Abstract
The single-stranded DNA- and RNA-binding protein, Purα, has been implicated in many biological processes, including control of transcription of multiple genes, initiation of DNA replication, and RNA transport and translation. Deletions of the PURA gene are frequent in acute myeloid leukemia. Mice with targeted disruption of the PURA gene in both alleles appear normal at birth, but at 2 weeks of age, they develop neurological problems manifest by severe tremor and spontaneous seizures and they die by 4 weeks. There are severely lower numbers of neurons in regions of the hippocampus and cerebellum of PURA−/− mice versus those of age-matched +/+ littermates, and lamination of these regions is aberrant at time of death. Immunohistochemical analysis of MCM7, a protein marker for DNA replication, reveals a lack of proliferation of precursor cells in these regions in the PURA−/− mice. Levels of proliferation were also absent or low in several other tissues of the PURA−/− mice, including those of myeloid lineage, whereas those of PURA+/− mice were intermediate. Evaluation of brain sections indicates a reduction in myelin and glial fibrillary acidic protein labeling in oligodendrocytes and astrocytes, respectively, indicating pathological development of these cells. At postnatal day 5, a critical time for cerebellar development, Purα and Cdk5 were both at peak levels in bodies and dendrites of Purkinje cells of PURA+/+ mice, but both were absent in dendrites of PURA−/− mice. Purα and Cdk5 can be coimmunoprecipitated from brain lysates of PURA+/+ mice. Immunohistochemical studies reveal a dramatic reduction in the level of both phosphorylated and nonphosphorylated neurofilaments in dendrites of the Purkinje cell layer and of synapse formation in the hippocampus. Overall results are consistent with a role for Purα in developmentally timed DNA replication in specific cell types and also point to a newly emerging role in compartmentalized RNA transport and translation in neuronal dendrites.
Purα is a nearly ubiquitous protein originally purified from mouse brain based on its ability to bind to the DNA fragment containing a GGCGGA sequence derived from the myelin basic protein (MBP) proximal regulatory region (21, 22). At that time, human Purα had been identified as a sequence-specific single-stranded DNA-binding protein with a potential role in DNA replication and it had been sequenced (2, 3). The sequence of mouse PURA cDNA (33) is nearly identical to that of humans (3), and the DNA binding region of the gene is strongly conserved throughout evolution. While Purα homologs are present in certain bacteria and plants, no counterparts have been detected in viruses or yeast. At present, there are four known Pur family members in humans and mice, including Purβ and two isoforms of Purγ (2, 27, 31, 32). Expression of Purα in mouse brain and its DNA binding activity are developmentally regulated and peak at 15 to 18 days after birth (46), a time at which many interneuronal connections, particularly in the cerebellum, are being established. In the brain, Purα has been reported to modulate transcription of the rat somatostatin (41) gene, the hamster neuron-specific FE65 gene (51), and the mouse BC1 gene (29), and it can interact with the promoter region of the rat nicotinic acetylcholine receptor β4 subunit (12). More recently, Purα has been revealed to be an RNA binding protein with potential effects on translation (28). Analysis of the predicted 322-amino-acid human Purα protein has revealed a modular structure with alternating 23- and 26-amino-acid repeats (3) which are important for Purα binding to DNA while other regions of the protein contribute by binding to several cellular regulatory proteins, including SP1 (47), YB-1 (42), E2F-1 (10), and pRb (25). Colocalizations of Purα with cyclin A/Cdk2 and cyclin B1/Cdk1 have been reported (1), and unpublished work suggests that these are mediated by binding to the Cdk component (S. Barr, H. Liu, and E. M. Johnson, unpublished data). While many actions of Purα occur in the nucleus, the protein is frequently located in the cytoplasm, depending at least in part upon the cell cycle phase. Purα localization to the nucleus depends on a complex set of motifs within the protein (1). The associations of Purα with pRb and colocalization with Cdk protein kinases suggest a role for Purα in control of the cell cycle and differentiation. The ability of Purα to inhibit replication of oncogenically transformed cells (1, 11) and the finding of deletions of the PURA gene in acute myelogenous leukemia (31) offer further support for such a role. Results from protein microinjection, followed by video time lapse monitoring, have provided direct evidence of Purα's role in cell cycle progression (44). More than 80% of cells injected with Purα were inhibited from passing through mitosis with cells blocked either in the G2 or G1 phase, depending on the time of injection. A mutant Purα protein lacking the Rb-binding and glutamine-rich domains had no effect upon cell cycle progression.
The ability of Purα to associate with the regulatory DNA sequence of MBP and several other cellular and viral genes led to the initial assumption that Purα may function as a transcription factor, modulating expression of genes during brain development and viral infection of brain cells (17). Purα can interact with the regulatory proteins of several viruses, including the human neurotropic JC virus (JCV) early protein, T antigen (16), and the human immunodeficiency virus type 1 (HIV-1) transactivator, Tat (18, 30), both of which are required for viral replication in infected cells of the central nervous system (CNS). Work on viruses has suggested a common mechanism in Purα interactions with proteins and nucleic acids. It was found that at low levels Purα stimulates initiation of JCV DNA replication, whereas at high levels Purα is inhibitory (9). Earlier studies have revealed that while Purα stimulates JCV early gene transcription, it suppresses the regulatory activity of T antigen when the two proteins are bound (15). The association of Purα with Tat, which is dependent on RNA molecules, increases its transcriptional activity upon the Tat-responsive HIV-1 promoters (16). In its effects on transcription of the HIV-1 genome (8) and the cellular BC1 gene (29), Purα both stimulates transcription and associates with the resulting RNA transcription product. The reported abilities of Purα to act in processes as diverse as DNA replication and RNA metabolism may now be evaluated in a mouse genetic model.
MATERIALS AND METHODS
Mice with PURA gene deletion.
Mice with targeted disruption of the PURA locus were generated by insertion of sequences coding for the selectable marker neomycin by homologous recombination in 129-derived embryonic stem (ES) cells. The targeting vector, pNTK-Purα, containing exons 1 and 2 of PURA with an insertion of a neomycin cassette resulting in the deletion of 418 nucleotides of the open reading frame immediately after the translation start site was utilized to introduce the disrupted allele into ES cells. After selection with neomycin, genomic DNA from ES cell clones was analyzed by genomic DNA blot hybridization. Cell lines containing a disrupted PURA allele were utilized for microinjection in blastocysts resulting in chimeric animals (Chrysalis/DNX). Chimeric animals were bred with PURA+/+ mice to generate animals that were PURA+/−.
Genomic DNA blotting.
Ten micrograms of genomic DNA was digested with XbaI, resolved by 1% agarose gel electrophoresis, and transferred to a nylon membrane (Hybond-N; Amersham). The blots were hybridized with 106 cpm of an [α-32P]dCTP-labeled 300-bp DNA fragment representing the 3′ end of the PURA gene in Ultrahyb (Ambion)/ml overnight at 55°C, washed with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) (two times for 6 min), and exposed to X-ray film at −70°C for 16 h.
Immunoblotting.
Immunoblotting was performed on whole-cell extract isolated from total brains of PURA−/−, PURA+/−, and PURA+/+ age-matched littermates homogenized in TNN buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, and 0.5% NP-40. The protein concentration was determined by the Bradford assay. Fifty micrograms of each extract was denatured by boiling at 95°C in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, and proteins were separated by SDS-10% PAGE and transferred to nylon-supported nitrocellulose (Hybond-P; Amersham) overnight at 4°C. Membranes were blocked in 1× Tris-buffered saline (TBS)-0.1% Tween 20 containing 10% nonfat dry milk for 1 h and incubated for 2 h or overnight in 1× TBS-0.1% Tween 20 containing 0.5% dry milk and specific primary antibodies (see below). After washing, membranes were incubated with secondary anti-mouse antibodies conjugated to alkaline phosphatase (1:10,000 dilution; Pierce) in 1× phosphate-buffered saline (PBS)-0.1% Tween 20 for 1 h. The membranes were then washed, equilibrated in 100 mM Tris (pH 9.5), incubated in CDP-Star (NEN-Perkin Elmer), and exposed to X-ray film.
Immunoprecipitation and immunoblotting.
Immunoprecipitation was performed by incubating 250 μg of whole-cell extract with 500 ng of antibody overnight at 4°C. Fifty microliters of Pansorbin (Calbiochem) was added, the samples were incubated at 4°C for 1 h, and the pellets were washed with TNN buffer. The pellets were denatured at 95°C in SDS-PAGE sample buffer and utilized for immunoblotting as described above. After immunoblotting, some membranes were stripped by incubation in buffer containing 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl (pH 6.7) for 20 min at 37°C. Following this incubation, blots were incubated with CDP-Star solution and exposed to film to ensure removal of antibodies. Then blots were equilibrated in 1× TBS-0.1% Tween 20, and immunoblotting proceeded as described above.
Immunohistochemistry.
