Abstract
The seven conserved enzymatic domains required for tryptophan (Trp) biosynthesis are encoded in seven genetic regions that are organized differently (whole-pathway operons, multiple partial-pathway operons, and dispersed genes) in prokaryotes. A comparative bioinformatics evaluation of the conservation and organization of the genes of Trp biosynthesis in prokaryotic operons should serve as an excellent model for assessing the feasibility of predicting the evolutionary histories of genes and operons associated with other biochemical pathways. These comparisons should provide a better understanding of possible explanations for differences in operon organization in different organisms at a genomics level. These analyses may also permit identification of some of the prevailing forces that dictated specific gene rearrangements during the course of evolution. Operons concerned with Trp biosynthesis in prokaryotes have been in a dynamic state of flux. Analysis of closely related organisms among the Bacteria at various phylogenetic nodes reveals many examples of operon scission, gene dispersal, gene fusion, gene scrambling, and gene loss from which the direction of evolutionary events can be deduced. Two milestone evolutionary events have been mapped to the 16S rRNA tree of Bacteria, one splitting the operon in two, and the other rejoining it by gene fusion. The Archaea, though less resolved due to a lesser genome representation, appear to exhibit more gene scrambling than the Bacteria. The trp operon appears to have been an ancient innovation; it was already present in the common ancestor of Bacteria and Archaea. Although the operon has been subjected, even in recent times, to dynamic changes in gene rearrangement, the ancestral gene order can be deduced with confidence. The evolutionary history of the genes of the pathway is discernible in rough outline as a vertical line of descent, with events of lateral gene transfer or paralogy enriching the analysis as interesting features that can be distinguished. As additional genomes are thoroughly analyzed, an increasingly refined resolution of the sequential evolutionary steps is clearly possible. These comparisons suggest that present-day trp operons that possess finely tuned regulatory features are under strong positive selection and are able to resist the disruptive evolutionary events that may be experienced by simpler, poorly regulated operons.
INTRODUCTION
It has become quite apparent from the recent flood of genomic data that dynamic gene reorganization is an ongoing process (albeit of unknown significance) that distinguishes even closely related genomes. Genes that stay together within operons must resist the gene-scrambling process. Operons that embrace a complete complement of pathway-specific structural genes (whole-pathway operons), such as the ones encoding all the enzymes of tryptophan (Trp) biosynthesis or histidine biosynthesis, have a classical status in both biochemistry and molecular genetics that extends far beyond understanding these pathways per se. Such whole-pathway operons are broadly distributed among prokaryotes. However, the pathway genes may be completely scattered in some organisms, and in yet other organisms, the pathway genes may be organized into two or more “split-pathway” operons. This raises intriguing questions about what the evolutionary relationship is between whole-pathway operons, split-pathway operons, and those cases where all pathway genes are unlinked. Is it possible to deduce whether a given whole-pathway operon was an ancient innovation and therefore that operon splitting and/or gene dispersal followed in some lineages? Or are whole-pathway operons relatively recent innovations that are derived from split-pathway operons? Or, since these two scenarios are not mutually exclusive, is it possible that both apply?
An ideal operon system for this analysis is the trp operon. We show that the trp operon must have been present in early prokaryote ancestors. In Bacteria but not in Archaea, sufficient genome representation exists to deduce an ancestral whole-pathway trp operon. The regulation of this operon may initially have been quite minimal since the first evolutionary step(s) probably would be to collect the structural genes together. Parsimony principles support a hypothesis developed in this paper of two major evolutionary events in Bacteria, one splitting the ancestral operon in two and the other rejoining it by gene fusion. We assert that a detailed analysis can recognize occasional events of lateral gene transfer (LGT) or paralogy. Both are likely to be associated with Trp pathway genes engaged in specialized metabolic pathways other than primary amino acid biosynthesis. We show that when two sister lineages differ in particular trp operon characteristics, it is possible to deduce which is the derived change and which reflects the state of the ancestral node.
Recently, Gogarten et al. (28) endorsed a “synthesis” that will acknowledge both the traditional tree-like behavior (vertical descent of genes) and web-like, reticulate behavior (horizontal gene transfer) of the evolutionary process. They leave it open whether or not “vertical descent remains the best descriptor of the history of most genes over evolutionary time.” Our overall analysis yields a very optimistic viewpoint that the evolution of the trp operon can be deduced as a vertical genealogy, with events of LGT and paralogy enriching the analysis as interesting features rather than undermining or obliterating the vertical trace of evolutionary history.
Biochemical Pathway of Tryptophan Biosynthesis
Nomenclature.
The inconsistencies of nomenclature for designations of genes involved in aromatic biosynthesis have created increasingly awkward problems for comparative analyses, and in order to cope with genomic comparisons, we have implemented a logical system of naming aro genes at a level corresponding to catalytic domains (13, 31, 44, 90, 91, 88). We have extended this nomenclature to the Trp pathway (89, 92) (see Fig. 1 and Table 1). The two overall enzyme reactions that utilize a complex of nonidentical subunits have been denoted with lowercase letters (TrpAa and TrpEa are α-subunits for anthranilate synthase and tryptophan synthase, respectively; TrpAb and TrpEb are β subunits for anthranilate synthase and tryptophan synthase, respectively). Capital letters are assigned according to the order of the enzyme reactions (or overall reactions, in the case of the two complexes). C. Yanofsky has expressed to us his preference (probably shared by most experimentalists working specifically with trp systems) for adherence to previous nomenclature schemes to minimize disruption of what is most familiar in the existing literature. Admittedly, the designations generally in use for the Trp branch do not generate as many problems of annotation errors as is the case for the rest of the aromatic pathway, but for consistency with our overall work with the aromatic pathway, we use the new naming system in this paper. Both sets of designations are shown in Table 1.
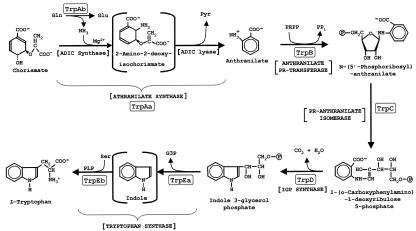
FIG. 1.
Biochemical pathway of tryptophan biosynthesis. The nomenclature used in this paper for the seven catalytic domains is in boxes. See Table 1 for the alternative designations used in the literature. Anthranilate synthase catalyzes the overall reaction from chorismate to anthranilate via the half-reactions shown, whereby 2-amino-2-deoxyisochorismate (ADIC) is an enzyme-bound intermediate (62). The TrpAa/TrpAb complex functions as an amidotransferase, utilizing glutamine as the source of the o-amino group of anthranilate. TrpAa can catalyze the overall reaction alone in the presence of NH3 (thereby functioning as an aminase). TrpAb alone in some cases may be able to function as a glutaminase. As shown by McDonald et al. (59), Pseudomonas and Streptomyces species form ADIC as the product of a reaction catalyzed by PhzE. PhzE has fused domains that are homologues of TrpAa and TrpAb, which we have denoted TrpAa•TrpAb_phz (93) (see Table 1). In these organisms, ADIC can be considered a branch point that proceeds to Trp on the one hand and to phenazine pigments on the other hand. Tryptophan synthase catalyzes a second overall reaction, converting indoleglycerol phosphate to Trp in a reaction path where indole is always an intermediate. The alpha (TrpEa) and beta (TrpEb) subunits catalyze the reactions shown in which the indole intermediate is processed through a tunnel (85). PR, phosphoribosyl; IGP, indoleglycerol phosphate; G3P, glyceraldehyde 3-phosphate.
TABLE 1.
Key to nomenclature conversions
| Function | Gene namea
|
Protein domain encoded | |
|---|---|---|---|
| Suggested | Conventional | ||
| Tryptophan biosynthesis | trpAa | trpE | Anthranilate synthase, aminase subunit (α) |
| trpAb | trpG | Anthranilate synthase, amidotransferase subunit (β) | |
| trpB | trpD | Anthranilate phosphoribosyl transferase | |
| trpC | trpF | Phosphoribosyl-anthranilate isomerase | |
| trpD | trpC | Indoleglycerol phosphate synthase | |
| trpEa | trpA | Tryptophan synthase, α subunit | |
| trpEb | trpB | Tryptophan synthase, β subunit | |
| trpAa•trpAbb | trpE(G) | Fusion of first two domains above | |
| Phenazine biosynthesis | trpAa•trpAb_phzb | phzE | ADIC synthase |
| Folate biosynthesis | pabAa | pabB | 4-Amino-4-deoxychorismate synthase, aminase subunit (α) |
| pabAb | pabA | 4-Aminobenzoate synthase, amidotransferase subunit (β) | |
| pabAc | pabC | 4-Amino-4-deoxychorismate lyase (γ) subunit | |
Nomenclature is at the level of catalytic domain in order of reaction steps in the pathway. Overall reactions of tight complexes are assigned one capital letter, and then α, β, and γ subunits are assigned lowercase letters. trpAa and pabAa are homologs, as are trpAb and pabAb. The convention of a bullet denotes a fusion. TrpAa•TrpAb catalyzes the overall reaction of anthranilate synthase (see Fig. 1). TrpAa•TrpAb_phz is a distinct and shortened subgroup of TrpAa•TrpAb that catalyzes only the first half-reaction, ADIC synthase.
Seven catalytic domains and two α/β-subunit complexes.
Trp is an essential amino acid among the assemblage of required amino acids in mammals. Trp is generally synthesized by free-living prokaryotes, lower eukaryotes, and higher plants. The Trp pathway is one of three amino acid branches diverging from a common flow route that produces chorismate. The apparent universal biosynthetic pathway for Trp biosynthesis that initiates with chorismate and l-glutamine is shown in Fig. 1. Seven catalytic domains are deployed to carry out the reactions shown. In a given organism these may be individually expressed, but a wide variety of gene fusions that encode single proteins carrying two or more catalytic domains are known.
TrpAa can function as an ammonia-utilizing aminase in the anthranilate synthase reaction. Although the aminase reaction can proceed with ammonia at unphysiologically high pH values, such reactions typically rely upon a glutamine-utilizing glutaminase subunit to deliver the ammonia at the active site (probably within a “tunnel”). Accordingly, TrpAb is a glutaminase homologue that forms a complex with TrpAa, thereby conferring an amidotransferase component to the overall anthranilate synthase reaction in the presence of glutamine. In either case, whether or not the overall anthranilate synthase reaction is carried out in the presence of TrpAb, 2-amino-2-deoxyisochorismate (ADIC) is an enzyme-bound intermediate. Interestingly, some species of Pseudomonas and Streptomyces produce an enzyme called PhzE (59), which carries out the ADIC synthase reaction but not the ADIC lyase reaction (see Fig. 1). ADIC is then converted ultimately to phenazine pigments. PhzE is a fusion of domains homologous to TrpAa and TrpAb (hence our designation TrpAa•TrpAb_phz in Table 1). TrpAa belongs to a protein superfamily that includes other chorismate-utilizing enzymes: PabAa converts chorismate to 4-amino-4-deoxychorismate (precursor of 4-aminobenzoate), and MenF and EntC are different homologue subgroups that convert chorismate to isochorismate (as precursors of ubiquinones and an iron siderophore, respectively).
Tryptophan synthase also exists as a complex of nonidentical subunits and is one of the best-understood examples of allosteric interaction exerted between subunits (97). Why indole should be sequestered to a tunnel in the α/β complex of tryptophan synthase is not known, but indole is volatile and rather toxic. Yanofsky has speculated that recent findings of a role for indole in quorum sensing and biofilm formation might suggest that indole either produced by tryptophanase or otherwise available in the environment may serve as a metabolite cue that might otherwise be disrupted if biosynthetic indole were not enzyme-bound (see reference 96 and references therein). It has been speculated (92) that some Archaea may not form a tryptophan synthase complex.
Relatives of Trp pathway catalytic domains.
The pathway of Trp biosynthesis is the first amino acid pathway for which the atomic structure of every catalytic domain has been determined (58), a circumstance of significance because evolutionary analysis can be greatly enhanced through insight gained at the structural level of protein folding. Consultation of the reference by Yanofsky et al. (97) is highly recommended for a definitive presentation of the detailed literature up to about 2000. Each catalytic domain belongs to a protein superfamily at the structural level of protein folding. Many of the catalytic domains exhibit clear homology on the criterion of amino acid identity with proteins that have different substrate specificities and which participate in different pathways. From an evolutionary perspective, this is of interest with respect to such questions as the extent to which the Trp pathway enzymes have been assembled (via gene duplication and substrate alteration) by recruitment of homologues from other pathways or the extent to which the Trp pathway has been the source of genes recruited for function in other pathways or a homologous gene with a recent history of function in another pathway has “crossed over” to replace a Trp pathway gene (or vice versa). This aspect is not addressed further in this article except indirectly (e.g., see the later section on the search for an elusive trpC gene).
Identical Trp pathways exist within varied metabolic contexts.
The Trp pathway is generally defined as an unbranched pathway that begins with chorismate and produces Trp as a substrate for general protein synthesis. The Trp pathway appears to have evolved only once. These aspects of universality are favorable for the task of deducing the evolutionary history. However, many aspects of biochemical individuality are not usually considered. In some cases, Trp biosynthesis does not compete with Phe and/or Tyr biosynthesis because one or both of these are absent. In other cases, as exemplified by the use of ADIC for phenazine biosynthesis in Pseudomonas and Streptomyces species, chorismate is no longer the last branch point, and if one starts with chorismate as a reference point, then the pathway is branched. The pathway does not necessarily end exclusively with the Trp end product supplying protein synthesis, e.g., in cases where Trp may be a component of an antibiotic (as in Streptomyces), or where it is converted to indoleacetic acid in plant symbionts such as Azospirillum. Eukaryotes (but no prokaryotes so far) deploy Trp as a precursor of niacin. In such cases, the pathway can be considered divergently branched at the end, with Trp being guided to different molecular fates.
Trp is the most biochemically expensive of the amino acid pathways, requiring the input of erythrose-4-phosphate, ATP, phosphoribosyl pyrophosphate (PRPP), two phosphoenolpyruvate molecules, l-glutamine, and l-serine. Thus, efficient regulation is generally expected, but these rules no longer apply in an endosymbiont such as Buchnera, which has abandoned Trp regulation. In this case, loss of regulation can be viewed as a positive selective step in order to satisfy the needs of its aphid host. In addition, some prokaryotes sustain different physiological or developmental states where the demands impacting the Trp pathway may be more complicated than just sensing the availability of Trp for protein synthesis. These often involve specialized pathways that coexist with primary Trp biosynthesis. These specialized pathways are encoded in part or entirely by divergent trp gene duplicates whose expression is triggered by a variety of temporal and environmental cues, e.g., to make a given pigment or antibiotic derived in part from the Trp pathway.
These are all interesting but complicating elements that we have tried to keep in mind. This is relevant to the task of sorting out and recognizing paralogues (or xenologues) that may be engaged in specialist pathways other than primary Trp biosynthesis. Appreciation of such complexity may also prove relevant to understanding the nature of split-pathway trp operons in many prokaryotes.
Operon Stability
Most molecular biologists who are familiar with the elaborate control features of well-studied operon systems would feel a strong conviction that once evolved, these would resist change (at least disruptive changes). Yet a bioinformatic analysis of the then-available genomes in 1999 (37) produced the conclusion that operon structures, such as the trp operon, are unstable, as inferred from observations of extraneous insertions, gene dispersal, and scrambling of gene order within operons. Characterization of operons as unstable connotes susceptibility to deterioration. If true, this implies that the selective advantages conferred by such operons must be weak.
The Itoh et al. study (37) was a broad-scope analysis of many operons that was necessarily limited with respect to in-depth consideration of any individual operon system. It should be noted that for these kinds of studies, operons have been considered simply as a collection of structural genes that are linked. The presence or absence of linked or unlinked regulatory elements has not usually been evaluated, undoubtedly because this is not easily done. In this paper we pursue in great detail the evolution of a single well-known operon system in the large number of prokaryote genomes now available. We found strong support for the hypothesis that the trp operon, minimally defined as the linked assemblage of structural genes for tryptophan (Trp) biosynthesis, is of ancient origin and has indeed followed a dynamic time course of change that includes several identifiable milestone events in Bacteria. Our study leads to the further hypothesis that the instability of early trp operons (and perhaps some modern ones) can be attributed to weak positive selection conferred by relatively undeveloped control mechanisms.
We suggest that since the time that operons evolved a variety of control mechanisms, the characterization of operons as dynamic (rather than unstable) yields better semantics to describe a positive ongoing process of fine-tuning. In modern free-living organisms, the variety of recently evolved trp operon systems which differ from one another and are endowed with intricate control features mediated by one or more unlinked regulatory genes may in fact be highly stable in the contemporary time frame. One caveat, however, is that this frequently will not apply to pathogenic or endosymbiotic relatives, where the rules dictating selective advantage have completely changed.
trp Operon and Its Regulation
The biochemical pathway of Trp biosynthesis is a classical system of biochemical genetics (95, 97). In Escherichia coli the component genes are organized into a single transcriptional unit to form the trp operon. (This is not strictly correct to the extent that a weak internal promoter exists.) The Trp pathway has become one of the most intensely studied systems in biology, thanks largely to the truly Herculean labors of C. Yanofsky and his many students and colleagues. This system has produced knowledge that extends well beyond the details of the Trp pathway per se, e.g., proof of codon and amino acid colinearity and an early precedent for attenuation mechanisms (95, 96). The individual reactions of Trp biosynthesis are invariant, but experimental work with a variety of organisms reveals substantial diversity with respect to gene fusion, gene organization, and mechanisms of regulation.
Known regulatory mechanisms.
At the bioinformatic level, the analysis of trp operons in the literature has been largely restricted to the structural genes. Consideration of regulatory features has been understandably limited, mainly because relatively little comparative information is available at the experimental level and also because analysis of alternative stem-loop structures, etc., is not a trivial task. Escherichia coli, Bacillus subtilis, Pseudomonas aeruginosa, and Lactococcus lactis represent clades for which detailed control mechanisms have been described, each of them entirely different. Importantly, each mechanism seems to be narrowly distributed, and therefore we infer that they are of recent origin. Note that in each case, unlinked genes exist that markedly decrease the probability that the total regulated operon system could be transferred by LGT in one event.
Regulation of Trp biosynthesis in E. coli, the most widely known system, is quite sophisticated (23, 94), being subject to the following multiple levels of control: (i) repression control via the Trp repressor (encoded by the unlinked trpR) which binds Trp as a corepressor moiety, (ii) an attenuation mechanism mediated by a Trp-rich leader peptide (encoded by trpL), and (iii) allosteric feedback inhibition of anthranilate synthase by Trp (95). The E. coli mechanisms of overall trp operon regulation are generally shared by the enteric lineage of Bacteria, defined by us as the clade that includes Shewanella putrefaciens as the outlying point of divergence from E. coli.
Bacillus subtilis has a different system of trp operon regulation (72, 80, 95, 96), whereby genes unlinked to the trp operon encode (i) a trp RNA-binding attenuation protein (TRAP) encoded by mtrB as well as (ii) an anti-TRAP gene product encoded by rtpA (80). Trp both feedback inhibits anthranilate synthase and activates TRAP for attenuator function, whereas uncharged tRNATrp induces synthesis of anti-TRAP. TRAP can also block translation of the trp operon through interference with the ribosome-binding site. The clade sharing the TRAP system of regulation includes Bacillus halodurans, Bacillus stearothermophilus, and Oceanobacillus iheyensis in addition to Bacillus subtilis. At this time it is not clear whether the anti-TRAP component is present throughout this clade.
A third finely tuned system of regulation has been documented in Lactococcus lactis (69). In this case uncharged tRNA can bind directly to the leader transcript, stabilizing an antiterminator configuration that promotes expression of the operonic genes. In Lactococcus lactis, unlinked, unknown genes involved in trp operon transcript processing and in transcription initiation have been suggested (69). The presence or absence of the Lactococcus lactis mode of trp operon regulation in close relatives, such as species of Streptococcus, has apparently not yet been investigated.
In Pseudomonas aeruginosa, the fourth well-documented system, the Trp pathway is represented by four operon entities: a free-standing trpAa, the trpAbBD operon, a free-standing trpC, and the trpEbTrpEa operon. The trpAa and trpAbBD operons are regulated by attenuation mechanisms employing leader peptides (67), whereas the trpEbtrpEa operon is controlled by an indoleglycerol phosphate-activated regulatory protein encoded by trpI (6). trpC is not known to be regulated in any way. The P. aeruginosa system is complicated by the presence of paralogues of trpAa and trpAb. These include genes of unknown physiological function (also known as phnA and phnB) expressed in stationary phase (57) as well as two copies of PhzE (trpAa•trpAb_phz), a gene that encodes ADIC synthase (Fig. 1), the initial reaction committed to phenazine biosynthesis. It is not entirely clear what physiological conditions exist in P. aeruginosa (and close relatives) that have resulted in its unusual use of indoleglycerol phosphate as a regulatory cue for the selective regulation of the trpEbTrpEa operon, but it is certainly evident that much has been committed to the overall regulation in this system. Close genomic neighbors of P. aeruginosa that possess identical split-pathway trp operons and trpI include Pseudomonas fluorescens, Pseudomonas syringae, and Azotobacter vinelandii.
Unknown regulatory systems awaiting discovery?
