Abstract
While the stress-responsive alternative sigma factor σB has been identified in different species of Bacillus, Listeria, and Staphylococcus, the σB regulon has been extensively characterized only in B. subtilis. We combined biocomputing and microarray-based strategies to identify σB-dependent genes in the facultative intracellular pathogen Listeria monocytogenes. Hidden Markov model (HMM)-based searches identified 170 candidate σB-dependent promoter sequences in the strain EGD-e genome sequence. These data were used to develop a specialized, 208-gene microarray, which included 166 genes downstream of HMM-predicted σB-dependent promoters as well as selected virulence and stress response genes. RNA for the microarray experiments was isolated from both wild-type and ΔsigB null mutant L. monocytogenes cells grown to stationary phase or exposed to osmotic stress (0.5 M KCl). Microarray analyses identified a total of 55 genes with statistically significant σB-dependent expression under the conditions used in these experiments, with at least 1.5-fold-higher expression in the wild type over the sigB mutant under either stress condition (51 genes showed at least 2.0-fold-higher expression in the wild type). Of the 55 genes exhibiting σB-dependent expression, 54 were preceded by a sequence resembling the σB promoter consensus sequence. Rapid amplification of cDNA ends-PCR was used to confirm the σB-dependent nature of a subset of eight selected promoter regions. Notably, the σB-dependent L. monocytogenes genes identified through this HMM/microarray strategy included both stress response genes (e.g., gadB, ctc, and the glutathione reductase gene lmo1433) and virulence genes (e.g., inlA, inlB, and bsh). Our data demonstrate that, in addition to regulating expression of genes important for survival under environmental stress conditions, σB also contributes to regulation of virulence gene expression in L. monocytogenes. These findings strongly suggest that σB contributes to L. monocytogenes gene expression during infection.
In several low-GC-content gram-positive bacteria, σB has been recognized as a general stress-responsive sigma factor. This alternative sigma factor contributes to the ability of organisms such as Listeria monocytogenes, Bacillus subtilis, and Staphylococcus aureus to survive under environmental and energy stress conditions (4, 10, 18, 19, 65). σB also contributes to biofilm formation in S. aureus and Staphylococcus epidermidis (37, 55). Biofilm formation may further enhance the survival of these organisms under conditions of environmental stress.
In L. monocytogenes, a food-borne pathogen capable of causing mild to severe infections in humans, σB confers stress resistance (e.g., under acid stress and osmotic stress) and contributes to pathogenesis. To illustrate, an L. monocytogenes σB null mutant survives less well than the wild-type parent at low pH (pH 2.5) and in a murine infection model (45, 67). σB has been demonstrated to contribute to transcription of prfA, which encodes the central virulence gene regulator PrfA in L. monocytogenes (45).
An increasing body of evidence suggests a broad role for σB-dependent genes in the virulence of gram-positive bacteria. For example, σB has been linked with regulation of virulence gene expression in S. aureus (6, 12). Specifically, σB contributes to transcriptional regulation of sarC in S. aureus. The sar locus partially controls expression of the agr locus; agr and sar are both global regulatory elements that control the synthesis of a variety of extracellular and cell surface proteins involved in the pathogenesis of S. aureus (12, 38). A total of 23 proteins showed increased expression in the presence of σB in a two-dimensional gel comparison of proteins isolated from both the wild-type S. aureus strain and its isogenic sigB null mutant (27). σB also has been shown to contribute to virulence in Bacillus anthracis. Specifically, a B. anthracis ΔsigB strain is less virulent than its wild-type parent (21). Links between environmental stress responses and virulence in these bacteria suggest a central role for σB in contributing to the ability of bacterial pathogens to survive environmental stress conditions, to direct expression of virulence genes, and to cause disease.
Identification of the genes regulated by σB is the first step in deciphering specific mechanisms through which this alternative sigma factor confers resistance to multiple environmental stresses and contributes to virulence. Previous efforts at defining the σB regulon have focused primarily on B. subtilis, a gram-positive, nonpathogenic model organism. Through a combination of in vitro transcription, reporter fusion transposon mutagenesis, two-dimensional protein gel electrophoresis, and RNA hybridizations (reviewed in reference 52), a total of 75 genes and proteins with σB-dependent expression patterns were identified in B. subtilis. Recently, full genome macroarray analyses enabled two research groups to independently define over 120 B. subtilis genes that showed σB-dependent expression profiles by comparing gene expression patterns of wild-type strains with those of isogenic sigB null mutants (51, 53). In addition, use of a full-genome B. subtilis array allowed Helmann et al. (30) to identify 44 heat shock-induced genes preceded by previously unidentified potential σB-dependent promoters. These transcriptome studies of the B. subtilis stress response illustrate the power of genome-wide, array-based expression studies.
The σB regulon in B. subtilis, as determined by Price et al. (53) and Petersohn et al. (51), includes genes with a wide variety of functions. Approximately one quarter of the B. subtilis σB-dependent genes encode proteins involved in metabolic functions, including glucose metabolism, protein degradation, and lipid metabolism. In addition, many genes regulated by σB in B. subtilis encode transporters, e.g., solute transporters, permeases, and ATP-binding cassette (ABC) transport systems. The broad range of gene functions associated with the σB regulon in B. subtilis suggests that, in addition to enhancement of fundamental cellular processes, σB-mediated stress resistance mechanisms are also responsible for targeted action against specific stresses.
To better understand the role of σB in stress resistance and virulence in L. monocytogenes, we combined promoter searches and DNA microarrays to identify genes directly activated by σB from experimentally defined or predicted σB-dependent promoter sequences. As some B. subtilis genes have shown apparent σB-dependent induction in the absence of well-conserved σB-dependent promoter sequences, some σB-mediated effects may also occur through indirect regulatory mechanisms (e.g., possibly through σB-dependent expression of additional transcriptional regulators [51]).
For the purposes of this study, we chose to focus specifically on defining L. monocytogenes genes that are transcribed from σB-dependent promoters. To that end, we first performed a hidden Markov model (HMM) similarity search for putative σB-dependent promoters against the published complete L. monocytogenes genome sequence (28). To allow a focused analysis of σB-dependent gene expression, a microarray was constructed with the genes identified by the HMM search. To more fully explore the role of σB in L. monocytogenes stress response and virulence, our microarray also included previously identified stress response and virulence genes that were not necessarily identified by HMM.