Tissues harvested from PURA−/− mice and their age-matched littermates were fixed in formalin, embedded in paraffin, and sectioned at 4 μM for immunohistologic analysis. Tissue sections were immunolabeled by using the avidin-biotin-peroxidase complex system according to the manufacturer's instructions (Vectastain Elite, ABC peroxidase kit; Vector Laboratories). Deparaffinization of sections in xylene followed by rehydration through graded ethanol was followed by nonenzymatic antigen retrieval at 95°C in 0.01 M citrate (pH 6.0) for 30 min, after which the sections were allowed to cool until they reached room temperature. Next, sections were incubated in methanol-3% H2O2 for 20 min to quench endogenous peroxidase. For detection of Purα, MBP, glial fibrillary acidic protein (GFAP), neurofilaments, calbindin, and class III β-tubulin, the mouse-on-mouse kit (MOM; Vector Laboratories) was used immediately before blocking in 5% normal serum for 2 h in order to reduce background when detecting mouse primary antibodies in mouse tissue. After blocking, sections were incubated with primary antibodies overnight at room temperature in a humidified chamber. The secondary antibody and avidin-biotin steps were then performed according to the manufacturer's instructions. For detection of MCM7, Cdk5, and Psd95, after deparaffinization, slides were heated to 100°C in 0.1 M sodium citrate buffer (pH 6.0) and rinsed with 0.14 M NaCl and 2.7 M KCl in 6 mM sodium phosphate buffer (pH 7.4) (PBS). Sections were then incubated at room temperature for 16 h with primary antibody in PBS containing 5% nonfat dry milk, 2% bovine serum albumin, and 5% serum from the species from which the secondary antibodies were derived. Sections were then rinsed with PBS and incubated for 2 min with 5% milk in PBS. Detection was performed by using the biotinylated anti-immunoglobulins and streptavidin-conjugated peroxidase of the Super Sensitive detection kit (BioGenex) according to the instructions of the manufacturer. Sections were then treated with 0.5% Triton X-100 in PBS for 10 min. All sections were developed with diaminobenzidine, counterstained with hematoxylin, and mounted with Permount.
Antibodies.
Antibodies utilized for immunoprecipitation and immunoblotting include monoclonal mouse anti-Purα antibody developed in the Johnson laboratory (clone 10B12), mouse anti-Grb2 (BD Biosciences), and mouse anti-Cdk5 (J-3; Santa Cruz). Antibodies utilized for immunohistochemistry include mouse anti-Purα antibody (clone 10B12), mouse anti-Cdk5 (H-291; Santa Cruz), mouse anti-MCM7 (141.2; Santa Cruz), mouse anti-MBP (no. 1118099; Roche), mouse anti-GFAP (6F2; Dako), mouse anti-total neurofilament H (SMI33; Sternberger Monoclonal Antibodies), mouse anti-phospho-neurofilament (SMI312 panaxonal cocktail; Sternberger Monoclonal Antibodies), mouse anticalbindin (KR6; NovoCastra), mouse anti-class III β-tubulin (SDL.3D10; Sigma), and goat anti-Psd95 (M-18; Santa Cruz).
RESULTS
Mice with targeted disruption of the PURA gene do not survive to maturity.
To gain insight into the functions of Purα, targeted disruption of the mouse Purα gene was performed through homologous recombination in ES cells in which a portion of the mouse Purα gene was replaced with sequences coding for the selectable marker neomycin (Fig. 1A). The altered ES cells were then microinjected into fertilized mouse embryos and implanted into pseudopregnant female mice. The resulting chimeric animals were utilized to create heterozygous and homozygous animals lacking one (PURA+/−) or both (PURA−/−) alleles of the Purα gene. The genotypes of the animals were confirmed by genomic DNA blotting (Fig. 1B), and expression of Purα protein in age-matched littermates was determined by immunoblot analysis with whole-cell protein extracts prepared from the brain (Fig. 1C). As expected, the PURA−/− mice showed no evidence of production of Purα protein, as the band corresponding to Purα is absent. Of interest, the heterozygous animals (PURA+/−) produce less Purα protein than their control PURA+/+ littermates. Since changes in Purα levels of about twofold can affect cell proliferation (1), this reduced level in the PURA+/− mice suggests haploinsufficiency of the Purα protein in certain tissues.
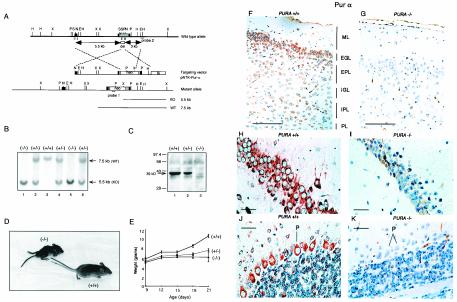
FIG. 1.
Creation of mice with targeted gene disruption of PURA, genotypic analysis, and absence of Purα protein in PURA−/− mice. (A) Strategy for inactivation of the PURA gene in ES cells. Sequences of the mouse PURA gene in ES cells were replaced with those encoding the eukaryotic selectable marker neomycin as described in Materials and Methods. The resulting alleles were 7.5 kb in size for the PURA+/+ mice and 5.5 kb for mice with the gene deletion. (B) Genomic DNA blotting demonstrates the genotype of mice which are homozygous (PURA−/−), heterozygous (PURA+/−), or wild-type (PURA+/+) for the PURA gene. (C) Immunoblot analysis of whole-cell mouse brain extract reveals a complete absence of the band representing Purα protein in the PURA−/− animal and reduced levels of Purα protein in PURA+/− animals in comparison to its level in wild-type, PURA+/+, littermates. Equal amounts of protein were loaded in each lane. (D) Homozygous PURA−/− mouse pictured next to a wild-type PURA+/+ littermate at 18 days of age. (E) Comparison of the total body weight of PURA mice between days 9 and 21 after birth. The growth of the PURA−/− mice is severely retarded while the PURA+/− mice show an intermediate phenotype. (F) Immunohistochemical analysis was performed on cortical brain sections from paraffin-embedded formalin-fixed tissue of day p19 PURA+/+ and PURA−/− mice with anti-Purα antibody. The +/+ mouse brains display abundant cytoplasmic immunoreactivity, particularly in the external granular layer (EGL). (G) Similar sections of brain tissue from PURA−/− mice exhibit no evidence of labeling; however, the cortical layers appear less cellular, the molecular layer (ML) appears broader, and the external granular layer is disorganized compared to those of the PURA+/+ mouse. EPL, external pyramidal layer; IGL, internal granular layer; IPL, internal pyramidal layer; PL, polymorphic layer. (H) The majority of hippocampal neurons in the horn of Ammon show cytoplasmic labeling for Purα in PURA+/+ mice. (I) A similar section within the hippocampus of PURA−/− mice shows an absence of immunoreactivity when labeled for Purα. (J) Cytoplasmic expression of Purα is seen in some cerebellar granular cells and all Purkinje cells (P) in PURA+/+ mice. (K) PURA−/− mice show an absence of labeling for Purα in the cerebellum. The granular and Purkinje cell layers are less populated than those of the PURA+/+ littermate. Panels F through K have a hematoxylin counterstain. Bars, 100 μm (F and G), 10 μm (H and I), and 20 μm (J and K). WT, wild type; KO, knockout.
Phenotypically, the PURA−/− mice are distinguishable from their PURA+/+ and PURA+/− littermates when they become mobile and leave the nest. Subtle changes in the mice such as tremor upon motion, infrequent mobility, slack posture, flaccid tail, and a waddling gait are first observed at approximately postnatal day 14 (p14). From the second week of life, PURA−/− mice are easily distinguished from their PURA+/+ age-matched littermates, as the animals do not gain weight normally. Figure 1D depicts a PURA−/− mouse and its PURA+/+ littermate at day p18, and Fig. 1E shows weight curves of PURA+/+, PURA−/−, and PURA+/− litters from day p9 to p21. The mice appear to be feeding properly, as milk can be found in the stomach of nursing animals and the older animals are observed to be eating rodent chow alongside their littermates. The severity of these features progressively increases from day 14 until death, which occurs between 18 and 28 days of age. None of the PURA−/− mice survived beyond 1 month. The PURA+/− animals display a delay in development similar to that of their PURA−/− siblings, with significantly reduced severity, although, by adulthood, the appearance of the PURA+/− animals is nearly indistinguishable from that of the PURA+/+ littermates. One exception is that the PURA+/− animals are seen to undergo occasional spontaneous seizures, usually following routine handling of the animals during cage changes.
Histopathological analysis was performed on the major organs of the body, including the brain, spinal cord, heart, kidney, liver, lung, spleen, gonads, thymus, pancreas, and gut in littermates of various ages. Examination of formalin-fixed paraffin-embedded cortical brain sections from wild-type (PURA+/+) and homozygous (PURA−/−) littermates showed distinct changes in the brain architecture of PURA−/− mice. As shown in Fig. 1, at 19 days after birth, strong cytoplasmic immunoreactivity upon incubation with anti-Purα antibody was seen in neurons of all six layers of the cortex, particularly the external granular layer in the PURA+/+ mouse (Fig. 1F). However, no labeling of neurons is evident in the PURA−/− mouse (Fig. 1G). In addition, the cortex of the PURA−/− mouse has fewer cells; reduced numbers of neurons overall can be seen and the molecular layer appears much broader while the external pyramidal layer appears not to be organized into a typically more compacted layer (Fig. 1G). Neurons in the hippocampus of PURA+/+ mice show robust cytoplasmic immunoreactivity to Purα in the majority of the cells within the horn of Ammon while labeling for Purα was noticeably absent in the PURA−/− mice (compare Fig. 1H and I). Likewise, in the cytoplasm of PURA+/+ mice, neurons in the cerebellar granular layer can be visualized positive for Purα, and the majority of the Purkinje cells show evidence of cytoplasmic immunoreactivity, whereas the PURA−/− mice are negative (compare Fig. 1J and K). Interestingly, reduced numbers of neurons can be discerned in both the cerebellar granular layer and the Purkinje cell layer of the PURA−/− mice (Fig. 1K and Table 1).
TABLE 1.
