Abstract
Gaucher disease is caused by mutations in the gene encoding the lysosomal enzyme glucocerebrosidase (GC). Three clinical types of Gaucher disease have been defined according to the presence (type 2 and 3) or absence (type 1) of central nervous system disease and severity of clinical manifestations. The clinical course of the disease correlates with the mutation carried by the GC gene. To produce mice with point mutations that correspond to the clinical types of Gaucher disease, we have devised a highly efficient one-step mutagenesis method—the single insertion mutagenesis procedure (SIMP)—to introduce human disease mutations into the mouse GC gene. By using SIMP, mice were generated carrying either the very severe RecNciI mutation that can cause type 2 disease or the less severe L444P mutation associated with type 3 disease. Mice homozygous for the RecNciI mutation had little GC enzyme activity and accumulated glucosylceramide in brain and liver. In contrast, the mice homozygous for the L444P mutation had higher levels of GC activity and no detectable accumulation of glucosylceramide in brain and liver. Surprisingly, both point mutation mice died within 48 hr of birth, apparently of a compromised epidermal permeability barrier caused by defective glucosylceramide metabolism in the epidermis.
Keywords: gene targeting, mouse model, sphingolipidosis, lysosomal storage disease
Gaucher disease, the most common lysosomal storage disease, is caused by mutations in the gene encoding the lysosomal enzyme, glucocerebrosidase (GC) (1–3). The deficiency of enzyme activity results in the accumulation of the GC substrate, glucosylceramide, leading to enlargement of liver and spleen, lesions in the bones, and, in the most severe cases, impairment of central nervous system function.
The disease has been classified into three clinical types based on the presence or absence of central nervous system involvement and disease severity. Type 1 Gaucher disease is the mildest form and is without neurologic involvement. Type 2, the most severe form of the disease, is characterized by an early onset of neurologic disease and acute course. The type 3 disease is of intermediate severity with a later onset of neurologic symptoms and a more chronic course. The clinical type of Gaucher disease, an autosomal recessive disorder, is dictated to a large extent by mutations carried in the GC gene. For instance, the N370S (substitution of serine for normal asparagine-370) mutation is diagnostic of type 1 disease. Homozygosity for the L444P (substitution of proline for leucine-444) mutation usually is associated with type 3 disease in the Norrbottnian population in Sweden. An allele termed RecNciI with two amino acid changes (substitution of proline for leucine-444 and proline for alanine-456) is found frequently in type 2 patients and causes perinatal lethality in homozygous form (4, 5).
In most cases studied, Gaucher patients retain residual GC enzyme activity and have at least one allele specifying an amino acid substitution in the GC coding sequence (2). Homozygosity for a GC null mutation has been shown to result in prenatal lethality in humans (6). GC knockout mice with a total enzyme deficiency die within 24 hr after birth with glucosylceramide storage in brain and liver and severe epidermal abnormalities (7, 8).
To establish more authentic mouse models of Gaucher disease we have devised a single insertion mutagenesis procedure (SIMP) to introduce mutations associated with type 2 (RecNciI) and type 3 (L444P) Gaucher disease into the murine GC gene in embryonic stem (ES) cells. SIMP uses an insertion-type targeting vector that results in duplication of the target sequence on homologous integration. The vector is carefully designed so that one genomic duplicant is an intact gene structure with introduced mutations and the other genomic duplicant is a truncated gene without function. The resulting point mutation mice had levels of GC activity and glucosylceramide storage that corresponded to the relative severity of the mutations. Unexpectedly, both mice died soon after birth—irrespective of the level of enzyme activity.
MATERIALS AND METHODS
Targeting Vector Construction and ES Cell Targeting.
Genomic clones from a 129/sv strain library (Stratagene; catalogue 946306) containing the mouse GC gene were isolated by using a human GC cDNA probe (ATCC; catalogue 65696). One clone that covers the entire GC locus, Φ8, was used to construct targeting vectors.
RecA-assisted restriction endonuclease (RARE) cleavage was used to introduce point mutations into this genomic clone (9). The 16-kb genomic DNA insert first was subcloned into the pBluescript SK vector. RARE cleavage then was carried out to specifically cut only the BamHI and PstI sites in exon 10 of the GC gene to release the 80-bp sequence delimited by the two restriction sites (Fig. 1 B and C). Two pairs of complementary oligonucleotides were synthesized to carry the desired mutations (L444P or RecNciI) and the diagnostic BamHI(−) site (Fig. 1C). Oligonucleotides were annealed and ligated into the BamHI–PstI plasmid to create pBL444P and pBRecNciI.
