Abstract
In a Xenopus egg replication system, the origin recognition complex (ORC) does not bind to CpG methylated DNA and DNA replication is inhibited. Insertion of low density CpG DNA of at least 1.2 kb into methylated plasmids rescues both replication and ORC binding. Using this pseudo-origin, we find that ORC binding is restricted to low-CpG-density DNA; however, MCM is loaded onto both weakly and highly methylated DNA and occupies at least ∼2 kb of DNA. Replication initiates coincident with MCM, and even the most distally bound MCM is associated with sites of replication initiation. These results suggest that in metazoans MCM is loaded onto and initiates replication over a large region distant from ORC.
During the G1 phase of the cell cycle proteins, including ORC the origin recognition complex (cdc6), and the MCM-helicase complex, assemble at multiple sites along DNA, forming prereplication complexes (Pre-RCs) (for a review, see reference 6). During the S phase of the cell cycle, chromatin-bound Pre-RCs function as sites for initiation of DNA replication. The assembly of Pre-RCs during G1 is an ordered process in which ORC recognizes and binds to DNA first and subsequently cdc6 associates with the DNA. The ORC-cdc6 complex then facilitates loading of the MCM-helicase onto DNA. The sequential nature of Pre-RC assembly implies that the initial association of ORC with DNA sites determines where Pre-RCs form along the DNA.
In the yeast, Saccharomyces cerevisiae, ORC binds specifically and with high affinity to well-defined DNA consensus sequences distributed along the genome (7, 62). As such, replication in this eukaryote initiates reproducibly from unique DNA sites. The specific association of the S. cerevisiae ORC with origin sites has proven to be extremely useful for defining both the compositional organization of a Pre-RC, as well as dynamic changes in this organization during initiation. For example, because ORC associates specifically with DNA, footprinting techniques can be used to demonstrate structural changes during the G1- to S-phase transition (21). Similarly, the “fixed” location of Pre-RCs in S. cerevisiae has allowed replication initiation to be mapped at the nucleotide level (9).
Although well-defined origins exist in S. cerevisiae, similar sequence-specific sites have remained elusive in other eukaryotic organisms. Rather, in metazoan somatic cells, sensitive PCR and genetic methods have identified large regions of DNA (2 to 50 kb) where initiation occurs (2, 22). Within these “initiation zones” or origins, initiation occurs at many sites. Similarly, strategies using plasmid maintenance assays in human cells have suggested that any human DNA of adequate length is sufficient for cell cycle-controlled autonomous DNA replication (35). Consistent with these observations is the finding that strict sequence-specific binding of purified Schizosaccharomyces pombe and Drosophila melanogaster ORC has not been detected (15, 16).
Moreover, in metazoans Pre-RC formation appears to be flexible and under developmental control. In early embryonic cells in both Xenopus and Drosophila, ORC binds DNA in a sequence-independent manner at Pre-RCs spaced ca. 10 kb apart (33, 58). However, as development progresses and cells acquire a somatic phenotype, Pre-RCs assemble at initiation sites 100 to 200 kb apart (32). Together, these observations indicate that the association of metazoan ORC with DNA may not depend strongly on DNA sequence.
Modifications of DNA or chromatin structure (e.g., DNA and histone methylation or acetylation) likely plays a critical role in determining where ORC can associate with DNA and assemble an initiation site or Pre-RC (for a review, see reference 27). For example, during early embryogenesis when DNA-chromatin structure is relatively simple and homogeneous, ORC may be able to associate with all DNA sites, while in somatic cells, as a result of multiple chromatin modifications, ORC may only be able to associate with a subset of potential binding sites (36).
DNA templates added to Xenopus egg extracts replicate completely, and replication is limited to a single round (33, 59). Plasmid DNA templates added to the extract initiate replication independent of sequence and a single initiation occurs per plasmid (37, 41). The use of the Xenopus membrane-free replication system has allowed for the detailed analysis and ordering of requisite steps for replication initiation. However, the sequence-independent nature of replication initiation in Xenopus egg extracts has limited the analysis of Pre-RC formation and its relationship to actual sites of replication initiation. To overcome this restriction, we have generated a template on which Pre-RC assembly is restricted to a defined zone.
In vertebrate cells, DNA methylation at CpG nucleotides is a prevalent modification (61). Moreover, there is some evidence that this modification may regulate initiation (47). We observe that methylated DNA templates fail to initiate replication when added to Xenopus extracts. Inhibition of replication is at the level of ORC recruitment; we find that ORC cannot bind to methylated DNA. Importantly, we found that both replication of a methylated template and ORC binding is rescued by inserting a low-density methylated sequence into the methylated template. By chromatin immunoprecipitation (ChIP) analysis, we observed that ORC binding is restricted to the insert. Thus, methylation can, in principal, serve to limit where Pre-RCs form along DNA.
Using this constructed “origin,” we investigated the organization and function of ORC and MCM. Most interestingly, we found that, although ORC binding occurs within the low-methyl insert, MCM binding occurs both within the insert and within the methylated regions flanking the insert, suggesting that multiple MCMs are loaded cooperatively onto DNA up to 1,000 bp on either side of ORC. Initiation of replication occurs throughout the region covered by the MCM protein, indicating that all of the bound MCMs are competent for initiating DNA replication. Our results provide insight about the formation and organization of Pre-RCs and provide a possible explanation for why specific initiation within metazoans has been difficult to observe.
MATERIALS AND METHODS
Preparation of Xenopus egg extracts.
Membrane-free egg cytosol and nucleoplasmic extract (NPE) were prepared as described previously (54, 59).
Preparation of methylated plasmids and lambda DNA.
Plasmid and lambda DNA were methylated in vitro by using the bacterial CpG methylase SssI (NEB, Beverly, Mass.) according to manufacturer's instructions. Plasmid DNA was then gel purified. Lambda DNA (Amersham-Pharmacia) was methylated as plasmid. Mock-methylated lambda was generated by excluding the methyl donor, SAM, from the methylation reaction. Both lambda DNA samples were heat inactivated at 65°C for 20 min. Methylation of both lambda and plasmid DNA was verified by restriction enzyme digestion with the methylation-sensitive enzymes ClaI, SmaI, and PvuI.
Generation of cdc45 deletions.
The inserts derived from cdc45 cDNA were a 146-bp EcoRI/KpnI fragment, a 441-bp HindIII/EcoRI fragment, a 1,263-bp EcoRI/HindIII fragment, and a 1,704-bp EcoRI fragment. The 1.8-kb bacterial plasmid DNA fragment was derived from pBSK− by restriction digestion with EcoRV and ScaI. All fragments were cloned into pCRII-TOPO at appropriate restriction sites by using the multiple cloning site.