Reduced levels of total and neurofilament-positive neurons in PURA−/− micea
| Day and PURA mouse type | Cerebral cortex
|
Hippocampus
|
Cerebellum
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total no. of neuronsb | No. of NF-positive neuronsc | NF labeling index (%) | Total no. of neuronsd | No. of NF-positive neuronse | NF labeling index (%) | Total no. of neuronsf | No. of NF-positive neuronsg | NF labeling index (%) | |
| 17 | |||||||||
| +/+ | 3,103 | 2,779 | 90 | 995 | 904 | 91 | 332 | 298 | 90 |
| −/− | 1,655 | 206 | 12 | 781 | 164 | 21 | 189 | 29 | 15 |
| 19 | |||||||||
| +/+ | 3,522 | 3,324 | 94 | 1,216 | 1,081 | 89 | 384 | 336 | 88 |
| −/− | 2,074 | 293 | 14 | 892 | 133 | 15 | 223 | 53 | 24 |
Calculation of numbers of neurons and neurofilament (NF) labeling index are as described in Materials and Methods.
Reductions of 47% on day 17 and 41% on day 19.
Reductions of 93% on day 17 and 91% on day 19.
Reductions of 22% on day 17 and 27% on day 19.
Reductions of 82% on day 17 and 88% on day 19.
Reductions of 43% on day 17 and 42% on day 19.
Reductions of 90% on day 17 and 84% on day 19.
PURA−/− mice show a decrease in cells undergoing DNA replication in multiple organs compared with PURA+/+ littermates.
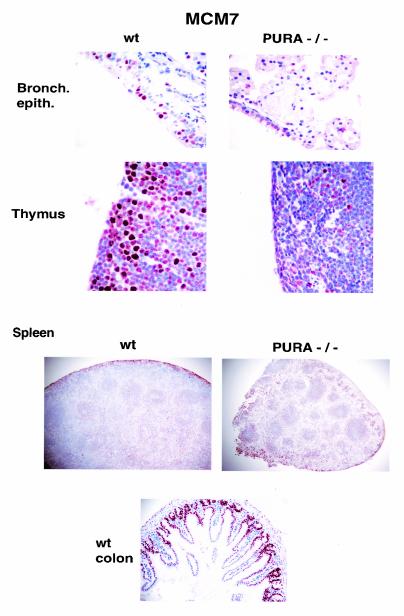
Detailed postmortem analysis of mouse tissues revealed that the organs of PURA−/− mice, with the exception of the brain, were smaller in size and weight than those of PURA+/+ mice. For example, the spleens appeared much smaller than those of the PURA+/+ mice (Fig. 2), and their average weights, presented in Table 2, were greatly reduced from 29.2 mg in PURA+/+ mice to 5.95 mg in the PURA−/− mice. We employed MCM7 as a protein marker for cell proliferation. MCM7 is reasonably well characterized. This protein is a component of a complex essential for initiation of DNA replication in eukaryotic cells (4, 39, 49), and its presence in nuclei is highly specific for the onset of DNA synthesis (13, 39, 43). MCM7 has previously been used as a marker for cell proliferation and as a diagnostic marker for malignant cells (14). Nuclear localization of MCM7 is highly specific for proliferating cells in S phase (14). Further evaluation of the spleen and other lymphoid tissues such as the thymus revealed fewer numbers of cells positive for MCM7 than in organs from PURA+/+ mice (Fig. 2), indicating a reduction in the number of proliferating cells. In addition to lymphoid organs, we examined nonlymphoid organs with highly proliferative epithelial cells for labeling with MCM7. As a positive control, it can be seen that MCM7 specifically labels nuclei of cells at the bases of colon crypts, which are the locations of proliferating cells in the normal colon (Fig. 2, lower panel). The bronchiolar epithelium and regions of the thymus and spleen of PURA−/− mice all exhibited reduced nuclear labeling with anti-MCM7 antibody when compared with normal tissues (Fig. 2). Furthermore, these changes were quantitated in the spleen, thymus, and peripheral blood, as summarized in Table 2. The total percentage of MCM7 positive cells within the thymic cortex showed a reduction of nearly 85% while the white and red pulp of the spleen showed reductions of 26 and 93%, respectively. The dramatic decrease in the red pulp is of note since this is where myeloid cells develop. It should be noted that the regions of the thymus and spleen where dramatic reductions in MCM7 positive cells are seen, namely the thymic cortex and the splenic red pulp, represent regions of these organs where cellular replication normally should be most active. Similar results to those obtained by labeling for MCM7 in various tissues were obtained by immunolabeling with anti-Ki67 antibody, another frequently used marker of proliferation (data not shown).
FIG. 2.
Decrease in number of cells undergoing DNA replication in a variety of tissues from PURA−/− mice. Paraffin-embedded tissues from littermate PURA+/+ and PURA−/− mice, taken at day 19 after birth, were sectioned, treated with anti-MCM7 antibody, and visualized as described in Materials and Methods. MCM7 is visualized as red-brown. The control colon section at the bottom confirms that cells undergoing DNA replication, at the bases of crypts, possess nuclei positively labeling for MCM7 while cells not replicating DNA, at the tops of crypts, do not. Bars, 50 μm. A hematoxylin counterstain was used in all panels. wt, wild type.
TABLE 2.
Quantitative changes in blood and lymphoid systems in PURA genetically altered mice
| PURA mouse type | No. of blood reticulocytesa | Spleen mass (mg) | % Replicating cellsb in:
|
||
|---|---|---|---|---|---|
| Thymic cortex (lymphoid) | Spleen, white pulp (lymphoid) | Spleen, red pulp (myeloid) | |||
| +/+ | 208 ± 27 | 29.2 ± 1.6 | 54.3 ± 2.5 | 11.56 ± 0.71 | 34.4 ± 1.74 |
| +/− | 184 ± 3.5 | 9.27 ± 1.0 | 13.55 ± 3.68 | 27.3 ± 8.06 | |
| −/− | 86 ± 14 | 5.85 ± 0.7 | 8.3 ± 09 | 8.53 ± 1.78 | 3.17 ± 0.66 |
Mean number of cells per 10 high-power microscopic fields ± standard deviation.
Percentage of cells staining positive for nuclear MCM7.
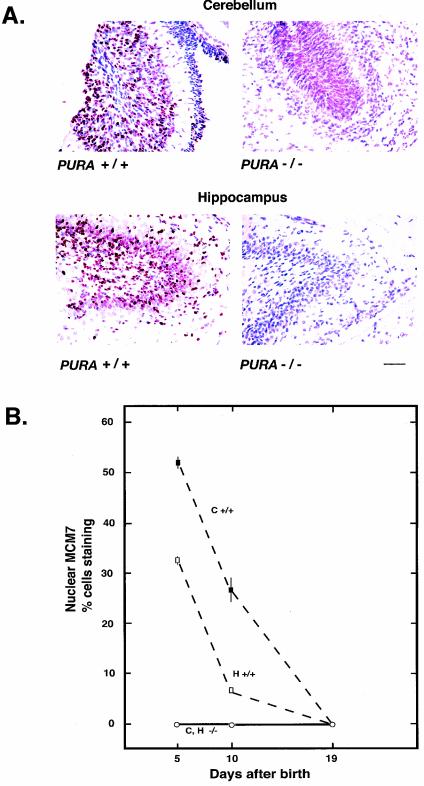
Decreased cellular proliferation in the cerebellum and hippocampus of PURA−/− mouse brains during development.
Examination of the granular layer of the cerebellum and the hippocampus of PURA−/− mice showed dramatic reductions in the percentage of MCM7-positive nuclei at day p10, as shown in Fig. 3A. During development, MCM7 labeling in PURA+/+ mice is high at day p5 and gradually reduces by p19; however, in PURA−/− littermates, levels of nuclear MCM7 labeling are extremely low at all time points studied (Fig. 3B). In addition, the data as presented in Table 2 and Fig. 3B provide further evidence of haploinsufficiency, as spleen, thymus, and brain tissue labeling with anti-MCM7 antibody shows percentages intermediate between those observed in PURA+/+ and PURA−/− mice. Overall results with anti-MCM7 raise the possibility that Purα may be affecting MCM7 expression. Therefore, levels of MCM7 protein were assayed by immunoblotting similar amounts of protein from brain extracts of PURA+/+ and PURA−/− mice taken at different times after birth (data not shown). No differences were seen in MCM7 expression levels. Therefore, any effect of Purα on DNA replication is not due to an effect on MCM7 expression.
FIG. 3.
Absence of cells undergoing DNA replication in the cerebellum and hippocampus of PURA−/− mice from 5 to 10 days after birth. Paraffin-embedded sections of brains from littermate PURA+/+ and PURA−/− mice, taken at day p10, were treated with antibody to MCM7, a marker for DNA replication, and visualized as described in Materials and Methods. (A) Sections from the most dorsal lobe of the cerebellum (top) and the horn of Ammon of the hippocampus (bottom). Bar, 50 μm. (B) Brains were obtained from littermate mice at 5, 10, and 19 days after birth. Microscopic fields from four sections from each time point labeled as described above, at ×200 magnification, were subjected to counting of total cell number and number of cells with nuclei labeling positive for MCM7. Presented are the averages and standard deviations (vertical bars) of the percentages of cells positively labeled for MCM7 in the cerebellum (C) and hippocampus (H) for each time point for either wild-type mice (+/+) or mice with genetically inactivated PURA (−/−).
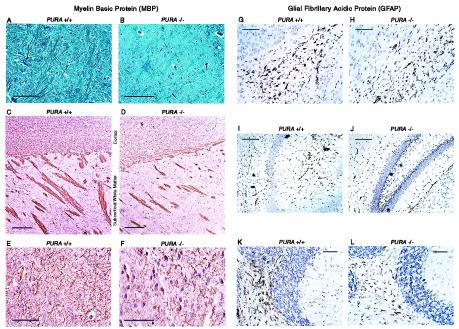
Glial cells in PURA−/− brains show reduced labeling for MBP and GFAP in white matter of the brain.