To be classified as being under direct σB control, genes had to meet the following two independent criteria: (i) significantly higher expression in the wild-type strain compared to the sigB null mutant under the conditions used for the microarray experiments and (ii) the presence of an identifiable σB-dependent promoter-like sequence upstream of the coding region. With these criteria, we identified 54 genes under direct σB control. Although we refer to these genes as σB dependent, it is important to recognize that the genes identified with this approach may not be exclusively σB dependent but may also be regulated by σB-independent mechanisms. Interestingly, while the genes that we found to be controlled by σB include many that had been identified previously as part of the B. subtilis σB regulon, several virulence and virulence-related genes were also identified (e.g., a gene encoding a bile salt hydrolase and genes encoding surface molecules with possible or confirmed roles in host cell attachment). Our data provide further evidence in support of the contributions of σB to L. monocytogenes stress resistance. Importantly, identification of multiple σB-regulated virulence genes strongly suggests that this alternative sigma factor also contributes to the ability of L. monocytogenes to infect and survive within a host.
MATERIALS AND METHODS
Bacterial strains and media.
L. monocytogenes strain 10403S (7) and the isogenic sigB null mutant (ΔsigB) FSL A1-254 (67) were used throughout this study. Cells were grown in brain heart infusion broth (BHI; Difco, Detroit, Mich.) at 37°C with shaking unless indicated otherwise.
HMM searches.
HMM searches were performed with HMMER (http://hmmer.wustl.edu) essentially as described by Price et al. (53). The training alignments included 29 B. subtilis σB-dependent promoters (described in reference 53) as well as four confirmed (prfA, rsbV, and opuCA) or likely (betL) L. monocytogenes σB-dependent promoter sequences. The models were searched against the complete L. monocytogenes strain EGD-e genome sequence (28). The output results were filtered, and only hits within 350 bp upstream of a coding region, as predicted by the ListiList web server (http://genolist.pasteur.fr/ListiList [44]), and with an E value of ≤0.006 were kept.
DNA and RNA isolation and treatment.
RNA was extracted from L. monocytogenes either grown to stationary phase or exposed to osmotic stress (as described below) with the RNeasy Protect Bacteria midi kit (Qiagen, Valencia, Calif.). The enzymatic and mechanical disruption protocol provided by the manufacturer was used, with the exception that cell lysis was performed with sonication at 21W three times for 20 s each (with cells iced between bursts) instead of bead beating. RNA was eluted from the column in RNase-free water and ethanol precipitated overnight at −20°C. Precipitated RNA was centrifuged, washed in 70% ethanol, and resuspended in RNase-free water. DNase treatment was performed with 30 U of RQ1 RNase-free DNase (Promega, Madison, Wis.) in the presence of 400 U of RNase inhibitor (RNasin; Promega) for 1 h at 37°C. The reaction mix was then extracted twice with an equal volume of 50% phenol-50% chloroform, followed by one extraction with an equal volume of 100% chloroform. RNA was ethanol precipitated from the aqueous layer and stored at −20°C in ethanol.
Cultures for RNA isolation were inoculated from single colonies and grown overnight at 37°C. The cultures were then diluted 1:200 in BHI and grown at 37°C to specified growth phases. Specifically, for harvest of stationary-phase cells, L. monocytogenes was grown for one additional hour after reaching an optical density at 600 nm (OD600) of 0.8. At this point, a 9-ml aliquot of culture was added to 18 ml of RNA Protect Bacteria reagent (Qiagen), and RNA isolation was performed as described above. For osmotic stress treatment, cells present in 40 ml of a culture grown to an OD600 of 0.4 were collected by centrifugation and resuspended in 8.25 ml of warm (37°C) BHI broth, to which 1.25 ml of 4 M KCl was added to yield a final concentration of 0.5 M KCl. After 7 min of incubation at 37°C, corresponding to the approximate peak of σB-induced transcriptional response reported in induction experiments in B. subtilis (64), 20 ml of RNA Protect Bacteria reagent was added, and RNA isolation was performed as described above.
Chromosomal DNA used for genomic DNA microarray control spots was isolated as described by Flamm et al. (20) from an overnight culture of L. monocytogenes. Briefly, cell walls were digested with lysozyme in 20% sucrose, followed by cell lysis with sodium dodecyl sulfate and proteinase K. DNA was purified by phenol-chloroform extractions and ethanol precipitated. DNA was then resuspended in spotting buffer consisting of 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% Sarkosyl) and quantified by UV spectroscopy.
Microarray construction.
DNA microarrays were constructed to include 208 L. monocytogenes genes spotted in triplicate, 30 mouse β-actin cDNA spots for nonhybridizing controls, and 60 spots of genomic DNA to aid in data normalization and analysis. The 208 L. monocytogenes genes included 166 of the 170 genes identified by HMM searches as including an upstream predicted σB-dependent promoter (PCR amplification failed for four genes) as well as 36 genes previously reported to be involved in virulence or stress response and six negative control genes (i.e., housekeeping genes predicted not to show σB-dependent expression). All 208 target genes as well as the PCR primers used for their amplification are detailed in the supplemental material available at http://www.foodscience.cornell.edu/wiedmann/Mark%20supplemental%20table%20 (4-15).doc.
PCR primers were designed with PrimeArray software (56) to amplify the complete open reading frames (ORFs) of the selected genes. For ORFs of >1 kb, primers were selected to amplify a 1-kb fragment at the 5′ end of each ORF. PCR was performed with AmpliTaq Gold (Applied Biosystems, Foster City, Calif.) with an L. monocytogenes 10403S cell lysate (prepared as described in reference 24) as template DNA. Appropriately sized products free of nonspecific amplification or extraneous bands, as determined by agarose gel electrophoresis, were purified by ethanol precipitation and reconstituted in spotting buffer (3× SSC-0.1% Sarkosyl) to a final concentration of 50 to 150 ng/μl. PCR was also used to generate a 1-kb PCR fragment of the mouse β-actin gene with mouse β-actin cDNA (Sigma, St. Louis, Mo.) as the template.
Arrays were spotted with a GMS 417 robotic arrayer (Affymetrix, Santa Clara, Calif.) on GAPS2 glass slides (Corning, Corning, N.Y.). Immediately before use, slides were blocked in a solution of 1-methyl-2-pyrrolidinone containing 1.44% succinic anhydride and 0.02 M boric acid (pH 8.0), as directed by the slide manufacturer.
cDNA labeling and microarray hybridization.
