for the 14 days before sacrifice at age 10 weeks. HFE+/+ mice showed an increase in transferrin saturation from 68 ± 3 to 94 ± 0.2%, a 6-fold increase in hepatic iron (370 ± 87 to 2,124 ± 149 μg/g dry wt, P < 0.001), and a 3.5-fold increase in splenic iron (1,244 ± 191 to 4,361 ± 905 μg/g dry wt, P < 0.004) in response to dietary iron loading. The HFE−/− mice had no increase in the already high transferrin saturation with dietary iron loading. However, their 1.5-fold increase in hepatic iron (from 1,660 ± 286 to 2,523 ± 262 μg/g dry wt) was significant (P < 0.03). Splenic iron was not significantly increased in response to iron loading in −/− mice (P = 0.15).
Measurement of Tissue Iron Content.
Tissues isolated from HFE+/+, HFE+/−, or HFE−/− littermates were analyzed for nonheme iron as described by Torrance and Bothwell (21). Liver and spleen samples were weighed dry, digested in acid digestion mixture (3 M hydrochloric acid, 10% trichloroacetic acid) at 65°C for 20 hr, and 400 μl of each acid extract was mixed with 1.6 ml of bathophenanthroline chromagen reagent. The absorbance at 535 nm was measured in a DU-65 spectrophotometer (Beckman).
Histology.
Six HFE+/+ and six HFE−/− mice fed standard basal diet, and six HFE+/+ and six HFE−/− mice fed control diet supplemented with carbonyl iron (2%, wt/wt) from week 8 to week 10 were sacrificed at age 10 weeks. Tissues fixed in 10% neutral buffered formalin for 18 hr were subjected to routine histologic processing, and the sections were stained for iron storage with Perls’ Prussian blue stain for the detection of storage iron.
Hematological Measurements.
Blood was obtained by retroorbital sampling, and hemoglobin, hematocrit, and mean corpuscular volume were determined by using a System 9110 plus hematology analyzer.
RESULTS
Generation of HFE-Deficient Mice.
To disrupt the HFE gene in mouse ES cells, we designed a replacement-type targeting vector by using fragments from two overlapping genomic clones. A 360-bp region, including the first 134 bp of the 275-bp exon 4, was deleted and replaced with the PGKneo cassette in the reverse orientation (Fig. 1A). The targeting vector had a total of 6.1 kb of homologous genomic sequence flanking the Neo cassette, with 3.5 kb upstream and 2.6 kb downstream. The construct was linearized and introduced into RW4 ES cells by electroporation. After selection with G418 and gangciclovir, doubly resistant clones were screened for homologous recombination by Southern blotting and hybridization with a 3′ external probe (Fig. 1A). Of 386 clones screened, 14 contained the 5.6-kb SacI band diagnostic of homologous recombination in one allele, in addition to the 7.6-kb fragment from the other wild-type allele. Targeted ES cells containing one disrupted HFE allele were injected into C57BL/6 blastocysts and chimeric males were derived from three independent clones (17, 23, and 124), all of which showed germ-line transmission of the disrupted allele. Heterozygous F1 offspring from ES clones 17 and 23 were independently intercrossed to generate F2 homozygous mice of 129/Sv × C57BL/6 hybrid strain background. Offspring were genotyped by Southern blotting of genomic tail DNA and hybridized with a 3′ external probe (Fig. 1B). Two independent mouse lines, derived from two separate ES cell clones, had identical, grossly normal phenotypes. All mice heterozygous and homozygous for the HFE gene disruption appeared healthy, grew and reproduced normally, and produced homozygous offspring in the number expected. Combined data from crosses between F1 heterozygous progeny derived from clone 17 and also between F1 progeny derived from clone 23 showed a distribution of 25% (+/+), 53% (+/−), and 22% (−/−) in 249 offspring analyzed.
The Disrupted HFE Allele Produces No Normal mRNA Transcript.
To confirm that HFE−/− mice do not express the HFE gene product, we performed Northern blot analyses on total RNA isolated from liver, kidney, and spleen of HFE +/+, HFE+/−, and HFE−/− littermates (Fig. 1C). An HFE transcript of 1.9 kb, present in multiple tissues from +/+ mice, was present in reduced amounts in +/− mice, and was not detectable in the tissues from HFE−/− mice. These results suggest that the targeting event resulted in a null allele. An additional 1.1-kb cross-hybridizing transcript was detected in RNA from some tissues from +/+ and +/− mice, and was more prominent in RNA from −/− mice. Both a 1.9-kb and a 1.1-kb transcript were also evident on the Northern blot of RNA of several tissues from normal mice reported by Hashimoto et al. (22).