Replication assays.
Replication reactions were performed as previously described (59). Briefly, DNA was incubated in 10 μl of extract for 30 min, and then 25 μl of NPE was added. [α-32P]dATP was included in all replication assays at a concentration of 0.1 μCi/μl of egg extract except for the two-dimensional (2-D) gel analysis, where it was used at 1 μCi/μl. Reaction products were visualized after they were run on a 1% agarose gel, which was then dried and exposed to a phosphorimaging screen (Molecular Dynamics).
Lambda and plasmid isolation and Pre-RC detection.
After incubation with membrane-free egg extract, lambda samples were diluted 10-fold in ELB (50 mM KCl, 2.5 mM MgCl2, 10 mM HEPES [pH 7.7], 250 mM sucrose) containing 2 mM ATP and 0.5% Triton X-100. Samples were spun at 16,000 × g for 2 min in a microfuge. Chromatin pellets were washed once in ELB, respun, resuspended in sodium dodecyl sulfate (SDS) loading buffer, and loaded on a 10% protein gel for analysis. Plasmid samples were diluted in 2 ml of ELB with 0.2 mM ATP, 0.5% Triton X-100, and spun at 100,000 × g for 10 min in an ultracentrifuge. Chromatin pellets were washed once in ELB-ATP-Triton X-100 and respun as previously described. Samples were resuspended in Tris-EDTA (TE) and split in half to determine ORC association and DNA recovery, respectively.
Western blot analysis.
Western blot analysis was performed with α-XORC2, α-MCM3, and α-RCC1 (19, 57), and α-Η2Α antibodies (Serotec). For detection of histone H2A, samples were subjected to SDS-15% polyacrylamide gel electrophoresis. After protein separation, the gel was incubated in 2.3% SDS-5% β-mercaptoethanol-67 mM Tris-HCl (pH 6.7) for 30 min at room temperature with gentle agitation. Gels were preincubated in transfer buffer (39 mM glycine, 48 mM Tris, 0.1% SDS, 25% methanol) for 5 min and then transferred for 2.5 h at 4°C and 70 V by using a Mini-Protean II (Bio-Rad) apparatus. Proteins were visualized by chemiluminesence (Pierce) with goat anti-rabbit immunoglobulin G-horseradish peroxidase. All incubations were performed at 1:5,000 dilutions. Western blot analysis was linear for protein concentration more than 2 orders of magnitude, as determined by using purified XOR2-His (58).
ChIP analysis.
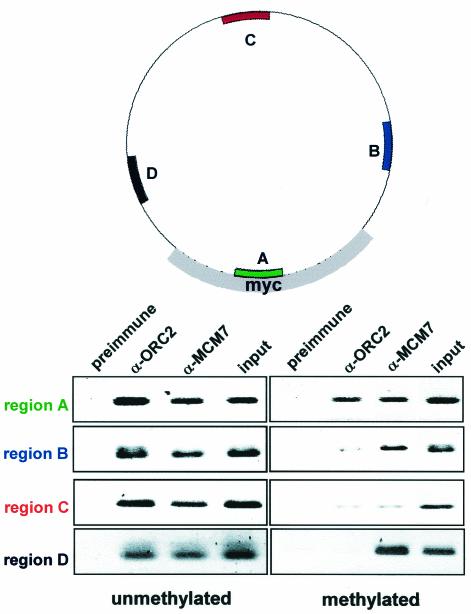
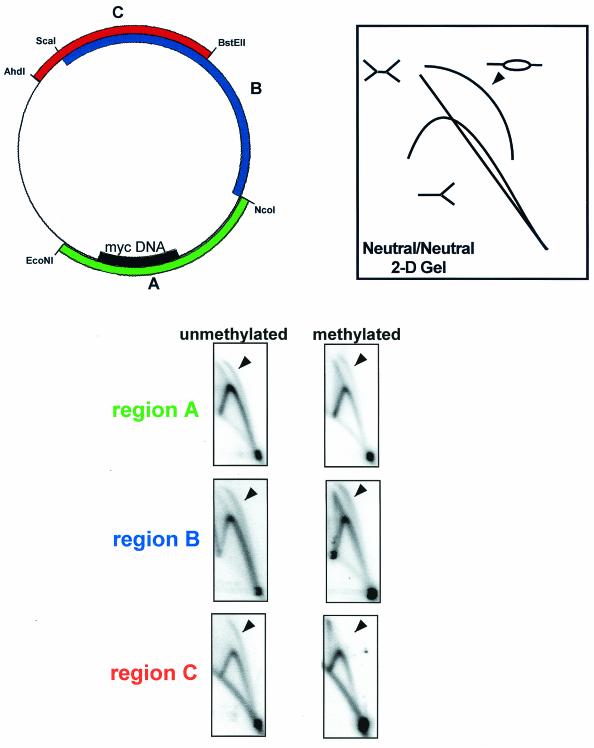
After assembly of 200 ng of plasmid in 100 μl of membrane-free egg cytosol, the reaction was diluted in ELB and 1% formaldehyde for 10 min. Fixed chromatin was isolated as described above, resuspended in 500 μl of immunoprecipitation buffer (20 mM Tris HCl [pH 7.7], 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 5 μg of aprotinin and leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride) and fragmented to <500 bp by sonication with a microprobe five times for 10 s at 6 W. Chromatin was immunoprecipitated with 2 μl of either rabbit α-ORC2, α-MCM7, or preimmune serum for 5 h at 4°C. Immune complexes were collected with protein A-Sepharose (Pharmacia) for 1 h, washed five times with immunoprecipitation buffer and twice with TE, and then resuspended in 100 μl of TE. DNA was recovered by incubation with 1 mg of proteinase K/ml and 0.5% SDS for 9 h at 37°C. The buffer was collected, and DNA was purified by phenol extraction and ethanol precipitation. PCR (20 cycles of denaturation at 94°C, annealing at 48°C, and extension at 72°C for 1 min each) was performed on 2 μl of each sample with primers specific for region A (5′-TCTCTTTTGGAGGTGGTG-3′ and 5′-CAGGCATTAATTTCCTAG-3′), region B (5′-TGGAGCGAACGACCTACA-3′ and 5′-ATAACGCAGGAAAGAACA-3′), and region C (5′-TGAGTATTCAACATTTCC-3′ and 5′-TGACTGGTGAGTACTCAA-3′) as depicted in Fig. 4. Amplified products were run on 2% agarose gels, stained with Sybergold, scanned, and inverted in Adobe Photoshop for clarity.