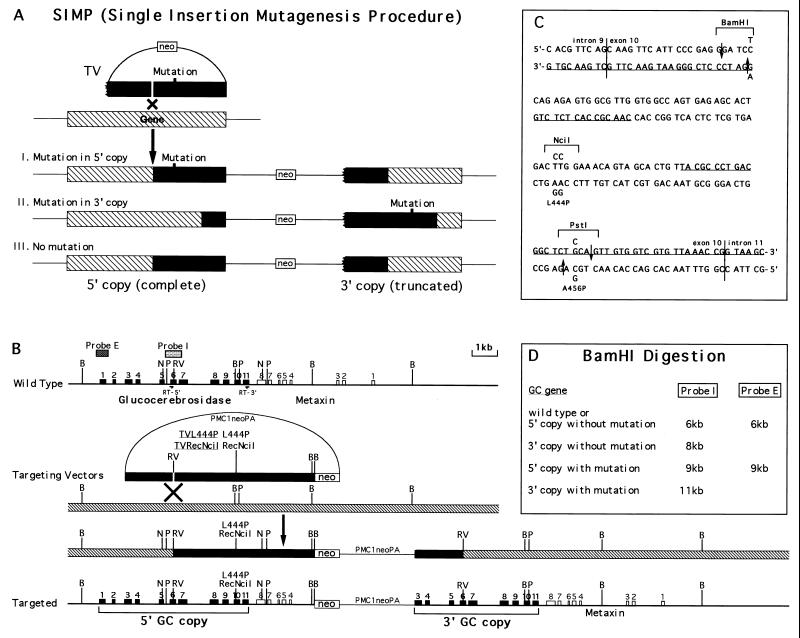
Figure 1.
Introduction of L444P and RecNciI human Gaucher disease mutations by the SIMP strategy. (A) A general scheme of the SIMP strategy. The ragged end of the gene in the targeting vector represents a truncation, filled bars represent the homology in the targeting vectors, hatched bars represent the endogenous allele. (B) The specific SIMP scheme for the introduction of the L444P and RecNciI mutations into the GC locus. The wild-type locus is shown on the top and the correctly targeted locus is shown on the bottom. The SIMP recombination step is shown in the middle. The filled boxes represent the exons of the GC gene, open boxes represent the exons of the nearby metaxin gene, stippled boxes represent probes used in the Southern analysis (probe I is a 0.7-kb KpnI fragment covering exon 6 and probe E is a 0.4-kb EcoRI fragment covering exon 1), filled bars represent the homology in the targeting vectors, hatched bars represent the endogenous allele. RT-5′, 5′ primer used in the reverse transcription (RT)-PCR primer (5′-CACGAATTCACATCACCCACTTGGCTCAAG-3′); RT-3′, 3′ primer used in the RT-PCR (5′-GTCGAATTCATGTCCATGCTAAGCCCAGGT-3′); B, BamHI; N, NciI; P, PstI; RV, EcoRV; Probe E, external probe; Probe I, internal probe. (C) The DNA sequence of the mouse GC exon 10 region. The vertical lines are exon/intron borders. The underlined sequences are oligonucleotides used in the RecA-assisted restriction endonuclease cleavage experiment. The mutations are indicated above and below the wild-type sequences. The mutated sequences between the two downward arrows were used as top strand of the mutation inserts. The mutated sequences between the two upward arrows were used as bottom strand of the mutation inserts. Both targeting vectors contained the change that eliminated the BamHI site and the L444P mutation. In addition, the RecNciI vector contain the L456P change. (D) The expected sizes of BamHI restriction fragments of wild-type and targeted GC genes detected by the I (internal) and E (external) probes.
The 7-kb HindIII fragments from pBL444P and pBRecNciI plasmids were purified and ligated into the HindIII site of the pMC1neopolyA vector (Stratagene; catalogue 213201) to generate targeting vectors TVL444P and TVRecNciI. In both vectors, the bacterial neomycin phosphotransferase (neo) cassette is in the opposite orientation to the GC gene.
The targeting vector (20 μg) was linearized with EcoRV endonuclease and electroporated into 5 × 106 J1 ES cells (in a volume of 0.8 ml) at 400 V and 25 μ (Bio-Rad). The G418-resistant ES cells were obtained and screened by Southern analysis as described (10). Chimeric mice were established from targeted ES cells as described (11).
Biochemical Analysis.
Acid β-glucosidase activity was determined in extracts of brain, liver, and skin. Aliquots (25 μl) of the extracts were incubated with 250 μl of a solution containing 5 mM 4-methylumbelliferyl-β-d-glucopyranoside (Toronto Research Chemicals, Downsview, ON, Canada), 0.125 mM sodium taurocholate, and 0.1 M sodium acetate (pH 4.0) at 37°C for 30 min. The reaction was stopped with 1 ml of a buffer containing 0.17 M glycine and 0.17 M sodium carbonate (pH 10). Fluorescence was determined on a Ratio-2 System fluorometer (Optical Technology Devices, Elmsford, NY).
Epidermis was separated from dermis after floating the skin of 1-day-old mice on Dispase (Boehinger Mannheim, grade II) diluted 1:1 in Hank’s buffer at 4°C overnight. Epidermal lipids were extracted in chloroform-methanol-water (1:2:0.6, vol/vol/vol) for 24 hr at 37°C. The total lipid extracts were applied to TLC plates, and the chromatograms were developed twice with chloroform-methanol-acetic acid (190:9:1, vol/vol/vol). After development, plates were air-dried, sprayed with 8% (wt/vol) H3PO4 containing 10% (wt/vol) CuSO4, charred, and quantitated by photodensitometry. Sphingolipids were isolated from brain and liver, separated by HPTLC and were quantitated as described (12).