Targeted Disruption of the HFE Allele Produces Iron Storage.
To determine the effect of the disrupted allele on iron homeostasis, we analyzed 18 F2 mice, six each of HFE+/+, HFE+/−, and HFE−/− genotypes, at 10 weeks of age, that were maintained on a standard diet (0.02% wt/wt iron) from weaning at age 3 weeks. The data in Table 1 show striking differences in transferrin saturation (96 ± 5% vs. 77 ± 3%, P < 0.007) and liver iron content (2,071 ± 450 vs. 255 ± 23 μg/g dry wt, P < 0.002) between the +/+ and −/− F2 mice. Although all six −/− mice had elevated liver iron levels, there was a considerable variation between individual −/− mice (800–3,400 μg/g dry wt), which might be caused by differences in C57BL/6 and 129/Sv genes segregating in the different F2 mice (23). The spleen iron contents showed considerable variation that did not correlate with HFE genotype. Heterozygote values for transferrin saturation and liver iron content both showed slight increases compared with +/+ littermates but the differences were not significant (P = 0.14 and 0.37, respectively).
Table 1.
Serum and tissue iron levels for mice on standard diet
| Mice | Serum
|
Tissue iron
|
|||
|---|---|---|---|---|---|
| Iron, μg/dl | TIBC, μg/dl | Transferrin saturation, % | Liver, μg/g dry wt | Spleen, μg/g dry wt | |
| HFE+/+ | 291 ± 9 | 379 ± 13 | 77 ± 3 | 255 ± 23 | 903 ± 153 |
| HFE+/− | 310 ± 21 | 358 ± 20 | 87 ± 5 | 280 ± 13 | 1,164 ± 232 |
| HFE−/− | 323 ± 24 | 338 ± 17 | 96 ± 5* | 2,071 ± 450* | 918 ± 120 |
Values are presented as mean ± SE for n = 6 mice from each group. TIBC, total iron binding capacity.
Significant differences were observed between HFE+/+ and HFE−/− mice for transferrin saturation (P < 0.007) and liver iron content (P < 0.002).
To verify the striking effect of the HFE gene disruption on iron homeostasis and to study its effect under conditions of iron loading, we placed 12 mice homozygous for the disrupted allele and 12 of their +/+ littermates on either a control diet (0.02% wt/wt iron) or an iron-enriched diet (2% wt/wt carbonyl iron) at age 8 weeks and sacrificed them for analysis at age 10 weeks. Fig. 2 summarizes the data on transferrin saturation, liver iron content, and spleen iron content of HFE knockout mice and their normal littermates. The left half of the figure shows the data from six mice of each group without iron supplementation. Again, the mice with the targeted disruption of the HFE allele showed profound differences in parameters of iron homeostasis. The HFE−/− mice fed with standard diet had a much higher transferrin saturation (Fig. 2A) than the HFE+/+ littermate mice (89 ± 2% vs. 68 ± 3%, P < 0.001). Their hepatic iron concentrations (Fig. 2B) were also much higher than those of the wild-type littermates (1,660 ± 290 vs. 370 ± 87 μg/g dry wt, P < 0.01). Fig. 2C shows the HFE−/− mice had spleen iron contents that did not differ significantly from those of their normal littermates. The right half of Fig. 2 shows the data from F2 HFE+/+ and HFE−/− mice that were fed the iron-supplemented diet. The +/+ mice showed increases in transferrin saturation (94 ± 0.2% vs. 68 ± 3%, P < 0.001), hepatic iron content (2,124 ± 149 vs. 370 ± 87 μg/g dry wt, P < 0.001), and spleen iron content (4,361 ± 905 vs. 1,244 ± 191 μg/g dry wt, P < 0.004) compared with +/+ mice on the control diet. The HFE−/− mice also showed an increase in liver iron content over the already elevated levels that were evident on the control diet in response to dietary iron loading (2,523 ± 262 vs. 1,660 ± 290 μg/g dry wt, P < 0.03). However, the −/− mice showed nowhere near the increase in iron loading of the spleen (2,305 ± 395 vs. 1,736 ± 339 μg/g dry wt, P = 0.15) that was seen in the +/+ mice on the iron-supplemented diet (4,361 ± 905 vs. 1,244 ± 191 μg/g dry wt, P < 0.004), suggesting that the HFE−/− mice are relatively resistant to iron deposition in reticuloendothelial cells as is the case in humans with HH (24–26).