Precipitated, DNase-treated RNA was centrifuged, washed once in 70% ethanol and once in 100% ethanol, and resuspended in RNase-free water. The RNA was quantified on a UV spectrophotometer and checked for quality by A260/280 ratio and formaldehyde-agarose gel electrophoresis. cDNA was generated from 10 μg of RNA with random hexamers and SuperScript II (Invitrogen, Carlsbad, Calif.) in the presence of indocarbocyanine (Cy3)-dUTP or indodicarbocyanine (Cy5)-dUTP (Amersham Biosciences, Piscataway, N.J.). cDNA synthesis and labeling were performed with a dye-swapping design (sometimes referred to as reverse labeling [62]); each RNA sample was used for two separate labeling reactions, one with Cy3 and one with Cy5. Completed reactions were incubated with 1.5 μl of 1 N NaOH at 65°C for 10 min to inactivate the enzyme and degrade the RNA. Reactions were then neutralized with 1.5 μl of 1 N HCl.
Wild-type and sigB mutant cDNA probes were mixed and purified with the QIAquick PCR purification kit (Qiagen) as described by the manufacturer, with the addition of a 750-μl 35% guanidine-HCl wash after binding. The purified probes were then dried and reconstituted in 20 μl of hybridization buffer (3.5× SSC, 0.25% sodium dodecyl sulfate, and 0.5 μg of salmon sperm DNA/μl; Invitrogen). Probes were boiled for 2 min, applied to arrays, and hybridized overnight in a 60°C waterbath. Before scanning, slides were washed in 2× SSC-0.1% sodium dodecyl sulfate at 60°C for 5 min, followed by room temperature washes with 2× SSC-0.1% sodium dodecyl sulfate, 2× SSC, and 0.2× SSC for 5 min each. Slides were centrifuged to dry them and scanned with a Perkin Elmer Scan Array 5000 confocal laser scanner (Perkin Elmer, Wellesley, Mass.).
Microarray replicates and data analysis.
For each stress condition described above (osmotic stress or stationary phase), two independent RNA isolations (for both wild-type and ΔsigB cells) were performed on separate days to provide true biological replicates. Each set of independent RNA samples was used to perform two microarray hybridizations with a dye-swapping design to correct for differences in Cy3 and Cy5 dye incorporation and to provide experimental duplicates. Briefly, both wild-type and ΔsigB cell RNAs from each of the two independent RNA isolations were labeled separately with either Cy5 or Cy3 as described above. Labeled cDNA was used to perform two independent microarray hybridizations, one with Cy3-labeled wild-type and Cy5-labeled mutant cDNA and one with Cy5-labeled wild-type and Cy3-labeled mutant cDNA. Thus, data for four microarray repetitions were collected for each stress condition.
Raw TIFF images from the scanner were loaded into ScanAlyze (http://rana.lbl.gov/EisenSoftware.htm) for analysis. Spot grids were manually fitted to the microarray images. Spots were flagged and eliminated from analysis only in obvious instances of high background or stray fluorescent signals.
Because our microarray contained a relatively small number of genes, most of which were expected to show differential expression, proper normalization of the raw data was critical. Equalizing the mean intensities of both channels of an array, the most common method used for microarray normalization, would thus have provided severely skewed data on an array, with the majority of spots appearing to be unequally expressed. Therefore, we normalized our data using the mean intensities of spots containing genomic DNA. Data normalization was performed simultaneously on all four array data sets for a given stress condition, as follows. For each channel (Cy3 or Cy5), the mean intensity of all genomic DNA control spots was calculated independently. A correlation factor comparing genomic means among channels was calculated and used to proportionally adjust all spot intensities (experimental and control) so that the means of genomic spot intensities were equal across all arrays. From these normalized values, a floor was calculated as the average intensity plus 2 standard deviations of all β-actin spots in a channel. Spots with values below the floor in both channels were eliminated from analysis; spots below the floor in only one channel had that channel intensity raised to the floor value, and the resulting data were included in the analyses (16).
Floored and normalized channel intensities were analyzed with the Significance Analysis for Microarrays (SAM) program (63). This statistical analysis involves factoring the change in expression of each gene relative to the standard deviation of all replicates for that gene. Therefore, genes with a low change will not be discounted if the ratios are consistent among repeats, effectively reducing false-negatives. False-positives are also avoided when genes have poor reproducibility between replicates. This method of statistical analysis maximizes both the quantity of genes found and the reliability of the results. Spot intensities for all channels were input in SAM as paired, unlogged values. All individual spots were considered repetitions, generating 24 data points for each gene (3 spots per gene × 4 arrays × 2 channels per array). Delta values were chosen according to the lowest false discovery rate. Only genes with expression ratios of >1.5 were considered biologically significant.
RACE-PCR.
Promoter regions were mapped with the 5′ rapid amplification of cDNA ends (RACE) system (Invitrogen) according to the manufacturer's protocol. Briefly, gene-specific first-strand cDNA was tailed with dCTP by using terminal transferase. The products were then amplified with a nested gene-specific primer and a poly(G/I) primer in a touchdown PCR with AmpliTaq Gold (Applied Biosystems). PCR products present in the wild-type L. monocytogenes but absent in the ΔsigB strain were purified with the Qiagen QIAquick PCR purification kit and sequenced with the Big Dye terminator system and an Applied Biosystems 3700 sequencer at the Cornell University Bioresource Center.
RESULTS
HMM search to identify potential σB promoters.
An HMM developed with 29 B. subtilis and four L. monocytogenes σB-dependent promoter sequences was used to search the L. monocytogenes EGD-e genome (28) to identify genes preceded by a σB consensus promoter and thus potentially regulated by σB. A total of 170 genes met the following criteria for putative σB-dependent genes: (i) location of a predicted σB consensus promoter sequence within 350 bp upstream of a predicted open reading frame and (ii) an E value of ≤0.006. The presence of a sequence adequately fitting the promoter consensus was confirmed visually at positions indicated by the HMM. The results of this search were used to create a 208-gene microarray to specifically study the σB-mediated stress response in L. monocytogenes. The array consisted of 166 of the 170 HMM-identified genes (PCR amplification failed for four genes), 36 genes involved in stress response or virulence, and six control genes.
Transcriptional analysis of potential σB-dependent genes.
Microarray analyses were performed with RNA isolated from wild-type L. monocytogenes and an isogenic ΔsigB strain grown to stationary phase or exposed to osmotic stress, two conditions shown to activate σB (4, 19, 22). We identified 55 genes that showed significant expression ratios, with >1.5-fold expression in the wild-type strain over that in the mutant strain in at least one of the two stress conditions tested (Table 1). Fifty-one of these genes displayed >2-fold induction; the highest relative induction was 27-fold.
TABLE 1.