It is known that Purα can affect MBP gene transcription and can upregulate late gene transcription of JCV, a virus involved in demyelination. We therefore examined oligodendrocytes, the myelin-producing cells in the CNS, and their neighboring astrocytes. Myelin tracts were analyzed by luxol fast blue staining and immunohistochemistry with the oligodendrocyte-specific protein MBP. As shown in Fig. 4A and B, myelin tracts (appearing dark blue) were more pronounced in PURA+/+ mice than in their PURA−/− littermates. Similarly, immunohistochemical labeling for MBP shows reduced white matter tracts in the subcortical white matter and in the cortex of PURA−/− mice compared with controls (Fig. 4C through F). However, electron microscopy was performed on sections of the spinal cord and optic nerve which revealed the presence of myelin sheaths, and immunoblotting for MBP in cellular lysates prepared from whole brains did not reveal changes in MBP expression (data not shown). It is possible that MBP expression may be altered in certain regions of the brain during development, but these differences may not be sufficient to be detectable in extracts prepared from whole brains.
FIG. 4.
Immunohistologic analysis of oligodendrocytes and astrocytes demonstrates a decrease in intensity of myelin and MBP immunolabeling as well as a reduction in GFAP expression in white matter tracts in the absence of Purα. Paraffin-embedded sections of brain cerebral cortex from day p19 PURA+/+ and PURA−/− mice were stained for myelin and immunolabeled for the oligodendrocyte-specific cellular marker MBP. Myelin staining patterns in the cerebral cortex and subcortical white matter of mice lacking Purα appeared less intense when compared with those of PURA+/+ littermates (A and B), and immunohistochemistry to detect MBP revealed similar patterns in the cortex and subcortical white matter tracts (C and D). Similar evaluation for the astrocytic marker GFAP revealed less-intense labeling of cellular processes of PURA−/− mice in the cortex (G and H), white matter adjacent to the hippocampus (I and J), and white matter adjacent to the granular layer of the cerebellum (K and L). Bars, 100 μm (A to F, K, and L) and 10 μm (G to J).
In parallel, labeling of the astrocyte-specific structural protein GFAP was performed in paraffin-embedded sections. Patterns similar to those observed with MBP labeling were observed, in which the labeling intensity and number of positive cells appeared slightly reduced in the PURA−/− mice in comparison to sections from matched PURA+/+ littermates of the subcortical white matter (Fig. 4G and H), hippocampus (Fig. 4I and J), and underlying white matter of the cerebellum (Fig. 4K and L).
Immunohistochemical analysis reveals reduction in the number of neurons and reduction in neurofilament labeling in the cerebellum, cortex, and hippocampus of PURA−/− mice.
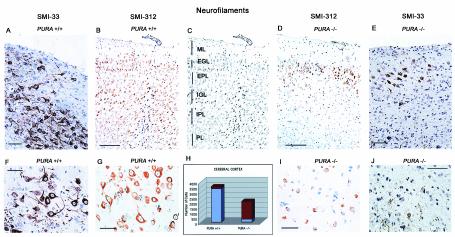
Next, we focused on neurons in specific regions of the brain by labeling with anti-neurofilament antibodies. Purα has been shown to associate with cyclin/Cdk complexes, and it is known that Cdk5 can phosphorylate neurofilaments. For this reason, we also labeled the brain sections with antibodies which recognize phosphorylated neurofilament proteins. Areas of the cerebral cortex, from age-matched PURA−/− and PURA+/+ littermates at day p19 were incubated with antibodies recognizing neurofilaments independent of phosphorylation (SMI33) (Fig. 5A, F, E, and J) or a cocktail specifically recognizing phosphorylated neurofilaments (SMI312) (Fig. 5B, G, D, and I). In the cortex of PURA+/+ mice, intense cytoplasmic immunoreactivity can be observed in neurons of all cortical layers with both antibodies while the number of neurons labeling positive in the PURA−/− mice was reduced for total neurofilaments as well as phosphorylated neurofilaments (compare Fig. 5A, B, F, and G with D, E, I, and J). Overall, levels of total neurons were reduced by approximately 41% in the cortex, and the level of phosphorylated neurofilaments dropped from 94 to 14% (compare Fig. 3A and B to D and E; also Fig. 3H and Table 1). Thus, while there are fewer total neurofilaments, further work will be necessary to determine if there is less phosphorylation of the neurofilaments present.
FIG. 5.
Reduction in number of neurons and in neurofilament-labeled neurons in the cortex of PURA−/− mice. Paraffin-embedded sections of cerebral cortex from day p19 PURA+/+ and PURA−/− littermates were immunolabeled with an antibody which recognizes total neurofilaments (SMI33) or a cocktail of antibodies specific for phosphorylated neurofilaments (SMI312). Intense immunopositivity of total (A and F) and phosphorylated (B and G) neurofilaments was observed in the PURA+/+ mice while reduced labeling of total (D and I) and phosphorylated (E and J) neurofilaments was seen in the PURA−/− mice. Panel C depicts the location of the six cortical layers, including the molecular layer (ML), external granular layer (EGL), external pyramidal layer (EPL), internal granular layer (IGL), internal pyramidal layer (IPL), and polymorphic layer (PL). As summarized in panel H, 20 microscopic fields from sections of two different mice labeled as described above, at ×200 magnification, were subjected to counting of total cell number and number of cells with positive cytoplasmic expression of phosphorylated neurofilaments using antibody SMI312. Presented are the average total number of neurons counted (red) and the fraction of those cells positively labeled for phosphorylated neurofilaments (blue). These cell counts expressed as percentages of neuronal loss and a calculated neurofilament labeling index are presented in Table 1. All panels contain a hematoxylin counterstain. Bars, 50 μm (A and E), 100 μm (B and D), and 20 μm (F, G, I, and J).
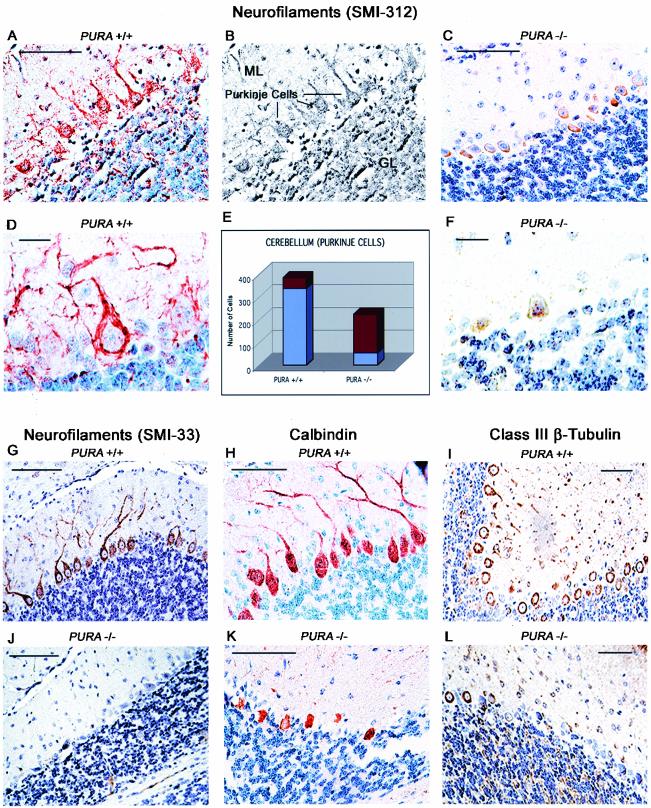
Within the cerebellum of PURA+/+ mice, labeling for total and phosphorylated neurofilaments was abundant throughout the foliae (Fig. 6A, D, and G). The molecular layer, Purkinje cell layer, and granular layer demonstrate strong nuclear immunoreactivity of phosphorylated neurofilaments of neurons (regions of the cerebellum are shown in Fig. 6B, where the molecular layer, Purkinje cells, and granular layer are depicted). In the granular and molecular layers, these neurofilaments are present in the majority of the cells, and strong cytoplasmic reactivity is detected in 9% of Purkinje cell bodies and within their processes (Fig. 6A and G). The molecular layer is known to specifically contain the dendrites of the Purkinje cells, and dendritic labeling for neurofilaments is prominent in the PURA+/+ mouse. However, in the PURA−/− mice, considerably fewer neurons are observed in all layers of the cerebellum, and immunoreactivity for phosphorylated neurofilaments is reduced from 88% of Purkinje cells to approximately 24% (Fig. 6C and F; also Fig. 6E and Table 1). Dendritic labeling is markedly reduced. Also, Purkinje cells of the PURA−/− mice were smaller in size than those of PURA+/+ mice and showed relatively little reactivity to total neurofilament antibody in the nucleus, cytoplasm, or within processes (compare Fig. 6G and J). For ease of identification, Purkinje cells were immunolabeled to detect the Purkinje marker protein, calbindin, and the neuronal marker, class III β-tubulin, which reveals their smaller size as well as their smaller numbers in the PURA−/− mice (compare Fig. 6H and I with K and L, respectively).
FIG. 6.
Reduction in number of neurons and neurofilament-labeled neurons in the cerebellum of PURA−/− mice. Wild-type mice display abundant immunolabeling of phosphorylated neurofilaments within the molecular, Purkinje, and granular layers (A and D), whereas in PURA−/− mice, this expression is severely reduced in all layers of the cerebellum (C and F). Panel B depicts the location of the molecular layer (ML), internal granular layer (IGL), and Purkinje cell layers of the cerebellum. As summarized in panel E, the total number of Purkinje cells and the number of Purkinje cells with positive labeling for phosphorylated neurofilaments were determined throughout a section containing all five foliae of thecerebellums (represented across approximately 50 microscopic fields) of two different mice labeled with antibody SMI312, at ×200 magnification. Presented are the average total number of neurons counted (red) and the fraction of those cells positively labeled for phosphorylated neurofilaments (blue). These cell counts expressed as percentages of neuronal loss and a calculated neurofilament labeling index are presented in Table 1. Immunolabeling with antibody recognizing total neurofilaments shows that Purkinje cell neurons are devoid of expression in the PURA−/− mice, whereas the Purkinje layer is intensely labeled in the PURA+/+ mice (G and J). The integrity of the Purkinje cells in the PURA−/− mice was examined by immunolabeling for the Purkinje cell marker calbindin or the neuronal cell marker class III β-tubulin. PURA−/− brain sections labeled with anticalbindin antibody demonstrate Purkinje cells with smaller and irregularly shaped cell bodies in reduced numbers compared with their PURA+/+ littermates (compare panels H and K). Immunohistochemistry for class III β-tubulin reveals robust cytoplasmic positivity of all Purkinje cells in sections of the PURA+/+ mouse brain while few Purkinje cells in the PURA−/− mice are positively labeled (I and L). All panels contain a hematoxylin counterstain. Bars, 100 μm (A, C, G, and H to L) and 10 μm (D and F).