Histopathology of HFE Knockout Mice Resembles That of HH.
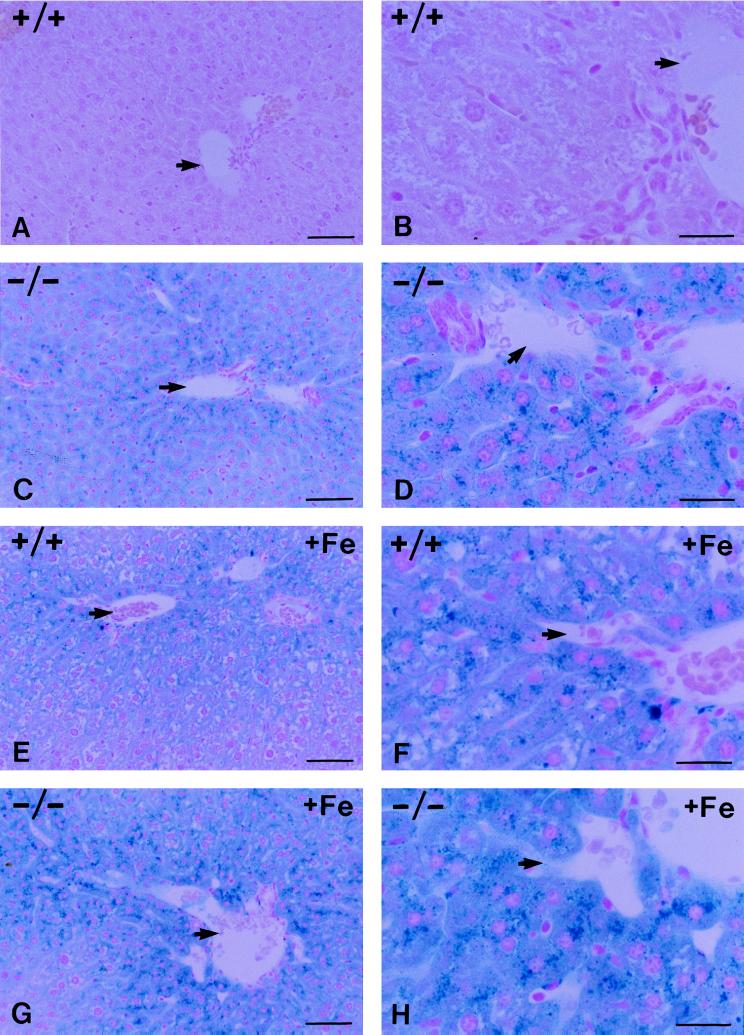
Histologic examination was done on the tissues from the HFE+/+ and HFE−/− mice sacrificed at age 10 weeks after 2 weeks on control diet with and without iron supplementation. Perls’ Prussian blue-stained sections of liver, spleen, heart, lungs, kidney, pancreas, and small intestine were examined. Iron deposition was seen only in liver, spleen, and small intestine. Fig. 3 presents low-power (A, C, E, and G) and high-power (B, D, F, and H) views of sections demonstrating the histopathology of liver. Fig. 3 A and B shows that there is no stainable iron in the liver from an HFE+/+ littermate on the control diet. Fig. 3 C and D shows a representative section of liver from an HFE−/− mouse fed with control diet. Stainable iron was prominent and was located predominantly in hepatocytes with a periportal to pericentral gradient. Fig. 3 E and F shows staining of a representative section from +/+ mice fed with iron-supplemented diet for 2 weeks before sacrifice. These mice showed a marked gradient of hepatocellular iron deposition with periportal predominance. Mild sinusoidal lining cell iron staining, presumably in Kupffer cells, was seen in the periportal area. Fig. 3 G and H shows a representative stained section from HFE−/− mice fed with the iron-supplemented diet. A zonal gradient with periportal predominance persisted in these mice. In all spleens, iron deposition was seen in varying amounts in scattered perisinusoidal cells. In the small intestine, stainable iron was present in the epithelial cells of the villi and not in the cryptal enterocytes (data not shown).