L. monocytogenes genes with significantly higher expression in the wild-type versus the ΔsigB strain under osmotic and stationary-phase stresses
| Changea (fold) | No. of genes with statistically significant higher expression under:
|
|||
|---|---|---|---|---|
| Stationary phase stress | Osmotic stress (0.5 M KCl) | Stationary-phase and osmotic stress | Stationary-phase or osmotic stress | |
| ≥1.5 | 41 | 51 | 37 | 55 |
| ≥2.0 | 37 | 44 | 30 | 51 |
Change was calculated by SAM based on microarray data (see Materials and Methods) and equals the average wild-type spot intensity divided by the average mutant spot intensity for two separate arrays from each of two replicate RNA isolations.
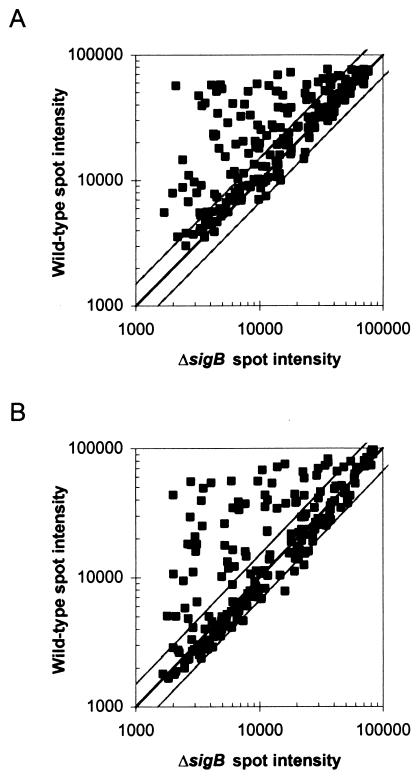
Figure 1 shows scatter plots of wild-type versus sigB mutant spot intensities. As the microarray predominantly targeted genes predicted to be positively regulated by σB, the distribution of the majority of the points above the diagonal, which indicates that a large proportion of the genes in the microarray were expressed more strongly in the wild type than in the mutant, was expected. As the presence of points falling far below the diagonal would signify increased gene expression in the absence of σB, none to few were expected. For the purpose of this study and in accordance with previous studies (31), we applied a cutoff of 1.5-fold induction to signify biological significance. Two genes included in the microarray fell below this cutoff (showing statistically significant expression ratios between 1.2- and 1.5-fold) and are not discussed further here. As shown in Fig. 1, a few genes showed higher expression in the ΔsigB mutant than in the wild-type strain; expression differences between strains for these genes were determined to be not statistically significant.
FIG. 1.
Scatter plots of normalized microarray spot intensities for wild-type and sigB mutant strains. The center line depicts equal expression in both strains. Outer lines depict 1.5-fold higher expression in either strain. Spot intensities were normalized as described in the text. Data for each gene represent the averages of three replicate spots from each of four hybridizations. RNA for microarray experiments was isolated from (A) stationary-phase cells or (B) osmotically stressed cells.
Overall, 44 of the 169 genes (26%) in our microarray that were either (i) predicted to be σB dependent by HMM or (ii) members of operons previously identified as being regulated by a σB-dependent promoter also showed σB-dependent expression in our microarray experiments. There was no apparent correlation between a promoter's HMM E value and the relative expression level of a corresponding gene, as determined by microarray (r2 < 0.01 for correlations between HMM E values and relative expression levels under either osmotic stress or stationary-phase conditions). In addition, 11 of the 36 currently recognized stress response and virulence genes that were included in the array even though they did not show an HMM-predicted σB-dependent consensus promoter displayed σB-dependent expression patterns. Subsequent visual inspection of the upstream regions for these 11 genes identified putative σB consensus promoter sequences for 10 of them (Table 2). Although inlD showed σB-dependent expression in our microarray experiments, no putative σB consensus promoter sequence was identified for this gene.
TABLE 2.
Summary of genes with σB-dependent expression patterns as identified by microarray analyses
| Category | Gene | Function or closest homologuea (E value) | Predicted σB promoter sequenceb | B. subtilis homologuec (E value) | Changed (fold)
|
|
|---|---|---|---|---|---|---|
| ST | OS | |||||
| Stress | lmo2230 | Arsenate reductase [Staphylococcus aureus] (le-08) | TTTAATGTTTCTAGTAATTTAAAAAGGGTAGATATTACTGT-N108-ATG | arsC (7e-10) | 27.0 | 21.9 |
| gadB | GadB glutamate decarboxylase (Lmo2434) | TGAACGGTTTGTCTCTGTGGTTTAATGGGTATTGGTGAGAGA-N32-ATG | 5.3 | 4.9 | ||
| sepA | Metallo-beta-lactamase superfamily protein [Shewanella oneidensis] (e-173) | CAAAAGGTTTTGAATAATTTTATGGAGGTATAAAAAGAAAGT-N33-ATG | 12.0 | 13.4 | ||
| ltrC | Low-temperature-requirement C protein | CCGTCCGTTTTTAGTTCGTGATGGCGGGGACTATGAACTTAT-N260-ATG† | yutG (le-53)‡ | 4.8 | 1.9 | |
| lmo1433 | Glutathione reductase (NADPH) [Methanosarcina acetivorans] (4e-91) | TTTTTCGTTTGAAAGTGAAATCAGACGGGAAAACAAGCTAAG-N14-GTG | 6.8 | 5.7 | ||
| ctc | B. subtilis general stress protein Ctc (le-27) | TCCTTTGTTTTGCTATTTTTCTAAAGGGTAGATATAATGTA-N35-ATG | ctc (le-33)‡ | 1.5 | 2.2 | |
| lmo1602 | General stress protein [Oceanobacillus iheyensis] (6e-11) | AAGAAAGTTTTAGAGGGGAATACTCAGGGTATAGAAAAAGGA-N18-ATG | ytxG (6e-13)‡ | 2.7 | 2.1 | |
| Virulence and virulence associated | bsh | Bsh bile salt hydrolase (Lmo2067) | AATTATGTTTTACTCCAAACTCCGAGGGTACTGGTATACAT-N31-ATG | 10.8 | 20.1 | |
| inlA | Internalin A | CCATTAGTGTTATTTTGAACATAAAGGGTAGAGGATAACAT-N437-GTG† | 6.2 | 3.9 | ||