Pathology.
Twelve newborn mice (two each of homozygotes for the L444P allele, RecNciI allele, the GC duplication without mutations and corresponding normal controls) were killed by ether inhalation and fixed by immersion in 0.1 M phosphate-buffered 4% paraformaldehyde. The tail and hind limb of some mice were removed before fixation for genotyping. The bodies of these mice were bisected sagittaly through the midline. The left half was embedded in paraffin and used for the light microscopic study. The right half was coronally sectioned, rostral to caudal, and pieces of cerebrum, cerebellum, brainstem, spinal cord, a portion of liver and skin from the back of the neck at the midline (nuchal region) were processed by plastic embedding for light and electron microscopic studies. Paraffin-embedded portions of the mice were sectioned (5 microns) and stained with solochrome and eosin and Luxol fast blue-periodic acid Schiff stains. Plastic embedded tissue was sectioned (1 micron) and stained with toluidine blue.
RESULTS
Scheme of SIMP.
Homologous integration of insertion-type targeting vectors results in a duplication of the entire genomic segment carried by the vector (13, 14) (Fig. 1A). In our experimental design, the genomic segment in the insertion-type targeting vector, containing the nucleotide changes to be introduced into the genome, is truncated at the 5′ end to render the gene nonfunctional. After homologous recombination, the resulting 5′ genomic duplicant is a complete gene with mutations transferred from the targeting vector; the resulting 3′ genomic duplicant is a truncated, nonfunctional gene fragment. In summary, this procedure (Fig. 1A) results in the introduction of desired mutations into the GC gene in one homologous integration step.
For introduction of the L444P and RecNciI mutations into the mouse GC gene, we constructed the TVL444P and TVRecNciI insertion-type targeting vectors (Fig. 1B). Each contained the pMC1neoPolyA cassette as a positive selection marker (15). TVL444P carried nucleotide changes that resulted in the substitution of proline for leucine-444 and in the elimination of a BamHI site 36 bp upstream with a neutral codon change to facilitate detection by Southern analysis (Fig. 1C). TVRecNciI carried the same changes as in TVL444P with an additional substitution of proline for alanine-456 that directly eliminated a PstI site (Fig. 1C). In both vectors, the L444P exchange created a novel NciI site. In patients with the RecNciI allele a third point mutation (G1597C) is present together with the L444P and A456P changes (2, 3). Because the G1597C mutation does not result in an amino acid change (V460V), it was not included in the RecNciI targeting vector.
The targeting vectors were linearized with EcoRV endonuclease at a single site within the GC gene, establishing a distance between the EcoRV cut and the mutations of ≈2.5 kb (Fig. 1B). The targeting vectors spanned the entire 3′ end of the GC gene from exon 3 through a portion of the downstream metaxin gene (Fig. 1B) (16). The predicted genomic structure resulting from homologous recombination is an intact 5′ GC gene copy with the introduced mutations, and a 3′ GC gene copy that is a truncated, nonfunctional gene fragment missing the first two exons and upstream promoter sequences (Fig. 1B). The downstream metaxin gene remains intact.
Identification of Targeting Events.
The linearized targeting vectors were transferred into ES cells by electroporation, and resulting G418r colonies were analyzed by Southern hybridization. On the basis of the double-strand break, gap repair, and mismatch repair models for homologous recombination of insertion-type vectors (17–19), three types of targeting events could be observed: (i) the correct transfer of the mutations to the 5′ GC gene copy as discussed above, (ii) the incorrect transfer of the mutations to the 3′ truncated GC gene copy caused by branch migration past the site of the mutations, and (iii) the repair of the mutations back to the wild-type sequence caused by gap repair or mismatch repair (Fig. 1A). An internal probe (probe I) and an external probe (probe E) were used to identify these three different targeting events (Fig. 1B).