Figure 3.
Perls’ Prussian blue staining of liver sections from HFE+/+ and HFE−/− mice fed control diet (A–D) or control diet supplemented with 2% (wt/wt) carbonyl iron (E–H). Shown are low-power views (A, C, E, and G) and high-power views (B, D, F, and H) of sections. A and B show the absence of stainable iron in the +/− mouse liver fed the control diet. C and D show prominent stainable iron in hepatocytes with periportal predominance in liver from HFE−/− mice fed the control diet. E and F show iron accumulation in HFE+/+ mouse liver in response to iron loading. G and H show the stainable iron in the HFE−/− mice after 2 weeks of feeding with the iron-supplemented diet. The arrows indicate the location of branches of the portal vein. (Bars: A, C, E, and G = 50 μm in the low-power views; B, D, F, and H = 20 μm in the high-power views.)
Erythroid Parameters.
To rule out the possibility that the increased iron storage in HFE−/− mice might result from anemia, we measured erythroid parameters on +/+ and −/− mice at age of 10 weeks. Data on red blood cells, hemoglobin concentration, hematocrit, and mean corpuscular volume are summarized in Table 2. These results demonstrate that anemia was not contributing to the increase in iron storage seen in HFE−/− mice.
Table 2.
Erythroid parameters
| Mice | n | RBC, 106/mm3 | Hb, g/dl | Hct, % | MCV, fl |
|---|---|---|---|---|---|
| HFE+/+ | 11 | 10.1 ± 0.5 | 16.7 ± 0.5 | 48.6 ± 2.1 | 48.8 ± 1.3 |
| HFE+/+ + iron | 6 | 9.9 ± 0.5 | 16.5 ± 0.4 | 48.2 ± 1.6 | 49.3 ± 1.5 |
| HFE−/− | 11 | 10.1 ± 0.4 | 16.7 ± 0.7 | 49.9 ± 1.7 | 49.2 ± 0.8 |
| HFE−/− + iron | 6 | 10.0 ± 0.6 | 17.0 ± 0.6 | 51.9 ± 2.4 | 51.8 ± 0.9 |
Red blood cells (RBC), hemoglobin concentration (Hb), hematocrit (Hct), and mean corpuscular volume (MCV) analyzed in male and female wild-type (HFE+/+) and mutant (HFE−/−) littermates at 10 weeks of age. HFE+/+ + iron and HFE−/− + iron are mice fed with an iron-supplemented diet containing 2% (w/w) carbonyl iron for 14 days before sacrifice. n, number of animals. Data are presented as mean ± SD.
DISCUSSION
Until now, HFE was considered a likely candidate gene for HH, but its role in iron metabolism was uncertain (25). The possibility remained that the C282Y mutation found in most patients with HH was a passenger mutation, present in most HH patients only because of tight linkage disequilibrium with a still unknown gene that was actually responsible for HH. Our results remove all doubt whether mutations that disrupt the function of the HFE gene product can produce HH. Even on a standard diet, the HFE gene knockout mouse described here exhibits abnormally high transferrin saturation and excessive iron accumulation in the liver. The iron deposition occurs predominantly in hepatocytes as is the case in HH. The HFE−/− mice on the iron-supplemented diet showed less loading of the spleen than the HFE+/+ mice on the iron-supplemented diet. This relative resistance of the spleen to iron loading is also similar to the findings in human HH (24, 25). Thus, the HFE gene knockout model appears to faithfully recapitulate both the biochemical abnormalities and the histopathology of HH. Like human HH, the murine HFE deficiency HH model shows autosomal recessive inheritance with little evident abnormality in the heterozygotes, at least at age 10 weeks. Given the fact that the homozygote shows such exaggerated iron storage at age 10 weeks, it will be interesting to follow the HFE−/− mice as they age to determine which of the usual complications of human HH such as liver fibrosis, diabetes, other endocrine problems, cardiomyopathy, and arthritis appear over time.