| inlB | Internalin B | CCTTTTGTTAGGGTTATTAGCAGTAGGAACTGCAATGGCTC-N113-GTG† | 4.6 | 2.1 | ||
| inlC2 | Internalin C2 | TTTTTTGTTAATTTGGTCTAAAAAAGGGTATCTATTATTAA-N55-ATG† | 11.4 | 7.3 | ||
| inlD | Internalin D | None found | 3.3 | 2.3 | ||
| inlE | Internalin E | TAAATCGTTAACAAGTCTAATTTTAGTGATTAAACGAAATC-N84-ATG† | 3.7 | 2.4 | ||
| Transporters | opuC operon | TAAAAAGTTTAAATCTATACTAGTTAGGGAAATTAGTTATCG-N48-GTG | ||||
| opuCA | Glycine betaine/carnitine/choline ABC transporter (ATP-binding protein) | opuCA (e-159) | 5.3 | 6.5 | ||
| opuCB | Glycine betaine/carnitine/choline ABC transporter (membrane protein) | opuCB (le-74) | 3.8 | 4.7 | ||
| opuCC | Glycine betaine/carnitine/choline ABC transporter (osmoprotectant-binding) | opuCC (le-92) | 4.1 | 5.2 | ||
| opuCD | Betaine/carnitine/choline ABC transporter (membrane protein) | opuCD (5e-66) | 5.0 | 5.8 | ||
| lmo0784 | PTS enzyme IIAB, mannose-specific [Yersinia pestis KIM] (5e-34) | AATTGCGTTTTCTGACTAATCTTTTAGGGTAATGTTGTATGT-N195-ATG | levD (le-21) | 7.5 | 5.0 | |
| lmo2602 | Cation-transporting ATPase [Oceanobacillus iheyensis] (2e-12) | TTGATTGTTTTGGTTTAATGCCAAAGGGAATATATTAACGT-N16-ATG | sapB (le-09) | 5.5 | 3.8 | |
| lmo0405 | PHO4, phosphate transporter family [Bacillus anthracis] (4e-76) | GATTCTTTTTATATTTGTATAAAAGGGGTATAGACAATAAA-N30-ATG | 3.9 | 4.9 | ||
| lmo2463 | Probable export protein (antibiotic) [Streptomyces coelicolor] (6e-80) | AATGATGTTTGGCATATGTAAAAAAGAGGTATAAATTAGCGTG | ydfJ (4e-30) | 3.0 | 5.2 | |
| lmo1539 | Glycerol uptake facilitator [Bacillus halodurans] (8e-66) | AAATGGGTTATAACTCTCGCGAATTGGGGTAAAAGTAAAGAG-N254-ATG | glpF (5e-81) | 2.4 | 2.3 | |
| lmo1421 | ABC transporter [Bacillus anthracis A2012] (8e-98) | AATTAGGAATATTTAGGGATGATTTAGGGTAATTGGATGATA-N74-ATG† | 2.5 | 2.1 | ||
| lmo0524 | Sulfate transporter [Mycobacterium tuberculosis CDC1551] (2e-50) | AGAGATGATTGTTCTATGTCAAAACGGGTAAATAGTATAAG-N65-ATG | 2.0 | 2.9 | ||
| lmo0593 | Formate/nitrite transporter family protein [Streptococcus agalactiae] (7e-37) | TTTTGTGTTTTAAGAGTTTGAAAACGGGGAAATTAACAACAT-N146-TTG | ywcJ (le-18) | 2.9 | 2.3 | |
| Metabolism | lmo0669 | Oxidoreductase [Oceanobacillus iheyensis] (3e-86) | TTAAACGTTTTAGCGTAAAACAGGAGGGAAGACATAAAGTA-N139-ATG | yhxD (3e-95)‡ | 14.2 | 9.5 |
| lmo1694 | Epimerase, NAD-dependent family [Bacillus anthracis] (2e-62) | AAAATGGTTTTAATACTACTAAAAAGGGAATAAACTAGTTA-N22-TTG | yfhF (5e-59)‡ | 11.5 | 11.7 | |
| lmo1883 | Chitinase [Serratia marcescens] (e-117) | TTTCTAGTTTTATTTTCACTATGTTGGGTATTTTCTAGTAG-N47-ATG | 4.1 | 2.1 | ||
| lmo2695 | Dihydroxyacetone kinase [Bacillus halodurans] (e-103) | AACCACGTTTTGACTTTCTAGTAAAGGGAAATTGAGGTAAG-N30-ATG | 2.8 | 4.3 | ||
| lmo0956 | N-Acetylglucosamine-6-phosphate deacetylase [Bacillus anthracis] (e-110) | TTTTTTGGTTATTTTACTTTTTTTCGGGTAAATAGGAAAAG-N71-ATG | nagA (le-72) | 1.8 | 1.4 | |
| lmo2205 | Phosphoglycerate mutase [Neisseria meningitidis] (4e-86) | AAAAAGGTTTGACACTTCACTTGAAAGGGAAAACTTTGGTTG-N44-ATG | 1.8 | 3.2 | ||
| pdhA | Pyruvate dehydrogenase (E1 alpha subunit) [Bacillus subtilis] (e-151) | ACCCATGTTTTCAATGAGGAATTTGTGGTAATAACTGGAGAG-N231-ATG | pdhA (e-162) | 1.7 | 1.6 | |
| lmo1933 | GTP cyclohydrolase I [Bacillus halodurans] (6e-67) | TTAGTGGTTTTCTGTTTTAGAAATAGGGAATATAATTACGA-N113-ATG | mtrA (7e-73)‡ | 2.0 | 2.0 | |
| Transcriptional regulation | rsbV operon | TTGAATGTTTTAATTTTATTTGTTAGGGTAAAATCGACAGT-N27-ATG† | ||||
| rsbV | Anti-anti-sigma factor (antagonist of RsbW) | rsbV (3e-24)‡ | 2.1 | 2.0 | ||
| rsbW | Sigma B activity negative regulator RsbW | rsbW (3e-42)‡ | 2.0 | 2.4 | ||
| sigB | RNA polymerase sigma-37 factor (sigma B) | sigB (7e-97)‡ | 2.2 | 2.3 | ||
| rsbX | Indirect negative regulation of sigma B (serine phosphatase) | rsbX (le-26)‡ | 2.0 | 2.2 | ||
| lmo2485 | Transcriptional regulator [Methanosarcina mazei Goel] (le-04) | GCAAAAGTAGGAAATGGAATTATCAAGGTTTCTGAACTCGA-N42-ATG | 1.8 | 3.9 | ||
| Cell wall | lmo2085 | Surface-anchored protein (LPXTG motif) [Listeria innocua] (le-18) | CTGGCTGTTTTCTTTTGCTGTTTTATGGGTATTTAATGATGT-N28-ATG | 14.7 | 10.7 | |
| lmo0880 | Surface-anchored protein (LPXTG motif) [Listeria innocua] (le-26) | TGTACGGTTTTTAACAAGCAAGTTGTGGGAACCTATAAAGATA-N26-ATG | 5.2 | 2.1 | ||
| Energy | qoxA | AA3-600 quinol oxidase subunit II | TTTTTTGTTTCGAATTTCACAATCTAGGGAATAGTGAACAGA-N265-GTG | qoxA (3e-91) | 1.5 | 1.9 |
| lmo2389 | Pyridine nucleotide-disulfide oxidoreductase [Bacillus anthracis] (e-123) | GATATAGTTTGTAAGCTGTGAAATAGGGAATATATTGGTAC-N127-ATG | yumB (e-128)‡ | 1.6 | 3.4 | |
| Translation | lmo1698 | Ribosomal protein (S5)-alanine N-acetyltransferase [Bacillus halodurans] (3e-) | TGGAAAGTTTATTTTTTAATAAAATGGGTATATAGAAAAAT-N72-ATG | yjcK (3e-55) | 3.1 | 1.9 |
| lmo2511 | Ribosomal_S30, sigma 54 modulation protein [Bacillus anthracis] (2e-53) | AAAGAGGTTTGCGGAAGCGGTATTAGTGGAATAAAATACTAA-N135-ATG | yvyD (e-162)‡ | 3.8 | 1.9 | |
| DNA translocase | lmo1606 | DNA translocase (spoIIIE) [Bacillus halodurans] (e-168) | TACCATGTTTAACCCCTCTATTACCAAGGTATTATAACGAAT-N97-ATG | ytpT (0.0) | 2.4 | 2.6 |