We do not know the extent to which the total network of regulatory elements governing the single trp operons in the E. coli, B. subtilis, and L. lactis clades or the multiple split-pathway operons of the P. aeruginosa clade might be more elaborate than that of most other organisms. Different lifestyles undoubtedly select mechanisms accommodating varied ranges of control responsiveness. A simple mode of Trp regulation may very well be appropriate in a cyanobacterium but not E. coli. A variety of alternative regulatory systems in other modern lineages probably remain to be elucidated. Transcriptional regulation has been reported in the whole-pathway operons of Methanobacterium thermoautotrophicum (26) and Pyrococcus kodakaraensis (77), but the exact mechanisms are unknown. The split-pathway operons of the clade represented by Rhizobium meliloti (7) and Azospirillum brasilense (21) exhibit an attenuation mechanism involving a Trp-rich leader peptide, upstream of the trpAa•trpAb fusion, but no regulation of the remaining two partial-pathway operons is known. Physically separated split-pathway trp operons may be of positive selective value per se for presently unknown reasons, whereby it might be of value to discoordinate the expression of some trp genes from others, or they may simply be the outcome of initially disrupted whole-pathway operons that subsequently recruited a refined control mechanism accommodating the gene separations.
Feasibility for Deduction of Evolutionary Histories
The current database for prokaryotes, at least for the Bacteria, now has sufficient genome representation to accommodate systematic attempts to deduce the evolutionary history of well-understood biochemical pathways. Such an effort requires the successful recognition and confrontation of complications such as (i) irregular genome expansions in the form of the unpredictable emergence of new paralogues or analogues, (ii) an erratic and differential phylogenetic loss of paralogue genes (often the cause of what has been termed unrecognized paralogy), and loss of analogue genes (could be termed unrecognized analogy), and (iii) lateral (horizontal) gene transfer (LGT). Errors and inconsistencies of database annotation as well as idiosyncrasies of nomenclature can create formidable hurdles for those who are not completely familiar with a given pathway and its scholarly literature. Global computational surveys to date are simply not very informative, and the algorithms employed for automated annotation have too many limitations. For example, a very recent effort at computational identification of operons in microbial genomes (98) chose to highlight the results of trp operon analysis as a prime example of the analysis. However, the results presented are not comprehensive and contain serious mistakes, most likely due to errors in annotation and confusing nomenclature issues that have been perpetuated in the databases.
As a first step toward deducing the evolutionary history of overall aromatic biosynthesis, we selected the Trp branch as a challenging but manageable metabolic segment for initial analysis. Trp pathway genes have sometimes been recruited for function in specialized biochemical pathways, and ancient paralogues or xenologues may coexist with the Trp pathway genes that are engaged in primary biosynthesis. We have shown (93) that detailed case-by-case analysis can distinguish ancient trp paralogues (or xenologues) from their homologues engaged in primary Trp biosynthesis. A comparable study in the literature produced a detailed analysis of homologues of ornithine carbamoyltransferase in which the challenges to tracking a vertical path of evolutionary descent that are caused by the complexities of xenology and ancient paralogy were sorted out (73). This study was preceded by an analysis (49) showing that ornithine carbamoyltransferases in turn belong to a larger protein family in which the ornithine and aspartate carbamoyltransferases are very ancient paralogues. The conclusions such comprehensive studies are consistent with the contentions of Glansdorff (27) and Woese (87) that complications of ancient paralogy, ancient analogy, and lateral gene transfer can be recognized sufficiently well to allow the events of vertical ancestry to be tracked.
Here we present results from an in-depth, manual analysis of Trp pathway genes in over 100 genomes. A limited amount of information is also given to illustrate the very important perspective that the evolutionary relationships of Trp biosynthesis will ultimately be best understood in its larger context as one branch of a highly divergent pathway responsible for the biosynthesis of aromatic amino acids as well as many other important metabolites.
GENOMIC DISTRIBUTION OF THE TRYPTOPHAN PATHWAY
Mapping of trp Gene Patterns to the 16S rRNA Tree
In most of the figures used in this paper, patterns of operonic gene arrangement in a given organism are mapped to the placement of that organism on a 16S rRNA tree. Table 2 keys each organism examined to the figures that show trp gene patterns in that organism. To what extent do the individual Trp protein trees parallel the 16S rRNA tree? It is well known that, unlike information-rich 16S rRNA, most individual proteins cannot be expected to yield robust phylogenetic trees in which the order of branching is well supported by high bootstrap values, at least not over wide phylogenetic ranges. However, in relatively narrow phylogenetic spans, we have found congruity between Trp protein trees and 16S rRNA trees (except for TrpAb, which is too small).
TABLE 2.
Cross-reference guide to organisms and figures
| Prokaryote | Figure(s) in which it appears |
|---|---|
| Aeropyrum pernix | 2A, 4, 5, 10 |
| Anabaena sp. | 2C, 4, 6A |
| Aquifex aeolicus | 2A, 4 |
| Archaeoglobus fulgidus | 2A, 4, 5, 10 |
| Azospirillum brasilense | 4, 9 |
| Azotobacter vinelandii | 9 |
| Bacillus anthracis | 2B, 4, 11 |
| Bacillus halodurans | 2B, 11 |
| Bacillus stearothermophilus | 2B, 4, 11 |
| Bacillus subtilis | 2B, 4, 6A, 11 |
| Bordetella pertussis | 2D, 4, 6B, 9 |
| Borrelia burgdorferi | 2C |
| Bradyrhizobium japonicum | 9 |
| Brucella melitensis | 2D, 4, 9 |
| Buchnera sp. | 8 |
| Buchnera aphidicola | 2D, 4 |
| Burkholderia fungorum | 2D, 6B, 9 |
| Burkholderia pseudomallei | 4 |
| Campylobacter jejuni | 2D, 4, 6A |
| Caulobacter crescentus | 2D, 4, 6B, 9 |
| Chlamydia muridarum | 2C |
| Chlamydia trachomatis | 2C |
| Chlamydophila pneumoniae | 2C |
| Chlamydophila psittaci | 2C, 4, 6A |
| Chlorobium tepidum | 2C, 4 |
| Chloroflexus aurantiacus | 4 |
| Clostridium acetobutylicum | 2B, 4, 6A |
| Clostridium difficile | 2B |
| Corynebacterium diphtheriae | 2B, 3, 4 |
| Corynebacterium glutamicum | 3, 4 |
| Coxiella burnetii | 2D, 6B, 9 |
| Cytophaga hutchinsonii | 2C, 6A |
| Dehalococcoides ethenogenes | 2A, 6A |
| Deinococcus radiodurans | 2A, 4, 6A |
| Desulfovibrio vulgaris | 2D, 4, 6A |
| Desulfitobacterium hafniense | 6A |
| Enterococcus faecalis | 2B, 7 |
| Escherichia coli | 2D, 4, 6B, 8 |
| Ferroplasma acidarmanus | 2A, 5, 10 |
| Geobacter sulfurreducens | 2D, 4, 6A |
| Haemophilus actinomycetemcomitans | 4, 8 |
| Haemophilus ducreyi | 2D, 8 |
| Haemophilus influenzae | 2D, 4, 8 |
| Halobacterium sp. | 2A, 4, 5, 10 |
| Helicobacter pylori | 2D, 4, 6A |
| Klebsiella pneumoniae | 4, 8 |
| Lactococcus lactis | 2B, 4, 7 |
| Legionella pneumophila | 2D, 4, 6B, 9 |
| Listeria monocytogenes | 2B, 7, 11 |
| Magnetospirillum magnetotacticum | 2D, 6B, 9 |
| Magnetococcus sp. | 6B, 9 |
| Mesorhizobium loti | 4 |
| Methanobacterium thermoautotrophicum | 2A, 4, 5, 10 |
| Methanococcus jannaschii | 2A, 4, 5, 10 |
| Methanosarcina barkeri | 2A, 5, 10 |
| Methylococcus capsulatus | 2D, 6B |
| Mycobacterium avium | 2B, 3, 4 |
| Mycobacterium bovis | 2B, 3 |
| Mycobacterium leprae | 2B, 3 |
| Mycobacterium smegmatis | 2B, 3 |
| Mycobacterium tuberculosis | 2B, 3, 4, 6A |
| Mycoplasma genitalium | 2B, 7 |
| Mycoplasma pneumoniae | 2B, 7 |
| Neisseria gonorrhoeae | 4, 9 |
| Neisseria meningitidis | 2D, 4, 6B, 9 |
| Nitrosomonas europaea | 2D, 6B, 9 |
| Nostoc punctiforme | 2C, 4 |
| Oceanobacillus iheyensis | 11 |
| Pasteurella multocida | 2D, 4, 8 |
| Porphyromonas gingivalis | 2C |
| Pseudomonas aeruginosa | 2D, 4, 6B, 9 |
| Pseudomonas fluorescens | 9 |
| Pseudomonas putida | 4 |
| Pseudomonas syringae | 2D, 4, 9 |
| Prochlorococcus marinus | 2C |
| Pyrobaculum aerophilum | 2A, 5, 10 |
| Pyrococcus abyssi | 2A, 4, 5, 10 |
| Pyrococcus furiosus | 2A, 4, 5, 10 |
| Pyrococcus horikoshii | 2A, 5, 10 |
| Ralstonia metallidurans | 2D, 6B, 9 |
| Rhizobium loti | 2D, 9 |
| Rhizobium meliloti | 4 |
| Rhodobacter capsulatus | 9 |
| Rhodobacter sphaeroides | 2D, 6B |
| Rhodopseudomonas palustris | 2D, 4, 6B, 9 |
| Rickettsia prowazekii | 2D |
| Salmonella enterica | 2D, 4, 8 |
| Shewanella putrefaciens | 2D, 4, 6B, 8 |
| Sinorhizobium meliloti | 2D, 9 |
| Sphingomonas aromaticivorans | 2D, 6B |
| Staphylococcus aureus | 2B, 4, 11 |
| Staphylococcus epidermidis | 2B, 5, 11 |
| Streptococcus equi | 2B, 7 |
| Streptococcus gordonii | 2B, 7 |
| Streptococcus mutans | 2B, 4, 7 |
| Streptococcus pneumoniae | 2B, 4, 6A, 7 |
| Streptococcus pyogenes | 2B, 7 |
| Streptomyces coelicolor | 2B, 3, 4 |
| Sulfolobus solfataricus | 2A, 4, 5, 10 |
| Synechococcus sp. | 2C |
| Synechocystis sp. | 2C, 4 |
| Thermomonospora fusca | 2B, 3, 4 |
| Thermoplasma acidophilum | 2A, 4, 5, 10 |
| Thermoplasma volcanium | 5, 10 |
| Thermotoga maritima | 2A, 4, 6A |
| Thiobacillus ferrooxidans | 2D, 4, 6B, 9 |
| Treponema denticola | 2C |
| Treponema pallidum | 2C |
| Ureaplasma urealyticum | 2B, 7 |
| Vibrio cholerae | 2D, 4, 8 |
| Wolbachia sp. | 2D |
| Xanthomonas campestris | 9 |
| Xanthomonas axonopodis | 9 |
| Xylella fastidiosa | 2D, 4, 6B, 9 |
| Yersinia pestis | 2D, 4, 8 |
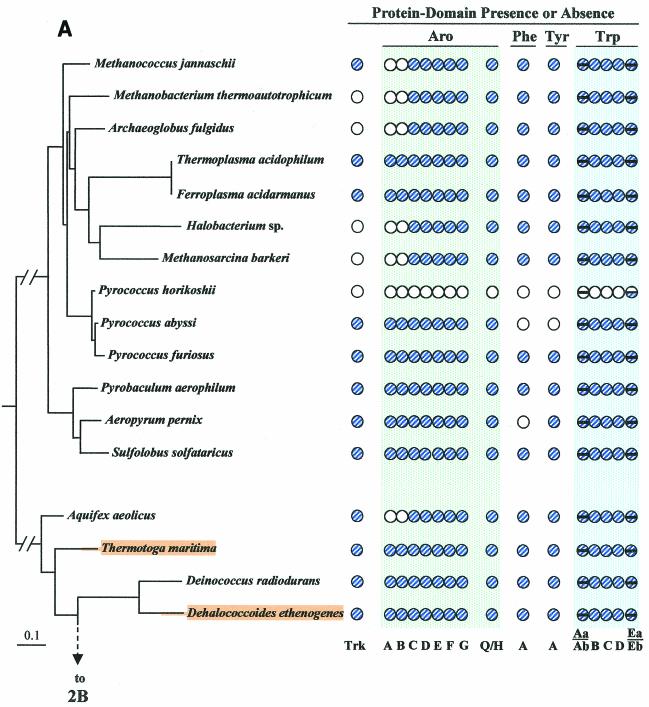
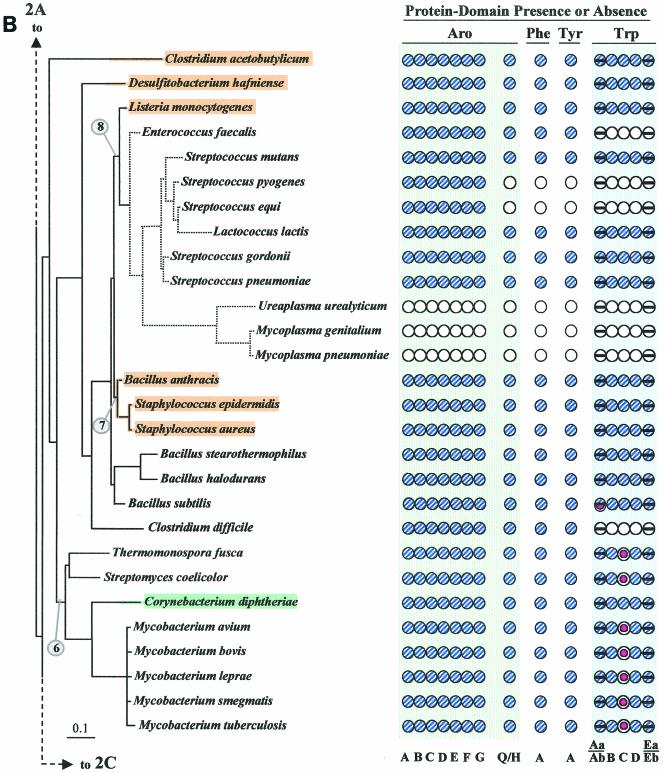
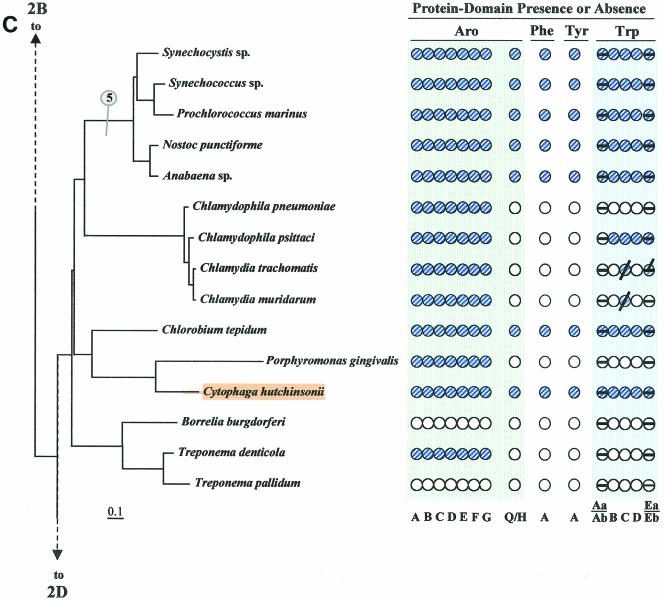
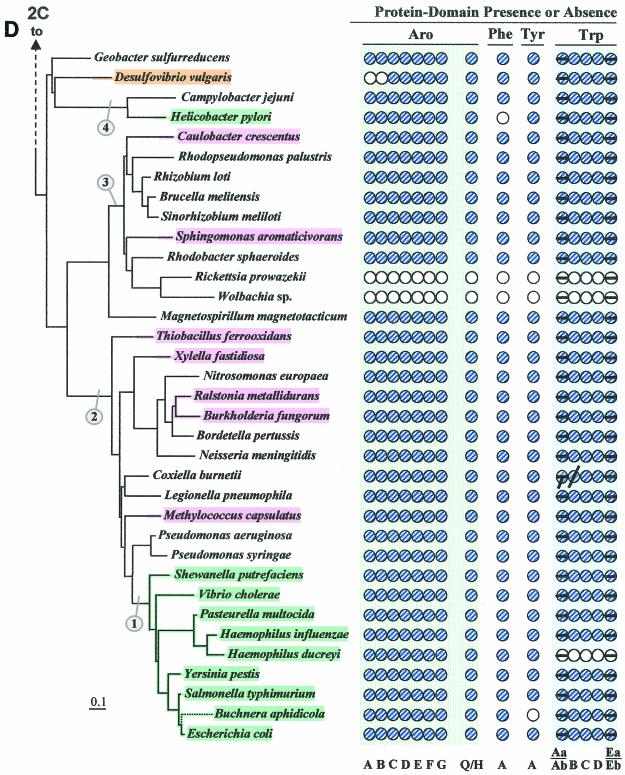
Figure 2 illustrates (see shaded and circled numbers) eight clades where reasonably good congruity is observed: a Listeria/Bacillus/Staphylococcus/Streptococcus grouping (Fig. 2B), B. subtilis, B. stearothermophilus, B. halodurans (Fig. 2B), actinomycete bacteria (Fig. 2B) cyanobacteria (Fig. 2C), Campylobacter/Helicobacter (Fig. 2D), Proteobacteria between Caulobacter crescentus and Rhodobacter sphaeroides in Fig. 2D, the clade between Thiobacillus ferrooxidans and Pseudomonas syringae (Fig. 2D), and the enteric lineage between Shewanella putrefaciens and Escherichia coli in Fig. 2D. These are all groups for which a sufficient number of closely related genomes have been sequenced. We expect that when genome sequences become available in more sparsely represented areas, e.g., around Chlorobium (Fig. 2C) or Thermotoga (Fig. 2A), additional phylogenetic spans will be congruent. Within a relatively narrow phylogenetic span, protein trees actually have the potential to discriminate branching order better than 16S rRNA trees. Our Trp protein trees (data available upon request), together with a variety of other aromatic-pathway information (see also the section on nested gene fusions), suggest that the Enterococcus/Streptococcus/Lactococcus grouping is outside the Listeria/Bacillus/Staphylococcus clade rather than within it, as shown on the 16S rRNA tree of Fig. 2B.
FIG.2.
Distribution of aromatic-pathway catalytic domains among prokaryotes. In each panel, 16S rRNA trees are shown at the left, and the presence (shaded circles) or absence (open circles) of domains is shown at the right. Note that only the presence or absence of genes, not gene order, is indicated. Catalytic domains of the common trunk of aromatic biosynthesis (Aro), the phenylalanine branch (Phe), the tyrosine branch (Tyr), and the Trp branch are labeled across the top right; the specific letter designation for a given domain is shown at the bottom. In the Trp grouping, split circles are used to indicate the presence or absence of TrpAa (top half-circle) and TrpAb (bottom half-circle) or TrpEa (top half-circle) and TrpEa (bottom half-circle). In panel A, the presence or absence of transketolase (Trk) is indicated by the left column of circles. The connecting point of a tree segment in any given panel (A, B, C, and D) with a tree segment(s) in another panel is marked with a broken line. The scale bar corresponds to substitutions per site. Dotted lines in the Streptococcus region (B) and the Buchnera region (D) indicate our suggestion that the 16S rRNA tree shown may not reflect exactly the correct order of branching, and perhaps these organisms branch from a slightly deeper position. See Fig. 8 for the suggested branching order of Buchnera. Circled numbers indicate eight node positions from which Trp protein trees are congruent with the 16S rRNA tree. The common trunk of aromatic biosynthesis is encoded by seven genes whose corresponding gene products are named AroA through AroG. The common-pathway genes are named in exact order of pathway reactions according to the precedent implemented in references 12, 31, 76, 90, and 91. The chorismate mutase block is represented by homologues of either AroQ (usually) or AroH (seldom) (12). PheA refers to prephenate dehydratase, the sequence of the relatively infrequent arogenate dehydratase being currently unknown. TyrA refers to a homologue family that includes prephenate dehydrogenase, arogenate dehydrogenase, or cyclohexadienyl dehydrogenase (9, 88). See Fig. 1 for details of Trp biosynthesis. The names of organisms retaining the putative ancestral whole-pathway trp operon are shaded orange, those having the two split-pathway operons are shaded magenta, and those having operons rejoined by fusion of trpD and trpC are shaded aqua. These correspond to the major evolutionary events portrayed in Fig. 12 and indicated with the same color-coding scheme. Probable pseudogenes in chlamydiae (C) and Coxiella (21) are indicated with heavy black slash marks. Genes that function in two pathways (trpAb in Bacillus subtilis and trpC in actinomycete bacteria) are marked with magenta bull's-eyes in B. Panel A includes the Archaea and a few of the deeper-branching Bacteria at the bottom. Panel B includes the gram-positive Bacteria. Panel C includes cyanobacteria, chlamydiae, and other organisms on the 16S rRNA tree between the gram-positive organisms in panel B and the organisms in panel D, which contains the gram-negative subdivisions of the Proteobacteria. Wolbachia sp. (panel D) is an endosymbiont of Brugia malayi. A cross-index of all organisms shown in both this figure and the remaining figures is given in Table 2.
Assembly of these protein trees is not a trivial task because divergent paralogues or xenologues engaged in specialized metabolic activities as well some genes originating by LGT must be recognized and sorted out. Examples are given in this paper.