The HFE knockout model we report here resembles the β2M knockout mouse, which also has excessive iron storage, and provided evidence that a major histocompatibility complex class I type gene might be involved in HH (15–17). The HFE knockout mice appear to have a more severe phenotype in that the iron accumulation in the liver on a standard diet develops more rapidly. Longer follow-up will be needed for further comparison of the two models. For studies of iron homeostasis, the HFE gene knockout mouse has the advantage that only the HFE gene function is disrupted. The β2M knockout mouse also has immunologic abnormalities because cell surface expression of all major histocompatibility complex class I gene products are affected by the deficiency of β2M (27).
The HFE knockout mouse should facilitate studies of the pathogenesis of HH, including the effect of the gene disruption on intestinal iron absorption. Because mice have marked strain differences in susceptibility to iron loading (23), it will be important for the HFE gene disruption to be bred onto a uniform genetic background. The individual differences in parameters of iron status in this study suggest that the F2 129/Sv × C57BL/6 mice may be segregating genes that affect the severity of the abnormal iron storage resulting from HFE gene disruption. Mapping and identifying these genes should be possible given the increasing density of DNA markers on the mouse map [The Jackson Laboratory, Mouse Genome Informatics, available at: http://www.informatics.jax.org/(accessed 12/11/97)]. The HFE knockout mouse also can be used to study the interaction with other mutations known to influence iron absorption such as those producing thalassemia (28), heme oxygenase 1 deficiency (29), and atransferrinemia (30). Additionally, it might be used to explore the functional significance of the recent observation that the normal HFE protein forms a stable complex with β2M and with the transferrin receptor (31, 32). The association with the transferrin receptor provides a link between the HFE gene product and a key protein involved in iron transport that could play a role in the pathogenesis of HH. Finally, the mutant mouse should be useful in testing experimental therapies for prevention and correction of the iron storage in HH and in other hematological disorders associated with iron overload.
Acknowledgments
We acknowledge Elizabeth Torno for editorial assistance and Joni Kneer for technical assistance. This work was supported by National Institutes of Health Grants DK40163 and GM34182 to W.S.S. and DK41816 to B.R.B. We also acknowledge the generous support from the Pediatric Research Institute, Cardinal Glennon Children’s Hospital.
ABBREVIATIONS
- HH
hereditary hemochromatosis
- β2M
β2-microglobulin
- ES cell
embryonic stem cell
Footnotes
A commentary on this article begins on page 2033.
References
- 1.Cartwright G E, Edwards C Q, Kravitz K, Skolnick M, Amos D B, Johnson A, Buskjaer L. N Engl J Med. 1979;301:175–179. doi: 10.1056/NEJM197907263010402. [DOI] [PubMed] [Google Scholar]
- 2.Borwein S T, Ghent C N, Flanagan P R, Chamberlain M J, Valberg L S. Clin Invest Med. 1983;6:171–179. [PubMed] [Google Scholar]
- 3.Edwards C Q, Griffen L M, Goldgar D, Drummond C, Skolnick M H, Kushner J P. N Engl J Med. 1988;318:1355–1362. doi: 10.1056/NEJM198805263182103. [DOI] [PubMed] [Google Scholar]
- 4.Bacon B R, Tavill A S. In: Hepatology: A Textbook of Liver Disease. Zakim D, Boyer T D, editors. Philadelphia: Saunders; 1996. pp. 1439–1472. [Google Scholar]
- 5.Barton J C, Bertoli L F. Nat Med. 1996;2:394–395. doi: 10.1038/nm0496-394. [DOI] [PubMed] [Google Scholar]
- 6.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 7.Beutler E, Gelbart T, West C, Lee P, Adams M, et al. Blood Cells Mol Dis. 1996;22:187–194. doi: 10.1006/bcmd.1996.0027. [DOI] [PubMed] [Google Scholar]