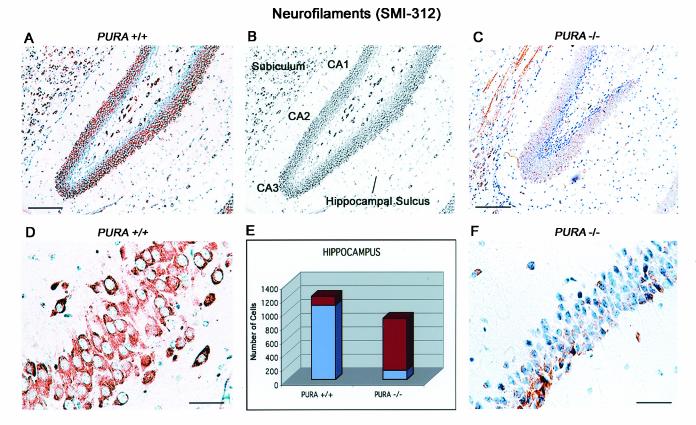
Immunolabeling for phosphorylated neurofilaments within the hippocampus of PURA+/+ and PURA−/− littermates reveals nearly 88% of the neurons within the horn of Ammon of PURA+/+ mice to be labeled with antibody (Fig. 7A and D), whereas in the PURA−/− mice, only 42% of neurons are seen in the horn, with only 24% of the neurons positive for phosphorylated neurofilaments (Fig. 7C and F; also Fig. 7E and Table 1). In particular, limited reactivity can be observed at CA3 in the PURA−/− mice (Fig. 3F). Thus, the cortex, hippocampus, and cerebellar Purkinje cells all demonstrate a reduction in total numbers of neurons as well as a reduction in the percentage of such cells which express neurofilaments. It should be noted that we also carefully examined brain sections for evidence of neuronal loss through programmed cell death. However, evaluation by light microscopy and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling revealed no signs of apoptotic bodies or fragmented DNA (data not shown), indicating that a lack of proliferation, rather than enhanced apoptosis, is responsible for the reduction of neurons observed in this mouse model.
FIG. 7.
Reduction in numbers of neurons and neurofilament-labeled neurons in the hippocampus of PURA−/− mice. Immunohistochemistry of sagittal sections with an antibody which recognizes phosphorylated forms of neurofilaments reveals intense cytoplasmic labeling in the majority of neurons within the horn of Ammon in PURA+/+ mice (A and D). Parallel immunolabeling of sections of PURA−/− mice revealed a reduction in the total number of neurons as well as a dramatic decrease in the number of neurons which are positive for neurofilaments (C and F). Panel B depicts the location of hippocampal landmarks, including the subiculum (S), hippocampal sulcus (HS), and the CA1, CA2, and CA3 regions. As summarized in panel E, the total number of hippocampal neurons and the number of hippocampal neurons showing cytoplasmic immunoreactivity with the phospho-specific neurofilament antibody SMI312 were counted throughout a section of the entire hippocampus (approximately 50 microscopic fields) from sections of two different mice labeled as above, at ×200 magnification. Presented are the average total number of neurons counted (red) and the fraction of those cells positively labeled for phospho-neurofilaments (blue). These cell counts expressed as percentages of neuronal loss and a calculated neurofilament labeling index are presented in Table 1. Bars, 100 μm (A and C) and 10 μm (D and F).
Purα and Cdk5 coimmunoprecipitate from whole-cell extracts from normal mouse brain.
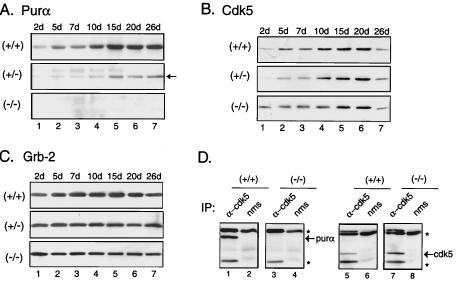
Purα levels were observed during development by immunoblot analysis of brain whole-cell extract from PURA+/+ and PURA−/− age-matched littermates at days p2, p5, p7, p10, p15, p20, and p26. As shown in Fig. 8A, Purα levels peak at approximately day p15 and remain at high levels in the PURA+/+ mice. As predicted, in mice with a targeted disruption of PURA, no protein can be detected. In PURA+/− animals, decreased levels of Purα protein are detectable and also peak at day p15, though much lower levels are seen than in the PURA+/+ mice. As mentioned above, Purα has been shown to associate with cyclin/Cdk complexes, and phosphorylation of neurofilaments is mediated by Cdk5 and its associated 35-kDa brain-specific protein (37, 45). We therefore compared the levels of Cdk5 in whole-cell protein extracts from PURA −/− and PURA+/+ mouse brains. A peak at day p15 was observed in all extracts, although total amounts of Cdk5 decreased by day p26 in all mice (Fig. 8B). However, some differences were detected at earlier time points. In the PURA+/+ mice, Cdk5 levels begin to rise at day p5, while Cdk5 levels are lower at days p5 and p7 in the PURA+/− mice than those observed in the PURA+/+ mice. Interestingly, levels of Cdk5 protein were higher at days p2 to p7 in the PURA−/− mice than those observed in the PURA+/+ and PURA+/− mice. The membranes used for Purα and Cdk5 immunoblotting were also stripped and reprobed with antibody to the nonspecific protein Grb2 as a control for equal protein loading, an example of which is shown in Fig. 8C.
FIG. 8.
Purα and Cdk5 coimmunoprecipitation and immunoblot analysis for levels of Purα and Cdk5 proteins during development in PURA+/+, PURA+/−, and PURA−/− mice. Immunoblotting of whole-cell extracts from mouse brain at 2, 5, 7, 10, 15, 20, and 26 days (d) postnatal shows a gradual increase in Purα levels during development (A). Reduced levels following a similar pattern of increase are seen in extracts from PURA+/− mice while the Purα protein is undetectable at any time point in PURA−/− mice (A). Levels of Cdk5 protein in the same extracts show a gradual increase during development which peak at day 20 in PURA+/+ mice, whereas extracts from PURA+/− mice show reduced amounts of Cdk5 protein at 2, 5, and 7 days compared to PURA+/+ littermates (B). While Cdk5 protein levels in PURA−/− mice appear greater at 2 days of age and are slightly reduced compared with PURA+/+ mice at day 10, levels peak at day 20 as seen in the PURA+/+ mice (B). For each gel shown, equal amounts of lysate were loaded in each lane. Immunoblotting for an unrelated protein, Grb2, is shown as a control for equal loading (C). Immunoprecipitation (IP) performed on whole-cell extracts from 26-day-old mice using anti-Cdk5 antibody or nms followed by immunoblotting with anti-Purα antibody demonstrates the association of Cdk5 and Purα proteins in extracts prepared from PURA+/+ but not PURA−/− mouse brains (D, left). The same blots, stripped and reblotted with anti-Cdk5 (α-cdk5) antibody, show the detection of Cdk5 protein in extracts from PURA+/+ and PURA−/− mouse brains but not in nms (D, right).
Since similar developmental patterns were seen for Purα and Cdk5 proteins and some alterations in Cdk5 levels were observed in the PURA+/− and PURA−/− mouse brain extracts, in the next experiment we performed immunoprecipitation of whole-brain extracts from mice at day p26 with antibody to Cdk5 followed by immunoblotting with antibody to Purα. As shown in Fig. 8D, a band corresponding to Purα was detected in immunocomplexes obtained by immunoprecipitation with anti-Cdk5 antibody from PURA+/+ mouse brain extracts but not in samples immunoprecipitated with serum from nonimmunized mice (nms). As expected, no band corresponding to Purα was obtained upon analysis of the Cdk5 complex in the extracts from the PURA−/− mice. These blots were stripped and reprobed with anti-Cdk5 antibody. These results illustrate the association of Purα with Cdk5 in mouse brain extract.
Cerebellar deficits and cellular localization of Purα protein, Cdk5, and neurofilaments at days p5 and p10 in PURA−/− mice.
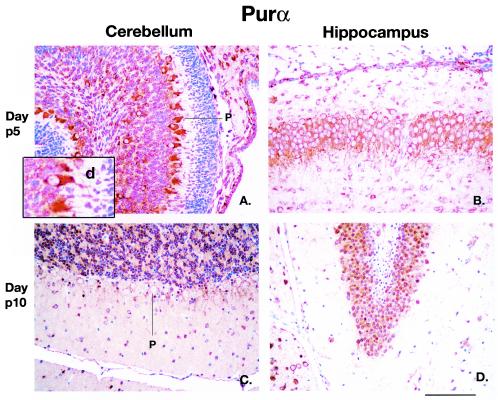
Immunohistochemistry labeling of cerebellar and hippocampal sections from day p5 normal mouse brains (Fig. 9A and B, respectively) show Purα localized in the cell bodies of Purkinje cells and granular cells of the cerebellum as well as the cytoplasm of hippocampal neurons at day p5. Note the appearance of labeling in the Purkinje cell dendrites in Fig. 9A. However, at day p10, Purα labeling is absent in the cerebellar Purkinje cells and is now present in the nuclei of cerebellar granular cells and hippocampal neurons (Fig. 9C and D, respectively). Sections of PURA−/− mice revealed an absence of labeling in all cells (data not shown).