Trp Biosynthesis in Its Larger Context of Aromatic Biosynthesis
Trp biosynthesis is usually described as the branch of aromatic biosynthesis (reference 10 is a comprehensive biochemical review) that begins with chorismate and l-glutamine as initial substrates. In view of the fact that chorismate is not generally available from the environment as a stable nutrient, Trp biosynthesis can be considered from an in vivo perspective to initiate further upstream via the enzymatic condensation of erythrose-4-phosphate and phosphoenolpyruvate. This step is common to the biosynthesis of all three aromatic amino acids and is positioned at a point of interface with carbohydrate metabolism.
The multipurpose Fig. 2 provides a summary of the presence or absence of Trp pathway genes in the larger context of the presence or absence of genes specifying the common aromatic trunk and the sister phenylalanine and tyrosine branches. The circles in Fig. 2 from left to right represent catalytic domains (specified at the bottom of each panel) corresponding to the seven common-pathway steps (aroA through aroG), chorismate mutase (aroQ or aroH) (which is common to the short Phe and Tyr branches), and the seven catalytic domains of the Trp pathway (Fig. 1 and Table 1).
The key enzyme of Phe biosynthesis is PheA, and the key enzyme of Tyr biosynthesis is TyrA. The Phe and Tyr branches each utilize an aminotransferase step, not shown as a circle because of bioinformatic difficulties associated with deducing the substrate specificity of multiple and ubiquitous broad-specificity aminotransferases (42). Most intermediary metabolites of aromatic biosynthesis are not likely to be available from the environment; only quinate, shikimate, and anthranilate, all abundant in nature (10), are feasible precursors of Trp. Although these metabolites are indeed readily utilized when available, no prokaryotes have yet been found to rely on an exogenous source of quinate, shikimate, or anthranilate as exclusive and obligatory beginning precursors. One interesting special-case exception is Chlamydophila psittaci, an obligate intracellular parasite that utilizes host-derived anthranilate as a required Trp precursor (89).
Implications of Missing Genes
Unidentified analogue genes.
The most obvious explanation for “missing” genes that leave a gap in an otherwise intact pathway is the existence of analogue genes, i.e., functionally equivalent genes that lack homology with the genes used to query the databases. The common pathway of aromatic biosynthesis is a good example of nonhomologous genes producing enzymes that catalyze the same reaction. These include the first step ([3-deoxy-d-arabino-heptulosonate-7-phosphate][DAHP] synthase) (31, 44), the third step (dehydroquinase) (13), and the fifth step (shikimate kinase) (13, 18). Chorismate mutase is represented in nature by three analogue genes (13). No analogue genes are presently known for Trp pathway genes except for trpC (9).
Alternative metabolic relationships.
In contrast to the apparent universality of the specific Trp branch, alternative enzyme steps appear to exist in nature for the Phe and Tyr branches as well as for the common trunk of aromatic biosynthesis. Some Archaea (Fig. 2A) and two widely spaced members of the Bacteria (Aquifex and Desulfovibrio, Fig. 2A and 2D) lack both AroA and AroB. Transketolase (Trk), required for generation of a substrate for AroA, is also shown in Fig. 2A because most (but not all) organisms that lack AroA and AroB also lack transketolase. (Desulfovibrio vulgaris [Fig. 2D] does have transketolase.) In the last six organisms, dehydroquinate, the substrate of AroC, presumably connects with carbohydrate metabolism in some unknown way that does not involve AroB or any of the known AroA homology groupings AroAIα, AroAIβ, or AroAII (31, 44, 76). Some support for this putative alternative metabolic connection, based on tracer methodology, exists in the literature (79). It is also possible that quinate, either from the environment or arising endogenously in some unknown way, could be the source of dehydroquinate via the action of a quinate dehydrogenase.
Although species of Chlamydophila and Chlamydia are very close phylogenetically, the presence of Trp pathway genes varies from complete absence in C. pneumoniae to almost all present in C. psittaci. It appears that the Trp pathway in C. trachomatis and C. muridarum is in a contemporary process of reductive evolution, and the few remaining genes may be remnants (25, 89). In contrast to these species, an “incomplete” trp operon in C. psittaci appears to play a role in the capture of host kynurenine derived from tryptophan (89). Although C. psittaci does lack trpAa and trpAb, the remaining five trp genes coexist in an operon into which two novel genes have been recruited. These encode kynureninase and PRPP synthase. This creates the ability to generate PRPP (needed for the TrpB step) and to intercept host kynurenine as a source of anthranilate, cycling host-catabolized Trp back to Trp in the intracellular parasite (89). Effectively, a host-pathogen metabolic mosaic has been created, and the variant operon generates a kynurenine-to-Trp flow route instead of the usual chorismate-to-Trp flow route.
As explained above, the absence of trpAa and trpAb in C. psittaci is by design, and the remaining Trp pathway is functional. The likelihood that aroA and aroB, which are absent in some organisms, will prove to reflect either a new metabolic connection or the existence of unknown analogue genes has already been mentioned. In a few cases tyrA or pheA was the only aromatic-pathway gene not found by homology search. The endosymbiont Buchnera (Fig. 2D), which lacks tyrA, may not need to synthesize tyrosine because the host has phenylalanine hydroxylase, which can convert phenylalanine to tyrosine. Aeropyrum pernix (Fig. 2A) and Helicobacter pylori (Fig. 2D), which both lack pheA, may very well possess arogenate dehydratase, an alternative pathway step for prephenate dehydratase (reference 39 and references therein). No gene encoding an arogenate dehydratase has yet been cloned and sequenced.
Reductive evolution.
Reductive evolution is descriptive of the process in which pathogens or symbionts decrease genome size by abandoning genes that are needed by their free-living relatives but dispensable because of the availability of resources from a host or symbiont partner. The genus Pyrococcus exhibits marked variation in the capability for aromatic biosynthesis. Pyrococcus horikoshii has experienced total reductive evolution. Only TrpEb remains in P. horikoshii, and the case has been made that this may have some other function, such as serine deaminase activity (92). P. abyssi possesses genes encoding common-pathway and Trp pathway steps but lacks the Phe and Tyr branches. Although chorismate mutase (aroQ) is present, it could have some other substrate specificity (13). Since P. abyssi lacks the competing Phe and Tyr branches, an unusual metabolic circumstance exists in which the representation of tryptophan biosynthesis can be collapsed to that of a linear pathway of 12 overall steps (corresponding to the seven common-pathway steps followed by the five overall steps that are specifically dedicated to Trp biosynthesis). In contrast to the foregoing two differentially auxotrophic species of Pyrococcus, P. furiosus possesses a complete assemblage of aromatic-pathway genes.
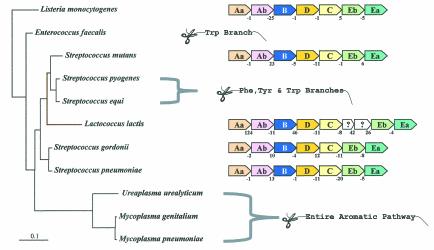
Organisms that lack the entire branched system of aromatic amino acid biosynthesis include P. horikoshii (Fig. 2A), Ureaplasma urealyticum and Mycoplasma species (Fig. 2B), Borrelia burgdorferi and Treponema pallidum (Fig. 2C), and Rickettsia prowazekii and Wolbachia spp. (Fig. 2D). These whole-pathway reductive evolutions are generally associated with intracellular parasitism or endosymbiosis, and they imply auxotrophic dependence upon the host not only for all three aromatic amino acids but also for end products of the vitamin-like branches (e.g., folate, vitamin K, and ubiquinones) that derive from chorismate. In the Bacteria, some organisms possess an otherwise intact aromatic pathway but the Trp branch is uniquely absent. Among gram-positive bacteria (Fig. 2B), this includes Enterococcus faecalis and Clostridium difficile, and this pattern is also seen in the gram-negative Haemophilus ducreyi (Fig. 2D).
Interestingly, some organisms lack all three of the terminal aromatic amino acid branches but possess an intact common pathway to chorismate: Streptococcus pyogenes (Fig. 2B), Streptococcus equi (Fig. 1B), chlamydial species (Fig. 2C), Porphyromonas gingivalis (Fig. 2C), and Treponema denticola (Fig. 2C). The implication is that the remaining common pathway still links to one or more of the vitamin-like pathways. In the chlamydiae, we could not detect (by use of homology searching) a single gene encoding any known chorismate-utilizing enzyme. However, this could easily be accounted for by the existence of analogue genes that have not yet been identified. For example, E. coli chorismate lyase, which catalyzes the initial step of ubiquinone biosynthesis, is encoded by a gene (66) that is of very limited distribution. Therefore, elucidation of presently unknown analogue genes encoding chorismate lyase surely must be forthcoming.
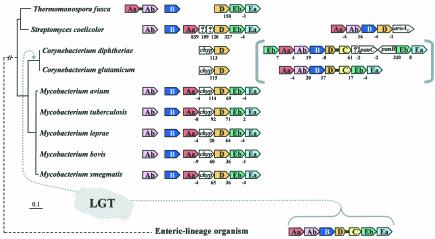
Search for an Elusive trpC Gene in Actinomycete Bacteria
A particularly challenging observation was that, aside from the fragmented presence of the Trp pathway genes already discussed for the chlamydiae, some organisms lacked a single gene of Trp biosynthesis (trpC in all cases). These organisms are restricted to a cohesive cluster of gram-positive actinomycete bacteria (Fig. 2B and Fig. 3) that includes Thermonospora fusca, Streptomyces coelicolor, Corynebacterium diphtheriae, Corynebacterium glutamicum, and five species of Mycobacterium. Since S. coelicolor can grow on defined minimal medium in the absence of Trp, it must possess an intact Trp pathway. Likewise, Mycobacterium smegmatis is a saprophytic species that can grow in a minimal medium in the absence of Trp. This therefore also indicates the presence of a functional pathway even though the presence of trpC in the genome is not apparent by homology searching.
FIG. 3.
Apparent absence of trpC and an event of LGT in a lineage of actinomycete bacteria. A broader phylogenetic context can be viewed in Fig. 2B and 6A. chyp denotes a conserved hypothetical membrane protein exhibiting about 28% identity in comparison of a given Mycobacterium species with a given Corynebacterium species. Color-coded boxes pointing in the direction of transcription represent genes of Trp biosynthesis. For clarity of presentation, trpAa is shown as Aa, etc. Open boxes with question marks denote hypothetical proteins. Intergenic spacing is shown, with negative values indicating gene overlap. trpD•trpC fusions are represented by short black linker bars. On the left are 16S rRNA-based phylogenetic trees of the genomes having the gene organizations shown on the right. Orthologues that match the mycobacterial trpAa/chyp/D/Eb/Ea operon genes are aligned vertically. Contemporary trp operons in coryneform species that originated in their common ancestor by LGT of trpAa/Ab/B/D•C/Eb/Ea from a source within the enteric lineage are shown within brackets. Except for the two coryneform species, all actinomycetes have a free-standing trpB gene. The Mycobacterium spp. and Streptomyces also have a free-standing trpAb gene. The corresponding TrpB and TrpAb proteins exhibit high identity with one another but not with TrpB and TrpAb of the coryneform species. Thermomonospora has dissociated trpAa from the typical clade operon and fused it with trpAb (as also shown in Fig. 4). The trpAa/Ab/B/D/aroAII operon of S. coelicolor is known to be specifically associated with antibiotic biosynthesis (see text).
One actinomycete exception is explained by LGT.
Within the actinomycete clade, the two species of Corynebacterium do possess a recognizable trpC (albeit fused to the trpD domain). However, this exception is explained by LGT displacement of not only trpC but also all trp genes in the Corynebacterium genus (except for a now-redundant trpD remnant) by the whole-pathway operon originating from an enteric bacterium (Xie and Jensen, unpublished data). Figure 3 shows that this actinomycete clade characteristically possesses a partial-pathway operon, trpAa/trpD/trpEb/trpEa, with gene insertions expanding the intergenic space between trpAa and trpD. In T. fusca, trpAa has not only dissociated completely from the trpD/Eb/Ea operon but has fused with trpAb. Only trpD and an associated conserved hypothetical gene denoted chyp remain in species of Corynebacterium as remnants of the original actinomycete genes. The remnants are pleasingly fortuitous because they show the Corynebacterium ancestor to be the recipient of LGT rather than the donor.
A comprehensive phylogenetic tree for trpD proteins (data not shown) reveals that all of the TrpD proteins in Fig. 3 exhibit cohesive clustering and an order of branching that is congruent with the corresponding genome positions on the16S rRNA phylogenetic tree except, of course, for the trpD domain of the trpD•trpC fusion protein in the two coryneform species. Thus, in C. diphtheriae and C. glutamicum, the free-standing trpD outside of the whole-pathway trp operon is more closely related to trpD inside the partial-pathway trp operons of all the other organisms. An inner-membrane protein of unknown function separating trpAa and trpD in all of the mycobacteria, encoded by chyp, also flanks the nonoperonic trpD of the two coryneform species. As expected for the suggested LGT scenario, trees of TrpAa, TrpEa, and TrpEb proteins that are encoded from the partial-pathway operons of mycobacterial species, Streptomyces, and Thermomonospora in Fig. 3 all cluster closely together with the exclusion of the corresponding LGT genes from the coryneform bacteria.
Post-LGT events of vertical descent can be tracked in C. diphtheriae.
Since the time that an alien trpAa/trpAb/trpB/trpD•trpC/trpEb/trpEa operon displaced the trp genes present in the common ancestor of coryneform bacteria, leaving behind only chyp and trpD as remnants, subsequent vertical evolutionary events in the C. diphtheriae genome are apparent. Thus, an insertion containing panB and panC occurred recently between trpD•trpC and trpEb in the C. diphtheriae lineage after its divergence from C. glutamicum. In C. glutamicum, closely related panB and panC orthologues (encoding ketopantoate hydroxymethyltransferase and pantothenate synthetase) comprise a characterized operon of d-pantothenate biosynthesis that is located elsewhere in the genome (71). In C. diphtheriae, the translocation of panB and panC into the trp operon is associated with an inversion event between these two genes. Hence, the opposite transcriptional direction of the inserted panC has now isolated trpEb/trpEa from its former operonic transcriptional continuity, presumably forcing it to become a separate transcriptional unit. It is interesting that the otherwise alien operon of C. diphtheriae now contains the native genes panB and panC, transposed from the resident genome. C. diphtheriae has also produced a gene duplicate of the gene encoding the alien TrpEb, which has then become the proximal member of the operon. This paralogue TrpEb is probably deficient in complex formation with TrpEa, because conserved residue K-167 (Salmonella enterica serovar Typhimurium numbering), which forms a salt bridge with residue D-56 of TrpEa, has been changed to S-167 (85). Also, the highly conserved residue 162-G has been changed to a charged residue, 162-E. Thus, after the LGT event, several subsequent vertical events of evolution that occurred in C. diphtheriae but not in C. glutamicum can be tracked.
The following approaches were taken in an attempt to locate the missing trpC genes in the above-mentioned actinomycete organisms.
Pattern and profile search.
TrpC is a short and relatively divergent sequence. Known TrpC homologues may have identities as low as 22%. In an initial Blast screening with E. coli TrpC as the query, for example, the Ferroplasma acidarmanus genome did not return any hits and appeared to lack TrpC by this criterion. However, the position of an unknown gene within the trp operon of F. acidarmanus strongly implicated its presence as a divergent trpC gene because it occupies the same relative position as trpC in two closely related Thermoplasma species. Indeed, identity as trpC (second iteration) was amply confirmed by use of PSI-Blast (5), as well as by the observed conservation in multiple alignments of critical residues established by structural studies of TrpC from E. coli. In addition, the use of TrpC query sequences from most of the Archaea did return positive Blast hits from the F. acidarmanus genome.
With this background in mind, the genomes of T. fusca, S. coelicolor, and the mycobacteria M. avium, M. tuberculosis, and M. bovis were subjected to a pattern and profile search that included a ProSite-like pattern based upon critical residues reported in the PDB summary, the use of TrpC domains as query sequences that were available from the closest relatives of the group missing TrpC, and the generation of a hidden Markov model based on a multiple sequence alignment of known TrpC sequences. No illuminating results were obtained with this approach.
Evaluation of an unknown gene inserted in the trp operon.
M. tuberculosis has a conserved hypothetical gene (Rv1610) inserted between trpAa and trpD (denoted chyp in Fig. 3). The absence of trpC coupled with the insertion of this unexpected gene within the trp operon invited careful scrutiny. This was, in fact, reminiscent of the previously mentioned situation with the operonic trpC of F. acidarmanus, which initially eluded detection as trpC. However, critical residues expected of TrpC could not be matched to Rv1610 by manual alignment. Furthermore, Rv1610 appears to encode an inner-membrane protein with three transmembrane segments. In addition, if Rv1610 were, in fact, a divergent TrpC, we would expect to find homologues in T. fusca and S. coelicolor. We did not.
Possible catalysis of the TrpC reaction by HisA.
TrpC catalyzes an intramolecular oxidoreduction (Amadori rearrangement) that parallels the isomerase reaction catalyzed by HisA. Both reactions involve isomerization of an identical phosphoribosyl moiety. TrpC and HisA each exhibit (βα)8 barrel structures. Jurgens et al. (46) in fact generated hisA mutants that could catalyze the TrpC reaction both in vivo and in vitro. One of these variants retained significant HisA activity. We therefore envisioned the possibility that an ancestor of the TrpC-deficient block of organisms might have duplicated hisA and recruited one copy to TrpC function. However, second copies of hisA were not found. We then further considered the possibility that HisA in these organisms might catalyze both reactions, since that potential had been established in vitro. However, the alignment of HisA sequences did not reveal any obvious variant residues common to the TrpC-deficient block of organisms that might suggest potential for TrpC activity.
Evolution of competence for TrpC catalysis by TrpD.
Altamirano et al. (3) recently reported the evolution of TrpC activity from the αβ barrel scaffold of TrpD following in vitro mutagenesis and recombination. Thus, one might envision an event of trpD gene duplication followed by divergence of one of the paralogues to TrpC function. Although a gene duplicate of trpD was found in S. coelicolor, other organisms of the trpC-“deficient” block do not have a trpD gene duplicate. In consideration of the additional possibility that a modified trpD might encode an enzyme capable of both reactions, a careful comparison of the multiple alignment for trpD sequences failed to reveal a variant subgroup that might be expected of an evolved dual-function trpC/trpD protein. This is perhaps not surprising in view of the recent retraction (4) of the results of Altamirano et al. (3).
Other possibilities.
Enzymes possessing triose phosphate isomerase (TIM) (βα)8 barrel-like folds are widespread and accommodate a particularly wide range of functions (15). Within this large grouping, TrpC, TrpD, TrpAa, and Rpe (d-ribulose 5-phosphate 3-phosphate epimerase) belong to the ribulose phosphate binding superfamily within the SCOP (structural classification of proteins) database (15, 86). Therefore, both TrpAa and Rpe were also evaluated as possible evolutionary sources of the missing TrpC, with the approaches described for HisA and TrpD. Suggestive evidence was not found.
The isomerase step catalyzed by TrpC is clearly a facile reaction, and although none of the foregoing possibilities considered produced the answer sought, they illustrate nicely the rationale and sorts of in silico strategies for gene discovery that can be anticipated in the near future. Until the time that this article was under review, the identity of trpC in the organisms included in Fig. 3 had remained a mystery. However, convincing evidence has been obtained recently that the HisA isomerase in these organisms does in fact catalyze the isomerase reaction in both pathways (9). The gene name, priA (phosphoribosyl transferase A), has been suggested to accommodate to its functional role in two pathways. Although this possibility was anticipated as outlined earlier, the natural bifunctional proteins of actinomycete bacteria did not resemble that obtained experimentally (46) in terms of amino acid sequence matches. Barona-Gómez and Hodgson (9) suggested that the bifunctional actinomycete isomerases represent an ancient evolutionary state that is in line with the recruitment hypothesis (38). If so, specialization in the gene duplicate that became trpC must have required more divergence than the gene duplicate that became hisA because the homology of PriA proteins with HisA is evident but not with TrpC proteins.
GENE FUSIONS
Phylogenetic Distribution of trp Gene Fusions
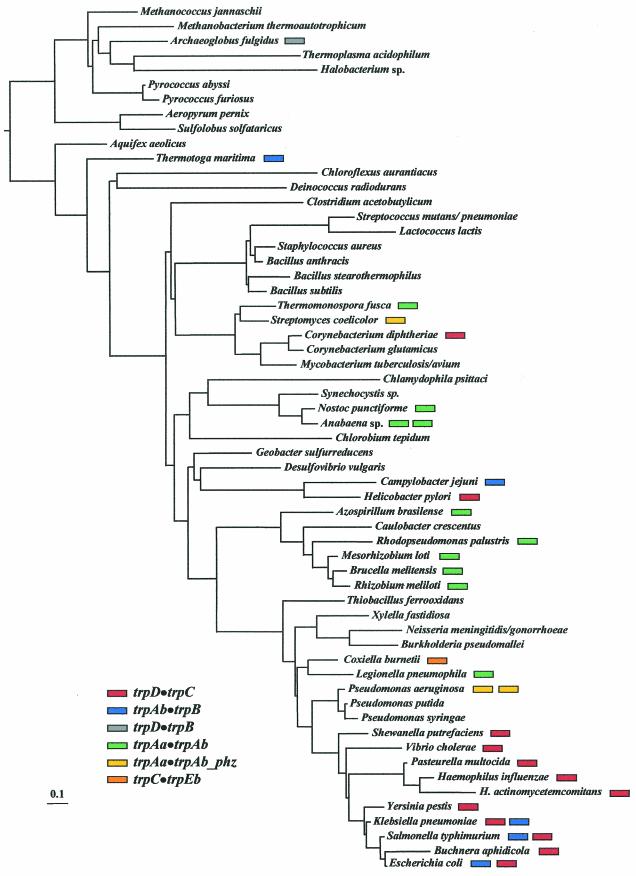
Each of the trp genes has been involved in various prokaryote fusion events except for trpEa. In some eukaryotes, however, trpEa and trpEb are fused (12, 16). Indeed, in Euglena, all of the trp genes except for trpAa and trpAb are fused together to form a pentafunctional protein (74). A trpD•trpB fusion is known in only a single instance (Archaeoglobus fulgidus), and a trpC•trpEb fusion is also thus far known in a single case (Coxiella burnetii). The remaining fusion types, all in the Bacteria, show an erratic distribution that is phylogenetically incongruous when mapped on the 16S rRNA tree (Fig. 4). Thus, the trpAb•trpB fusion is present not only in a small subcluster of the enteric bacteria, but also in the remote taxa Thermotoga maritima and Campylobacter jejuni. The trpD•trpC fusion, present throughout most of the enteric lineage (gamma proteobacteria), is also present in the widely separated Helicobacter pylori and in species of Corynebacterium. (In this case, we have already mentioned that a single origin followed by LGT events is likely.) Two distinct types of trpAa•trpAb fusions have occurred, one dedicated to primary biosynthesis (denoted trpAa•trpAb) and the other to phenazine pigment synthesis (denoted trpAa•trpAb_phz). The considerable extent of amino acid changes in TrpAa•TrpAb_phz has resulted in a shortened protein which no longer allows the ADIC product to continue through the ADIC lyase reaction to yield anthranilate, as is the case with anthranilate synthase (Fig. 1).