| Hypothetical | lmo2570 | Hypothetical protein YvaZ [Bacillus subtilis] (.023) | CAACAGGTGTTCTAGAATATTTACGGGAACAAAAAGTCGA-N234-ATG | yvaZ (5e-09) | 5.3 | 5.9 |
| lmo2269 | YhzC of Listria monocytogenes (9e-26) | TTTTAAGATTGTAATAAATATGAGTGAAGGAGTGGTAGTCGTG | 4.0 | 5.3 | ||
| lmo0911 | BH2119-unknown [Bacillus halodurans] (le-06) | CATCTTGTTTTAACTTGCCCTCAGGCGGGTATTTATTATATG-N53-ATG | 2.2 | 1.9 | ||
| lmo0794 | Conserved hypothetical protein YwnB [Bacillus subtilis] (2e-50) | ATAAATGTTTCCCAGTCCCCTCTTTCGGGAATAATTCTTCTA-N45-ATG | ywnB (5e-54) | 5.2 | 5.8 | |
| lmo1580 | Conserved hypothetical ATP-binding domain [Bacillus anthracis] (2e-40) | TTTTATGGTTCTTTTAGGAAAAAGAGGGTAAAATATAAGTA-N35-ATG | yxiE (4e-11) | 2.0 | 2.7 | |
| lmo2673 | Conserved hypothetical ATP-binding domain [Bacillus anthracis] (4e-28) | AATCATGCTTCTTTCTTTTATTTATGGGTATTAAGTAAATA-N30-ATG | 3.7 | 6.9 | ||
| lmo2386 | Conserved hypothetical protein YuiD [Bacillus subtilis] (le-37) | GAATAGGTTTTTAATAAGCTCATTGTGGTAAAATAAGAATAG-N22-ATG | yuiD (le-50) | 1.8 | 2.4 | |
| lmo2399 | Hemolysin homolog YhdP [Bacillus subtilis] (2e-89) | CTTGTGGTTTAAACGCTAAGAACGGGTATAGAATGGGGA-N15-ATG | 2.0 | 2.5 | ||
| lmo0438 | No similarity | ACTAACGAATCGCGTGGATGCGCTTCGGGAATTGTTGAAATA-N157-GTG | 2.7 | 3.4 | ||
| lmo0994 | No similarity | GAAAGAGGTTATTTTTCACTAAATGGGGTAAAAATACGTTA-N257-ATG | 6.1 | 6.5 | ||
The function of the protein encoded is listed if it has been supported by genetic or biochemical data. Otherwise, the nearest homologue of the protein with known function is listed. The E values in parentheses were obtained with Blast 2.0 searches with the BLOSUM62 matrix against the nr database. PTS, phosphotransferase system.
The promoter sequence as predicted by HMM is listed. For genes without an HMM-predicted promoter sequence (marked with †), a putative promoter identified by visual inspection is indicated; “none found” indicates that no suitable promoter could be found by HMM or visual inspection. The expected −35 and −10 regions are in bold, and the last nucleotide triplet is the presumed translational start codon.
The B. subtilis genome was searched for ORFs encoding proteins homologous to the given L. monocytogenes protein by Blast 2.0 on the SubtiList web server (http://genolist.pasteur.fr/SubtiList). The closest homologue is listed, if one was found, along with the E value in parentheses. Homologues that have been determined to be σB dependent in B. subtilis (51,53) are marked with ‡.
Change was calculated with SAM based on microarray data (see the text) and equals the average wild-type spot intensity divided by the average mutant spot intensity for two separate arrays from each of two replicate RNA isolations. Numbers in italics indicate ratios that were determined by SAM to not be statistically significantly different from a ratio of 1.0. ST, stationary-phase cells; OS, osmotically stressed cells (0.5 M KCl).
Overall, we identified 54 σB-dependent L. monocytogenes genes through combined application of HMM and microarray analyses. Based on identification of the predicted proteins, the genes were grouped into 10 functional groups (Table 2). The two most highly represented categories were transport and metabolism proteins, which together comprised 20 of the 54 genes (Table 2). Statistical analysis of microarray data (SAM [63]) showed that, in addition to high expression ratios, the putative oxidoreductase gene lmo0669 also had the highest significance score for differential expression of all genes represented in the microarray (Table 2). The known σB-dependent betaine/carnitine transporter operon opuC also showed σB-dependent expression, along with eight other solute transport genes. The large proportion of metabolism and transport genes regulated by σB underscores the importance of maintaining proper cellular functions during exposure to stress conditions and suggests that σB provides protection by enhancing the operation of a specialized subset of basic cellular processes, which may help to produce a more stress-resistant state for the cell.
Two other notable functional categories of σB-dependent genes are stress response and virulence genes (Table 2). The stress response genes, which have apparent functions in bacterial stress resistance but do not encode transporters or proteins from other recognized categories included in Table 2, represented some of the most highly differentially expressed genes. For example, in the SAM output, which ranks differentially expressed genes based on the statistical significance associated with the expression ratios, four of the seven stress response genes ranked among the 16 genes with the highest significance values for differential expression, for RNA isolated from cells exposed to osmotic stress. The stress response genes identified include gadB, which was ranked as the third or fifth most significantly differentially expressed gene in cells grown to stationary phase or exposed to osmotic stress, respectively.