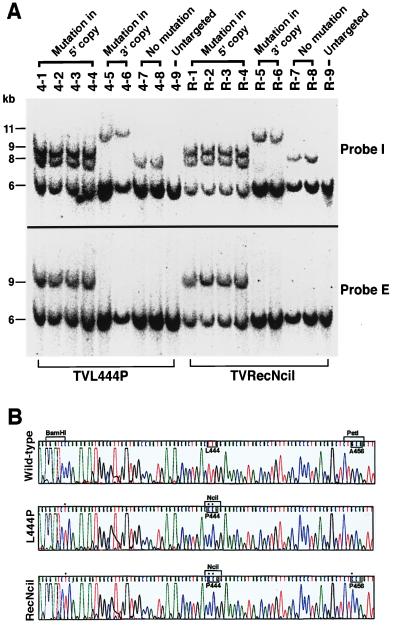
Genomic DNA from TVL444P transfected clones (4-1 → 4-9) and TVRecNciI transfected clones (R-1 → R-9) were analyzed by Southern blotting after digestion with BamHI restriction enzyme. The correspondence of band size to GC genomic segment is shown is Fig. 1D. The blot first was hybridized with the internal probe (Fig. 2A; probe I, Top). The correctly targeted clones (4-1 → 4-4 and R-1 → R-4) showed three bands of equal intensity: a 6-kb band from the wild-type (untargeted) allele, a 9-kb band from the 5′ GC copy of the targeted allele caused by the elimination of the BamHI site in exon 10, and an 8-kb band from the truncated 3′ GC copy. The clones that had mutations incorrectly transferred into the truncated 3′ copy (4-5 and 4-6; and R-5 and R-6) showed two bands of unequal intensity: a 6-kb band of double intensity derived from both the wild-type allele and the 5′ GC copy, and an 11-kb band from the 3′ GC copy caused by the elimination of the BamHI site by the transferred mutations. The clones where repair of mutations occurred (4-7 and 4-8, and R-7 and R-8) showed two bands of unequal intensity: a double-intensity 6-kb band derived from both the wild-type allele and the 5′ GC copy, and a single-intensity 8-kb band from the truncated 3′ GC copy. In clones 4-9 and R-9 the targeting vectors were randomly integrated into the genome. In these cases, the ends of the vectors are likely lost during insertion because of exonuclease activity and the lack of a repair function. Because the probe I hybridizes to these ends, nontargeted clones show either weak bands of random sizes or no band other than the 6-kb band from the wild-type allele.
Figure 2.
Analysis of three different homologous insertion events by the two targeting vectors: TVL444P and TVRecNciI. (A) ES cell DNAs were digested with BamHI, and the filter was probed with probe I (Upper) and probe E (Lower), respectively. Clones 4-1 → 4-9, ES cell clones from TVL444P targeting experiment; Clones R-1 → R-9, ES cell clones from TVRecNciI targeting experiment. (B) Sequence analysis of the mouse GC transcripts from ES cells. cDNA clones, obtained by reverse transcription–PCR, of wild-type and targeted alleles were sequenced with an automated DNA sequencer (Applied Biosystems model 373A). Only mutated amino acids are labeled. Nucleotide mutations are indicated by black dots.
The same blot of BamHI-digested genomic DNA was stripped and rehybridized with the external probe (Fig. 2A; probe E, Lower) derived from a region around exon 1. Probe E detects fragments of the wild-type gene (6 kb) and the 5′ GC copy fragment with the mutation (9 kb). It does not detect the truncated 3′ copy of the targeted allele caused by the absence of exons 1 and 2. Correctly targeted clones (4-1 → 4-4 and R-1 → R-4) showed 6-kb and 9-kb bands as expected. Other clones, such as those with mutations in the 3′ duplication, with mutations repaired and with a untargeted GC gene, showed only the 6-kb wild-type band and, thus, cannot be distinguished from one another with this probe.
Table 1 provides a summary of the three targeting events from experiments using the two vectors. Both vectors gave similar frequencies for the desired genomic changes (31% for TVL444P and 26% for TVRecNciI) and for total targeting (45% for TVL444P and 50% for TVRecNciI). For a 2.5-kb distance between mutation and the cutting site in our vectors, the percentages for the correct transfer of the mutation among all the targeting events was 71% for TVL444P (17/24) and 50% for TVRecNciI (14/27), a reasonable frequency to enable successful transfer of mutations.
Table 1.
Summary of the three targeting events by using the TVL444P and TVRecNciI vectors
| Targeting vectors | Total ES cell clones | Targeted clones
|
|||
|---|---|---|---|---|---|
| Mutation in 5′ copy | Mutation in 3′ copy | No mutation | Total targeted | ||
| TVL444P | 54 | 17 (31%) | 4 | 3 | 24 (45%) |
| TVRecNciI | 54 | 14 (26%) | 7 | 6 | 27 (50%) |
The existence of human mutations in the GC transcripts was confirmed by cloning and sequencing the reverse transcription–PCR products derived from the targeted GC transcripts in ES cells. The sequencing results showed that the desired changes (Fig. 1C), carried by the L444P and RecNciI targeting vectors, were precisely transferred into the GC gene transcripts (Fig. 2B). In addition, no alterations were detected around the EcoRV cleavage site (data not show). This is consistent with previously reported efficient double-strand gap repair in gene targeting (19).
Mice with Type 2 and 3 Gaucher Disease Mutations.
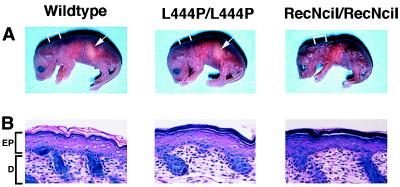
The L444P- and RecNciI-targeted ES clones were injected into C57BL/6 mouse blastocysts, and highly chimeric male mice were produced that transmitted the targeted alleles through the germ line. The heterozygotes were intercrossed to obtain mice homozygous for each mutated gene. Homozygotes for both genotypes were obtained in the expected Mendelian ratio. Both types of homozygous mice appeared abnormal at birth (Fig. 3). The RecNciI‖ mouse was the most severely affected with small size, poor turgor and an extremely red, wrinkled appearance. These mice appeared not to feed as evidenced by the absence of milk in their stomachs (Fig. 3A). The skin of the L444P mice was also abnormally red and wrinkled, especially around the torso area, but was less severely affected than the RecNciI mouse. The skin turgor of L444P mice was much improved compared with the RecNciI mouse. The improved hydration may be caused by the ability of the L444P mice to feed as evidenced by milk in the stomach (Fig. 3A). Both the RecNciI and L444P mice died within 24–48 hr after birth.