FIG. 4.
Mapping of the distribution of Trp pathway gene fusions to the 16S rRNA tree. The presence of fusion subtypes is color-coded as indicated in the legend. Although Buchnera aphidicola maps near E. coli on the 16S rRNA tree, as shown, its true point of divergence is probably prior to Yersinia, as portrayed by dotted lines in Fig. 8.
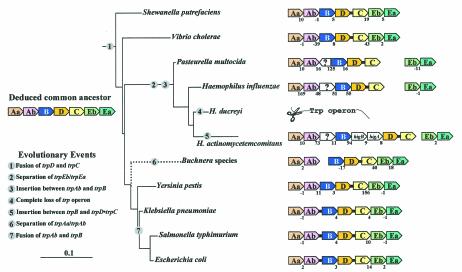
A priori, the scattered phylogenetic distribution of these gene fusions could be attributed to (i) LGT, (ii) an initial ancestral fusion (of rare occurrence) followed by numerous events of gene loss in different lineages, or (iii) independent gene fusions (therefore being of relatively frequent occurrence). Table 3 shows that all of the gene fusions exhibit a GC content that is similar to that of the resident genome. Thus, either these did not originate by LGT, the donor genome fortuitously had a similar GC content, or the LGT event occurred sufficiently long ago that amelioration has masked LGT. Unpublished data (Xie and Jensen, unpublished data) support the occurrence of many of the gene fusions as independent events of evolutionary innovation. Although the trpD•trpC fusions in coryneform bacteria and in Helicobacter pylori originated from the enteric lineage by LGT, the comparison of parametric data, e.g., GC content, does not reflect this, probably due to amelioration.
TABLE 3.
Comparison of GC content in gene fusions and cognate genomes
| Fusion | Organism | GI no.a | % G+C
|
|
|---|---|---|---|---|
| Gene fusion | Ge- nome | |||
| trpAb•trpB | Escherichia coli | 2506459 | 56 | 51 |
| Salmonella enterica | 1351306 | 57 | 55 | |
| Thermotoga maritima | 6226721 | 50 | 47 | |
| Campylobacter jejuni | 11268541 | 31 | 31 | |
| trpD•trpB | Archaeoglobus fulgidus | 11499197 | 52 | 49 |
| trpD•trpC | Escherichia coli | 136292 | 53 | 51 |
| Vibrio cholerae | 9655647 | 50 | 46 | |
| Haemophilus influenzae | 1574224 | 39 | 39 | |
| Buchnera sp. | 10038954 | 24 | 27 | |
| Pasteurella multocida | 12720848 | 41 | 38 | |
| Helicobacter pylori | 7227935 | 37 | 40 | |
| Salmonella enterica | 136301 | 55 | 55 | |
| Vibrio parahaemolyticus | 136302 | 47 | 46 | |
| Shewanella putrefaciens | N/A | 48 | 46 | |
| Yersinia pestis | 16122433 | 51 | 50 | |
| Corynebacterium diphtheriae | N/A | 57 | 53 | |
| Corynebacterium diphtheriae | 136291 | 58 | 56 | |
| trpAa•trpAb | Brucella melitensis | 13487153 | 58 | 56 |
| Rhizobium meliloti | 136328 | 63 | 62 | |
| Azospirillum brasilense | 1717765 | 74 | 68 | |
| Anabaena sp. | 17227910 | 41 | 44 | |
| Anabaena sp. | 17230725 | 46 | 44 | |
| Nostoc punctiforme | N/A | 42 | 44 | |
| Thermomonospora fusca | N/A | 68 | 69 | |
| Rhodopseudomonas palustris | N/A | 67 | 65 | |
| Mesorhizobium loti | 13472468 | 64 | 57 | |
| Legionella pneumophila | N/A | 38 | 40 | |
| Agrobacterium tumefaciens | 15889565 | 60 | 60 | |
| trpAa•trpAb_phz | Streptomyces coelicolor | 21220595 | 75 | 72 |
| Pseudomonas aeruginosa | 15597100 | 70 | 67 | |
| Streptomyces violaceus | 7481909 | 74 | 70 | |
| Pseudomonas chlororaphis | 6572982 | 63 | 62 | |
| Pseudomonas fluorescens | 2494756 | 62 | 62 | |
| Pseudomonas aureofaciens | 2494755 | 63 | 62 | |
GI, gene identification; N/A, not available.
Nested Gene Fusions
Jensen and Ahmad (1, 41) proposed that a series of nested gene fusions could be exploited as markers of phylogenetic branch points in prokaryotes. Thus, any organism that belongs to the enteric lineage (shaded green in Fig. 2D) shown in Fig. 4 would be expected to possess the trpD•trpC fusion, provided that the pathway has not been lost. At a more narrow hierarchical level, any organism belonging to the E. coli/S. enterica serovar Typhimurium/Klebsiella pneumoniae clade would be expected to possess the trpAb•trpB fusion as well. Thus, the clade defined by the trpAb•trpB fusion is nested within the more ancient clade defined by the trpD•trpC fusion. The presence of an AroQ•AroAIβ (chorismate mutase•DAHP synthase) fusion in Listeria, Bacillus species, and Staphylococcus but not in Enterococcus, Streptococcus, or Lactococcus is consistent with the suggestion made earlier that, contrary to the 16S rRNA tree, the order of branching is slightly different, so that these group diverged at a deeper tree position.
The ultimate analysis of the total inventory of fused genes in any given genome should provide an excellent phylogenetic tool for deducing the order of branching. This approach should be greatly enhanced by the rapid increase in the number of sequenced genomes coupled with the enormous advantage of being able to identify gene fusions with bioinformatic methods. However, it was not expected at the time that fusions could occur independently at such frequencies or that LGT should be taken seriously. Therefore, application of the approach of nested gene fusions will require sufficient background work to recognize and discriminate fusion clusters that have independent origins on the vertical tree as well as ones that might have been spread in the horizontal direction by LGT.
Trp PATHWAY GENE ORGANIZATION IN THE ARCHAEA
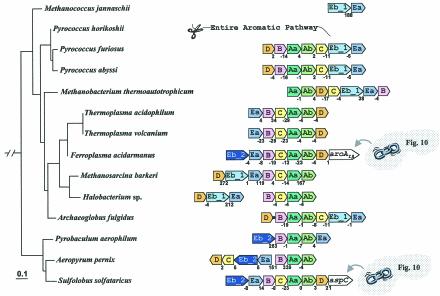
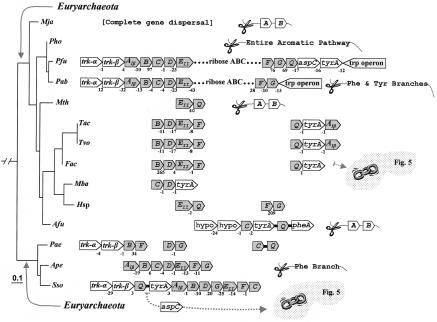
In general the Archaea deploy the Trp pathway genes as whole-pathway operons or as partial-pathway operons (Fig. 5). A very limited amount of experimental work provides data supporting the qualitative existence of regulation at the transcriptional level (26, 77). Rearrangements of gene order following events of inversion, translocation, and gene loss have been sufficiently dynamic that it is currently not possible to deduce the gene order of the common ancestor without more closely spaced genome representation. The only certainty would appear to be the existence in the archaeal ancestor of the partial gene orders →trpAa→trpAb and → trpEb_1→trpEa.
FIG. 5.
Organization of trp operon genes in the Archaea. Each trp gene is color coded differently, including the two subtypes of trpEb (Eb_1 and Eb_2) (92). trp genes that exist in the genome unlinked to any other trp genes are not shown. Archaeoglobus fulgidus has a trpD•trpB gene fusion (see Fig. 4). Intergenic spacing is shown, with negative values indicating gene overlap. Genes that are not specific trp pathway genes are in white boxes. F. acidarmanus possesses a gene encoding the aroAIβ subclass (44) of DAHP synthase. aspC in S. solfataricus is an aromatic aminotransferase of the Iγ aspartate aminotransferase type (42). This gene insertion corresponds to genes that appear to have escaped from the aro operons shown in Fig. 10. The gene order shown for Methanosarcina barkeri is the same as those in Methanosarcina acetivorans and Methanosarcina mazei. The gene order shown for S. solfataricus is the same as that for Solfolobus tokodaii.
In the compact Pyrococcus genus, P. horikoshii has lost the entire pathway. Although the trp operons of P. abyssi and P. furiosus are virtually identical, great variation can be seen for the remainder of aromatic biosynthesis (see Fig. 2A). In the Crenarchaeota grouping (Pyrobaculum, Aeropyrum, and Sulfolobus; lowest clade of Fig. 5), dramatic scrambling of gene order is apparent. This group has replaced trpEb_1 with trpEb_2. trpEb_2 is a distinct subgroup of trpEb that is present mainly in Archaea and that may often (but not always, as indeed exemplified by the Crenarchaeota) have a separate stand-alone function (92).
Usually, the pair of genes encoding the two subunits of tryptophan synthase are adjacent in prokaryotes. In the case of P. aerophilum, trpEa and trpEb_2 have been separated from one another within the operon. This may reflect the inability of trpEb_2 to form a complex with trpEa. In P. aerophilum, trpC and trpD have become separately dissociated from the operon. trpC and trpD are adjacent in the operon of Aeropyrum pernix, but separated in the operon of Sulfolobus solfataricus. Although all of the trp genes in A. pernix are adjacent, they are organized as two divergently transcribed groups, trpEa/trpEb_2/trpC/trpD and trpB/trpAa/trpAb. The A. pernix →trpEa→trpEb order is very unusual, the →trpEb→trpEa gene order being one of the most highly conserved gene couples in all prokaryotic genomes (17). Methanosarcina barkeri and Halobacterium spp. have identical gene orders, but the intact operon currently seen in M. barkeri corresponds to a splitting into two separate operons in Halobacterium spp. In some cases, other aromatic-pathway genes have been inserted into the trp operon. Thus, the trp operon of F. acidarmanus has aroAIβ as its most distal gene, whereas S. solfataricus has aspC (encoding aromatic aminotransferase) as the most distal gene of its trp operon.
Trp PATHWAY GENE ORGANIZATION IN THE BACTERIA
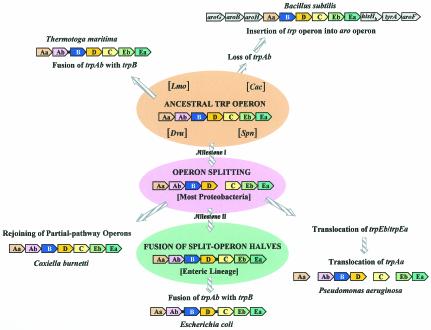
Whole-Pathway trp Operons
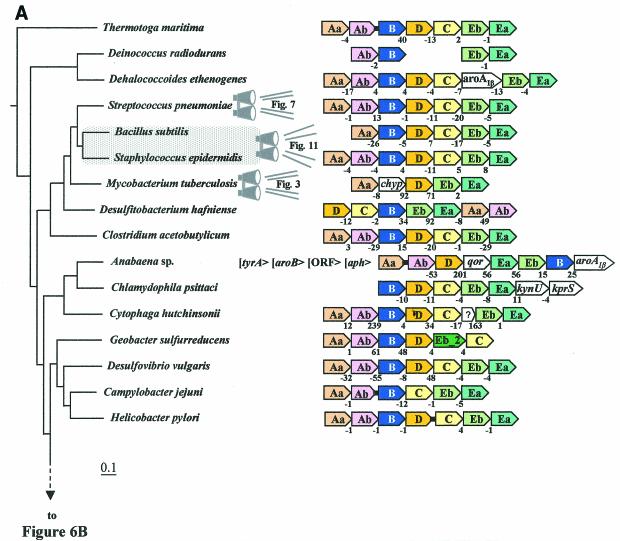
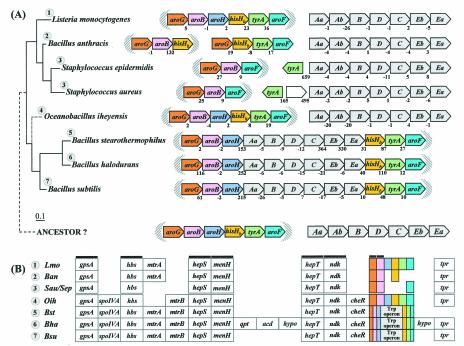
Unlike the domain Archaea, a consensus gene order can be discerned within the domain Bacteria, trpAa/Ab/B/D/C/Eb/Ea (Fig. 6A and 6B). The overall trace of organisms having the consensus gene order can be followed by noting the orange highlighting on the 16S rRNA tree of Fig. 2. A reasonable deduction is that this gene order (operon) was already present in the common ancestor of Bacteria, perhaps similar to the compact operons still present in the contemporary organisms Thermotoga maritima, Listeria monocytogenes, Streptococcus pneumoniae, Staphylococcus epidermidis, Staphylococcus aureus, and Coxiella burnetii. In contrast to the compact operon structure of these organisms, where intergenic spacing is <70 nucleotides and indeed often exhibits negative values (gene overlap), other whole-pathway operons exhibit significantly greater intergenic spacing, sometimes in the form of insertions of seemingly irrelevant hypothetical genes. For example, although both Lactococcus lactis and Cytophaga hutchinsonii maintain a consensus operon gene order, L. lactis exhibits an intergenic space of 124 bp between trpAa and trpAb, whereas both L. lactis and C. hutchinsonii have inserted a hypothetical gene between trpC and trpEb. Dehalococcoides ethenogenes possesses a very compact but expanded operon that includes an insertion of aroAIβ between trpC and trpEb. In this case, the insertion is probably physiologically relevant because AroA catalyzes the initial step of aromatic biosynthesis and thus forms the beginning precursor of chorismate.
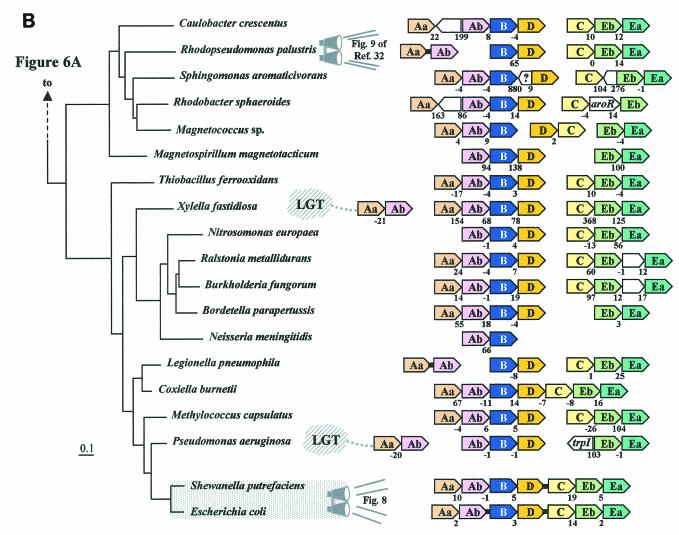
FIG. 6.
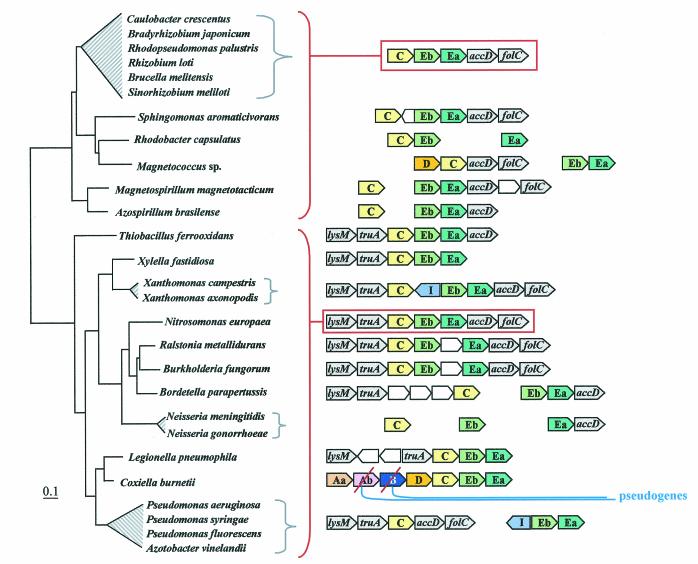
Organization of trp operon genes in the Bacteria. Each trp gene is color coded differently, including the two subtypes of trpEb. (trpEb in this figure refers to the major trpEb_1 subtype.) The tree sections in A and B join as indicated by the dashed line. Intergenic spacing is shown, with negative values indicating gene overlap. Separations showing white space and no intergenic spacing values indicate that the gene clusters are not linked to one another. Insertions of hypothetical genes and known genes are shown as white boxes. Short black bars connecting arrows denote gene fusions. Links to zoom-in expansions of particular lineages in other figures of this paper are indicated by binoculars. In B, the gene organization shown for Rhodopseudomonas palustris is identical to those of the closely related Agrobacterium tumefaciens, Rhizobium loti, Brucella melitensis, and Sinorhizobium meliloti; that shown for Burkholderia fungorum is identical to that of Burkholderia pseudomallei and Burkholderia mallei; that shown for Bordetella parapertussis is identical to that of Bordetella pertusis and Bordetella bronchiseptica; that for Neisseria meningitidis is identical to that of Neisseria gonorrhoeae; and that for Pseudomonas aeruginosa is identical to that of Pseudomonas putida, Pseudomonas fluorescens, and Pseudomonas syringae. The apparent supraoperon of Anabaena sp. (A) has been discussed in reference 93. kynU and kprS on the Chlamydophila psittaci line (A) refer to genes encoding kynureninase and PRPP synthase, respectively (89). The linked trpAa/trpAb genes shown for P. aeruginosa (B) were named phnA/phnB by Essar et al. (24). because they were thought to be dedicated to phenazine biosynthesis, a conclusion shown to be incorrect by Mavrodi et al. (57). This gene pair is not within the vertical line of descent (see later section), as indicated by the LGT notation. The trpAaAb operon shown on the left for Xylella is also outside the vertical line of descent (i.e., origin by LGT) (93). Shewanella putrefaciens (B) has the newly proposed name of Shewanella oneidensis (81).
Dispersal of trp Operon Genes
Gene dissociation has disrupted the trp operon of Bacteria in occasional lineages. For example, as illustrated at the bottom of Fig. 6A, Campylobacter jejuni possesses an extremely compact trp operon, but trpD has become dissociated, leaving behind a six-member partial-pathway trp operon. In Deinococcus radiodurans the separate dissociations of trpAa, trpD, and trpC have resulted in the retention of two small operons (each with overlapping genes) that are remnants of the ancestral whole-pathway operon. In these figures, isolated trp genes are not shown in order to conserve space, but they are present in the genome unless their absence is indicated in Fig. 2, e.g., trpAa and trpAb are missing in species of Chlamydophila psittaci (Fig. 2C).
In some cases, bacteria possess two chromosomes (19, 54). It is interesting that, in Rhodobacter sphaeroides, not only has the ancestral trp operon been split apart, but also the resulting partial-pathway operons (trpAa/yibQ/trpAb/trpB/trpD and trpC/aroR/trpEb) now reside on separate chromosomes. TrpEa has become completely dissociated from these operons (54). The closest available genomic neighbor of R. sphaeroides that is available on the 16S rRNA tree is Sphingomonas aromaticivorans, and it possesses the same split-pathway arrangement as R. sphaeroides except that trpEb and trpEa have remained together (i.e., trpC/trpEb/trpEa). The intriguing partitioning of the trp split-pathway operons between two chromosomes is typical of a 16S rRNA grouping of organisms that includes Rhodopseudomonas palustris, Rhizobium loti, Brucella melitensis, Sinorhizobium meliloti, Rhodobacter sphaeroides, and Sphingomonas aromaticivorans. Most of these organisms are not shown in Fig. 6, but a detailed breakdown of Trp pathway gene organization in this part of the tree is given in reference 93.