FIG. 4.
The four regions of the myc origin construct that are amplified after ChIP are depicted. Region A (green) contains the putative ARS consensus sequences and DUE within the myc origin. Region B (blue) and region D (black) are 969 bp upstream and downstream from region A. Region C (red) is 2,444 bp from region A. ChIP analysis of unmethylated (left) and methylated (right) plasmids containing the myc origin.
Neutral/neutral 2-D gel analysis.
A total of 25 ng of methylated or unmethylated DNA construct was incubated with 10 μl of cytosolic egg extract for 30 min and then supplemented with 2.5 volumes of NPE in the presence of [α-32P]dATP. After 15 min, 150 μl of stop buffer (20 mM Tris HCl [pH 8], 200 mM NaCl, 5mM EDTA, 0.5% EDTA) and 100 μg of RNase A/ml were added, and the samples were incubated at 37°C for 1 h. After a further 1 h of incubation in the presence of 1 mg of protease K/ml, samples were phenol extracted twice and ethanol precipitated. Restriction digestion for region A (EcoNI/NcoI), region B (NcoI/ScaI), or region C (BstEII/AhdI) was carried out for 2 h, and samples were phenol extracted and ethanol precipitated again. Samples were loaded onto a 0.4% agarose gel and run at 1 V/cm for 18 h in the first dimension. Electrophoresis on a 1% agarose gel in the second dimension was carried out for 8 h at 4°C at 6 V/cm in the presence of 0.3 μg of ethidium bromide/ml. Gels were dried and evaluated with the phosphorimager.
RESULTS
Methylation of DNA inhibits replication.
As a model for investigating how DNA-chromatin modifications might limit initiation sites in somatic cells, we assessed how DNA methylation, a prevalent epigenetic modification in human and frog genomes (56, 61), affected DNA replication in embryonic extracts. To do this, both plasmid and lambda DNA were methylated in vitro by using the bacterial CpG methylase SssI. Methylation was verified by using methylation-sensitive restriction enzymes and was stable during all subsequent incubations in egg cytosol (Fig. 1A and data not shown).
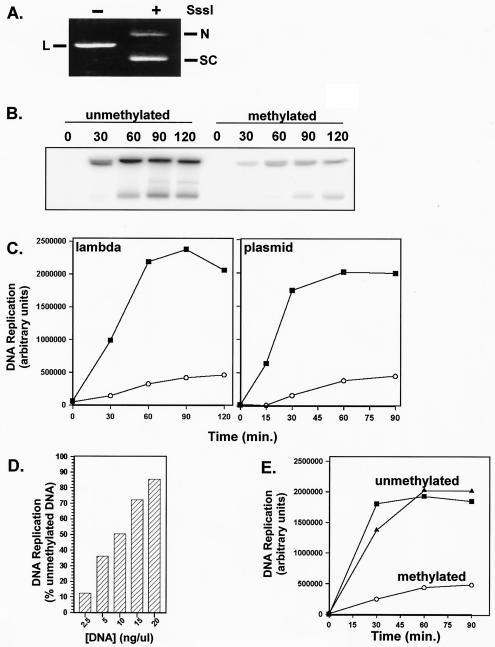
FIG. 1.
In vitro replication of CpG-methylated DNA. (A) Methylation status was determined after incubation of 200 ng of either unmethylated or SssI-methylated pBSK− in DNA Xenopus egg extract for 30 min. Isolated and purified DNA was then subjected to restriction digestion with ClaI. N, nicked plasmid; S, supercoiled plasmid; L, linear plasmid. (B) Replication was determined after incubation of 25 ng of either unmethylated or SssI-methylated lambda DNA in 10 μl of membrane-free Xenopus egg extract for 30 min, followed by the addition of 2.5 volumes of NPE. [α-32P]dATP was included in all replication assays at a concentration of 0.1 mCi of egg extract/μl. Then, 3-μl portions of the replication reaction products were visualized after being loaded onto a standard agarose gel. Time refers to number of minutes after the addition of the NPE. (C) Quantitation of replication of unmethylated (▪) and methylated (○) lambda DNA or a 2.9-kb plasmid, pBSK−, as determined by using ImageQuant software. (D) Quantitation of replication of increasing concentrations of pBSK− methylated plasmid after 60 min in NPE. DNA replication is expressed as the percent replication relative to replication of unmethylated plasmid at an equivalent concentration. (E) Quantitation of replication of an unmethylated 5.4-kb plasmid (pET-28a) in the absence (▪) or presence (▴) of the 2.9-kb pBSK− methylated plasmid (○).
It has already been established that when DNA is incubated in membrane-free egg cytosol, Pre-RCs, including ORC, Cdc6, Cdt1, and MCM, assemble on DNA (14, 17, 42, 49, 50). After Pre-RC formation, the addition of a concentrated NPE, derived from in vitro-assembled nuclei, induces these assembled Pre-RCs to initiate replication (59). In this system, CpG methylation resulted in a 90% inhibition of replication from either linear bacteriophage DNA (Fig. 1B and C) or circular plasmid DNA (Fig. 1C).
Interestingly, the methylation-dependent inhibition of replication can be inactivated by adding increasing amounts of methylated DNA into the reaction (Fig. 1D). At 2.5 ng of plasmid DNA per μl of extract, the inhibition of replication is 90%. However, the inhibition is lost as the concentration of DNA is increased. At a methylated DNA concentration of 20 ng/μl, replication of methylated plasmid approaches the level of unmethylated DNA. This result suggests that inhibition is due to a factor present in egg extracts and that the inhibitory activity of this factor can be “titrated out” by increasing the concentration of methylated DNA. Furthermore, an unmethylated 5.4-kb plasmid (pET-28a) replicated normally in the presence of a methylated 2.9-kb plasmid (pBSK−) (Fig. 1E). Thus, methylation-dependent inhibition of replication only occurs in cis and is not due to a soluble signal generated by methylated DNA. In addition, methylation-dependent inhibition of replication is not due to the generation of an inter-S phase ATM/ATR-dependent checkpoint signal (18, 23), since caffeine, an inhibitor of these checkpoints, does not rescue replication of methylated plasmids (data not shown).
Low-CpG-density DNA rescues replication of methylated DNA.