FIG. 9.
Localization of Purα protein in cells of the cerebellum and hippocampus of wild-type PURA+/+ mice at postnatal days 5 and 10. Sections of paraffin-embedded brain tissue were treated with anti-Purα antibody and visualized as described in Materials and Methods. Sections taken at day p5 from the cerebellum and hippocampus are shown in panels A and B, respectively. The inset in panel A shows a higher magnification in which labeling with anti-Purα antibody in Purkinje cells can be visualized in the dendrites (d). Sections taken at day p10 from the cerebellum and hippocampus are shown in panels C and D, respectively. Cells in the cerebellar Purkinje cell layer, intensely labeling at day p5 but not at day p10, are labeled P. The darker brown labeling for Purα in panels C and D results from largely nuclear labeling. All panels are at the same magnification. Bar, 100 μm (D).
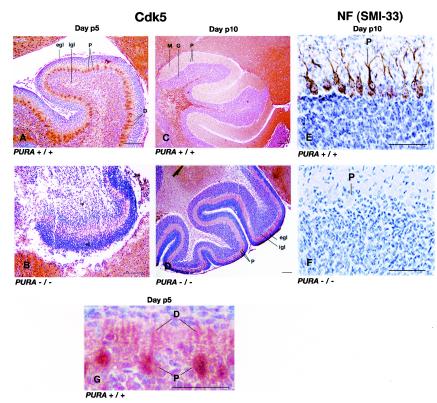
Immunohistochemistry was performed on parallel sections of cerebellum from PURA+/+ and PURA−/− mice with antibodies specific for Cdk5 and total neurofilaments (SMI33) at day p5 or p10. Robust immunolabeling for Cdk5 was observed in the Purkinje cells at day p5 while this labeling disappears by day p10 in the wild-type mice (Fig. 10A and C). The Cdk5 labeling in PURA+/+ mice extends into the Purkinje cell dendrites (Fig. 10G). Note that in PURA+/+ mice, the external granular layer is present at day p5, the Purkinje cell layer is formed, and the internal granular layer has begun to organize (Fig. 10A). By day p10, the external granular layer has disappeared in PURA+/+ mice, the molecular layer has appeared, the Purkinje cell layer is still present, and the granular layer is well organized (Fig. 10C). Sections from PURA−/− mice at days p5 and p10 were incubated in parallel (Fig. 10B and D). A lack of cerebellar organization and lack of intense labeling in the cells with anti-Cdk5 were observed at days p5 and p10. The external granular layer and internal granular layer are still present in the day p10 PURA−/− mouse cerebellum. Sections labeled for total neurofilaments at day p10 from PURA+/+ and PURA−/− mice show the presence of strong cytoplasmic positivity of cell bodies and dendrites in the PURA+/+ mice while this labeling is absent in the PURA−/− mouse sections. Thus, it appears that the absence of Purα correlates with a lack of Cdk5 and neurofilament labeling and improper cerebellar organization during development.
FIG. 10.
Defects in Cdk5 localization and neurofilament development in cerebellar Purkinje cells of mice with genetically inactivated PURA. Paraffin sections of mouse brains taken at the indicated postnatal times were treated with antibodies for either Cdk5 (A to D and G) or with SMI33, an antibody which recognizes total neurofilaments (E and F), and visualized as described in Materials and Methods. Sections from wild-type PURA+/+ mice taken at days p5 and p10 are shown in panels A and C, respectively. Labeled at day 5 are the external granular layer (egl), internal granular layer (igl), and Purkinje cell layer (P) as well as the Purkinje cell dendritic layer (D). Note the intense immunolabeling of the Purkinje cells with anti-Cdk5. At day p10 the external granular layer is not present, and the molecular (M) and granular layers have developed but the Purkinje cells at day p10 are devoid of Cdk5. Sections from PURA−/− mice at days p5 and p10 are shown in panels B and D, respectively. Note the lack of cerebellar organization and lack of intense immunolabeling of cells with anti-Cdk5 at days p5 and p10. Labeled are the external and internal granular layers, which are still observed in the day p10 PURA−/− mouse cerebellum. Sections labeled for total neurofilaments at day p10 from PURA+/+ and PURA−/− mice are shown in panels E and F, respectively. Cells corresponding to the Purkinje cell layer are labeled P. Purkinje cells (P) labeled with anti-Cdk5 are also shown at a higher magnification labeling in the PURA+/+ mouse at day p5, showing labeling extending to the dendrites (D). Bars, 100 μm.
Defective synapse formation in the hippocampus of the PURA−/− mouse.
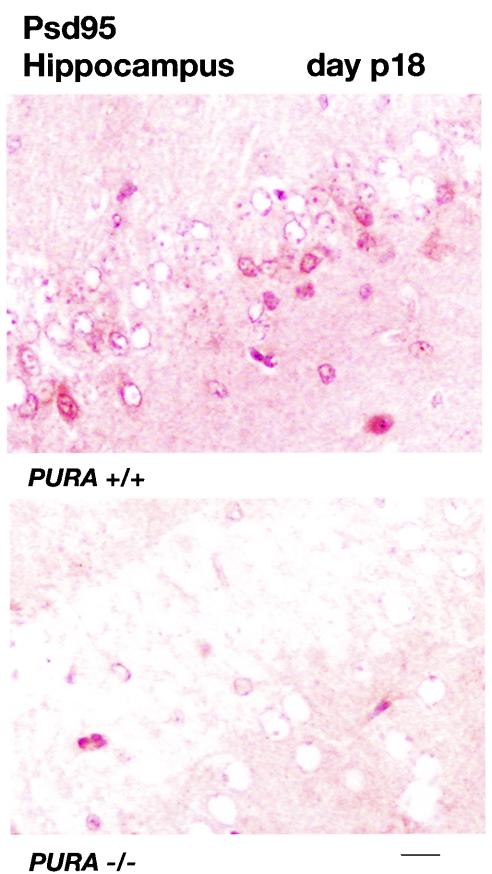
To examine functional consequences of the absence of Purα in the brain, sections of hippocampus from the CA3 region from mice at day p18 were incubated with antibody specific for the postsynaptic density protein 95 (Psd95). Psd95 has previously been employed as a marker to visualize synapses in the CA3 region of the hippocampus (38). Synapses are visualized as dense foci of Psd95 at the outer membranes of neurons. To facilitate visualization of these foci, no counterstain was employed in the experiments shown in Fig. 11. As seen in Fig. 11, immunolabeling in the PURA−/− mice (lower panel) is reduced compared with sections from the PURA+/+ mice (upper panel), indicating a reduction in the formation of synapses in the absence of Purα. By counting foci in multiple high-powered fields of similar sections of the hippocampus in PURA+/+ and PURA−/− mice, it could be estimated that synapse formation in the PURA−/− hippocampus is reduced by approximately 69%.
FIG. 11.
Defective synapse formation in the hippocampus of the PURA−/− mouse. Brain sections from littermate PURA+/+ (upper panel) and PURA−/− (lower panel) mice at day p18 were treated with antibody specific for the postsynaptic density protein 95 (Psd95) and visualized as described in Materials and Methods. No counterstain was employed. Synapses on neuronal cell bodies can be visualized within the CA3 region as intensely labeling foci at the outer cell membranes. Bar, 10 μm.
DISCUSSION
Purα is a sequence-specific DNA- and RNA-binding protein with local helix-unwinding capacity (for reviews, see references 17 and 24). Purα has been detected in virtually all mammalian tissues thus far examined. Studies of functions of this protein over the last few years have revealed three major areas in which its specific interaction with nucleic acids could be important. First, Purα was initially identified through its association with a sequence element present in initiation zones of DNA replication, and it has been demonstrated to interact with several proteins to influence replication, cell cycle progression, and oncogenic transformation (17, 24). Recently, deletions of the PURA gene have been recorded at a high frequency in myelodysplastic syndrome and acute myelogenous leukemia (31). Second, Purα is a transcription factor reported by many laboratories to regulate transcription of a variety of genes through interaction with promoter sequences. In particular, Purα has been reported to regulate both viral (6, 8, 30) and cellular (22, 47, 48) genes in human glial cells. Third, Purα has been reported to bind to RNA transcripts of certain genes under its control and to subsequently alter transcriptional elongation (8), intracellular transport (36), or mRNA translation (17, 36). In particular, Purα reportedly binds to an RNA element involved in transport to neuronal dendrites (26, 29) and influences compartmentalized translation in dendrites (24, 36). The present analyses of PURA gene inactivation shed considerable light on each of these putative Purα functions.