Three confirmed virulence genes were expressed at a higher level in the wild-type L. monocytogenes strain compared to the ΔsigB mutant (Table 2). bsh exhibited 20.1-fold higher mRNA levels in the wild type compared to the ΔsigB strain under osmotic stress (Table 2); this expression ratio was determined to be highly significant by SAM analysis (ranked as the second most significant ratio for all the genes in the array). The virulence genes inlA and inlB showed significantly higher expression under osmotic and/or stationary-phase stress in the wild type compared to the mutant strain, with expression ratios consistently >2.0 (Table 2). In addition to these confirmed virulence genes, three other internalins (inlC2, inlD, and inlE) also showed significant expression ratios of >2.0 under osmotic and/or stationary-phase stress (Table 2).
PCR mapping showed that the L. monocytogenes strain used in our study (10403S) bears an inlC2DE operon rather than an inlGHE operon, which is present in the L. monocytogenes EGD-e strain (57). Each ORF of the inlC2DE gene cluster is potentially transcribed independently of the others (13). We identified a σB promoter consensus sequence upstream of both inlC2 and inlE but not inlD. Only one previously described internalin gene (inlC) that was included in our array and is present in strain 10403S did not show differential expression between the wild-type and the ΔsigB mutant strains.
Microarray identification of operons and known σB-dependent genes.
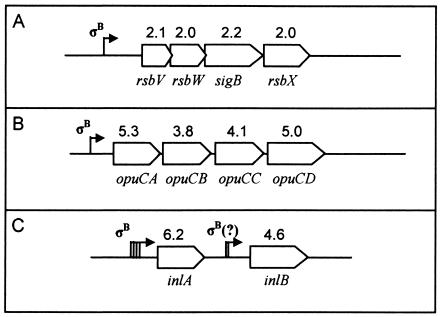
For putative σB-regulated genes that were identified only by HMM, only PCR products from the genes most proximal to given HMM-predicted promoters were spotted on the microarray. Therefore, downstream genes that may have been cotranscribed from a given promoter were not tested for σB dependence in this array. However, the microarray did include genes comprising two operons (opuC and rsbV) that were known a priori to be σB dependent (4, 22). All genes in these two operons were expressed at significantly higher levels in the wild-type compared to the ΔsigB strain. Furthermore, expression ratios for each gene within these operons were approximately equal (Fig. 2A and B). The inlAB operon was not previously known to be regulated by σB, but inlA and inlB were among the virulence genes included on the microarray. We found that both genes in this operon were expressed at higher levels in the presence of σB and that inlA and inlB displayed similar expression ratios (Fig. 2C).
FIG. 2.
Graphic depiction of three operons represented on the microarray. Numbers above genes indicate wild-type-to-mutant expression ratios in stationary-phase cells. Arrows indicate transcriptional start sites. (A) The four genes of the rsbV operon are transcribed from a σB-dependent promoter (7). Transcription can also occur from a σA-dependent promoter further upstream, which also transcribes the four upstream genes rsbR, rsbS, rsbT, and rsbU. (B) The four genes of the opuC operon are transcribed from a σB-dependent promoter (22). (C) Transcription upstream of inlA can be initiated from one of four promoters (40), one of which is σB dependent (as confirmed by RACE). Cotranscription of inlB can occur from at least one of the inlA promoters (40). Transcription of inlB can also occur independently from promoters directly upstream of inlB (i.e., not including inlA), including one promoter that does not have a σB consensus sequence (40) and potentially from one with a visually predicted σB-dependent promoter.
Confirmation of predicted σB-dependent promoters by RACE-PCR.
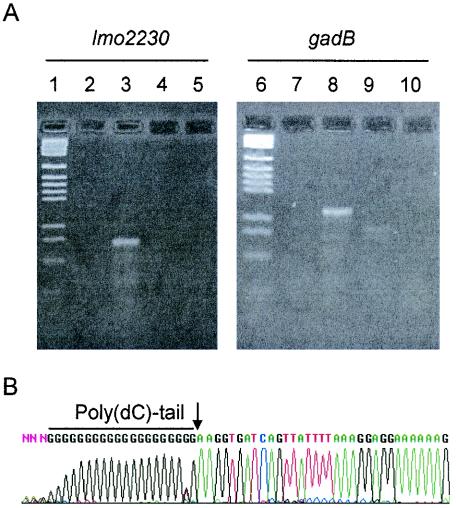
To confirm that predicted σB-dependent promoters were in fact responsible for the differential gene expression seen in the microarrays, we performed RACE-PCR on eight selected genes. These genes were chosen to represent highly differentially expressed genes and genes with high significance scores in SAM as well as known stress response or virulence genes. Gene-specific first-strand cDNA was generated for each gene, and a 3′ poly(dC) tail was added to each cDNA product with terminal transferase. The tailed cDNA product was amplified by touchdown PCR with a poly(G/I) primer (Invitrogen) and a nested gene-specific primer.
PCR bands from wild-type cDNA reactions that met the following conditions were purified and sequenced (Fig. 3): (i) bands that must have been generated by reactions with wild-type cDNA and (ii) equivalent bands that must have been absent in sigB mutant cDNA reactions. For all genes tested, the transcriptional start site determined by RACE was 10 nucleotides (± 2 nucleotides) downstream of the σB-dependent promoter −10 region that had been predicted by HMM or by visual inspection (Fig. 4). The genes selected for promoter confirmation by RACE were bsh and inlA, both virulence genes; lmo0669, a putative metabolic gene; lmo1421 and opuCA (which encode putative and known compatible solute transporter proteins, respectively); and lmo2230, lmo1433, and gadB, all putative or proven stress response genes.
FIG. 3.
Determination of σB-dependent transcriptional start sites by RACE-PCR. (A) Agarose gel electrophoresis of touchdown-PCR amplified poly(dC)-tailed cDNA. DNA was stained with ethidium bromide and visualized under UV light. Two gene-specific experiments (lmo2230 and gadB) are shown. Lanes 1 and 6, pGEM DNA size marker (Promega). Lanes 2 and 7, negative control PCR of untailed wild-type cDNA. Lanes 3 and 8, PCR of poly(dC)-tailed wild-type cDNA. Lanes 4 and 9, PCR of poly(dC)-tailed ΔsigB cDNA. Lanes 5 and 10, negative control PCR of untailed ΔsigB cDNA. (B) A typical chromatogram from sequencing the wild-type RACE-PCR product (gadB shown). As a reverse-oriented primer was used in sequencing reactions, the reverse complement of the sequence obtained is shown in order to depict the actual transcript sequence. The poly(dC) tail is shown, and the transcript beginning is marked with an arrow.