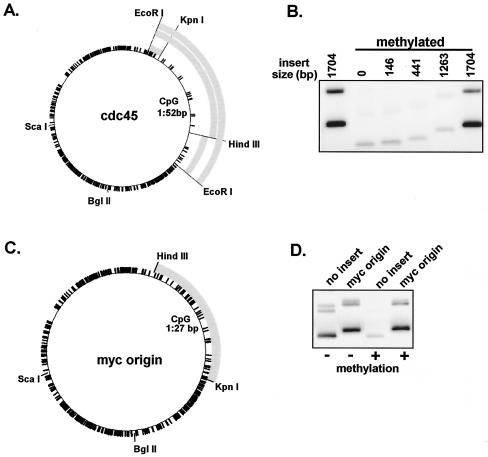
Due to the relatively high CpG content characteristic of both lambda and bacterial plasmid DNA (one CpG dinucleotide every 14 bp), we determined the effect on replication of different sized inserts of DNA with lower CpG content. Inserts derived from the Xenopus cdc45 cDNA display a low density of CpG dinucleotides (one every 52 bp) compared to the bacterial vector (Fig. 2A). Strikingly, a 1,704-bp EcoRI fragment from the cdc45 cDNA rescued replication of methylated plasmid (Fig. 2B). Rescue of replication by the 1,704-bp insert is not due to the increased size of the plasmid because a 1,825-bp fragment derived from the bacterial vector, pBSK−, did not rescue replication (data not shown). Dividing the 1,704-bp fragment into 1,263-bp and 441-bp fragments resulted in a loss of replication rescue (Fig. 2B). The 1,704-bp fragment does not contain any apparent consensus ARS (autonomous replicating sequence) or putative DNA unwinding elements (DUE), suggesting that rescue of replication of methylated plasmids simply requires the inclusion of a low-CpG-density insert longer than ∼1.2 kb. We have observed that any DNA characterized by low CpG density of sufficient length can rescue the replication of methylated plasmids (unpublished data).
FIG. 2.
Low-CpG-density DNA insert and human myc origin DNA rescue replication of methylated plasmid. (A) A 3.9-kb plasmid, pCRII-TOPO, containing fragments derived from a cdc45 cDNA with low CpG density relative to the bacterial vector DNA. Black tick marks represent CpG dinucleotides. (B) Replication of methylated and unmethylated plasmids which harbor cdc45 inserts. Replication products were visualized after 120 min in NPE. (C) A 1,271-bp HindIII/KpnI fragment from the 5′ region of the human myc origin (gray) has a low CpG content relative to the bacterial vector DNA. (D) Replication of unmethylated and methylated plasmids containing myc origin DNA or no insert (detected as for panel B).
A number of mapped mammalian origins are coincident with unmethylated stretches of CpG motifs associated with 5′ transcriptional regulatory regions (4, 5, 20, 47). However, these “origins” have remained impervious to further molecular dissection; this is due, in part, to the fact that initiation within these regions occurs over a broad range of sequence. The origin associated with the upstream regulatory region of the c-myc proto-oncogene is one such origin. Initiations from this origin take place within a 2.4-kb, XhoI/HindIII fragment just upstream from the first intron (60). Furthermore, this fragment of DNA can function as an origin when moved to an ectopic location (43) and has been reported to function in plasmid maintenance assays (44). The 5′ half of this 2.4-kb region contains putative ARS consensus sites and an A/T rich DUE, as well as binding sites for transcription factors (24). Like cdc45 cDNA, the 5′ region of the myc origin is lower in CpG density than the bacterial plasmid (Fig. 2C). We determined the ability of a 1,271-bp fragment containing the 5′ portion of the myc origin to rescue replication of methylated plasmids. Indeed, the fragment completely rescued replication of methylated plasmid (Fig. 2D). In contrast, vector without insert DNA or with an 1,825-bp bacterial DNA insert did not replicate under these conditions (Fig. 2D and data not shown).
ORC cannot bind to methylated DNA.
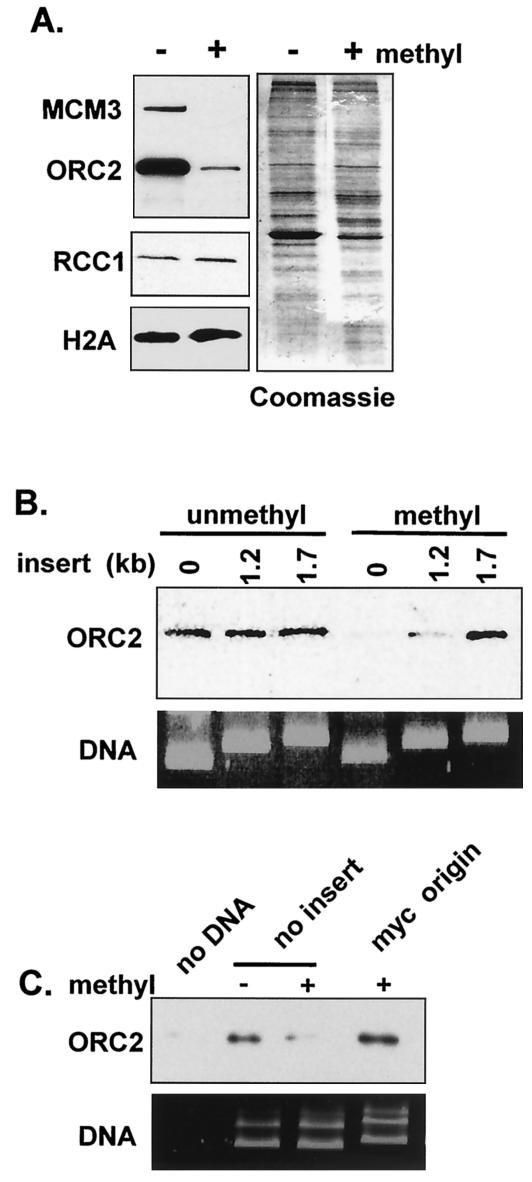
To determine how CpG methylation inhibits replication, pre-RC formation on linear bacteriaphage lambda DNA was examined. Unmethylated or SssI methylated DNA was first incubated in cytosol and then isolated and assayed for pre-RC formation based on the association of ORC2 and MCM3 proteins. The amount of chromatin-associated ORC on methylated lambda was <5% of the level detected on unmethylated lambda, and no MCM was detected (Fig. 3A). In contrast, CpG methylation has no effect on nucleosome assembly (46), as evidenced by the incorporation of the core histone H2B and the binding of a well-characterized chromatin-binding protein, RCC1 (Fig. 3A). Similarly, when the total protein associated with methylated and unmethylated DNA isolated from extract is compared by Coomassie blue staining, few differences in protein content are observed (Fig. 3A, right panel). Together, these observations suggest that methylation of DNA greatly reduces the affinity of ORC for DNA. This reduction in affinity appears to be specific to ORC in that many other proteins bind to methylated DNA normally.