Figure 3.
Phenotypes of RecNciI and L444P mice after birth. (A) Photographs of the mice within 12 hr after birth. The arrow denotes the presence of milk in the stomach. Note the smaller size and wrinkled skin of the RecNciI/RecNciI mouse. The skin of the L444P/L444P mouse is less severely wrinkled. The bracket indicates the sites where skin samples were taken for the photomicrographs in B. (B) Section of epidermis (×150). Stratum corneum (SC) of RecNciI/RecNciI and L444P/L444P mice are compacted and less eosinophillic compared with the loosely packed eosinophillic SC in the wild-type mouse. No significant differences are noted in the three other layers of the epidermis (EP) (stratum basale, stratum spinosum, and stratum granulosum) or the dermis (D).
Light microscopic examination of the brain, spinal cord, and visceral organs of the RecNciI and L444P mice failed to demonstrate abnormal macrophages (Gaucher cells). In the skin of point mutation mice and controls, four layers—basal (stratum basale), spinous (stratum spinosum), granular (stratum granulosum), and cornified (stratum corneum)—all were identified in the epidermis. All except the cornified layer appeared similar to the corresponding layers of control mice. In control mice, the cornified layer consisted of loosely stacked eosinophillic membranous structures, whereas in the point mutation mice the cornified layer was compact and basophilic; the eosinophillic membranous structures were scarce (Fig. 3B). The abnormal cornified layer appeared slightly thicker in RecNciI mice than in L444P mice. On Luxol fast blue-periodic acid Schiff sections (PAS) (not shown), PAS-positive granules in the granular and spinous layers were more prominent in the point mutation mice than in control mice. These PAS-positive granules, indicative of glycocojugate accumulation, appeared to be more conspicuous in the RecNciI mice than in the L444P mice.
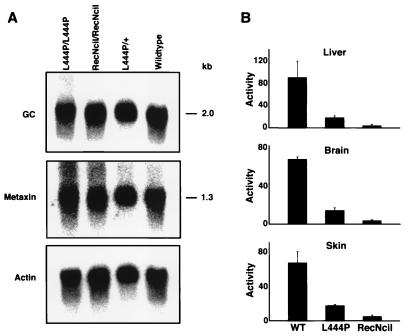
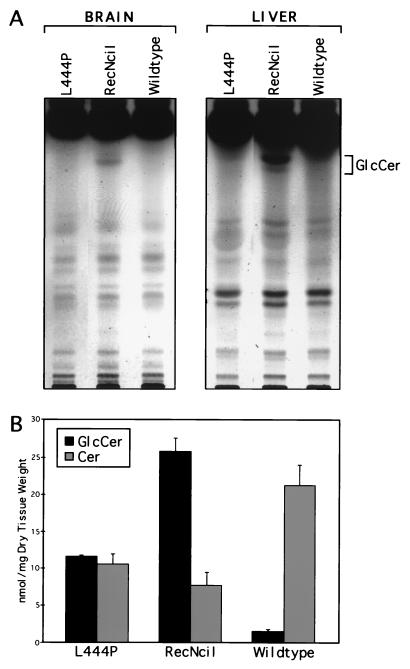
Northern analysis revealed that levels of brain GC mRNA in both the RecNciI and L444P mice were similar to wild type (Fig. 4A). The expression of the downstream gene, metaxin, also was found to be unaffected by the genomic changes in the GC allele. A large reduction in GC enzyme activity was found in extracts from liver, brain, and skin from both the L444P mouse and RecNciI mouse (Fig. 4B). The activity in the L444P mouse tissues was about 20% of normal activity compared with 4–9% residual activity found in the RecNciI mouse tissues. Analysis of the sphingolipid fractions of brain and liver from the point mutation mice demonstrated glucosylceramide accumulation in the RecNciI mouse but not in the L444P mouse (Fig. 5A). In epidermis, abnormally high glucosylceramide accumulation was found in both mutant mice together with a lower than normal level of ceramides. The glucosylceramide and ceramide levels were most severely affected in the RecNciI mice (Fig. 5B).
Figure 4.
Expression of GC mRNA and enzyme in RecNciI and L444P mice. (A) A Northern blot with 5 μg of brain poly(A)+ RNA from a RecNciI homozygote mouse, a L444P homozygote mouse, a L444P heterozygote mouse, and a wild-type mouse. The blot first was probed with a mouse GC cDNA. After stripping, it was reprobed with a mouse metaxin cDNA. Finally, it was stripped once more and probed with a human actin cDNA. (B) Acid β-glucosidase activity was determined in extracts of liver, brain, and skin from homozygote L444P and RecNciI mice. Tissues from three mice of each genotype were assayed. The assays were repeated three times. The activity (±SD) is expressed as nmol substrate cleaved/hr per mg of protein.