At one extreme of gene dissociation, gene dispersal has completely eliminated any linkage of trp pathway genes, as observed in Aquifex, unicellular cyanobacteria (Synechocystis, Synechococcus, and Prochlorococcus), and Chlorobium tepidum. (Only organisms possessing at least some linked trp genes are shown in the various figures of this paper.) One might reasonably consider whether these organisms simply manifest retention of a “preoperon” ancestral state, but this seems untenable with respect to the application of parsimony principles because they represent distinctly separate, widely spaced lineages.
All cyanobacteria possess a common phylogenetically congruous set of completely dispersed genes for tryptophan biosynthesis. However, Nostoc and Anabaena possess in addition some redundant trp genes that are linked to one another. The assemblage of linked trp genes in Anabaena spp. (shown in the middle of Fig. 6A) is very similar to that of the closely related Nostoc punctiforme (not shown, but see Fig. 2 of reference 93 for details) and seems to be part of a larger gene assemblage (possible supraoperon) that includes several other aromatic-pathway genes. Nostoc and Anabaena (large-genome, filamentous, and heterocystous cyanobacteria) possess these linked genes in addition to copies of all of the dispersed trp pathway genes found in the unicellular cyanobacteria. Hence, the redundant set of linked genes that are uniquely present in Nostoc and Anabaena seemed to be obvious candidates for origin by LGT. However, no support for LGT was found, and it has been suggested (93) that ancient paralogues have been retained in the Nostoc/Anabaena lineage, whereas the set of linked paralogue genes has been lost in the unicellular cyanobacteria.
Gene Scrambling
Examples of extreme gene scrambling can be found in Bacteria, e.g., in Desulfitobacterium hafniense (middle of Fig. 6A). However, gene scrambling seems to have been generally less pronounced in the Bacteria than in the Archaea. One of the most bizarre trp operons of the Bacteria is that of Geobacter sulfurreducens (Fig. 6A). Not only have trpEa and trpEb_1 dissociated from the operon and from one another, but trpEb_2 (rarely found in Bacteria) has also been inserted into the operon between trpD and trpC. trpEb_2 is an “alternative” β subunit of tryptophan synthase whose usual functional role has been speculated to be catalysis of the serine deaminase reaction (92). The gene fusions designated by short connecting black bars in Fig. 6A and Fig. 6B have already been discussed (Fig. 4).
Brown and Doolittle (11) made the correct observations, as long ago as 1997, even with vastly less data, that the consensus gene order seemed to be trpAa/Ab/B/D/C/Eb/Ea in Bacteria, that archaeal gene orders seem to be more variable than in Bacteria, and that the trpAa/trpAb and trpEb/trpEa linkage groups might be ancestral.
RETENTION OF THE ANCESTRAL OPERON AT SPACED PHYLOGENETIC NODES IN BACTERIA
The gene order trpAa/Ab/B/D/C/Eb/Ea of trp operons generally persists in the phylogenetic section of Bacteria shown between Thermotoga maritima and Helicobacter pylori of Fig. 6A, and a rationale of parsimony supports the thesis that this operon was already present in the common ancestor of modern Bacteria. This is not immediately obvious to casual inspection because of the dynamics of gene dissociation, gene loss, gene scrambling, and gene dispersal. However, a progression of conserved ancestral operons can be identified from the deepest phylogenetic position to the point of operon splitting between trpD and trpC (see orange highlighting in Fig. 2).
At the deepest branching position shown in Fig. 6A, T. maritima possesses a compact ancestral operon, differing only in that trpAb and trpB have fused. This fusion is rare, having occurred elsewhere only in the distant E. coli/S. enterica serovar Typhimurium/K. pneumoniae subgrouping (Fig. 4). At the next phylogenetic node in Fig. 6A, D. ethenogenes has retained the ancestral trp operon, albeit with an aroAIβ insertion between trpC and trpEb. In the gram-positive organisms shown in the Fig. 2B tree, ancestral operons are present in the following organisms from the deepest to more shallow phylogenetic nodes: Clostridium acetobutylicum > Desulfitobacterium hafniense > Listeria monocytogenes, Bacillus anthracis, and species of Staphylococcus. The ancestral trp operon has not survived in most of the phylogenetic groupings shown in Fig. 2C. In many cases, some or all of the Trp pathway genes have been lost by reductive evolution in pathogenic Bacteria. In other cases (cyanobacteria and Chlorobium tepidum), the trp genes have all been dispersed. Cytophaga hutchinsonii is the sole organism shown in Fig. 2C that has retained a complete trp operon with the ancestral gene order. Finally, in the top node illustrated in Fig. 2D, Desulfovibrio vulgaris has retained the compact ancestral trp operon, as shown near the bottom of Fig. 6A.
TWO MAJOR EVENTS UNDERLIE THE DYNAMICS OF trp OPERON CHANGE IN BACTERIA
Operon Scission Yields Two Half-Pathway Operons
In the common ancestor of the section of the 16S rRNA tree between Caulobacter crescentus and Pseudomonas aeruginosa (Fig. 6B), the ancestral operon was split into two, trpAa/Ab/B/D and trpC/Eb/Ea. The resulting two partial-pathway operons have exactly the gene organization as the contemporary Thiobacillus ferooxidans, Xylella fastidiosa, and Methylococcus capsulatus. An additional four genomes (Caulobacter crescentus, Sphingomonas aromaticivorans, Ralstonia metallidurans, and Burkholderia fungorum) exhibit the same two operons, but they are less compact. These partial-pathway operons are colored magenta in Fig. 2D. This split-operon pattern is very similar throughout this phylogenetic block of organisms, with some variations of gene insertion, gene dissociation, and gene fusion (see Fig. 9 in reference 93 for more detail).
FIG. 9.
Conserved genes flanking the trpC/trpEb/trpEa operon of organisms within the split-operon portion of the 16S rRNA tree. Organisms in the upper grouping are α-Proteobacteria; the cluster between Thiobacillus and Neisseria are β-Proteobacteria; and the bottom cluster is that fraction of the γ-Proteobacteria that diverged prior to the trpD•trpC fusion event. lysM and truA, conserved at the flanking gene position at the left throughout the β- and γ-Proteobacteria, are shaded grey, as are accD and folC (conserved in the flanking gene position at the right throughout the phylogenetic span portrayed in this figure). The deduced gene order of the common ancestor for each of the two major 16S rRNA clades is the same as shown for the two contemporary organisms Rhodopseudomonas palustris and Nitorosomonas europaea, as indicated by outlining in orange. Intervening genes, either hypothetical or known, are shown as open block arrows.
Only Magnetococus sp. and Coxiella burnetii in this section of the 16S rRNA tree exhibit trpD and trpC in contiguous positions. Presumably trpD and trpC were rejoined in these organisms. Zheng et al. (98) recently noted the frequency of split-pathway operons, and they made the intuitively reasonable conclusion that these are evolutionary forerunners of the complete trp operon. (In one sense, this is correct as a special case for the aforementioned Coxiella burnetii.) However, an inspection of the comprehensive set of data now available, with parsimonious principles applied, can only lead to the conclusion that a previously intact operon has undergone fragmentation.
Fusion of trpD with trpC Restores a Whole-Pathway Operon
In the common ancestor of the phylogenetic block of organisms sandwiched between Shewanella putrefaciens and Escherichia coli (Fig. 4, Fig. 6B, and Fig. 8), trpD and trpC were joined by fusion to restore an intact operon having the original ancestral gene order. All organisms within this “enteric” lineage possess the trpD•trpC fusion (green shading in Fig. 2D). Protein domain trees of the TrpD•TrpC proteins from the enteric lineage are highly congruent with 16S rRNA trees, as indeed are all seven protein domains. These protein trees (all seven) include out-of-position Trp proteins from Helicobacter pylori, Corynebacterium glutamicum, and Corynebacterium diphtheriae, the consequence of LGT (see next section).
FIG. 8.
Zoom-in from Fig. 6B showing Trp pathway gene organization in a range of Proteobacteria defined by the presence of the trpD• trpC fusion. Deduced phylogenetic events described on the left are identified by number on the 16S rRNA tree at the evolutionary times indicated. The actual position of Buchnera on the 16S rRNA tree (as shown in Fig. 2D and Fig. 4) is closest to E. coli. However, the long branch (Fig. 4) is consistent with the more likely order of branching depicted by the dotted line for Buchnera in this figure.
It should be kept in mind that our perception of milestone evolutionary events is biased by the relatively unbalanced selection of complete genomes currently available, and this will undoubtedly be altered as genome representation in parts of the evolutionary tree that are currently sparsely represented expands and yields more balanced representation. Thus, we readily see splitting of the ancestral operon as a milestone event in the Proteobacteria because many proteobacterial genomes have been sequenced. Likewise, we see the fusion of the split-operon halves as another milestone event because organisms of the enteric lineage have received high priority for genome sequencing. In other words, the current biases in genome selection have favored the deduction of evolutionary events in those lineages. Thus, in the future one can expect an expansion of milestone events recognized in other lineages.
LATERAL GENE TRANSFER OF trp OPERONS
Lateral Gene Transfer of Whole-Pathway Operons
From the vantage point of primary Trp biosynthesis, free-living prokaryotes will already have a reasonably integrated Trp pathway, and it seems unlikely that any selective advantages would come from displacing the native pathway with an alien one which evolved in a different metabolic context. It is possible that organisms that have lost the Trp pathway might occasionally reacquire it in one LGT event (from a whole-operon pathway donor). One could envision displacement of a native operon by an alien one if these possessed a more effective regulation mechanism (provided that advanced regulation really meshes with the needs of the recipient). However, the most sophisticated regulatory systems thus far described seem to utilize unlinked regulatory genes, such as trpR in E. coli and mtrB in B. subtilis. Thus, it would be difficult to transfer the entire operon system of structural genes and one or more unlinked regulatory genes via LGT.
It also is worthwhile to consider whether what is effective regulation for one organism would be appropriate for organisms that have a completely different lifestyle. E. coli, for example, experiences regular episodes of feast and famine in the gut of humans, and the ability of E. coli to regulate Trp enzymes over a large range of expression confers rapid response and efficiency. On the other hand, cyanobacteria generally grow in a nutritionally dilute environment and synthesize most of their amino acids most of the time. Under these conditions, possession of an operon system that is responsive over several orders of magnitude may not confer selective advantages.
There are a number of well-spaced genomes that possess the putative ancestral operon of Bacteria, highlighted orange in Fig. 2, e.g., trpAa/Ab/B/D/C/Eb/Ea is present in species of Listeria, species of Streptococcus, species of Staphylococcus, Clostridium acetobutylicum, and Desulfovibrio vulgaris. We considered the possibility that the trp operons in these organisms are related to one another by LGT rather than by vertical descent. However, we did not find that the trp operon proteins in any of these organisms clustered together when comprehensive trees for all seven proteins were inspected (data not shown), as would be expected for relationships of LGT. Therefore, we conclude that in these lineages, the exact ancestral operon was simply retained without gene dispersal, gene insertion, or gene fusion.
On the other hand, the fusion-containing trp operon (trpAa/Ab/B/D/•C/Eb/Ea) in the enteric lineage is related to those of coryneform bacteria and Helicobacter pylori by LGT. We know that coryneform bacteria must have been the recipient rather than the donor because they retain remnants of the original host. We conclude that H. pylori was also a recipient of LGT from the ancestral lineage because the Helicobacter/Campylobacter node of divergence is more recent than the root of divergence for the enteric lineage. Therefore, if Helicobacter had been the trp operon donor, one would expect Campylobacter to also have the fusion-containing trp operon. As pointed out before, the modern Helicobacter operon lacks repression control by trpR, presumably because trpR of the alien enteric lineage donor was unlinked to the transferred operon. It would be interesting to know how the regulation of the modern H. pylori trp operon compares to that of the modern Campylobacter jejuni trp operon, which presumably would be similar to the original H. pylori trp operon that was displaced.
Lateral Gene Ttransfer of Partial-Pathway trp Operons
Trp pathway enzymes can have metabolic roles other than to serve protein synthesis as a primary source of Trp. Specialized pathways leading to pigments, antibiotics, etc., have already been mentioned, and many unknown specialized pathways probably exist. Both partial-pathway and whole-pathway operons can be associated with specialized pathways. Cases in which one or more enzymes can serve the needs of both primary biosynthesis and some specialized pathway are known. It is quite common in such cases that the specialized pathway will possess key paralogues or analogues of Trp-regulated enzymes of the primary pathway. Such paralogues or analogues differ in the absence of the usual regulatory properties in order to abolish Trp as a regulatory cue (see below for examples). Operons encoding specialized pathways are more likely to confer immediate selective advantages to a recipient if a novel capability is transferred. Lawrence has asserted (51) that the selfish-operon model “predicts that operons are unstable as genes associate and disperse between transfer events.” This scenario probably would be more applicable to trp operons associated with some metabolic specialization than to those associated with primary amino acid biosynthesis.
Figure 6B shows two different partial-pathway trpAa/trpAb operons that were acquired by LGT in Xylella fastidiosa and in Pseudomonas aeruginosa, as discussed previously by Xie et al. (93). In Xylella it has been speculated (93) that trpAa/trpAb coexists within an operon with acl, which encodes an aryl-coenzyme A ligase that might have the specificity of an anthranilate-coenzyme A ligase. This might then be a point of divergence, whereby coenzyme A-activated anthranilate proceeds to an antibiotic, siderophore, etc. This anthranilate synthase appears to be resistant to feedback inhibition by Trp, consistent with the absence of Trp as an end product of the putative specialized pathway. The P. aeruginosa trpAa/trpAb operon shown in Fig. 6B was originally denoted phnA/phnB (phn for phenazine) because their expression in stationary phase, unregulated by Trp, was thought to be a mechanism to produce anthranilate precursor for phenazine synthesis in the presence of Trp. Although it is now known (57) that this operon is not part of the phenazine pathway and that anthranilate is not a phenazine precursor, it would appear to constitute a system designed for production of anthranilate in an unknown functional role in stationary-phase metabolism.
Streptomyces coelicolor possesses an operon (trpAa/trpab/trpB/trpD/aroAII) (Fig. 4) that is nested within a large cluster of genes that dictate synthesis of a calcium-dependent antibiotic (CDA) (70). This antibiotic contains Trp. The origin of this operon by LGT has been mentioned (70), but a detailed analysis has not yet been done. However, even if it originated via ancient paralogy instead, it is a good example of a contemporary operon that could confer a specialized ability to make Trp in the presence of fully charged tryptophanyl-tRNA via LGT. The key aspects are an operon free of any mode of regulation by Trp and inclusion of the gene encoding a homologue of DAHP synthase (AroAII) that is not inhibited by amino acids, hence ensuring an unrestrained supply of chorismate. Thus, normal restraints in place for primary biosynthesis at the branch point levels of both DAHP synthase and anthranilate synthase have been removed in order to accommodate the secondary synthesis of antibiotic. Note also in these examples with S. coelicolor that the primary and secondary pathways are not entirely separate.
The antibiotic-oriented operon system lacks trpEa and trpEb. Therefore, the tryptophan synthase that is utilized for primary biosynthesis must also be used to make Trp molecules destined for incorporation into antibiotic molecules. In view of the recent revelation (9) that priA fulfills the isomerase function in both the histidine and tryptophan pathways in S. coelicolor, as discussed earlier, it would appear that priA must also have a functional role in a third pathway to the CDA antibiotic. S. coelicolor has four paralogues of trpAa: one engaged in primary Trp biosynthesis (undoubtedly sensitive to feedback inhibition), a free-standing trpAa of unknown function, one dedicated to antibiotic biosynthesis (probably not sensitive to feedback inhibition), and another (not shown in Fig. 3) that is a domain component of trpAa•trpAb_phz and dedicated to phenazine biosynthesis.
FINE-TUNED EVOLUTIONARY DEDUCTIONS
At the exponentially increasing rate of genome sequencing, it is becoming feasible to examine, or at least to anticipate, the examination of organisms that are sufficiently close in a phylogenetic progression to facilitate refined evolutionary conclusions.
Single Change in a Common Ancestor versus Multiple Independent Changes in Descendants
Figure 7 illustrates the state of the trp operon in a gram-positive lineage containing Listeria, Enterococcus, Streptococcus, Lactococcus, Ureaplasma, and Mycoplasma. Some of these organisms have become auxotrophic following loss of the Trp branch (Enterococcus), loss of all three branches of aromatic biosynthesis (Streptococcus pyogenes and Streptococcus equi), or loss of the entire aromatic pathway (Ureaplasma and Mycoplasma). These events of gene loss had to be quite recent, undoubtedly linked to a relationship between the pathogenic lifestyles of these organisms and the relinquishing of selective pressure to retain the Trp pathway.
FIG. 7.
Zoom-in from Fig. 6A showing instances of Trp pathway reductive evolution and expansion of intergenic space in one phylogenetic section of some gram-positive bacteria whose 16S rRNA tree relationships are shown at the left. Loss of various metabolic capabilities is indicated by scissors. Note that the order of branching of Lactococcus lactis (shown in orange) has been altered from that shown in the 16S rRNA tree of Fig. 2B. The gene order and compact spacing of Listeria innocua is the same as that shown for Listeria monocytogenes.
If the 16S rRNA tree (Fig. 2B) reflects an exactly correct order of branching, then reductive evolution led to the loss of the Trp pathway independently on four occasions: in Enterococcus, S. pyogenes, S. equi, and the common ancestor of the Ureaplasma and Mycoplasma genera. However, the resolution power of 16S rRNA for determination of exact branching order can be imperfect for closely related organisms, and other character states can help fine-tune branching orders. Figure 7 illustrates a suggested modification of the 16S rRNA branching order (as shown in Fig. 2B) so that S. pyogenes and S. equi have a common ancestor (Fig. 7), rather than a common ancestor for S. equi and L. lactis (Fig. 2B). This modified tree yields a parsimonious loss (one event) in the common ancestor of S. pyogenes and S. equi. Note that this alteration of branching order is conservative, considering that the distance between S. pyogenes and S. equi is distinctly less than the distance between S. equi and L. lactis on the16S rRNA tree of Fig. 2B.
Distinguishing Derived States from Ancestral States
Listeria and three species of the divergent Streptococcus genus have retained a compact operon of the ancestral order trpAa/Ab/B/D/C/Eb/Ea. In contrast, although Lactococcus exhibits gene overlap between gene pairs at three positions, expansion of intergenic spacing is evident between trpAa and trpAb, between trpB and trpD, and especially between trpC and trpEb. Evolutionary direction can be deduced; that is, it is more parsimonious to conclude that the intergenic expansions within the L. lactis operon are a derived state rather than the ancestral state. Other examples are given in the next two sections, and the ability to distinguish derived evolutionary states from ancestral states in sister lineages is key to the ability infer evolutionary character states at given phylogenetic nodes.
Deducing Ancestral Character States at Phylogenetic Node Positions
Figure 8 provides a zoom-in picture of the gene organization exhibited by the enteric lineage of Bacteria (gamma proteobacteria). This entire group had a common ancestor that acquired the landmark fusion of trpD and trpC (trpD•trpC), restoring a whole-pathway operon. In the Pasteurella/Haemophilus grouping, dynamic changes occurred, including both the separation of trpEb/trpEa from the original operon and expansion of the intergenic space between trpAb and trpB by insertion. After the divergence of Pasteurella from the common ancestor of the Haemophilus lineage, the entire assemblage of Trp pathway genes was discarded in Haemophilus ducreyi. On the other hand, in Haemophilus actinomycetemcomitans, the intergenic space between trpB and trpD•trpC was expanded by insertion of two hypothetical genes. In the lower cluster of organisms in Fig. 8, trpAa/trpAb became separated from the rest of the operon in the outlying lineage that is represented by Buchnera sp. In the common ancestor of E. coli, S. enterica serovar Typhimurium, and Klebsiella pneumoniae, a fusion between trpAb and trpB occurred at a very recent time. trpAb•trpB and trpD•trpC exemplify one of the sets of nested gene fusions discussed by Jensen and Ahmad (41) that can be exploited for hierarchical ordering of taxa.
Value of Flanking-Gene Context
Figure 9 shows the bacterial organisms in the section of the 16S rRNA tree whose common ancestor possessed the two split-pathway trp operons that resulted from the separation of the ancestral trpAa/Ab/B/D/C/Eb/Ea operon between trpD and trpC. These organisms include the α-Proteobacteria (top major grouping), the β-Proteobacteria, and some of the γ-Proteobacteria. We did not find the trpAa/Ab/B/D partial-pathway operon to be flanked by conserved genes, but the trpC/Eb/Ea partial-pathway operon did exhibit flanking conserved genes. On the right in Fig. 9 are shown the positions of conserved genes that do flank the trpC/Eb/Ea operon. Genes encoding the β subunit of acetyl-coenzyme A carboxylase (accD) and folylpolyglutamate synthase/dihydrofolate synthase (folC) follow trpEa in most cases. Occasionally folC appears to have been translocated away from trpEa/accD, as exemplified in Bordetella parapertussis and Neisseria meningitides and Neisseria gonorrhoeae. For the lower group of organisms (from Thiobacillus through the Pseudomonas/Azotobacter cluster), the trpC/Eb/Ea operon is additionally flanked on the left by genes encoding fimbria V protein (lysM) and tRNA pseudouridine synthase A (truA). The top group of organisms shown in Fig. 9 exhibit the gene order trpC/trpEb/trpEa/accD/folC (boxed) that likely mirrors the ancestral gene order of the alpha-Proteobacteria, whereas it is reasonable to suggest that the gene order of Nitrosomonas europeae represents the ancestral gene order of the remaining organisms in the tree.