FIG. 3.
myc origin DNA rescues Pre-RC assembly on CpG-methylated DNA. (A) ORC2, MCM3, RCC1, and H2A association with unmethylated and methylated lambda DNA assembled in membrane-free egg extracts. A total of 25 ng of methylated and unmethylated lambda was incubated for 30 min in 10 μl of membrane-free cytosol. Western blot analysis (left) or total protein staining with Coomassie blue (right) is shown. (B) ORC2 association with methylated and unmethylated plasmids containing either no insert, a 1,263-bp cdc45 insert, or a 1,704-bp cdc45 insert. A total of 125 ng of unmethylated and methylated plasmid was assembled in 50 μl of membrane-free egg cytosol for 30 min. DNA recovery was monitored by staining a standard agarose gel with Sybergold reagent. (C) ORC2 association was determined as in panel B except that plasmid containing either no insert or myc origin DNA was used.
To determine whether ORC is recruited to methylated plasmid, DNA was incubated in membrane-free egg extract for 30 min and then isolated by ultracentrifugation through a sucrose cushion. Based on Sybergold staining of gels, plasmid recovery using this method was identical for the different templates used (Fig. 3B and C). Western blot analysis revealed that binding of ORC to methylated plasmids was reduced 7- to 10-fold compared to unmethylated DNA (Fig. 3B and C). Samples that did not contain plasmid showed no ORC signal (Fig. 3C). In contrast, methylated plasmids containing either the 1,704-bp cdc45 insert (Fig. 3B) or the 5′ myc origin (Fig. 3C) showed no reduction in chromatin-associated ORC, suggesting that both of these DNA inserts rescue replication of methylated plasmids through the recruitment of ORC to chromatin. The observation that the 1,263-bp cdc45 insert, which failed to rescue replication, does not rescue ORC binding (Fig. 3B) is consistent with the premise that metazoan origins, in the context of methylated DNA, must be at least ∼1.2 kb in length.
MCM can be loaded at a distance from ORC onto methylated DNA.
Using ChIP analysis on plasmids that contain the 1.2-kb myc origin, we directly determined whether methylation could target pre-RC formation to specific regions of DNA. These experiments showed that in the absence of methylation both ORC and MCM bound without specificity to all regions of the plasmid tested (Fig. 4, left panel). However, when the construct was methylated, ORC failed to bind to the heavily methylated B, C, and D regions. Rather, ORC binding was restricted and occurred exclusively within the undermethylated A region (Fig. 4, right panel). This result demonstrates that ORC is indeed targeted to the low-methyl insert.
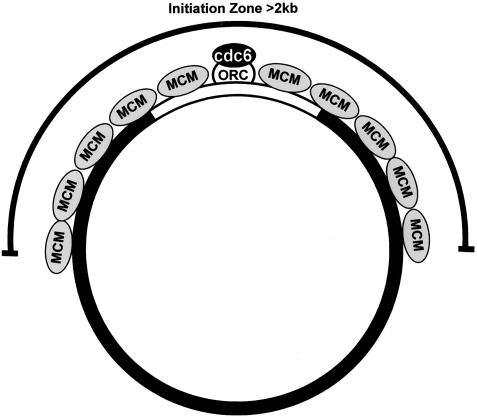
Surprisingly, ChIP analysis showed that MCM loading, unlike ORC, was not restricted to this undermethylated region. Like ORC, MCM is loaded onto the undermethylated region A. However, unlike ORC, MCM is also loaded onto regions B and D, which are heavily methylated and located 969 bp upstream and downstream from region A (Fig. 4, right). Interestingly, like ORC, MCM is not loaded onto region C, which is also heavily methylated but is 2,444 bp away from region A (Fig. 4, right). Based on previous estimates of the amounts of ORC and MCM bound to sperm chromatin (40, 58), we calculate that origin-associated MCMs cover at least 2 kb of DNA at a density of one MCM complex every one to two nucleosomes. Thus, in egg extracts at least, Pre-RCs may consist of an ORC with multiple MCM complexes bound at least 969 bp laterally both upstream and downstream of the chromatin-bound ORC.
The ChIP analysis shows that methylation restricts ORC, but not MCM, to regions of low methylation on the plasmid. To determine the location of actual initiation relative to the location of ORC and MCM, we used neutral/neutral 2-D gel electrophoresis. The detection of a distinct bubble arc above the prominent Y arc is diagnostic of initiation within the restriction fragment being analyzed (12). When analysis was carried out on an umethylated construct containing the myc origin (Fig. 5), prominent Y and bubble arcs were detected in fragments containing region A, B, or C, a finding consistent with previous work demonstrating random initiation of replication on plasmids in Xenopus eggs. Likewise, fragments from methylated samples representing regions A and B also showed prominent Y and bubble arcs. However, in contrast to unmethylated samples, bubble arcs were never detected in region C when methylated samples were used (Fig. 5). Thus, methylation can restrict replication initiation. In methylated constructs, region C is negative for both ORC and MCM binding as well as initiation; and region A is positive for both ORC and MCM binding, as well as initiation. Importantly, on these templates region B is negative for ORC but positive for MCM binding and initiation. The ChIP and 2-D gel analysis suggest that sites of replication initiation are coincident with MCM occupation, but not ORC, and even the most distant MCM complexes are associated with sites of initiation.
FIG. 5.
Methylation restricts replication initiation events to sequences within or immediately adjacent to the myc origin. Neutral/neutral 2-D gel electrophoresis of unmethylated or methylated replication intermediates after restriction digestion to generate region A (green), region B (blue), and region C (red). Three typical replication intermediates are depicted: the Y arc, the double Y arc, and the bubble arc (8, 27). Arrowheads indicate the positions of predicted bubble arcs.
DISCUSSION
In Xenopus, the same set of proteins function to establish Pre-RCs in both embryonic and somatic cells. Presumably, in embryonic cells these proteins can form Pre-RCs at all locations along the genome resulting in random replication initiation (10, 30, 32). However, as development progresses and cells take on somatic functions, Pre-RC formation may become restricted to certain zones, resulting in the detection of regions of specific replication initiation (32). This restriction process most likely involves, in part, multiple developmentally controlled DNA and or chromatin template modifications that limit where Pre-RCs can assemble along the genome (27). To date, these postulated “restrictive modifications” are poorly understood and characterized and, as a result, the compositional organization of metazoan Pre-RCs is not well defined. In the present study we report that DNA methylation, a modification prevalent in vertebrates but largely absent in invertebrates, can contribute to restricting Pre-RC formation and describe how the ORC and MCM components of the Pre-RC are organized under these restricted conditions.