Figure 5.
Sphingolipid accumulation in tissues from RecNciI and L444P mice. (A) Lipid profiles from brain and liver. Each lane represents a portion of the material derived from a single mouse (L444P, RecNciI, or wild type). The position of the glucosylceramide standard is indicated. (B) The levels of total ceramides (Cer) and glucosylceramides (GlcCer) in epidermis from RecNciI and L444P mice.
To demonstrate that the severe phenotypes of the RecNciI and L444P mice were the result of point mutations introduced into the GC gene and not the result of a locus rearrangement induced by the targeting procedure, we established mice by using ES cells with the GC duplication but without human point mutations (clone 4-7; Fig. 2A). Mice homozygous for this GC configuration appear normal at birth, are viable into adulthood, are fertile, have normal levels of GC activity, and have a normal epidermal structure (not shown).
DISCUSSION
The introduction of point mutations into the mouse genome is an important refinement of the original gene targeting technique. Several strategies have been developed based on various vector designs. Generally, they can be divided into either one-step or two-step procedures. The two-step procedures involve either an insertion-type of homologous recombination event followed by a single reciprocal intrachromasomal recombination event (20, 21) or two replacement-type of homologous recombination events (22–24). The one-step procedure (cotransfer) uses a replacement vector with a mutated gene linked to a selectable marker placed into a neutral site to substitute for the wild-type gene (25). Neither the two-step procedure nor the one-step cotransfer procedure have been successful thus far in producing mice with point mutations in the GC gene. We were not able to introduce point mutation into the GC locus with a two-step procedure because of low second-step targeting efficiency (Y.L. and R.L.P., data not shown). The one-step cotransfer procedure was also unsatisfactory; in an attempt to transfer the N370S mutation, the neo marker placed at the end of the GC gene unexpectedly disrupted the downstream metaxin gene, resulting in embryonic lethality (16).
To overcome difficulties associated with introducing point mutations into the GC gene, we devised SIMP. SIMP has some attractive features. First, the targeting vector contains a single selectable marker and an uninterrupted segment of genomic homology making vector construction simple. Second, the frequency of homologous recombination is relatively high because of the effectiveness of insertion-type targeting vectors (26); our SIMP vectors showed very high targeting frequency (≈50%) when compared with a similar-sized replacement targeting vectors used in the previous mouse GC gene knockout experiment (7). Finally, only one step of ES cells culturing and drug selection is required, increasing the likelihood that the targeted ES cell will contribute to the germ line. The SIMP methodology may be particularly useful in situations where homologous recombination frequency is low, there is difficulty in obtaining germ-line transmission, or the locus is crowded, as is the case with the GC gene (27).
After homologous recombination, a properly configured SIMP vector should produce a complete version of the targeted gene with mutations in tandem with an inactive gene fragment. For this to occur, SIMP vector must encompass the 5′ or 3′ end of the gene to be targeted. Subsequently the targeted locus should be examined to exclude clones with unwanted events such as repair of the mutations or transfer of the mutations into the inactive gene duplication. Another consideration in constructing the SIMP vector is the distance between the linearization site and the mutation. On the basis of the double-strand break repair theory, mutations very close to the double-strand break will tend to be lost or transferred to the truncated duplication. In our experiments, the 2.5-kb distance between the linearization site and the mutation resulted in a high frequency of faithful transfer of the mutation into the GC gene.
The locus duplication occurring as a result of SIMP was found not to influence the expression of either GC or the closely adjacent gene, metaxin. To prove that the phenotypes of the mutant mice were not the result of the genomic rearrangement, we established a mouse with the same duplication as in the L444P and RecNciI mice but without the point mutations. The ES cells used to produce the mice were obtained as one of the targeting events obtained by using SIMP (Fig. 1A, event III). The mice with the GC duplication without point mutations were phenotypically normal—without neonatal lethality, with wild-type levels of GC enzyme, and with no epidermal pathology—demonstrating that the introduced point mutations were responsible for the striking phenotype of the L444P and RecNciI mice.
The RecNciI mouse had a severe deficiency in GC enzyme activity, did not feed, and died within 24 hr. The mouse showed epidermal abnormalities and glucosylceramide storage in the brain, liver, and skin. These features of the RecNciI mouse appear virtually identical to the previously described GC knockout mouse, suggesting that the RecNciI mutation results in a null allele. Consistent with this conclusion is the perinatal lethal phenotype of Gaucher disease infants homozygous for the RecNciI allele (4, 5), and the finding that human RecNciI mutant enzyme has little or no GC activity (28, 29).