The link between Purα activities and regulation of DNA replication is now strengthened, and its role in development is highlighted. In the PURA−/− mice there is virtually no DNA replication in subsets of neuronal precursor cells in the hippocampus and cerebellum as revealed by lack of nuclear MCM7, a protein essential for initiation of DNA replication in the S phase. Our results indicate a high rate of replication in these cells in the PURA+/+ mice at day p5, and previous reports note a high rate of proliferation of these cells in the cerebellar cortex between days p3 and p10 (20, 40). Similar aberrant absence or reduction of DNA replication is seen in several tissues of the PURA−/− mice. Nonetheless, many cells do undergo DNA replication, and the mouse does develop past birth, indicating that Purα is not universally required for replication. Rather, Purα may be required for the developmental replication of selective classes of cells at specific times. Previous studies have described a stimulation of JCV DNA replication at low Purα levels (9) and an inhibition of replication at higher Purα levels (5, 9). Ectopic overexpression of Purα in cells has been reported to inhibit DNA replication (1), cell cycle progression (1, 44), and oncogenic transformation (1, 11). The present results are highly consistent with this dual nature of the Purα effect on replication. Purα is required for replication in the specific neuronal precursor cells at day p5, a time when Purα levels in the PURA+/+ mouse are rapidly increasing. When Purα levels are at their peak, however, after day 19, DNA replication is negligible in the brain. These results suggest that the dual effect of Purα on replication is not due simply to a binding or unwinding effect at a PUR element but that interactions of Purα with partner proteins are paramount. In this regard, reports of interactions of Purα with cell cycle regulatory proteins such as Rb (25) or cyclin/Cdk complexes (1, 23) warrant further investigation. At this time, it remains to be determined whether effects of Purα on cellular DNA replication are through interaction with the replication apparatus, as they are in the JCV model (5, 7, 9), or whether they are mediated by effects on transcription or translation.
It has been reported that Purα can influence transcription of the MBP gene in oligodendroglia (22, 47, 48). In the present analysis, MBP and GFAP immunolabeling reveals aberrant expression of these structural proteins in oligodendrocytes and astrocytes, respectively, in PURA−/− mice. In this mouse model, however, data not shown indicate that myelin sheaths of the spinal cord and optic nerve are present. Therefore, Purα is likely not absolutely required for MBP gene transcription. Myelin sheaths in the PURA−/− mouse may, however, bear subtle differences from their PURA+/+ littermates, and further work is being conducted to determine whether myelin sheath formation is altered in these mice.
Deletions in the PURA gene in humans, at chromosome band 5q31 in acute myelogenous leukemia potentially implicate Purα as a critical factor in myeloid development (31, 34). Present analysis strongly supports such a role for Purα. Replication of DNA in myeloid cells in the spleen is dramatically reduced in the PURA−/− mice (Table 2). It must be noted, however, that lack of Purα does not cause a block in differentiation at any developmental step. All classes of fully differentiated myeloid cells could be seen in marrow smears from the PURA−/− mice (data not shown). Thus, again, Purα appears to be required for proliferation of a particular class of cells, in this case myeloid cells, at a specific developmental time. It is notable that the absence of Purα results in decreased proliferation of cells in the brain and myeloid tissue. This is nearly the opposite effect of that observed in mice with genetically inactivated RB. In such mice lacking the retinoblastoma protein Rb, ectopic mitoses were observed in differentiated brain and hematopoietic tissue, indicating a lack of cessation of proliferation. Rb is a reported binding partner of Purα (23, 25). It is conceivable that interaction of these proteins provides a regulatory balance for cell proliferation in certain cell types. Purα may also interact with E2F1, a transcription factor downstream of Rb that may be involved in the upregulation of apoptosis, although as mentioned above, we did not observe any signs that reduced cell numbers are the result of apoptosis.
Whereas PURA is frequently disrupted in acute myelogenous leukemia, RB is only rarely so. Only one allele of PURA is disrupted in acute myelogenous leukemia. When that is the case in mice, i.e., in PURA+/− mice, replication of myeloid cells in the spleen is strongly affected, although less than in PURA−/− mice (Fig. 2 and Table 2). This supports the notion that a haploinsufficiency effect at the PURA locus could be involved in progression to acute myelogenous leukemia. This may be further addressed by observing whether PURA+/− mice are more susceptible to cancer. Future studies of the heterozygotes will address these questions.
It has been reported that Purα binds BC1 DNA, which possesses an identity element targeting RNA to neuronal dendrites in mice (29). It has been further reported that Purα associates with several proteins involved in dendritic mRNA transport, including Staufen, which has now been implicated in RNA transport to dendritic translation sites (35, 36). Several observations in the present study support a role for Purα in dendritic protein synthesis. In PURA+/+ mice, Purα is at high levels in dendrites of Purkinje cells at a time when cerebellar development is occurring. In the PURA−/− mice at this time, hippocampal neurons that normally have high levels of Purα are failing to form synapses (Fig. 11). We have shown that Purα associates with Cdk5 (Fig. 8). Mice deficient in p35, an activator of Cdk5, exhibit dysplasia and heterotopia of principal neurons in the hippocampal formation (50), a phenotype similar to one reported here for Purα-deficient mice. Mice deficient in Cdk5 display inversion of cerebral cortical lamination (19), a phenotype not observed in the PURA−/− mice, although lamination is affected. It is conceivable that altered timing of neurofilament phosphorylation by Cdk5 in the PURA−/− mice could influence aspects of brain development. The observation that Purα levels in the brain remain nearly maximal after cell proliferation has largely ceased, and that Purα levels are high in dendrites, indicates that the role of the protein extends beyond replication, transcription, or cell cycle control. A unifying theme in Purα functions is that it may selectively recruit regulatory proteins, such as Cdk enzymes, to specific nucleic acid sequence elements. The attractiveness of such a theme is enhanced by the present genetic analysis, and further work will be conducted with the PURA genetically altered mice to address this.
If Purα is critical for processes such as timing of DNA replication, gene transcription, and compartmentalized translation, and if it is present in so many cell types, one might hypothesize that PURA−/− animals would not survive as long as they do. It may be that overlapping or redundant functions of Pur family members are responsible for their prolonged survival. The amino acid sequences of the DNA-binding repeat regions of Purα, Purβ, and Purγ are nearly 90% identical, and these sequences have evolved little since bacteria (24). Whereas Drosophila melanogaster has only one Pur protein, humans have four known family members, including two splice forms of Purγ (32). Purβ has been reported to act in a complex with Purα in certain effects on transcription (28). Further evidence for an interaction between the two proteins may lie in the observation that PURA, at chromosome band 5q31, and PURB, at 7p13 in humans, are simultaneously deleted much more frequently than expected in acute myelogenous leukemia (31). In the developing brain, Purα levels increase dramatically after birth and peak at approximately the time the PURA−/− mice die. Timing of Purα expression in various tissues may explain why effects of its absence in the brain are so predominant at the time of death. It is expected that the simultaneous inactivation of multiple PUR family genes in the future will further elucidate their roles in cellular proliferation and CNS development.
Acknowledgments
We thank past and present members of the Center for Neurovirology and Cancer Biology for insightful discussion and sharing of ideas and reagents, particularly Sidney Croul for effort and guidance in the early stages of the neuropathological studies. We thank C. Schriver for editorial assistance. We thank Andrew D. Bergemann for helpful discussion. Jane A. Strauchen provided valuable pathologic analyses of mouse blood and lymphoid systems.
This work was made possible by grants awarded by the NIH to K.K., J.G., E.M.J., and S.A.
REFERENCES
- 1.Barr, S. M., and E. M. Johnson. 2001. Ras-induced colony formation and anchorage-independent growth inhibited by elevated expression of Puralpha in NIH3T3 cells. J. Cell. Biochem. 81:621-638. [DOI] [PubMed] [Google Scholar]
- 2.Bergemann, A. D., and E. M. Johnson. 1992. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol. Cell. Biol. 12:1257-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergemann, A. D., Z. W. Ma, and E. M. Johnson. 1992. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol. 12:5673-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blow, J. J., and B. Hodgson. 2002. Replication licensing-origin licensing: defining the proliferative state? Trends Cell Biol. 12:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, C.-F., G. Gallia, V. Muralidharan, N. N. Chen, P. Zoltick, E. Johnson, and K. Khalili. 1996. Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Pur α. J. Virol. 70:4150-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, N. N., and K. Khalili. 1995. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur α in glial cells. J. Virol. 69:5843-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, N. N., C.-F. Chang, G. L. Gallia, D. A Kerr, E. M. Johnson, C. P. Krachmarov, S. M. Barr, R. J. Frisque, B. Bollag, and K. Khalili. 1995. Cooperative action of cellular proteins YB-1 and Purα with the tumor antigen of the human JC polymovirus determines their interaction with the viral lytic control element. Proc. Natl. Acad. Sci. USA 92:1087-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chepenik, L. G., A. P. Tretiakova, C. P. Krachmarov, E. M. Johnson, and K. Khalili. 1998. The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene 210:37-44. [DOI] [PubMed] [Google Scholar]