Using a soluble egg extract replication system we find that methylation of DNA strongly inhibits DNA replication (Fig. 1). In addition, we show that the affinity of ORC for methylated DNA is very low (Fig. 3). These two observations alone suggest a simple model in which ORC binding to DNA is blocked by DNA methylation, resulting in inhibition of Pre-RC assembly and suppression of replication initiation. We assume that the residual replication and ORC binding detected on methylated plasmid and lambda DNA may be due to incomplete methylation. Because methylated DNA retains its affinity for many of the other proteins present in the extract (Fig. 3A), including RCC1, histone H2B, and the MCM-helicase (Fig. 4), this decreased affinity seems specific for ORC. Moreover, we observe that replication is rescued when a DNA sequence with low methylation potential is inserted into a highly methylation-restricted template (Fig. 2). Under these conditions we observe that ORC is targeted, by default, to the low-methylation insert (Fig. 4). Together, these observations demonstrate in principle that methylation of DNA can, in part, serve to restrict where Pre-RCs form along the genome in vertebrates. The mechanism of inhibition is not clear at this time. However, the fact that the inhibitory effect can be titrated with methylated DNA suggests that a soluble factor that specifically binds to methylated DNA may be present at low levels in egg extract. Consistent with this is the observation that binding of ORC to methylated plasmid is not inhibited when concentrations of plasmid DNA are raised to 20 ng/μl (unpublished data). Certainly, candidates for an inhibitory factor include methyl CpG-binding proteins.
MCM binds DNA at sites distant from ORC.
The stable targeting of ORC to low-methylation regions provides a rare opportunity to examine the compositional organization of a Pre-RC in a “restricted” situation. In particular, we have investigated the relationship between the binding of ORC and the MCM-helicase under this condition. Using ChIP analysis we observed that ORC is found exclusively bound to the low methyl insert (Fig. 4). In contrast, the MCM-helicase is located both within the low-methylation insert and in the highly methylated restricted zone (Fig. 4). The data strongly supports a model whereby ORC, along with cdc6 and cdt1, facilitates the loading of MCM onto a 1-kb region of DNA on either side of where ORC is bound (Fig. 6). As for how MCM might be loaded onto DNA at a distance from ORC, several possibilities exist. First, we considered the possibility that the results observed were due to methodological artifact. Specifically, if ORC binds with lower or with modest affinity to methylated DNA, then it might be able to load MCM at methylated sights but not cross-link efficiently and therefore be undetectable in a ChIP assay of the methylated regions. However, if this were the case, then MCM should be detected at all regions of the methylated DNA and not just regions flanking the low-methylation insert. This does not occur. Rather, our results show that although MCM can bind to methylated DNA up to 963 bp away from ORC (regions B and D), it does not bind to methylated DNA that is at a distance of 2,444 bp away from ORC (region C). Thus, the ability of ORC to load MCM onto DNA at distant sites appears to be limited.
FIG. 6.
Replication initiation sites are coincident with MCM but not ORC. ORC (white) binding to DNA is promiscuous, and a mechanism for selective binding of ORC to DNA relies on differences in chromatin structure (e.g., methylation) that exclude ORC from chromatin (black plasmid DNA). Once ORC binds to accessible chromatin (white insert DNA), multiple MCMs (gray) are loaded in a cdc6 (black)- and cdt1 (not shown)-dependent manner and occupy DNA sites at least 1 kb distal to ORC. Sites of replication initiation are detectable in a broad region and, like MCM binding, are detectable at distant regions of DNA that are negative for ORC binding.
Second, we considered a model in which DNA-bound ORC can come into contact with and load MCM onto distant sites as a result of DNA bending or looping. Thus, it is possible that although ORC cannot bind to methylated DNA directly it is able to contact such sites as a result of DNA bending or looping and load MCM onto these sites while remaining bound to low-methylation DNA. Although we cannot rigorously rule this model out, the constrained nature of local bending over a 1-kb (four to five nucleosomes) region argues against this possibility (48).
A third model suggests that multiple MCMs bind cooperatively to DNA flanking ORC and that this cooperative loading extends to regions at least ∼1 kb away from ORC. Such a mechanism would predict that loading of the first MCM would be ORC dependent but that the loading and propagation of subsequent MCMs onto flanking DNA might depend on MCM-MCM interactions and be independent of ORC. In support of this, we observed that initiation of MCM loading onto DNA was ORC dependent. However, once ORC-dependent loading of MCM has occurred, further loading of MCM onto highly methylated DNA may be independent of ORC. If cooperative loading of MCM is occurring, our results suggest that it propagates at least 1 kb but less than 2.5 kb away from ORC. The cause for the restriction of MCM spreading to less than 2.5 kb of the 5.1-kb construct used in the present study is not clear. It is certainly possible that the use of larger plasmid constructs reduces torsional stress related to the assembly of plamids in egg extracts, resulting in increased spreading of MCM.
A cooperative model is also consistent with current measurements of the ratio of ORC to MCM bound at a metazoan origin. In Xenopus, early studies suggested a level of 5 to 6 MCMs per ORC (40), whereas more recent studies suggest that there are 30 MCMs loaded per ORC (references 26 and 58 and unpublished data). Although these stoichiometric studies could be interpreted as supporting a model in which MCM is distributed at a distance from ORC, no spatial data to support this conclusion were presented, and alternative mechanisms are equally likely. For example, if ORC repeatedly loaded MCM and then dissociated from DNA, high ratios of MCM to ORC would be observed. Whether MCM can bind cooperatively and, if so, whether this cooperative binding requires known interacting proteins such as cdt1 and MCM10 require further study.
MCM, not ORC, determines where initiation occurs.
With respect to where DNA replication initiates, our findings strongly suggest that initiation occurs where MCM is positioned along the DNA rather than where ORC is located. Specifically, we observe initiation occurring both within poorly methylated regions containing both MCM and ORC and in highly methylated regions that contain MCM but lack ORC (Fig. 4 and 5, region B). Moreover, initiation does not occur in highly methylated regions lacking MCM (Fig. 4 and 5, region C). Thus, initiation is MCM dependent and MCMs located both near to and at a distance from ORC can initiate replication. This suggests that all of the bound MCMs can initiate replication. This conclusion is also supported by a recently published report demonstrating that multiple MCMs bound to a small fragment of DNA can all be phosphorylated by the initiation-specific Cdc7 kinase (26).