These conserved flanking genes provide information that can help guide fine-tuned evolutionary deductions. For example, the clade that includes Pseudomonas aeruginosa, P. syringae, P. fluorescens, and Azotobacter vinelandii possesses a trpC gene that has become separated from trpEb/trpEa. Was trpC or trpEb/Ea transposed away from the original trpC/Eb/Ea operon? The answer clearly is trpEb/trpEa, since trpC is flanked on the left by lysM/truA and on the right by accD/folC. Likewise, in the Magnetococcus sp., trpC is flanked on the right by accD and folC, and therefore the trpEb/trpEa operon must have been translocated away from trpC. Also, trpD in Magnetococcus must have migrated from the trpAa/Ab/D operon to its anomalous contemporary position near trpC.
Both Magnetospirillum and Azospirillum resemble the Magnetococcus sp. in that trpC has separated from trpEb/trpEa. However, in contrast to Magnetococcus sp., in which trpEb/trpEa has been transposed, in both Magnetospirillum and Azospirillum it is trpEb/trpEa that is linked to accD, and therefore it is clearly trpC that has been transposed away.
In Bordetella parapertussis trpC has separated from trpEb/trpEa in such a way that trpC retains linkage with lysM/truA on the left and trpEb/trpEa retains linkage with accD on the right. This could be consistent with a very large insertion (49,000 bp) between trpC and trpEb, or more likely trpEb/trpEa/aacD were jointly transposed. In Neisseria meningitides and Neisseria gonorrhoeae, trpC, trpEb, and trpEa have all become separated from one another. In this case, trpEa has retained its linkage with accD.
EXPANDED METABOLIC CONTEXT
Biochemical pathways are complexly interlinked in a net-like fashion, as any wall chart reveals, and it is of interest to examine the organization of Trp pathway genes in the larger metabolic context of aromatic amino acid biosynthesis (90). While even this is a relatively elementary metabolic expansion, a comprehensive analysis of this is well beyond the scope of this paper. However, two examples are given below (one from the Archaea and one from the Bacteria) which illustrate that the evolution of Trp biosynthesis has not necessarily occurred in isolation from its immediate biochemical connections. It is important to appreciate that a fuller future understanding of Trp biosynthesis will ultimately extend to the larger scope of interlocking metabolic ties that exist. A fuller appreciation of the varied interlocking ties of Trp biosynthesis with its metabolic context should be quite relevant to understanding the selective pressure favoring or disfavoring LGT. In addition, one can expect that conservation of an existing operon system would be significantly strengthened by a full repertoire of integrated metabolic ties.
Pyrococcus and Its Archaeal Relatives
Convergent trp and giant aro operons of Pyrococcus.
Pyrococcus furiosus possesses a truly remarkable array of linked genes for general aromatic biosynthesis (Fig. 10). These include not only genes encoding every common-pathway step, but also all genes specifically tied to phenylalanine, tyrosine, and Trp biosynthesis except for pheA. This even includes a gene encoding aromatic aminotransferase (denoted aspC). Incredibly, ties to the pentose phosphate pathway (the source of erythrose-4-phosphate) are reflected by the presence of linked genes for transketolase and for an ABC system of ribose transport. All of the genes encoding common-pathway steps are in the exact order of the reactions in the pathway. Shikimate kinase, encoded by aroEII (our designation), is an analogue class of kinase that is specific to Archaea (18). If one orients to tyrosine biosynthesis, all of the genes, beginning with transketolase (which generates erythrose-4-phosphate), are present in exact order through the final tyrA step. aroQ (chorismate mutase) and aspC (aromatic aminotransferase) are used for both phenylalanine and tyrosine biosynthesis. The P. furiosus aspC gene product has been shown experimentally to be utilized specifically for phenylalanine and tyrosine biosynthesis (84). The adjacent trp operon (see Fig. 4 for detail), with its many overlapping genes, is transcribed convergently from genes of the large aro operon.
FIG. 10.
Linkage relationship of genes within the larger context of aromatic amino acid biosynthesis in Archaea. The tree is the same as that shown in Fig. 5, where the full organism names corresponding to the acronyms used can be viewed. Common-pathway genes are shaded and designated by the gene letter, e.g., Q = aroQ. Hypothetical genes are denoted as hypo. Genes are labeled within block arrows that point in the direction of transcription. Copies of genes encoding transketolase are designated trk-α and trk-β. Short black bars connecting arrows indicate gene fusions. Deleted genes, pathway branches, and entire pathways are indicated with scissors.
Pyrococcus abyssi possesses exactly the same array of linked genes except that the aroQ/aspC/tyrA segment is absent from the genome (84). These genes are specific for tyrosine and phenylalanine biosynthesis. P. furiosus possesses a stand-alone copy of pheA, whereas P. abyssi lacks pheA altogether.
Dynamics of archaeal gene shuffling.
It is suggestive that the gene orders within the largest archaeal linkage groups that represent either Crenarchaeota (P. furiosus) or the Euryarchaeota (S. solfataricus) show some similarities, and we speculate that the ancestral gene order might have resembled that of the P. furiosus aro operon. This speculation is influenced by the gene order (aroAIβ/aroB/aroC/aroD/aroE/aroF/aroG) of the closest neighbor of S. solfataricus, Aeropyrum pernix. The altered order of aroC and aroG in S. solfataricus may reflect derived transposition events. If P. furiosus does represent the ancestral order, deletion of aspC could have resulted in the aroQ•tyrA fusion in S. solfataricus, which must then have been inserted between the ancestral trk-β and aroAIβ. If so, the deleted aspC gene was then inserted into the trp operon of S. sulfataricus (see Fig. 5) to become the distal gene member of the operon. Whether the trp operon became associated with the convergently transcribed aro operon in the Pyrococcus lineage or whether the trp operon dissociated from the aro operon in the Euryarchaeota seem to be equally possible alternatives that await resolution with the advent of more closely spaced genome representation.
The two Thermoplasma species (T. acidophilum and T. volcanium) and the closely related Ferroplasma acidarmanus have two identical aro operons except that aroAIβ is missing in Ferroplasma in comparison with the aroQ/tyrA/aroAIβ operon of the Thermoplasma species (Fig. 10). It is quite intriguing that this aroAIβ gene has been inserted into the trp operon of F. acidarmanus at the distal gene position (Fig. 5).
It is apparent that genes of both Trp biosynthesis (Fig. 5) and overall aromatic biosynthesis (Fig. 10) have been atypically dispersed in Methanococcus jannaschii. This is reminiscent of the tendencies toward gene dispersal seen in some but relatively few of the Bacteria (species of cyanobacteria, Aquifex, and Chlorobium). Methanopyrus kandleri, a relatively close relative of M. jannaschii, also has dispersed Trp pathway genes, with only trpAa and trpAb (20-bp gene overlap) being adjacent (data not shown).
Bacillus/Staphylococcus Clade
The entire clade shown in Fig. 11A is distinguished by having an aroQ•aroA fusion that is the basis for the novel allosteric pattern of sequential feedback inhibition of DAHP synthase by intermediary metabolites (43). The lack of this fusion in Enterococcus, Streptococcus, and Lactococcus is one of a number of reasons for our exclusion of these groupings from the Bacillus/Staphylococcus clade. The aro operons shown within shaded brackets in Fig. 11A exist within a general genomic region between gpsA (encoding glycerol-3-phosphate dehydrogenase) and a conserved gene (tpr) encoding a TPR repeat-containing protein (shown in Fig. 11B). Throughout the entire clade, the gene order gpsA/hbs/hepS/menH/hepT/ndk/aroG/aroB is conserved.
FIG. 11.
Zoom-in from Fig. 6A showing a conserved gram-positive region containing the six-gene aro operon (or remnants of it) and the trp/aro supraoperon of the B. subtilis/B. halodurans/B. stearothermophilus subgroup. (A) The aro and trp operons are mapped on a 16S rRNA tree at the far left. (The exact branching order of Oceanobacillus iheyensis has not been determined.) The Enterococcus/Streptococcus/Lactococcus grouping branches off between Listeria and the B. anthracis subgroup on a 16S rRNA tree (not shown, but see Fig. 2B and Fig. 7), but we believe from a variety of observations that it belongs just outside of the lineage shown in this figure. Shaded bracketed regions around aro operons and trp/aro supraoperons can be related to the presence of a context of conserved, flanking genes, as shown in part B. The separate aro and trp operons of a putative common ancestor are shown at the bottom of A. aro genes in B are color coded to match the genes shown in A. The conserved region to the left of aro operon genes includes eight genes (gps, hbs, hepS, menH, hepT, ndk, aroG, and aroB) that are conserved in every organism shown (heavy black overbars). Gene abbreviations: gpsA, glycerol 3-phosphate dehydrogenase; spoIVA, sporulation protein IVA; hbs, nonspecific DNA-binding protein; mtrA, GTP cyclohydrolase I; mtrB, TRAP; hepS, heptaprenyldiphosphate synthase (component I); menH, heptaprenyl naphthoquinone methyltransferase; qpt, quinone polyprenyltransferase; acd, aromatic acid decarboxylase; hypo, hypothetical gene; hepT heptaprenyldiphosphate synthase (component II); ndk, nucleoside diphosphate kinase; cheR, chemotaxis protein methyltransferase, tpr, tetratricopeptide repeat-containing protein (COG0457).
B. subtilis subgroup.
Bacillus subtilis, a member of the lower of the two major subgroups shown on the left in Fig. 11A, possesses a well-studied (35) supraoperon in which the trp operon is nested within a larger transcriptional unit. The B. subtilis trp operon has lost trpAb but pabAb (originally called trpX [47]) has been shown to support the amidotransferase function for both anthranilate synthase and p-aminobenzoate (PABA) synthase, i.e., the TrpAa/PabAb complex functions as an anthranilate synthase and PabAa/PabAb/PabAc functions as a PABA synthase. The six-gene B. subtilis trp operon is very compact, with four points of translational coupling. It is flanked on the N-terminal side with three aromatic-pathway genes (aroG, aroB, and aroH) and on the C-terminal side with three additional aromatic-pathway genes (hisHb, tyrA, and aroF).
hisHb (subscript denotes broad specificity) encodes a subgroup of imidazole acetyl aminotransferase that is widespread and functions as an aromatic aminotransferase (42). The other subgroup, HisHn (subscript denotes narrow specificity) functions in the pathway of histidine biosynthesis. Interestingly, the hisHb/tyrA/aroF gene combination is part of another supraoperon (serC/aroQp•pheA/hisHb/tyrA/aroF/cmk/rpsA) which has been characterized in Pseudomonas stutzeri and P. aeruginosa (90, 91). aroH is a relatively rare analogue class of chorismate mutase, thus far known to be present only in cyanobacteria and in a scattered distribution of gram-positive Bacteria, including, in addition to the lower group of Bacillus in Fig. 11A, Desulfitobacterium hafniense, Carboxydothermus hydrogenoformans, Clostridium botulinum (but not other Clostridium species), Thermoanaerobacter tengcongensis, Streptomyces coelicolor, Thermomonospora fusca, and Heliobacillus mobilis. The gene organizations of the Bacillus halodurans and Bacillus stearothermophilus supraoperons are essentially identical to that of B. subtilis. However, note that in B. stearothermophilus a conspicuous expansion of intergenic space between trpC and trpEb and between trpEb and trpEa is evident (Fig. 11A). We can be fairly sure, because of parsimony principles applied to the comparative data, that this intergenic expansion is a derived evolutionary state rather than an ancestral one.
Upstream of the B. subtilis supraoperon is the mtrA/mtrB operon, encoding GTP cyclohydrolase I and the TRAP regulatory protein, respectively (Fig. 11B). mtrB is uniquely present within the lower subgroup. B. stearothermophilus has conserved the general region shown in Fig. 11B between gpsA and the supraoperon, but tpr and its flanking region to the right have been transposed away. B. halodurans exhibits a number of unique insertions in the conserved region shown in Fig. 11B.
Listeria subgroup.
In spite of the current generic naming, Bacillus anthracis is closer on the 16S rRNA tree to species of Staphylococcus and Listeria (upper group in Fig. 11A) than to the other Bacillus species of the lower group. Members of this upper group all possess a complete seven-gene trp operon, including trpAb, which is absent in the lower Bacillus grouping of Fig. 11A. The Staphylococcus/B. anthracis group lacks the tryptophan RNA-binding attenuator protein (TRAP) encoded by mtrB (29), which is present throughout the lower group. The Staphylococcus/B. anthracis group also differs from the lower group and Listeria in the absence of aroH.
The aroH gene may be in a general process of displacement by aroQ, which is by far the most ubiquitous gene encoding chorismate mutase (13). Indeed, even within the lower group, one widely used strain of B. subtilis (strain 168) has lost aroH and relies exclusively on aroQ (48). The strain 168 genome, which has been sequenced and reported to possess aroH (as shown in Fig. 11A), is actually a hybrid prototrophic transformant with B. subtilis strain 23, the donor of aroH and linked trp genes (48). In Staphylococcus species of the upper group of Fig. 11A, the presumptive ancestral hisHb/tyrA/aroF linkage group has been disrupted, and aroF is now linked to aroG/aroB, whereas tyrA is now linked (divergently) with an intact trp operon. In contrast, B. anthracis retains the hisHb/tyrA/aroF linkage, but this has been expanded by addition of a gene duplicate of aroG at the 3′ end. In addition, the putative ancestral aroG/aroB has acquired a duplicate of hisHb at the 5′ end.
Note that we can distinguish which paralogues of aroG and hisHb in B. anthracis have remained in flanking gene context and which have been transposed away, i.e., the bracketed aroG/aroB/hisHb operon of Fig. 11A exists within the context shown in Fig. 11B. If aroH was present in the common ancestor of the clade, as speculated at the bottom of Fig. 11A, then it was lost in the common ancestor of the upper group. Otherwise, it arrived in the lower group either as a newly evolved innovation or by LGT. The first alternative may be more likely, considering that some fairly close relatives outside of the clade shown (e.g., Clostridium botulinum and Thermoanaerobacter tengcongensis) possess aroH.
Interconnectivity of the trp, aro, pab, and his operons.
Figure 11 illustrates that organisms like Listeria and Oceanobacillus possess six-gene aro and seven-gene trp operons that are located in widely spaced parts of their genomes. They also have pab operons and his operons (not shown) that altogether constitute four separately spaced and seemingly unrelated operons. This presumably represents the straightforward ancestral state of the clade. In the B. subtilis clade, however, these separate operon systems have become integrated via the following events. (i) The trp operon was inserted into the aro operon to produce the well-studied supraoperon. (ii) hisHn, a substrate-specific imidazole acetol phosphate aminotransferase, was deleted from the his operon, making the histidine pathway dependent upon HisHb, a broad-specificity imidazole acetol phosphate aminotransferase encoded by the aro portion of the supraoperon. (iii) trpAb was deleted from the trp operon, leaving the Trp pathway dependent upon the dual-function PabAb encoded from the pab operon. A metabolic basis for integration of the aro, trp, and pab operons is readily apparent in that the component genes are all part of the divergently branched pathway of aromatic biosynthesis. A metabolic relationship between the aromatic and histidine pathways is not as straightforward. However, both have a precursor relationship with pentose phosphate metabolism, both utilize a glutamine amidotransferase reaction, and both utilize PRPP as a key early substrate.
Evolutionary information derived from flanking-gene context.
Figure 11B shows a conserved region between gpsA and tpr that is the location of the six-gene aro operon in Listeria and Oceanobacillus. Upstream between the highly conserved hbs and hepS are mtrA and mtrB (if present). The shaded brackets in Fig. 11A indicate the genes that are present within the flanking gene context detailed in Fig. 11B. In the major upper group, the trp operon has no consistent pattern of flanking genes. In B. subtilis and B. halodurans, the supraoperon genes are ordered within the region shown on the bottom line of Fig. 11B. In B. anthracis, an aroG/aroB/hisHb segment of the original six-gene aro operon has remained in the original context of flanking genes. Paralogues of aroG and hisHb, now associated with tyrA and aroF, have migrated to a new genomic position. In Staphylococcus the remnant of the original aro operon, aroG/aroB/aroF, has remained in the original context of flanking genes; aroH has been lost from the genome; and hisHb and tyrA have separately been moved elsewhere. In the case of tyrA, it has now been divergently positioned directly upstream of the trp operon.
Thus, both this analysis and the analysis represented by the data shown in Fig. 9 illustrate how flanking-gene context in relatively close sister lineages can help sort derived evolutionary events from ancestral ones.
Deducing the likely common ancestor of the clade.
Thus, the major upper and lower groups of Fig. 11A differ in the gene organization of the trp operon (presence or absence of trpAb), in the regulation of the operon (presence or absence of mtrB), and in the particular context of association with other aromatic-pathway genes (Fig. 11B). The most conserved gene order arrangements overall, in addition to the trp operon, are aroG/aroB and hisHb/tyrA/aroF. One can be fairly certain that the common ancestor possessed the complete trpAa/Ab/B/D/C/Eb/Ea, aroG/aroB, and hisHb/tyrA/aroF gene orders. This is because the linkage of aroG/aroB persists throughout the organisms shown in Fig. 11A and because the hisHb/tyrA/aroF linkage is well conserved, even at a deeper level, in the Bacteria. Deduction of a convincing common ancestor will require the genome sequences of additional organisms that will present a more finely spaced phylogenetic progression. A case in point that illustrates the process was our recent consideration of the new genome sequence for Thermoanaerobacter tengcongensis in this connection. When the Blast similarities of proteins from T. tengcongensis were scored against the overall genomic database (8), the highest score was for B. halodurans. Had this reflected membership of T. tengcongensis in the Fig. 11A clade, as we anticipated, it might have assisted deduction of evolutionary events in the clade. However, T. tengcongensis does not have the clade-conserved aroQ•aroA fusion, and its position on the 16S rRNA tree also places it outside the clade.
Given the tentative deduced ancestral linkages shown at the bottom of Fig. 11A, evolution of the supraoperon of the lower group must have involved loss of trpAb and the connection of aroG/aroB/aroH at the 5′ end of the operon, as well as joining of hisHb/tyrA/aroF at the 3′ end of the operon. If the common ancestor possibly possessed aroG/aroB/aroH/hisHb/tyrA/aroF as a single linkage group (as seems probable in view of the presence of this six-gene aro cluster in Listeria and Oceanobacillus), a single event of insertion of the trp operon between aroH and hisHb would account for the contemporary supraoperon. We propose that the gene organization of the common ancestor of the clade shown in Fig. 11A was very similar to that of the modern Listeria monocytogenes.
OVERVIEW PERSPECTIVES
Lineage-Specific Evolutionary Trends
There may be lineage-specific forces at work that have favored processes of gene dispersal, operon fragmentation, and gene insertion for reasons that are currently unappreciated. When considering in a comparative context the intact and highly compact Trp and His operons of E. coli, we noticed that various trp operon features (which are comprehensively documented in this paper) seem to exhibit parallel differences with respect to histidine operon features. Thus, it seems more than coincidence (i) that both the Trp pathway genes and the histidine pathway genes are dispersed in Aquifex and in the unicellular cyanobacteria, (ii) Campylobacter jejuni has an intact his operon except for the dissociation of hisC, reminiscent of its otherwise intact trp operon which exhibits dissociation of only trpD (Fig. 6A), (iii) P. aeruginosa exhibits fragmentation into four partial-pathway operons of histidine biosynthesis (hisGDC, hisBHAF, and hisIE) reminiscent of its fragmentation into partial-pathway operons for Trp biosynthesis (Fig. 6B), and (iv) Lactococcus lactis exhibits seemingly extraneous multiple gene insertions in its complete his operon (2), similar to insertions observed in its complete trp operon.
The comparative analysis of the histidine operon is well beyond the scope of this paper, but dynamics of gene scrambling similar to those seen with the trp operon are evident, e.g., the E. coli gene order hisG/D/C/Bd•Bpx/H/A/F/I•E compared to the Sulfolobus solfataricus gene order hisC/G/A/Bd/F/D/E/H/I/Bpx. A preliminary assessment indicates that the histidine pathway gene organization exhibits some intriguing parallels to Trp pathway gene organization. Different events of gene scrambling, gene dispersal, gene fusion, intergenic expansion, and operon fragmentation exist in both Bacteria and Archaea. Similar to what seems to be the case for the Trp pathway gene organization, gene scrambling also seems to be more frequent for histidine pathway gene organization in the Archaea than in the Bacteria.
Individual Divergences Unmasked in the Larger Genomic Context
Figure 12 portrays the relationship of a few selected organisms with respect to the overall deduced evolutionary histories of the trp operon. The three major trp operon gene organizations are displayed within color-coded ovals that correspond to the highlighting of specific organisms in Fig. 2. Subsequent evolutionary events deduced for selected organisms emerging from each group are shown. The intent is to illustrate how detailed case-by-case analyses can elucidate evolutionary histories that would not be at all apparent otherwise. Thus, the trpAa/Ab/B/D/C/Eb/Ea operon of Coxiella burnetii (lower left) at first inspection appears to have experienced no evolutionary change because it is identical to the deduced ancestral operon. However, our analysis indicates the intervention of two evolutionary events, one producing the two “split-pathway operons” present in most Proteobacteria and the second rejoining the two previously separated operons.
FIG.12.
Schematic of the major evolutionary events (milestone I and milestone II) following the ancient establishment of a trp operon in the domain Bacteria. The ancestral trp operon has been retained by organisms such as Listeria monocytogenes (Lmo), Clostridium acetobutylicum (Cac), Streptococcus pneumoniae (Spn), and Desulfovibrio vulgaris (Dvu). The emergence of selected contemporary organisms is shown. The three stages highlighted with an orange oval, a magenta oval, and a green oval correspond to the color coding used in Fig. 2 to designate the particular contemporary organisms that have retained the exact gene organization illustrated within one of the three ovals.