- 8.Jouanolle A M, Gandon G, Jézéquel P, Blayau M, Campion M L, et al. Nat Genet. 1996;14:251–252. doi: 10.1038/ng1196-251. [DOI] [PubMed] [Google Scholar]
- 9.Jazwinska E C, Cullen L M, Busfield F, Pyper W R, Webb S I, Powell L W, Morris C P, Walsh T P. Nat Genet. 1996;14:249–251. doi: 10.1038/ng1196-249. [DOI] [PubMed] [Google Scholar]
- 10.Carella M, D’Ambrosio L, Totaro A, Grifa A, Valentino M A, Piperno A, Girelli D, Roetto A, Franco B, Gasparini P, Camaschella C. Am J Hum Genet. 1997;60:828–832. [PMC free article] [PubMed] [Google Scholar]
- 11.Borot N, Roth M-P, Malfroy L, Demangel C, Vinel J-P, Pascal J-P, Coppin H. Immunogenetics. 1997;45:320–324. doi: 10.1007/s002510050211. [DOI] [PubMed] [Google Scholar]
- 12.Feder J N, Tsuchihashi Z, Irrinki A, Lee V K, Parkkila S, Sly W S, Schatzman R C. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 13.Waheed A, Parkkila S, Zhou X Y, Tomatsu S, Tsuchihashi Z, Feder J N, Schatzman R C, Britton R S, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:12384–12389. doi: 10.1073/pnas.94.23.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkkila S, Waheed A, Britton R S, Feder J N, Tsuchihashi Z, Schatzman R C, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:2534–2539. doi: 10.1073/pnas.94.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Sousa M, Reimao R, Lacerda R, Hugo P, Kaufmann S H, Porto G. Immunol Lett. 1994;39:105–111. doi: 10.1016/0165-2478(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 16.Rothenberg B E, Voland J R. Proc Natl Acad Sci USA. 1996;93:1529–1534. doi: 10.1073/pnas.93.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos M, Schilham M W, Rademakers L H P M, Marx J J M, de Sousa M, Clevers H. J Exp Med. 1996;184:1975–1985. doi: 10.1084/jem.184.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iancu T C. Adv Vet Sci Comp Med. 1993;37:379–401. [PubMed] [Google Scholar]
- 19.Zhou X Y, Morreau H, Rottier R, Davis D, Bonten E, et al. Genes Dev. 1995;9:2623–2634. doi: 10.1101/gad.9.21.2623. [DOI] [PubMed] [Google Scholar]
- 20.Fielding J. Methods Hematol. 1980;1:15–43. [Google Scholar]
- 21.Torrance J D, Bothwell T H. Methods Hematol. 1980;1:90–115. [Google Scholar]
- 22.Hashimoto K, Hirai M, Kurosawa Y. Biochem Biophys Res Comm. 1997;230:35–39. doi: 10.1006/bbrc.1996.5889. [DOI] [PubMed] [Google Scholar]
- 23.Leboeuf R C, Tolson D, Heinecke J W. J Lab Clin Med. 1995;126:128–136. [PubMed] [Google Scholar]
- 24.Anderson G J, Halliday J W, Powell L W. Hepatology. 1987;7:967–969. doi: 10.1002/hep.1840070529. [DOI] [PubMed] [Google Scholar]
- 25.Cox T. Nat Genet. 1996;13:386–388. doi: 10.1038/ng0896-386. [DOI] [PubMed] [Google Scholar]
- 26.Cairo G, Recalcati S, Montosi G, Castrusini E, Conte D, Pietrangelo A. Blood. 1997;89:2546–2553. [PubMed] [Google Scholar]
- 27.Steinmetz M, Haas W. BioEssays. 1993;15:613–615. doi: 10.1002/bies.950150907. [DOI] [PubMed] [Google Scholar]
- 28.Yang B, Kirby S, Lewis J, Detloff P J, Maeda N, Smithies O. Proc Natl Acad Sci USA. 1995;92:11608–11612. doi: 10.1073/pnas.92.25.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poss K D, Tonegawa S. Proc Natl Acad Sci USA. 1997;94:10919–10924. doi: 10.1073/pnas.94.20.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craven C M, Alexander J, Eldridge M, Kushner J P, Bernstein S, Kaplan J. Proc Natl Acad Sci USA. 1987;84:3457–3461. doi: 10.1073/pnas.84.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkkila S, Waheed A, Britton R S, Bacon B R, Zhou X Y, Tomatsu S, Fleming R E, Sly W S. Proc Natl Acad Sci USA. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feder J N, Penny D M, Irrinki A, Lee V K, Lebron J A, Watson N, Tsuchihashi Z, Sigal E, Bjorkman P J, Schatzman R C. Proc Natl Acad Sci USA. 1998;95:1475–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]