The observation that initiation occurs in methylated regions where ORC is absent suggests that initiation is independent of ORC position. This conclusion is consistent with observations demonstrating that after MCM is loaded onto DNA, ORC can be dissociated from the Pre-RC without affecting later S-phase events leading to initiation of replication (25, 31, 51). Importantly, if multiple MCMs are cooperatively bound at a Pre-RC, then this would account for the observed 2-kb zone of initiation (Fig. 6). Because initiation within this zone would occur randomly, mapping a specific site for initiation in metazoans, as has been carried out in S. cerevisiae (9), may be impossible for metazoan origins.
Although a few origins of replication have been mapped to discrete start sites in metazoans, these may represent unique cases. In the case of the lamin B origin in humans, assembly of a specific replication-associated protein at this site has not been demonstrated (1). In contrast, initiation within the amplification region II/9A of Sciara has been mapped to a precise start site immediately adjacent to an ORC binding site (8). However, amplification in flies represents an exceptional case of replication initiation that functions in the absence of normal cell cycle controls. Indeed, during normal mitotic divisions before the stage of amplification, replication initiates from multiple sites within a broad 8-kb region (38). It is not clear weather ORC binding determines these developmental changes in origin usage. Most well-described origins in vertebrates are characterized by large zones of initiation (28). For example, at the large 55-kb zone of initiation within the dihydrofolate reductase locus, replication bubbles, as well as MCM protein, are detected across the whole intergenic region (3). Origins of this type may better represent the paradigm for cell cycle regulated origins found in metazoans and especially vertebrates.
Overall, our results suggest that methylation may be one of the contributing elements that define where Pre-RCs form in vertebrate cells. As is the case in prokaryotes, origins of replication, like transcriptional regulatory regions, are differentially methylated (20), and late-replicating origins are associated with imprinted genes due to differential methylation (53). Since it seems that ORC is recruited to chromatin in a sequence-independent manner, one method of restricting pre-RC assembly from regions of the genome may be CpG methylation. For example, although the density of CpG motifs around the dihydrofolate reductase locus in humans does not approach the densities in bacterial or lambda DNA, Rein et al. report that hypomethylation of this region does not inhibit replication in general but correlates with a loss of specific initiation from this locus (47). During Xenopus development, the relationship between DNA methylation and the change in replicon size during the mid-blastula transition remains to be determined. At a gross level the Xenopus genome is in a hypermethylated form throughout development although subregions are maintained in a hypomethylated state (56).
Although DNA methylation is qualitatively and quantitatively different in the Drospohila genome (29, 39), origin selection may be influenced by chromatin modifications as well. For example, the DNA-binding proteins E2F and DP regulate ORC recruitment and origin usage during chorion gene amplification (11, 52). These transcription factors have both positive and negative effects on origins. E2F mutants unable to bind Rb-like proteins, and in all probability associated repressive histone deacetylases (HDACs) result in overamplification of the chorion locus (52). Undoubtedly recruitment of HDACs by Rb results in a repressive, compact, chromosomal structure in accordance with current models of transcriptional regulation (34). It is likely that this compact chromatin structure is inhibitory for replication initiation as well. Consistent with this idea is the observation that exogenous human Rb inhibits replication in Xenopus extracts in an MCM-dependent manner (55).
Analogously, methylation of CpG motifs is inhibitory for E2F binding and transcription in mammalian cells (13). Presumably, this occurs via the recruitment of CpG-binding proteins, such as MeCP2, which recruit repressive HDACs (34, 45). In Xenopus extracts, replication of methylated plasmid and ORC recruitment is not rescued by inclusion of the HDAC inhibitor trichostatin A (unpublished data), suggesting that CpG methylation inhibits ORC recruitment independently of histone acetylation. However, we find that inhibition due to methylation is severely reduced by the addition of increasing amounts of methylated DNA. At low levels of plasmid DNA, the inhibition of replication is 90%. In contrast, at high concentrations of methylated DNA the replication of methylated plasmid approaches the level for unmethylated plasmid. This result suggests that egg extracts contain a limited amount of a CpG-binding protein and that the binding of this protein competitively inhibits ORC from interacting with methylated regions.
The system we describe here with Xenopus extracts might prove useful for examining other modifications, such as histone acetylation and phosphorylation, suspected to be involved is restricting where Pre-RCs can assemble in metazoan somatic cells. By using methylation as a means for targeting ORC to a specific region of DNA and thereby, in effect, creating an targeted origin of replication, we have found that neither the MCM-helicase nor initiation of replication are spatially coincident with the position of ORC. Rather, our results suggest that the MCM-helicase is loaded onto DNA, perhaps cooperatively, at a distance up to 1 kb away from ORC and that initiation of replication occurs at these distant sites. These results suggest that there may be fundamental differences in the compositional organization of origins between different eukaryotic organisms reminiscent of the variability in composition and organization of eukaryotic centromeres.
Acknowledgments
We thank Michael Leffak and John Casper for providing myc origin DNA constructs and Ryuji Yamaguchi, Zhongsheng You, and Samantha Zeitlin for helpful discussion regarding the preparation of the manuscript.