As a second example, a comparison of the E. coli trp operon with the ancestral trp operon reveals only two differences in structural gene organization, fusion of trpD with trpC and fusion of trpAb with trpB. However, we have shown that the ancestral operon must have split into the two halves shown in Fig. 12 prior to the trpD•trpC fusion. In this connection, the trp operon of Thermotoga maritima differs from the ancestral operon only in having the trpAb•trpB fusion. With limited information, one might have predicted that the T. maritima operon was directly intermediate between the ancestral state and the E. coli state, i.e., the ancestral state, followed by the Thermotoga state (trpAb•trpB fusion), followed by the E. coli state (trpAb•trpC fusion). However, we have found that the two trpAb•trpB fusions occurred independently. The contemporary operons of T. maritima and E. coli do not show any common steps of operon change, and a much richer evolutionary history exists than would be evident from superficial inspection.
The well-studied trp operons of Pseudomonas aeruginosa and Bacillus subtilis are illustrated in Fig. 12 as examples of operons from organisms that are not representative of the deeper phylogenetic node. Since the time of the landmark splitting of the ancestral operon, a history of additional fragmentations in P. aeruginosa has left trpAa isolated from trpAb/B/D and trpC isolated from trpEb/Ea. Likewise, the well-studied trp operon of Bacillus subtilis is not representative of the broader Listeria/Bacillus/Staphylococcus node. In relatively recent events, trpAb has been discarded, and the remaining trp operon appears to have been inserted into an aro (aroG/aroB/aroH/hisHb/tyrA/aro/F) operon. (see Fig. 11 and the attending discussion in this text). Since the dual use of pabAb in the lower group for both anthranilate and PABA synthesis is isolated to this lineage, the seven-gene trp operon of Bacillus anthracis and Staphylococcus species is more representative of the node of Fig. 11 organisms than is the six-gene B. subtilis operon.
Analysis of the Ancestral State at Phylogenetic Nodes
Our study illustrates how one can avoid errors due to LGT and ancient paralogy and identify the most likely common ancestor that represents a phylogenetic node. If nodes at the bottom of the tree are sufficiently well represented to deduce the state of the trp operon at those nodes, one can deduce the likely common ancestor at progressively more ancient nodes, working backwards in evolutionary time up the tree. This is illustrated by zoom-in figures in relationship to the mapping of Trp pathway genes on the 16S rRNA tree for Bacteria in Fig. 6. Thus, to give some examples of cases where evolutionary differences in closely related members of a clade can be distinguished as ancestral states or derived states, we have seen (i) that the trp operon of Listeria monocytogenes but not B. subtilis is representative of the node position of the common ancestor for the Listeria/Lactococcus/Staphylococcus/Bacillus clade (Fig. 7 and 11), (ii) that the two partial-pathway operons of Thiobacillus ferooxidans and Methylococcus capsulatus but not the trp gene arrangements of Pseudomonas aeruginosa or Neisseria meningitidis are representative of the ancestral state at the node representing those Proteobacteria (Fig. 6B) that diverged after the major event of operon splitting (Fig. 12), (iii) that Shewanella putrefaciens is more representative of the phylogenetic node for enteric bacteria than Haemophilus influenzae (which is probably undergoing an early phase of reductive evolution) or E. coli (which has experienced a recent additional gene fusion); and (iv) that Campylobacter jejuni is more representative of its common node with Helicobacter pylori because the native trp operon of H. pylori was displaced by an alien trp operon via LGT.
The clade of actinomycete Bacteria shown in Fig. 3 offers a particularly apt example of how genes representing the ancestral state of Trp biosynthesis can be sorted out from genes originating by LGT or ancient paralogy. The Thermomonospora, Streptomyces, Corynebacterium, and Mycobacterium genera each exhibit substantial differences from one another. Mycobacterium lacks paralogue copies of trp genes, and therefore its trpAa/D/Eb/Ea operon plus dispersed copies of trpAb, trpB and the missing trpC that are present can reasonably be assumed by default to specify the primary pathway of Trp biosynthesis. The situation is the same in Thermomonospora except that trpAa and trpAb are fused. Streptomyces possesses several trp operons, but the primary trpAa/D/Eb/Ea biosynthetic operon can be identified by phylogenetic analysis. Thus, proteins encoded by each of the trpAa/D/Eb/Ea operons as well as the free-standing copies of trpAb and trpB in the organisms shown in Fig. 3 all cluster together on phylogenetic trees to the exclusion of other paralogues present in the Streptomyces genome.
The trpAa/Ab/B/D/aroAII operon is known to have a specialized role in antibiotic production that is unique to Streptomyces. The free-standing TrpD of S. coelicolor specifically clusters in the phylogenetic tree with TrpD proteins encoded by the trpAa/D/Eb/Ea operons in the rest of the clade. It is a trpD remnant, since all other genes have been otherwise replaced by a whole-pathway operon via LGT. Thus, with all of this information, we can reasonably predict that the common ancestor at the node position for actinomycete bacteria (as depicted in Fig. 6) possessed a trpAa/D/Eb/Ea operon, with the remaining trp genes dispersed.
Intellectual Dilemma Addressed
Does trp gene reorganization necessarily imply functional deterioration?
A central dilemma that merits consideration was posed in the Introduction. trp operons of model organisms such as E. coli and B. subtilis are elegantly geared for the efficient regulation of what is the most biochemically expensive of the 20 amino acids. As such, one might think that once evolved, forces of selection would enforce stability of the first order. Therefore, the variety and frequency of trp operon rearrangements, which have involved events of gene shuffling, gene fusion, operon splitting, total gene dispersal, and insertion of seemingly unrelated genes, is a dilemma that underlies this study. Since the overwhelming majority of modern prokaryote lineages maintain whole-pathway or at least partial-pathway trp gene organizations, the operon surely must generally constitute a selective advantage. To what extent do all of these changes imply operon disruption as opposed to fine-tuned improvement (or neutrality) of the operon system? Among all of the types of change, only total gene dispersal, as occurred, for example, in unicellular cyanobacteria, clearly constitutes an event of operon disruption. The cleavage of whole-pathway operons to yield two or more partial-pathway operons would seem disadvantageous, but this may reflect an evolutionary strategy of which we are presently unaware. Certainly the multiple control mechanisms used to control three different trp transcriptional units in P. aeruginosa hint at this.
Events of gene insertion and gene shuffling are not necessarily events of gene disruption. The reshuffled deck of trp genes in an operon such as that of Desulfitobacterium hafniense (Fig. 6A) seems curious, indeed, but there is no reason to believe that this compact operon is any less efficient for the shuffling. Perhaps the shuffling reflects nature's continuing experimentation to test for different orders of translationally coupled genes that produce different protein-protein interactions. When previously compact operons are altered by expansion of intergenic spacing, perhaps this is a necessary evolutionary step for successful gene fusion. Sufficient intergenic space would seem to be necessary for evolution of a linker region that does not intrude on the catalytic domains being fused.
Are there any clear examples of efficient operons systems that have been disrupted?
We do not know the extent to which the high efficiency of regulation that is fully documented in only a few organisms such as E. coli and B. subtilis is typical of other trp operons. The regulatory features of E. coli and B. subtilis are distributed within rather narrow clades, and it may be that these exemplify relatively recent advanced operon systems that will in fact strongly resist future disruptive events in all of the free-living descendants. It would be most informative to know the details of regulation in a well-spaced phylogenetic progression of other modern whole-pathway operons (such as the operons carried by the orange-highlighted organisms in Fig. 2). For example, a two-component response regulator gene is positioned only 17 bp upstream of trpAa in Thermotoga maritima. Might this reflect the presence of a completely different mode of control?
It is possible that many trp operons in nature are relatively primitive and only have the advantages conferred by a common promoter and (in the case of overlapping genes) either translational coupling or protection from mRNA degradation. It seems quite probable that many free-living organisms have no use for the huge range of trp gene expression that is typical of a feast-and-famine organism such as E. coli. For example, cyanobacteria probably make most or all Trp endogenously and thus may require regulation over a minimal range. One could envision that simple feedback inhibition of anthranilate synthase might constitute the main regulation in operation. This is consistent with the results of two studies of cyanobacteria in which exogenous Trp transport was two orders of magnitude less than in B. subtilis (32), anthranilate synthase is 100% inhibited at 10 μM Trp (36), and the range of enzyme expression varies only two- to threefold except for a 20-fold range in the case of tryptophan synthase (36).
There are distinct examples where operon disruption has followed acquisition of a finely tuned trp operon system, e.g., dissociation of trpEb/trpEa in Pasteurella multocida in the enteric lineage (see Fig. 8). However, these are special cases in organisms that have become pathogens or intracellular symbionts. There is ample evidence that evolved interorganismal relationships can produce completely new selective conditions that no longer require an efficient operon. In the extreme case, many pathogenic organisms undergo reductive evolution and abandon the pathway altogether because the host provides Trp. Since eukaryote hosts (such as humans) are relatively recent, such processes are likely to be in an ongoing state. In these cases, events of gene insertion and gene dissociation may not be selectively disadvantageous. Indeed, they may be steps in the selected process of genome reduction. In this connection it might be instructive to consider the recent disruptive events that have occurred in the pathogenic Corynebacterium diphtheriae but not in the free-living sister species Corynebacterium glutamicum since the LGT-mediated acquisition of the trp operon in their common ancestor (see Fig. 3 and attending discussion).
In a completely different context of interorganismal relationship, Buchnera aphidicola is an endosymbiont that produces Trp for the host. In this case, one can pinpoint a fairly recent time of selection against efficient regulation of Trp biosynthesis. Here the endosymbiont cells have been challenged to overproduce Trp for export to the host. This is primarily accomplished by translocation of trpAa/trpAb to a plasmid, with the result of giving a 16-fold amplification of the rate-limiting first step of Trp biosynthesis (50). It is very important to keep in mind that genomic sequencing has been heavily biased in favor of organisms that directly impact humans, and genomic representation of free-living organisms is still relatively weak.
The answer to the question raised in the heading is then yes and no. Pathogens (especially obligate pathogens) are in the process of abandoning the trp operon altogether. Endosymbionts, such as Buchnera, may abandon the regulation altogether in order to engineer themselves to saturate the needs of the host. However, there is thus far no evidence that a free-living organism equipped with a highly evolved and efficiently regulated trp operon experiences instability with respect to that operon.
Elaborate regulation seems to be fairly recent.
Primitive trp operons may have been regulated by relatively simple schemes. Consistent with this is that all elaborate control systems for trp operons are restricted to marrow clades. The advanced trp operon of E. coli differs from that of the putative common ancestor of Bacteria in having two pairs of structural gene fusions (Fig. 4), the trpR repressor, and a leader peptide (trpL) for attenuation. The distribution of trpR is limited to the enteric lineage except for Coxiella burnetii, Xylella fastidiosa, and some species of Chlamydia (89). Regulation by attenuation mechanisms seems to be distinctly more widespread than repression control by trpR (7, 53, 75). However, particular attenuation mechanisms can be distinctly different. Thus, the mechanism in E. coli that relies on the trpL leader peptide (95) is quite distinct from the Bacillus subtilis mechanism that utilizes a Trp-activated RNA-binding protein (TRAP) (29) as well as an anti-TRAP protein whose synthesis is induced by uncharged tRNATrp (80).
Does the enteric clade (see Fig. 8), with its multiple mechanisms of control, perhaps possess a relatively superior trp operon that would resist future events of operon disruption? It may very well be that the enteric lineage (as represented in Fig. 8) currently has a very highly conserved operon system in its free-living members. Exceptions in pathogenic organisms that are undergoing reductive evolution are easily understood (e.g., Haemophilus species), as are exceptions in intracellular symbionts such as Buchnera.
The L. lactis tRNA-directed transcription termination mechanism might prove to be the most broadly distributed mechanism, since various gram-positive organisms utilize this mechanism for a number of different amino acid biosynthetic pathways (34). The loss of trpAb from the trp operon of the B. subtilis clade and reliance upon the broad-specificity homologue in the folate pathway for dual function in anthranilate and 4-aminobenzoate synthesis may have favored an even more advanced regulatory system that integrates folate and Trp biosynthesis. In accord with this, TRAP also regulates the transcript levels in the B. subtilis folate operon (20).
Regulation of Trp biosynthesis in organisms lacking the whole-pathway operon may be relatively undeveloped aside from the widespread sensitivity of anthranilate synthase to feedback inhibition by Trp. Several partial-pathway operons are known to possess only a degree of regulation. Thus, in Rhizobium meliloti, the trpAa•trpAb operon is regulated by transcription attenuation but not the trpBD operon or the trpCEbEa operon (7). However, such a generalization may not be justified in consideration of P. aeruginosa and its close relatives P. putida, P. fluorescens, P. syringae, and Azotobacter vinelandii, in which transcription of the trpEbEa operon is activated by trpI (6, 14) and the free-standing trpAa and the trpAbBD operon are regulated by attenuation (67).
Given the variety of trp operon regulatory mechanisms that are known to have evolved and others undoubtedly yet to be discovered (30), one might think that selection for the most efficient operons would have proceeded rapidly via LGT. This may be an oversimplification in that different levels of efficiency may be selected for different lifestyles. Feast-and-famine organisms such as E. coli may be most suited to relatively large ranges of control modulation. In any event, only the LGT relationship of whole-operon transfer between Helicobacter pylori, coryneform bacteria, and enteric bacteria is evident at present. An obvious roadblock to LGT of at least some complexly regulated operons is the presence of regulatory genes at unlinked loci with respect to the operonic structural genes. It may very well be (see following section) that what is efficient in the metabolic context of one lineage is not so efficient in the metabolic context of another lineage (40).
Regulation extending beyond the Trp pathway.
From the vantage point of operon stability, we think that it is very important to consider how deeply some modern trp operon systems have become integrated into a broader metabolic network. The first example is trpR in E. coli. Not only the trp operon but also four additional transcription units belong to the trpR regulon (68). Other members of the regulon include the trpR gene itself (which is therefore autoregulated), mtr (encodes a Trp-specific transporter), aroL (encodes shikimate kinase II), and aroH (a paralogue of the DAHP synthase AroAIα homology group that is also feedback inhibited by Trp). aroL is also a member of the tyrR regulon. Thus, fine-tuned regulation by trpR is not only focused upon the specific Trp branch, but also influences the broader aromatic pathway, which generates precursor molecules. There is a certain integrant relationship in which the presence of trpR correlates with multiple, differentially regulated isoenzymes of DAHP synthase. It may be relevant here that there is a correlation between disruption of the whole-pathway trp operon in Haemophilus influenzae and the loss of genes encoding two of the three differentially regulated isoenzymes of DAHP synthase that are typically present in enterics.
The second example is that of mtrB in B. subtilis, which encodes TRAP. Here again, TRAP exerts regulatory influences across metabolic pathways, in this case between the Trp and folate pathways. TRAP not only regulates the trp operon by both transcription attenuation and a translational control mechanism, it also regulates the translation of pabAb (required for both Trp and folate biosynthesis), yhg (a putative Trp transporter), and ycbk (encoding a protein of unknown function). Thus, Trp and folate biosynthesis are coordinated via the regulatory abilities of TRAP. An organism such as Oceanobacillus possesses mtrB, a seven-gene trp operon that contains trpAb, and a folate operon that contains pabAb. Thus, it seems likely that once the dual regulatory role of mtrB in both pathways was established, integration was further elevated in the B. subtilis/B. halodurans/B. stearothermophilus clade by loss of trpAa and reliance upon pabAb to form alternative complexes with either trpAa or pabAa.
Does Regulation Power Evolutionary Dynamics?
In view of the foregoing points, we offer the following broad perspective. In ancient free-living prokaryotes, trp structural genes had already become organized as whole-pathway operons. The selfish-operon model of operon origin promoted by Lawrence and Roth (52) might apply to these early stages. Presumably, coordinate expression from a common promoter, overlapping genes (perhaps protecting from mRNA degradation), and translational coupling (perhaps accommodating protein-protein interactions) have been of selective benefit. If, however, these are relatively weak benefits, then persistent gene scrambling may have been tolerated prior to the eventual acquisition of operons having the ultimate detail of regulation seen in the contemporary E. coli, B. subtilis, and L. lactis. An intermediate stage of regulation (possibly still persisting in some contemporary lineages) might have been a simplified form of transcription control involving small molecules that can bind directly to RNA and regulate attenuation. Attenuation mechanisms may have evolved in an RNA world (45), and a number of recent articles (63, 61, 82) describing the ability of small molecules to interact directly with nascent RNA suggest that this mechanism for influencing transcription might be widespread.
Aspects of regulation that may merit increased attention are the factors that influence the rate of mRNA decay. It is generally accepted that the differential stability of mRNA plays an important role in determining the steady-state levels of gene expression. Individual mRNA decay rates can vary more than 100-fold. In contrast to the level of knowledge about initiation of trp gene transcription, little is known about the specificity, precision, and regulatory role of mRNA decay. New capabilities for the systematic measurement of mRNA decay rates (83) should enhance our understanding of this important aspect of regulation.
One can envision that such mechanisms might have preceded the commitment of genetic material to the elaboration of regulatory proteins. Consider the relative contribution of attenuation (relatively weak) and trpR-mediated repression (relatively strong) in E. coli. Repression is designed to detect Trp, whereas attenuation is designed to detect uncharged tTNATrp. Under many growth conditions, the free Trp concentration in the cell may be fairly low but still sufficient to keep tRNATrp largely charged. Thus, trpR-mediated repression is responsible for a large range of expression, and only after maximal derepression does relief from attenuation ensue. Consider also that the repressor binds not only to the trp operator but also to operators relevant to DAHP synthase and trpR itself (autoregulation). The modern whole-pathway operon systems that do possess efficient control features should be highly stable, barring any evolutionary transitions to pathogenic or symbiotic relationships. This would not preclude presumably desirable changes such as gene fusions. Simple, unregulated operons (both ancient and modern) or weakly regulated operons can be expected to be relatively unstable compared to complex, regulated operon systems that can sense a variety of different cues with a good range of sensitivity. To the extent that these deploy unlinked regulatory elements, intergenomic transfer should be relatively unlikely due to the necessity for cotransfer of unlinked genes in order to obtain the complete operon system.
FUTURE PROSPECTS FOR ELEVATED KNOWLEDGE OF Trp PATHWAY EVOLUTION
The comparative organization of the seven structural genes responsible for Trp biosynthesis has been analyzed in comprehensive detail. We have asserted that the vertical trace of descent with respect to the primary pathway can be sorted from paralogy that leads to specialized pathways and from occasional events of LGT. We have shown how relatively nonconserved contexts of flanking genes in relatively narrow organismal clades can be used to elucidate which of two evolutionary states is derived and which is ancestral. We have given examples of how the ancestral state at a given phylogenetic node can be determined.
Thus, we are beginning to get a fairly good picture of the evolutionary progressions that have taken place with respect to the organization of trp genes as whole-pathway operons, partial-pathway operons, and dispersed genes. However, a rationale for what driving forces exist to power the evolutionary dynamics that we can describe is not so clear. This limitation can probably be attributed to the relatively small amount of information about Trp pathway regulation that is available in the broad comparative context. To completely describe trp operon systems, one needs to evaluate any linked or unlinked regulatory elements that may exist. Two widely spaced organisms may have identical whole-pathway trp operons but may have evolved completely different control systems, or one of the two may be quite complex and the other simple. It seems significant that the current systems of trp operon regulation that can be described as elaborate are present in narrow bacterial clades and therefore must be of relatively recent origin. Comparative bioinformatics data to elucidate the range of regulatory mechanisms in place for trp operons in modern organisms is an initiative that is only beginning (60) and should be most informative.
Complexly regulated Trp systems are likely to involve the integration of Trp biosynthesis with other pathways, as has been elucidated between Trp and folate (mediated by TRAP) in B. subtilis or between Trp and the greater aromatic pathway (mediated by TrpR) in E. coli. One could envision a yet-to-be discovered metabolic relationship between Trp and serine or between Trp and histidine.
A second aspect of complexity involves the variety of multiple pathways that can exist within a single organism in which Trp or Trp intermediates can have different fates. For example, Streptomyces coelicolor has four TrpAa/TrpAb homologues that compete to direct chorismate to the specific alternative fates of phenazine biosynthesis, antibiotic biosynthesis, siderophore (coelibactin) biosynthesis, and primary Trp substrate for protein synthesis. All of these competing systems would be expected to respond to entirely different regulatory cues. In some cases, a given trp gene product may be shared by more than one pathway. Larger genomes can be expected to more frequently exhibit this kind of paralogy/xenology complexity, and indeed we have seen examples for the Trp pathway in large-genome organisms such as Nostoc sp., Pseudomonas aeruginosa, and Streptomyces coelicolor.
In this article, a strong foundation has been developed that should help guide the selection of key organisms for studies designed to gain insight into how Trp pathway regulation is related to the driving forces of evolution.
Acknowledgments
Gary Xie was partially supported in this work through the STDGEN project at Los Alamos National Laboratory (NIH/NIAID IAG Y1-A1-8228-05). Some of the preliminary sequence data were obtained from the Institute for Genomic Research (TIGR) (http://www.tigr.org), National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov.Microb_blast/unfinishedgenome.html), and Integrated Genomics, Inc. (http://wit.integratedgenomics.com/ERGO).