By contrast, the L444P mouse had higher levels of GC activity—about 20% of normal—with no detectable storage of glucosylceramide in brain and liver at 1 day of age. Expression studies with human GC carrying the L444P change had shown that the mutant enzyme retains partial activity (28, 29). Compared with the RecNciI mouse, the L444P mouse had an improved physical condition with better hydration and an ability to feed. Unexpectedly, this mouse also did not survive past 24–48 hr after birth. Previous work with the GC knockout mouse demonstrated an altered skin permeability barrier resulting from elevated glucosylceramide and reduced ceramide in the epidermis (30). Similar skin changes—increased glucosylceramide storage, lowered ceramide content, and abnormal stratum corneum—were present in both point mutation mice. With the lack of neuronal and visceral glucosylceramide storage in the L444P mice, their rapid demise seems likely a consequence of severe stratum corneum abnormalities and compromised skin barrier function. Although neurologic dysfunction cannot be ruled out in the RecNciI and GC knockout mice because both have glucosylceramide storage in brain—abnormal skin barrier function must be a major factor limiting their life span.
Ceramides are essential components of the epidermal permeability barrier in the stratum corneum and may be derived in part from glucosylceramides by the action of GC (30). The reaction presumably takes place in the extracellular spaces of the stratum corneum after extrusion of the glucosylceramides from the epidermal lamellar bodies. The ceramides and other lipids then are assembled into lamellar layers which function as the permeability barrier. That GC activity is essential for this process was clearly illustrated by the altered barrier function and epidermal glucosylceramide storage in the completely enzyme deficient GC knockout mouse (30). An altered permeability barrier in the RecNciI mouse also might be expected because the mutation effectively produces a null allele. The abnormal skin manifestation in the L444P mice, however, was unexpected in light of their residual level of GC activity that is sufficient to prevent glucosylceramide storage in brain and liver. Epidermal abnormalities are not observed in Gaucher patients homozygous for the L444P mutation or in patients carrying other mutations resulting in residual GC activity. This difference between rodents and humans may reflect the differences in skin barrier formation during fetal development. In rodents a competent skin permeability barrier forms very late in gestation—1–2 days before birth (31, 32). A GC-mediated conversion of glucosylceramide to ceramide occurs during this period to produce a competent barrier (31). The residual level of GC activity in the L444P mice may not be sufficient to completely process the epidermal glucosylceramide in this short time period during gestation. In humans, the permeability barrier normally forms well before birth at between 30 and 34 weeks of gestation (33). The levels of residual GC activity in most forms of Gaucher disease are apparently sufficient to process epidermal glucosylceramide to ceramide before term birth at 40 weeks. An exception is seen with the Gaucher disease “collodion babies” exhibiting congenital ichthyosis and essentially no residual enzyme activity (8, 34). Infants homozygous for the RecNciI mutation have been described to have a severe skin phenotype (4, 5).
Another factor contributing to the glucosylceramide storage in epidermis but not in brain and liver in the L444P mouse could be related to biochemical differences of the glucosylceramides found in different tissues. Skin contains glucosylceramides with additional hydroxyl groups and with very long chain fatty acids, in addition to common types of glucosylceramides like those found in brain and liver (35). The L444P mutation may render the GC enzyme less active against the hydroxylated glucosylceramides with long chain fatty acids than against the common types of glucosylceramides, resulting in storage restricted to epidermis.
Establishment of a Gaucher disease model with a longer life span will be important for both investigating the complex pathogenesis of the disorder and evaluating new therapeutic approaches such as gene therapy and the use of glycolipid synthesis inhibitors. Our results show that the differences in skin biology between mice and humans must be taken into account in any strategy to produce a model of late onset disease. Possible approaches include preservation of epidermal GC activity by a transgene in the context of an enzyme-deficient animal or conditional gene disruption after the formation of the epidermal permeability barrier.
Acknowledgments
We thank Lance Ferrin for help with the RecA-assisted restriction endonuclease technique, and Cyndi Tifft, Chuxia Deng, and Dan Camerini-Otero for helpful comments on the manuscript. We are grateful to John T. Woosley who helped with the interpretation of the histopathology of the point mutation mice. This work was supported in part by U.S. Public Health Service Grants RO1-NS 24453 and P30-HD 03110 and the Deutsche Forschungsgemeinschaft (SFB 284).
ABBREVIATIONS
- ES
embryonic stem
- GC
glucocerebrosidase
- SIMP
single insertion mutagenesis procedure
Footnotes
RecNciI and L444P mice refer to homozygotes for the RecNciI and L444P targeted alleles, respectively.