- 9.Daniel, D. C., M. J. Wortman, R. J. Schiller, H. Liu, L. Gan, J. S. Mellen, C. F. Chang, G. L. Gallia, J. Rappaport, K. Khalili, and E. M. Johnson. 2001. Coordinate effects of human immunodeficiency virus type 1 protein Tat and cellular protein Puralpha on DNA replication initiated at the JC virus origin. J. Gen. Virol. 82:1543-1553. [DOI] [PubMed] [Google Scholar]
- 10.Darbinian, N., G. L. Gallia, M. Kundu, N. Shcherbik, A. Tretiakova, A. Giordano, and K. Khalili. 1999. Association of Purα and E2F-1 suppresses transcriptional activity of E2F-1. Oncogene 18:6398-6402. [DOI] [PubMed] [Google Scholar]
- 11.Darbinian, N., G. L. Gallia, J. King, L. Del Valle, E. M. Johnson, and K. Khalili. 2001. Growth inhibition of glioblastoma cells by human Pur(alpha). J. Cell. Physiol. 189:334-340. [DOI] [PubMed] [Google Scholar]
- 12.Du, Q., A. E. Tomkinson, and P. D. Gardner. 1997. Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. A possible role for the DNA-binding protein, Purα. J. Biol. Chem. 272:14990-14995. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, M. C., A. V. Tutter, C. Cvetic, C. H. Gilbert, T. A. Prokhorova, and J. C. Walter. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277:33049-33057. [DOI] [PubMed] [Google Scholar]
- 14.Freeman, A., L. S. Morris, A. D. Mills, K. Stoeber, R. A. Laskey, G. H. Williams, and N. Coleman. 1999. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin. Cancer Res. 5:2121-2132. [PubMed] [Google Scholar]
- 15.Gallia, G. L., M. Safak, and K. Khalili. 1998. Interaction of the single-stranded DNA-binding protein Purα with human polyomavirus JC virus early protein T-antigen. J. Biol. Chem. 273:32662-32669. [DOI] [PubMed] [Google Scholar]
- 16.Gallia, G. L., N. Darbinian, A. Tretiakova, S. A. Ansari, J. Rappaport, J. Brady, M. J. Wortman, E. M. Johnson, and K. Khalili. 1999. RNA-dependent interaction between the cellular protein Purα and the HIV-1 Tat protein. Proc. Natl. Acad. Sci. USA 96:11572-11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallia, G. L., E. M. Johnson, and K. Khalili. 2000. Purα: a multifunctional single-stranded DNA and RNA-binding protein. Nucleic Acids Res. 28:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallia, G. L., N. Darbinian, N. Jaffe, and K. Khalili. 2001. Single-stranded nucleic acid-binding protein, Pur alpha, interacts with RNA homologous to 18S ribosomal RNA and inhibits translation in vitro J. Cell. Biochem. 83:355-363. [DOI] [PubMed] [Google Scholar]
- 19.Gilmore, E. C., T. Ohshima, A. M. Goffinet, A. B. Kulkarni, and K. Herrup. 1998. Cyclin-dependent kinase 5-deficient mice demonstrate novel developmental arrest in cerebral cortex. J. Neurosci. 18:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldowitz, D., and K. Hamre. 1998. The cells and molecules that make a cerebellum. Trends Neurosci. 21:375-382. [DOI] [PubMed] [Google Scholar]
- 21.Haas, S., J. Gordon, and K. Khalili. 1993. A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol. Cell. Biol. 13:3103-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas, S., P. Thatikunta, A. Steplewski, E. Johnson, K. Khalili, and S. Amini. 1995. A 39 kD DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytic cells. J. Cell Biol. 130:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, H., M. J. Wortman, M. Kanovsky, R. R. Uson, R. E. Gordon, N. Alfano, and E. M. Johnson. 1998. Alterations in Pura levels and intracellular localization in the CV-1 cell cycle. Cell Growth Differ. 9:651-665. [PubMed] [Google Scholar]
- 24.Johnson, E. M. 2003. The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res. 23:2093-2100. [PubMed] [Google Scholar]
- 25.Johnson, E. M., P.-L. Chen., C. P. Krachmarov, S. Barr, Z. W. Ma, and W.-H. Lee. 1995. Association of human Purα with the retinoblastoma protein, Rb, regulates binding to the Purα single-stranded DNA recognition element. J. Biol. Chem. 270:24352-24360. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, E. M., M. J. Wortman, K. Khalili, and D. C. Daniel. 2002. Mechanisms of regulation of replication of HIV-1 and JC virus in the CNS provide a model for regulation of cellular gene expression by sequence-specific single-stranded DNA- and RNA-binding protein Pur-alpha. J. Neurovirol. 8(Suppl. 1):26. (Abstract.) [Google Scholar]
- 27.Kelm, R. J., Jr., P. K. Elder, A. R. Strauch, and M. J. Getz. 1997. Sequence of cDNAs encoding components of vascular actin single-stranded DNA-binding factor 2 establish identity to Purα and Purβ. J. Biol. Chem. 272:26727-26733. [DOI] [PubMed] [Google Scholar]
- 28.Kelm, R. J., Jr., J. G. Cogan, P. K. Elder, A. R. Strauch, and M. J. Getz. 1999. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter. J. Biol. Chem. 274:14238-14245. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi, S., K. Agui, S. Kamo, Y. Li, and K. Anzai. 2000. Neural BC1 RNA associates with Purα, a single-stranded DNA and RNA binding protein, which is involved in the transcription of the BC1 RNA gene. Biochem. Biophys. Res. Commun. 277:341-347. [DOI] [PubMed] [Google Scholar]
- 30.Krachmarov, C. P., L. G. Chepenik, S. M. Barr-Vagell, K. Khalili, and E. M. Johnson. 1996. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein, Purα. Proc. Natl. Acad. Sci. USA 93:14112-14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lezon-Geyda, K., V. Najfeld, and E. M. Johnson. 2001. Deletions of PURA, at 5q31, and PURB, at 7p13, in myelodysplastic syndrome and progression to acute myelogenous leukemia. Leukemia 15:954-962. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., and E. M. Johnson. 2002. Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res. 30:2417-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, Z. W., A. D. Bergemann, and E. M. Johnson. 1994. Conservation in human and mouse Purα of a motif common to several proteins involved in initiation of DNA replication. Gene 149:311-314. [DOI] [PubMed] [Google Scholar]
- 34.Ma, Z. W., T. Pejovic, N. Najfeld, D. C. Ward, and E. M. Johnson. 1995. Localization of PURA, the gene encoding the sequence-specific single-stranded-DNA-binding protein Pur alpha, to chromosome band 5q31. Cytogenet. Cell Genet. 71:64-67. [DOI] [PubMed] [Google Scholar]
- 35.Mallardo, M., A. Deitinghoff, J. Muller, B. Goetze, P. Macchi, C. Peters, and M. A. Kiebler. 2003. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. USA 100:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohashi, S., K. Koike, A. Omori, S. Ichinose, S. Ohara, S., Kobayashi, T. A. Sato, and K. Anzai. 2002. Identification of mRNP complexes containing pur alpha, mStaufen, fragile X protein and myosin Va, and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 277:37804-37810. [DOI] [PubMed] [Google Scholar]
- 37.Qi, Z., D. Tang, X. Zhu, D. J. Fujita, and J. H. Wang. 1998. Association of neurofilament proteins with neuronal Cdk5 activator. J. Biol. Chem. 273:2329-2335. [DOI] [PubMed] [Google Scholar]
- 38.Qin, L., G. S. Marrs, R. McKim, and M. E. Dailey. 2001. Hippocampal mossy fibers induce assembly and clustering of PSD95-containing postsynaptic densities independent of glutamate receptor activation. J. Comp. Neurol. 440:284-298. [DOI] [PubMed] [Google Scholar]
- 39.Romanowski, P., M. A. Madine, and R. A. Laskey. 1996. XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc. Natl. Acad. Sci. USA 93:10189-10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruppert, C., D. Goldowitz, and W. Wille. 1986. Proto-oncogene c-myc is expressed in cerebellar neurons at different developmental stages. EMBO J. 5:1897-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadakata, T., C. Kuo, H. Ichikawa, E. Nishikawa, S. Y. Niu, E. Kumamaru, and N. Miki. 2000. Puralpha, a single-stranded DNA binding protein, suppresses the enhancer activity of cAMP response element. Brain Res. Mol. Brain Res. 77:47-54. [DOI] [PubMed] [Google Scholar]
- 42.Safak, M., G. Gallia, and K. Khalili. 1999. Reciprocal interaction between two cellular proteins, Purα and YB-1, modulates transcriptional activity of human JCVCY in glial cells. Mol. Cell. Biol. 19:2712-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaarschmidt, D., E. M. Ladenburger, C. Keller, and R. Knippers. 2002. Human MCM proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 30:4176-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stacey, D. W., M. Hitomi, M. Kanovsky, L. Gan, and E. M. Johnson. 1999. Cell cycle arrest and morphological alterations following microinjection of NIH3T3 cells with Pur alpha. Oncogene 18:4254-4261. [DOI] [PubMed] [Google Scholar]
- 45.Sun, D., C. L. Leung, and R. K. H. Leim. 1996. Phosphorylation of the high molecular weight neurofilament protein (NF-H) by Cdk5 and p35. J. Biol. Chem. 271:14245-14251. [DOI] [PubMed] [Google Scholar]
- 46.Tretiakova, A., G. L. Gallia, N. Shcherbik, B. A. Jameson, E. M. Johnson, S. Amini, and K. Khalili. 1998. Association of Purα with RNAs homologous to 7 SL determines its binding ability to the MBP promoter DNA sequence. J. Biol. Chem. 273:22241-22247. [DOI] [PubMed] [Google Scholar]
- 47.Tretiakova, A., A. Steplewski, E. M. Johnson, K. Khalili, and S. Amini. 1999. Regulation of myelin basic protein gene transcription by Sp1 and Purα; evidence for association of Sp1 and Purα in brain. J. Cell. Physiol. 181:160-168. [DOI] [PubMed] [Google Scholar]
- 48.Tretiakova, A., J. Otte, S. E. Croul, J. H. Kim, E. M. Johnson, S. Amini, and K. Khalili. 1999. Association of JC virus large T antigen with myelin basic protein transcription factor (MEF-1/Purα in hypomyelinated brains of mice transgenically expressing T antigen. J. Virol. 73:6076-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tye, B. K. 1999. MCM proteins in DNA replication. Annu. Rev. Biochem. 68:649-686. [DOI] [PubMed] [Google Scholar]
- 50.Wenzel, H. J., C. A. Robbins, L. H. Tsai, and P. A. Schwartzkroin. 2001. Abnormal morphological and functional organization of the hippocampus in a p35 mutant model of cortical dysplasia associated with spontaneous seizures. J. Neurosci. 21:983-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zambrano, N., S. De Renzis, G. Minopoli, R. Faraonio, V. Donini, A. Scaloni, F. Cimino, and T. Russo. 1997. DNA-binding protein Purα and transcription factor YY1 function as transcription activators of the neuron-specific FE65 gene promoter. Biochem. J. 328:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]