We thank A. Osterman of Integrated Genomics, Inc. (Chicago, Ill.) for provision of access to ERGO. Most importantly, we thank Charles Yanofsky for his extraordinary generosity in providing an almost continuous critique of the early version of this work and for a number of suggestions, among them the idea that an ancient attenuation mechanism might have involved a simple and direct association of small molecules with nascent RNA.
APPENDIX
Analysis of Raw DNA Sequence Data
Raw DNA contig sequences available from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgemome.html) and TIGR unfinished microbial genomes database (http://www.tigr.org/tdb/ufmg/) were screened with the built-in Blast service. The protein sequences from GenBank were used as query entries. The Blast 2.0 (5) and the open reading frame finder (ORF Finder) offered by NCBI were used to locate open reading frames and to confirm the similarity search result of the raw sequence.
Deduced amino acid sequences were analyzed for N-terminal signal sequences and transmembrane domains with Psort (http://psort.ims.u-tokyo.ac.jp/) (64).
Hidden Markov model and Prosite pattern search. Multiple alignments were obtained with the ClustalW program (78) included in the BioEdit (version 5.0.9) multiple alignment tool (33). A hidden Markov model based upon a multiple sequence alignment of known TrpC sequences was generated by version 2.2g of the HMMER program (22). A Prosite-like regular expression pattern was generated manually, and this hidden Markov model and Prosite pattern were further searched against the genomes that are missing trpC.
16S rRNA Tree Construction
16S rRNA subtrees were obtained from the Ribosomal Datebase site (http://rdp.cme.msu.edu/html/) (55).
DNA Composition
The GC percentages for individual genes were computed with the GEECEE program, which was written by R. Bruskiewich at the Sanger Centre (Cambridge, United Kingdom). The whole-genome GC value was obtained from the codon usage database (http://www.kazusa.or.jp/codon/) (65).
Fusion Protein and Linker Region Analyses
All the fusion protein sequences from the GenBank and NCBI Microbial Genomes Blast databases (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) werescreened by use of Blast (5) service. The known fusion protein sequences were used as query entries. A multiple alignment was obtained by input of single-domain and fusion protein sequences into the ClustalW (78) program (version 1.4). The linker region was defined by comparing the multiple sequence alignment of fusion proteins and monofunction proteins. Then the conserved domain database result (56) was used as the reference guide to find the boundary of the fusion protein (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
Footnotes
Florida Agricultural Experiment Station Journal series no. R-09160.
REFERENCES
- 1.Ahmad, S., and R. A. Jensen. 1989. Utility of a bifunctional tryptophan pathway enzyme for the classification of the Herbicola-Agglomerans complex of bacteria. Int. J. Syst. Bacteriol. 39:100-104. [Google Scholar]
- 2.Alifano, P., R. Fani, P. Lio, A. Lazcano, M. Bazzicalupo, M. S. Carlomagno, and C. B. Bruni. 1996. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 60:44-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altamirano, M. M., J. M. Blackburn, C. Aguayo, and A. R. Fersht. 2000. Directed evolution of new catalytic activity with the α/β-barrel scaffold. Nature 403:617-622. [DOI] [PubMed] [Google Scholar]
- 4.Altamirano, N., J. M. Blackburn, C. Aguayo, and A. R. Fersht. 2002. Directed evolution of new catalytic activity with the α/β-barrel scaffold: retraction. Nature 417:468. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auerbach, S., J. Gao, and G. N. Gussin. 1993. Nucleotide sequences of the trpl, trpB, and trpA genes of Pseudomonas syringae: positive control unique to fluorescent pseudomonads. Gene 123:25-32. [DOI] [PubMed] [Google Scholar]
- 7.Bae, Y. M., and I. P. Crawford. 1990. The Rhizobium meliloti trpE(G) gene is regulated by attenuation, and its product, anthranilate synthase, is regulated by feedback inhibition. J. Bacteriol. 172:3318-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao, Q., Y. Tian, W. Li, Z. Xu, Z. Xuan, S. Hu, W. Dong, J. Yang, Y. Chen, Y. Xue, Y. Xu, X. Lai, L. Huang, X. Dong, Y. Ma, L. Ling, H. Tan, R. Chen, J. Wang, J. Yu, and H. Yang. 2002. A complete sequence of the T. tengcongensis genome. Genome Res. 12:689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barona-Gómez, F., D. A. Hodgson. 2003. Occurrence of a putative ancient-like isomerase involved in histidine and typtophan biosynthesis. EMBO reports 4:296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley, R. 1990. The shikimate pathway − a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25:307-384. [DOI] [PubMed] [Google Scholar]
- 11.Brown, J. R., and w. F. Doolittle. 1997. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 61:456-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns, D. M., V. Horn, J. Paluh, and C. Yanofsky. 1990. Evolution of the tryptophan synthetase of fungi. Analysis of experimentally fused Escherichia coli tryptophan synthetase alpha and beta chains. J. Biol. Chem. 256:2060-2069. [PubMed] [Google Scholar]
- 13.Calhoun, D. H., C. A. Bonner, W. Gu, G. Xie, and R. A. Jensen. 2001. The emerging periplasm-localized subclass of AroQ chorismate mutases, exemplified by those from Salmonella typhimurium and Pseudomonas aeruginosa. Genome Biol. 2:30.1-30.16. [DOI] [PMC free article] [PubMed]
- 14.Chang, M., and I. P. Crawford. 1990. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 18:979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Copley, R. R., and P. Bork. 2000. Homology among (βα)(8) barrels: implications for the evolution of metabolic pathways. J. Mol. Biol. 303:627-641. [DOI] [PubMed] [Google Scholar]
- 16.Crawford, I. P., M. Clarke, M. van Cleemput, and C. Yanofsky. 1987. Crucial role of the connecting region joining the two functional domains of yeast tryptophan synthetase. J. Biol. Chem. 262:239-244. [PubMed] [Google Scholar]
- 17.Dandekar, T., B. Snel, M. Huynen, and P. Bork. 1998. Conservation of gene order. Fingerprint of proteins that physically interact. Trends Biochem. Sci. 23:324-328. [DOI] [PubMed] [Google Scholar]
- 18.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Vecchio, V. G., Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, and A. Lykidis. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Saizieu, A., P. Vankan, C. Vockler, and A. P. van Loon. 1997. The trp RNA-binding attenuation protein (TRAP) regulates the steady-state levels of transcripts of the Bacillus subtilis folate operon. Microbiology 143:979-989. [DOI] [PubMed] [Google Scholar]
- 21.de Troch, P., F. Dosselaere, V. Keijers, P. deWilde, and J. Vanderleyden. 1997. Isolation and characterization of the Azospirillum brasilense trpE(G) gene, encoding anthranilate synthase. Curr. Microbiol. 34:27-32. [DOI] [PubMed] [Google Scholar]
- 22.Eddy, S. R. 1998. Profile hidden Markov models for sequence analysis. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 23.Elf, J., O. G. Berg, and M. Ehrenberg. 2001. Comparison of repressor and transcriptional attenuator systems for control of amino acid biosynthetic operons. J. Mol. Biol. 313:941-954. [DOI] [PubMed] [Google Scholar]
- 24.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehlner-Gardiner, C., C. Roshick, J. H. Carison, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-268903. [DOI] [PubMed] [Google Scholar]
- 26.Gast, D. A., U. Jenal, A. Wasserfallen, and T. Leisinger. 1994. Regulation of tryptophan biosynthesis in Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 176:4590-45906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glansdorff, N. 2000. About the last common ancestor, the universal life-tree and lateral gene transfer: a reappraisal. Mol. Microbiol. 38:177-185. [DOI] [PubMed] [Google Scholar]
- 28.Gorgarten, J. P., W. F. Doolittle, and J. G. Lawrence. 2002. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19:2226-2238. [DOI] [PubMed] [Google Scholar]
- 29.Gollnick, P. 1994. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol Microbiol. 11:991-997. [DOI] [PubMed] [Google Scholar]
- 30.Gollnick, P., and P. Babitzke. 2002. Transcription attenuation. Biochim. Biophys. Acta 1577:240-250. [DOI] [PubMed] [Google Scholar]
- 31.Gosset, G., C. A. Bonner, and R. A. Jensen. 2001. Microbial origin of plant-type 2-keto-3-d-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and tryptophan-regulated enzyme from Xanthomonas campestris. J. Bacteriol. 183:4061-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall, G., M. B. Flick, and R. A. Jensen. 1980. Approach to recognition of regulatory mutants of cyanobacteria. J. Bacteriol. 143:981-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor program for Windows 95/98/NT. Nucleic Acid Symp. Ser. 41:95-98. [Google Scholar]
- 34.Henkin, T. M. 1994. tRNA-directed transcription antitermination. Mol. Microbiol. 13:381-387. [DOI] [PubMed] [Google Scholar]
- 35.Henner, D., and C. Yanofsky. 1993. Bacillus subtilis and other gram-positive bacteria, p. 269-280. In A. L. Sonenshein, J. Hoch, and R Losick (ed.), Biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 36.Ingram, L. O., D. L. Pierson, J. F. Kane, C. Van Baalen, and R. A. Jensen. 1972. Documentation of auxotrophic mutation in blue-green bacteria: characterization of a tryptophan auxotroph in Agmenellum quadruplicatum. J. Bacteriol. 111:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itoh, T., K. Takemoto, H. Mori, and T. Gojobori. 1999. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 16:332-346. [DOI] [PubMed] [Google Scholar]
- 38.Jensen, R. A. 1976. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30:409-425. [DOI] [PubMed] [Google Scholar]
- 39.Jensen, R. A. 1992. An emerging outline of the evolutionary history of aromatic amino acid biosynthesis, p. 204-236. In R. P. Mortlock (ed.), The evolution of metabolic function. CRC Press, Inc., Boca Raton, Fl.
- 40.Jensen, R. A. 1996. Evolution of metabolic pathways in enterics, p. 2649-2662. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium. ASM Press, Washington, D.C.
- 41.Jensen, R. A., and S. Ahmad. 1990. Nested gene fusions as markers of phylogenetic branch points in prokaryotes. Trends Ecol. Evol. 5:219-224. [DOI] [PubMed] [Google Scholar]
- 42.Jensen, R. A., and W. Gu. 1996. Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J. Bacteriol. 178:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen, R. A., and E. W. Nester. 1965. The regulatory significance of intermediary metabolites: control of aromatic acid biosynthesis by feedback inhibition in Bacillus subtilis. J. Mol. Biol. 12:468-481. [DOI] [PubMed] [Google Scholar]
- 44.Jensen, R. A., G. Xie, D. H. Calhoun, and C. A. Bonner. 2001. The correct phylogenetic relationship of KdsA (3-deoxy-d-manno-octulosonate 8-phosphate synthase) with one of two independently evolved classes of AroA (3-deoxy-d-arabino-heptulosonate 7-phosphate synthase). J. Mol. Evol. 54:416-423. [DOI] [PubMed] [Google Scholar]
- 45.Joyce, G. F. 2002. The antiquity of RNA-based evolution. Nature 418:214-221. [DOI] [PubMed] [Google Scholar]
- 46.Jürgens, C., A. Strom, D. Wegener, S. Hettwer, M. Wilmanns, and R. Sterner. 2000. Directed evolution of a (beta alpha)8-barrel enzyme to catalyze related reactions in two different metabolic pathways. Proc. Natl. Acad. Sci. USA 97:9925-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kane, J. F., W. M. Holmes, and R. A. Jensen. 1972. Metabolic Interlock: the dual function of a folate pathway gene as an extra-operonic gene of tryptophan biosynthesis. J. Biol. Chem. 247:1587-1596. [PubMed] [Google Scholar]
- 48.Kane, J. F., S. L. Stenmark, D. H. Calhoun, and R. A. Jensen. 1971. Metabolic interlock: the role of the subordinate type of enzyme in the regulation of a complex pathway. J. Biol. Chem. 246:4308-4316. [PubMed] [Google Scholar]
- 49.Labedan, B., A. Boyen, M. Baetens, D. Charlier, P. Chen, R. Cunin, V. Durbeco, N. Glansdorff, G. Herve, C. Legrain, Z. Liang, C. Purcarea, M. Roovers, R. Sanchez, T. L. Toong, M. Van de Casteele, F. Van Vliet, Y. Xu, and Y. F. Zhang. 1999. The evolutionary history of carbamoyltransferases: a complex set of paralogueous genes was already present in the last universal common ancestor. J. Mol. Evol. 49:461-473. [DOI] [PubMed] [Google Scholar]
- 50.Lai, C. Y., L. Baumann, and P. Baumann. 1994. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc. Natl. Acad. Sci. USA 91:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence, J. G. 1999. Gene transfer, speciation, and the evolution of bacterial genomes. Curr. Opin. Microbiol. 2:519-523. [DOI] [PubMed] [Google Scholar]
- 52.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, C., A. S. Paradkar, and L. C. Vining. 1998. Regulation of an anthranilate synthase gene in Streptomyces venezuelae by a trp attenuator. Microbiology 144:1971-1980. [DOI] [PubMed] [Google Scholar]
- 54.Mackenzie, C., A. E. Simmons, and S. Kaplan. 1999. Multiple chromosomes in bacteria: The yin and yang of trp gene localization in Rhodobacter sphaeroides. Genetics 153:525-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marachler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayans, O., A. Ivens, L. J. Nissen, K. Kirschner, and M. Wilmanns. 2002. Structural analysis of two enzymes catalyzing reverse metabolic reactions implies common ancestry. EMBO J. 21:3245-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDonald, M. D. V. Mavrodi, L. S. Thomashow, and H. G. Floss. 2001. Phenazine biosynthesis in Pseudomonas fluorescens: branch point from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J. Am. Chem. Soc. 123:9459-9460. [DOI] [PubMed] [Google Scholar]
- 60.Merino, E., and C. Yanofsky. 2002. Regulation by termination-antitermination: a genomic approach, p. 323-336. In A. L. Sonenshein (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 61.Mironov, A. S., I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov, and E. Nudler. 2002. Sensing small molecules by nascent RNA. A mechanism to control transcription in bacteria. Cell 111:747-756. [DOI] [PubMed] [Google Scholar]
- 62.Morollo, A. A., and R. Bauerle. 1993. Characterization of composite aminodeoxyisochorismate synthase and aminodeoxyisochorismate lyase activities of anthranilate synthase. Proc. Natl. Acad. Sci. USA 90:9983-9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nahvi, A., N. Sudarsan, M. S. Ebert, X. Zou, K. L. Brown, and R. R. Breaker. 2002. Genetic control by a metabolite binding mRNA. Chem. Biol. 9:1043-1049. [DOI] [PubMed] [Google Scholar]
- 64.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 65.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nichols, B. P., and J. M. Green. 1992. Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase. J. Bacteriol. 174:5309-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olekhnovich, I., and G. N. Gussin. 2001. Effects of mutations in the Pseudomonas putida miaA gene: regulation of the trpR and trpGDC operons in P. putida by attenuation. J. Bacteriol. 183:3256-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pittard, A. J. 1996. Biosynthesis of the aromatic amino acids, p. 458-484. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium. ASM Press, Washington, D.C.
- 69.Raya, R., J. Bardowski, P. S. Andersen, S. D. Ehrlich, and A. Chopin. 1998. Multiple transcriptional control of the Lactococcus lactis trp operon. J. Bacteriol. 180:3174-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryding, N. J., T. B. Andreson, and W. C. Champness. 2002. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184:794-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sahm, H., and L. Eggeling. 1999. D-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding l-valine synthesis for d-pantothenate overproduction. Appl. Environ. Microbiol. 65:1973-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis operon containing genes of unknown function senses tRNA-Trp charging and regulates the expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. USA 97:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sawada, H., S. Kanaya, M. Tsuda, F. Suzuki, K. Azegami, and N. Saitou. 2002. A phylogenomic study of the OCTase genes in Pseudomonas syringae pathovars: the horizontal transfer of the argK-tox cluster and the evolutionary history of OCTase genes on their genomes. J. Mol. Evol. 54:437-457. [DOI] [PubMed] [Google Scholar]
- 74.Schwarz, T., K. Uthoff, C. Klinger, H. E. Meyer, P. Bartholmes, and M. Kaufmann. 1997. Multifunctional tryptophan-synthesizing enzyme. The molecular weight of the Euglena gracilis protein is unexpectedly low. J. Biol. Chem. 272:1061-1063. [DOI] [PubMed] [Google Scholar]
- 75.Stulke, J. 2002. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch. Microbiol. 177:433-440. [DOI] [PubMed] [Google Scholar]
- 76.Subramaniac, P. S., G. Xie, T. Xia, and R. A. Jensen. 1998. Substrate ambiguity of 3-deoxy-d-manno-octulosonate 8-phosphate synthase from Neisseria gonorrhoeae in the context of its membership in a protein family containing a subset of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthases. J. Bacteriol. 180:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang, X.-F., S. Ezaki, S. Fujiwara, M. Takagi, H. Atomi, and T. Imanaka. 1999. The tryptophan biosynthesis gene cluster trpCDEGFBA from Pyrococcus kodakaraensis KODI is regulated at the transcriptional level and expressed as a single mRNA. Mol. Gen. Genet. 262:815-821. [DOI] [PubMed] [Google Scholar]
- 78.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tumbula, D. L., Q. Teng, M. G. Bartlett, and W. B. Whitman. 1997. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis. J. Bacteriol. 179:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valbuzzi, A., P. Gollnick, P. Babitzke, and C. Yanofsky. 2002. The anti-trp RNA-binding attenuation protein (anti-TRAP), anti-TRAP, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J. Biol. Chem. 277:10608-10613. [DOI] [PubMed] [Google Scholar]
- 81.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 82.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang, Y. C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed]
- 84.Ward, D. E., W. M. De Vos, and V. D. Oost. 2002. Molecular analysis of the role of two aromatic aminotransferases and a broad-specificity aspartate aminotransferase in the aromatic amino acid metabolism of Pyrococcus furiosus. Archaea 1:133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weber-Ban, E., O. Hur, C. Bagwell, U. Banik, L. H. Yang, E. W. Miles, and M. F. Dunn. 2001. Investigation of allosteric linkages in the regulation of tryptophan synthase: the roles of salt bridges and monovalent cations probed by site-directed mutation, optical spectroscopy, and kinetics. Biochemistry 40:3497-3511. [DOI] [PubMed] [Google Scholar]
- 86.Wilmanns, M., C. C. Hyde, D. R. Davies, K. Kirschner, and J. N. Jansonius. 1991. Structural conservation in parallel β/α-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry 30:9161-9169. [DOI] [PubMed] [Google Scholar]
- 87.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie, G., C. A. Bonner, and R. A. Jensen. 1999. Cyclohexadienyl dehydrogenase from Pseudomonas stutzeri exemplifies a widespread type of tyrosine-pathway dehydrogenase in the TyrA protein family. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 125:65-83. [DOI] [PubMed] [Google Scholar]
- 89.Xie, G., C. A. Bonner, and R. A. Jensen. 2002. Dynamic diversity of the tryptophan pathway in the chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol. 3:005.1-005.17. [DOI] [PMC free article] [PubMed]
- 90.Xie, G., C. A. Bonner, and R. A. Jensen. 1999. A probable mixed-function supraoperon in Pseudomonas exhibits gene organization features of both intergenomic conservation and gene shuffling. J. Mol. Evol. 49:108-121. [DOI] [PubMed] [Google Scholar]
- 91.Xie, G., T. S. Brettin, C. A. Bonner, and R. A. Jensen. 1999. Mixed-function supraoperons that exhibit overall conservation, albeit shuffled gene organization, across wide intergenomic distances within eubacteria. Microb. Comp. Genomics 4:5-28. [DOI] [PubMed] [Google Scholar]
- 92.Xie, G., C. Forst, C. A. Bonner, and R. A. Jensen. 2001. Significance of two distinct types of tryptophan synthase beta chain in Bacteria, Archaea and higher plants. Genome Biol. 3:5.1-5.13. [DOI] [PMC free article] [PubMed]
- 93.Xie, G., N. O. Keyhani, C. A. Bonner, T. S. Brettin, R. Gottardo, and R. A. Jensen. 2003. Lateral gene transfer and ancient paralogy of operons containing redundant copies of tryptophan-pathway genes in Xylella and in heterocystous cyanobacteria. Genome Biol. 4:14.1-18. [DOI] [PMC free article] [PubMed]
- 94.Xiu, Z. L., Z. Y. Chang, and A. P. Zeng. 2002. Nonlinear dynamics of regulation of bacterial trp operon: model analysis of integrated effects of repression, feedback inhibition, and attenuation. Biotechnol. Prog. 18:686-693. [DOI] [PubMed] [Google Scholar]
- 95.Yanofsky, C. 2001. Advancing our knowledge in biochemistry, genetics, and microbiology through studies on tryptophan metabolism. Annu. Rev. Biochem. 70:1-37. [DOI] [PubMed] [Google Scholar]
- 96.Yanofsky, C. 2003. Reflections: with studies on tryptophan metabolism to answer basic biological questions. J. Biol. Chem. 278:10859-10878. [DOI] [PubMed] [Google Scholar]
- 97.Yanofsky, C., E. Miles, R. Bauerle, and K. Kirschner. 1999. Trp operon, p. 2676-2689. In T. E. Creighton (ed.), Encyclopedia of molecular biology, vol. 4. John Wiley and Sons, Inc., New York, N.Y.
- 98.Zheng, Y., J. D. Szustakowski, L. Fortnow, R. J. Roberts, and S. Kasif. 2002. Computational identification of operons in microbial genomes. Genome Res. 12:1221-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]