References
- 1.Beutler E, Grabowski G A. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw-Hill; 1995. pp. 2641–2670. [Google Scholar]
- 2.Balicki D, Beutler E. Medicine (Baltimore) 1995;74:305–323. doi: 10.1097/00005792-199511000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Horowitz M, Zimran A. Hum Mutat. 1994;3:1–11. doi: 10.1002/humu.1380030102. [DOI] [PubMed] [Google Scholar]
- 4.Strasberg P M, Skomorowski M A, Warren I B, Hilson W L, Callahan J W, Clarke J T. Biochem Med Metab Biol. 1994;53:16–21. doi: 10.1006/bmmb.1994.1052. [DOI] [PubMed] [Google Scholar]
- 5.Sidransky E, Tayebi N, Stubblefield B K, Eliason W, Klineburgess A, Pizzolato G P, Cox J N, Porta J, Bottani A, DeLozier-Blanchet C D. J Med Genet. 1996;33:132–136. doi: 10.1136/jmg.33.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tayebi N, Cushner S R, Kleijer W, Lau E K, Damschroder-Williams P J, Stubblefield B K, Den Hollander J, Sidransky E. Am J Med Genet. 1997;73:41–47. doi: 10.1002/(sici)1096-8628(19971128)73:1<41::aid-ajmg9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 7.Tybulewicz V L J, Tremblay M L, LaMarca M E, Willemsen R, Stubblefield B K, et al. Nature (London) 1992;357:407–410. doi: 10.1038/357407a0. [DOI] [PubMed] [Google Scholar]
- 8.Sidransky E, Sherer D M, Ginns E I. Pediatr Res. 1992;32:494–498. doi: 10.1203/00006450-199210000-00023. [DOI] [PubMed] [Google Scholar]
- 9.Ferrin L J, Camerini-Otero R D. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka S, Johnson M, Grinberg A, Westphal H, Crawley J N, Tanike M, Suzuki K, Proia R L. Proc Natl Acad Sci USA. 1994;91:9975–9979. doi: 10.1073/pnas.91.21.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Hoffmann A, Grinberg A, Westphal H, McDonald M P, Miller K M, Crawley J N, Sandhoff K, Suzuki K, Proia R L. Proc Natl Acad Sci USA. 1997;94:8138–8143. doi: 10.1073/pnas.94.15.8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sango K, Yamanaka S, Hoffmann A, Okuda Y, Grinberg A, et al. Nat Genet. 1995;11:170–176. doi: 10.1038/ng1095-170. [DOI] [PubMed] [Google Scholar]
- 13.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 14.Bronson S K, Smithies O. J Biol Chem. 1994;269:27155–27158. [PubMed] [Google Scholar]
- 15.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 16.Bornstein P, McKinney C E, LaMarca M E, Winfield S, Shingu T, Devarayalu S, Vos H L, Ginns E. Proc Natl Acad Sci USA. 1995;92:4547–4551. doi: 10.1073/pnas.92.10.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, Treco D, Szostak J W. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 18.Szostak J W, Orr-Weaver T L, Rothstein R J. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 19.Valancius V, Smithies O. Mol Cell Biol. 1991;11:4389–4397. doi: 10.1128/mcb.11.9.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valancius V, Smithies O. Mol Cell Biol. 1991;11:1402–1408. doi: 10.1128/mcb.11.3.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasty P, Ramirez-Solis R, Krumlauf R, Bradley A. Nature (London) 1991;350:243–246. doi: 10.1038/350243a0. [DOI] [PubMed] [Google Scholar]
- 22.Askew G R, Doetschman T, Lingrel F B. Mol Cell Biol. 1993;13:4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stacey A, Schnieke A, McWhir J, Cooper J, Colman A, Melton D W. Mol Cell Biol. 1994;14:1009–1016. doi: 10.1128/mcb.14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Liu X, Jaenisch R. Proc Natl Acad Sci USA. 1994;91:2819–2823. doi: 10.1073/pnas.91.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng C, Thomas K R, Capecchi M R. Mol Cell Biol. 1993;13:2134–2140. doi: 10.1128/mcb.13.4.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasty P, Rivera-Perez J, Chang C, Bradley A. Mol Cell Biol. 1991;11:4509–4517. doi: 10.1128/mcb.11.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winfield S L, Tayebi N, Martin B M, Ginns E I, Sidransky E. Genome Res. 1997;7:1020–1026. doi: 10.1101/gr.7.10.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grace M E, Newman K M, Scheinker V, Berg-Fussman A, Grabowski G A. J Biol Chem. 1994;269:2283–2291. [PubMed] [Google Scholar]
- 29.Pasmanik-Chor M, Madar-Shapiro L, Stein E O, Aerts H, Gatt S, Horowitz M. Hum Mol Genet. 1997;6:887–895. doi: 10.1093/hmg/6.6.887. [DOI] [PubMed] [Google Scholar]
- 30.Holleran W M, Ginns E I, Menon G K, Grundmann J U, Fartasch M, McKinney C E, Elias P M, Sidransky E. J Clin Invest. 1994;93:1756–1764. doi: 10.1172/JCI117160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanley K, Jiang Y, Holleran W M, Elias P M, Williams M L, Feingold K R. J Lipid Res. 1997;38:576–584. [PubMed] [Google Scholar]
- 32.Hanley K, Devaskar U P, Hicks S J, Jiang Y, Crumrine D, Elias P M, Williams M L, Feingold K R. Pediatr Res. 1997;42:610–614. doi: 10.1203/00006450-199711000-00010. [DOI] [PubMed] [Google Scholar]
- 33.Evans N J, Rutter N. Biol Neonate. 1986;49:74–80. doi: 10.1159/000242513. [DOI] [PubMed] [Google Scholar]
- 34.Fujimoto A, Tayebi N, Sidransky E. Am J Med Genet. 1995;59:356–358. doi: 10.1002/ajmg.1320590315. [DOI] [PubMed] [Google Scholar]
- 35.Wertz P W. Semin Dermatol. 1992;11:106–113. [PubMed] [Google Scholar]