This work was supported by NIH postdoctoral fellowship F32GM20633-01 to K.J.H. and NIH grant GM33523 to J.N.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrow, M. G., M. Ritzi, A. Pemov, and J. L. Hamlin. 2002. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277:2702-2708. [DOI] [PubMed] [Google Scholar]
- 4.Antequera, F., and A. Bird. 1999. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol. 9:R661-R667. [DOI] [PubMed] [Google Scholar]
- 5.Araujo, F. D., J. D. Knox, M. Szyf, G. B. Price, and M. Zannis-Hadjopoulos. 1998. Concurrent replication and methylation at mammalian origins of replication. Mol. Cell. Biol. 18:3475-3482. (Erratum, 19:4546, 1999.) [DOI] [PMC free article] [PubMed]
- 6.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 7.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 8.Bielinsky, A. K., H. Blitzblau, E. L. Beall, M. Ezrokhi, H. S. Smith, M. R. Botchan, and S. A. Gerbi. 2001. Origin recognition complex binding to a metazoan replication origin. Curr. Biol. 11:1427-1431. [DOI] [PubMed] [Google Scholar]
- 9.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 10.Blow, J. J., and R. A. Laskey. 1986. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47:577-587. [DOI] [PubMed] [Google Scholar]
- 11.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 12.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in Saccharomyces cerevisiae. Cell 51:463-472. [DOI] [PubMed] [Google Scholar]
- 13.Campanero, M. R., M. I. Armstrong, and E. K. Flemington. 2000. CpG methylation as a mechanism for the regulation of E2F activity. Proc. Natl. Acad. Sci. USA 97:6481-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter, P. B., P. R. Mueller, and W. G. Dunphy. 1996. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature 379:357-360. [DOI] [PubMed] [Google Scholar]
- 15.Chesnokov, I., D. Remus, and M. Botchan. 2001. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. USA 98:11997-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang, R. Y., L. Chretien, J. Dai, and T. J. Kelly. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 277:16920-16927. [DOI] [PubMed] [Google Scholar]
- 17.Coleman, T. R., P. B. Carpenter, and W. G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87:53-63. [DOI] [PubMed] [Google Scholar]
- 18.Costanzo, V., K. Robertson, C. Y. Ying, E. Kim, E. Avvedimento, M. Gottesman, D. Grieco, and J. Gautier. 2000. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol. Cell 6:649-659. [DOI] [PubMed] [Google Scholar]
- 19.Dasso, M., H. Nishitani, S. Kornbluth, T. Nishimoto, and J. W. Newport. 1992. RCC1, a regulator of mitosis, is essential for DNA replication. Mol. Cell. Biol. 12:3337-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado, S., M. Gómez, A. Bird, and F. Antequera. 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 17:2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diffley, J. F., J. H. Cocker, S. J. Dowell, and A. Rowley. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303-316. [DOI] [PubMed] [Google Scholar]
- 22.Dijkwel, P. A., S. Wang, and J. L. Hamlin. 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 22:3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitrova, D. S., and D. M. Gilbert. 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2:686-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobbs, D. L., W. L. Shaiu, and R. M. Benbow. 1994. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 22:2479-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donovan, S., J. Harwood, L. S. Drury, and J. F. Diffley. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 94:5611-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards, M. C., A. V. Tutter, C. Cvetic, C. H. Gilbert, T. A. Prokhorova, and J. C. Walter. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding ORC in Xenopus egg extracts. J. Biol. Chem. 277:33049-33057. [DOI] [PubMed]
- 27.Gerbi, S. A., and A. K. Bielinsky. 2002. DNA replication and chromatin. Curr. Opin. Genet. Dev. 12:243-248. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gowher, H., O. Leismann, and A. Jeltsch. 2000. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 19:6918-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harland, R. M., and R. A. Laskey. 1980. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell 21:761-771. [DOI] [PubMed] [Google Scholar]
- 31.Hua, X. H., and J. Newport. 1998. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 140:271-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyrien, O., C. Maric, and M. Méchali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 33.Hyrien, O., and M. Méchali. 1993. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 12:4511-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 35.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawlis, S. J., S. M. Keezer, J. R. Wu, and D. M. Gilbert. 1996. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. J. Cell Biol. 135:1207-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucas, I., M. Chevrier-Miller, J. M. Sogo, and O. Hyrien. 2000. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 296:769-786. [DOI] [PubMed] [Google Scholar]
- 38.Lunyak, V. V., M. Ezrokhi, H. S. Smith, and S. A. Gerbi. 2002. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol. Cell. Biol. 22:8426-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyko, F., B. H. Ramsahoye, and R. Jaenisch. 2000. DNA methylation in Drosophila melanogaster. Nature 408:538-540. [DOI] [PubMed] [Google Scholar]
- 40.Mahbubani, H. M., J. P. Chong, S. Chevalier, P. Thömmes, and J. J. Blow. 1997. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol. 136:125-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahbubani, H. M., T. Paull, J. K. Elder, and J. J. Blow. 1992. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 20:1457-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiorano, D., J. Moreau, and M. Méchali. 2000. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404:622-625. [DOI] [PubMed] [Google Scholar]
- 43.Malott, M., and M. Leffak. 1999. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 19:5685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McWhinney, C., and M. Leffak. 1990. Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res. 18:1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 46.Nightingale, K., and A. P. Wolffe. 1995. Methylation at CpG sequences does not influence histone H1 binding to a nucleosome, including a Xenopus borealis 5S rRNA gene. J. Biol. Chem. 270:4197-4200. [DOI] [PubMed]
- 47.Rein, T., T. Kobayashi, M. Malott, M. Leffak, and M. L. DePamphilis. 1999. DNA methylation at mammalian replication origins. J. Biol. Chem. 274:25792-25800. [DOI] [PubMed] [Google Scholar]
- 48.Rippe, K. 2001. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 26:733-740. [DOI] [PubMed] [Google Scholar]
- 49.Romanowski, P., M. A. Madine, A. Rowles, J. J. Blow, and R. A. Laskey. 1996. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6:1416-1425. [DOI] [PubMed] [Google Scholar]
- 50.Rowles, A., J. P. Chong, L. Brown, M. Howell, G. I. Evan, and J. J. Blow. 1996. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell 87:287-296. [DOI] [PubMed] [Google Scholar]
- 51.Rowles, A., S. Tada, and J. J. Blow. 1999. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 112:2011-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Royzman, I., R. J. Austin, G. Bosco, S. P. Bell, and T. L. Orr-Weaver. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, I., T. Tenzen, B. E. Reubinoff, D. Hillman, J. R. McCarrey, and H. Cedar. 1999. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401:929-932. [DOI] [PubMed] [Google Scholar]
- 54.Smythe, C., and J. W. Newport. 1991. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 35:449-468. [DOI] [PubMed] [Google Scholar]
- 55.Sterner, J. M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J. M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18:2748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veenstra, G. J., and A. P. Wolffe. 2001. Constitutive genomic methylation during embryonic development of Xenopus. Biochim. Biophys. Acta 1521:39-44. [DOI] [PubMed] [Google Scholar]
- 57.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 58.Walter, J., and J. W. Newport. 1997. Regulation of replicon size in Xenopus egg extracts. Science 275:993-995. [DOI] [PubMed] [Google Scholar]
- 59.Walter, J., L. Sun, and J. Newport. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1:519-529. [DOI] [PubMed] [Google Scholar]
- 60.Waltz, S. E., A. A. Trivedi, and M. Leffak. 1996. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 24:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolffe, A. P., and M. A. Matzke. 1999. Epigenetics: regulation through repression. Science 286:481-486. [DOI] [PubMed] [Google Scholar]
- 62.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in Saccharomyces cerevisiae: high-resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]