FIG. 4.
Promoter sequences of genes with σB-dependent transcriptional start sites confirmed by RACE-PCR. The −35 and −10 regions are underlined and in bold. Diamonds indicate transcriptional start sites determined by RACE. All promoter sequences displayed were predicted by HMM except for those of inlA and lmo1421, which were identified by visual inspection.
L. monocytogenes σB promoter consensus sequence resembles that of B. subtilis.
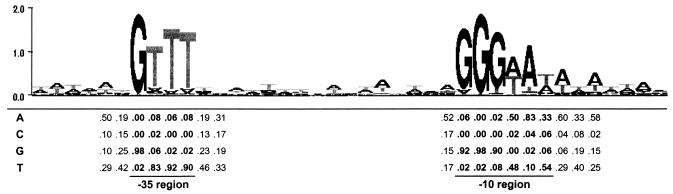
Successful identification of specific σB-dependent genes in L. monocytogenes provided new information needed to facilitate further refinement of the predicted consensus sequence for σB-dependent promoters in L. monocytogenes. The 54 σB-dependent promoter sequences described above were aligned and used to generate a new consensus recognition site (Fig. 5). The −35 region (GTTT) is separated from the −10 region (GGGWAT) by 13 to 17 nucleotides, most frequently by 15 or 16. The guanidine in the −35 box is almost completely conserved (98%). Overall, the L. monocytogenes σB consensus sequence resembles the consensus sequence for B. subtilis σB, which was reported by Petersohn et al. (50, 51) as GTTTAA-N12-15-GGGWAW. This finding is not surprising, as the majority of the sequences used to train the HMM used in this study were obtained from B. subtilis genes. However, the predicted L. monocytogenes σB consensus sequence does differ slightly from the B. subtilis consensus promoter sequence. Specifically, the two adenosines at the 3′ end of the B. subtilis −35 region are not conserved in L. monocytogenes. The different consensus sequences suggest that σB may vary in its ability to recognize certain promoters in these two species.
FIG. 5.
L. monocytogenes consensus sequence logo generated with the GENIO/logo RNA/DNA and Amino Acid Sequence Logos web server (http://genio.informatik.uni-stuttgart.de/GENIO/logo). Predicted promoter sequences of the 54 σB-dependent genes were aligned manually and entered into the program. The vertical axis is information content in bits. The height of a nucleotide represents its frequency at that location. Letters displayed upside down indicate a nucleotide frequency of less than 25%. Numbers below selected residues indicate nucleotide frequencies at that position. Conserved −35 and −10 regions are indicated by bold numbers.
Effect of induction conditions on gene expression patterns.
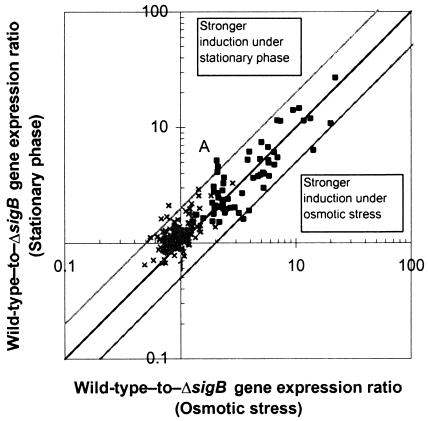
As σB-dependent gene expression patterns were determined under two different stress conditions, we also analyzed whether the magnitude of σB dependence for individual genes varied with the condition. Comparison of the microarray data generated under different stress conditions indicated that the relative magnitude of σB-dependent expression was similar under the two conditions for the genes tested. The scatter plot in Fig. 6 shows the wild-type-to-ΔsigB mutant gene expression ratios obtained for stationary-phase cells plotted against the gene expression ratios for osmotically stressed cells.
FIG. 6.
Comparison of expression ratios for two stress conditions. Wild-type-to-sigB mutant gene expression ratios for stationary-phase and osmotic stress are plotted on the two axes. Genes that are not expressed at a significantly higher rate in the wild type (×) cluster near the origin. Significantly σB-dependent genes (▪) lie near the diagonal, indicating similar expression ratios under both conditions. Outer lines indicate twofold differences in expression ratios among conditions. The point labeled A represents lmo0880, which showed the largest difference in expression ratio.
The majority (67%) of the σB-dependent genes defined as described above showed significantly higher mRNA levels, with expression ratios of >1.5 in the wild-type L. monocytogenes strain under both stress conditions (Table 1). For five genes, the expression ratios between the two stress conditions varied from each other by more than twofold; the largest difference was 2.5-fold (lmo0880, point A in Fig. 6). However, the majority of the genes (39 out of 55) had expression ratios that differed by less than 1.5-fold under the two stress conditions. These results suggest that the majority of genes comprising the σB regulon are induced to a similar extent in relation to each other regardless of the specific environmental stress. A subset of σB-dependent genes (e.g., lmo0880) may also be subject to additional transcriptional regulation by other stress-specific mechanisms. This is not to say, however, that all stress conditions activate σB to the same degree. For example, while our specific conditions resulted in similar induction levels among the targeted genes (slope of regression = 0.89), primer extension data suggest that induction of transcription from the σB-dependent L. monocytogenes rsbV promoter may vary under the different stress conditions (4).
DISCUSSION
To improve our understanding of the specific roles of the alternative sigma factor σB in regulating gene expression and bacterial stress response, we used a combination of whole-genome HMM similarity searches and microarray-based strategies to identify members of the σB regulon in the facultative intracellular pathogen L. monocytogenes. A 208-gene microarray, which included 166 HMM-identified L. monocytogenes genes located downstream of putative σB-dependent promoter sequences as well as selected additional virulence and stress response genes, allowed us to identify 54 genes as being directly regulated by L. monocytogenes σB. Transcriptional start site mapping on selected genes confirmed the functionality of putative σB-dependent promoters at the locations predicted by the HMM or by visual inspection.
Similar to the σB regulon previously defined for B. subtilis, a gram-positive soil bacterium closely related to the genus Listeria, members of the L. monocytogenes σB regulon were shown to encode a variety of protein functions involved in metabolic pathways, transport, and other fundamental cellular functions. As expected, many genes identified in this study encode proteins directly associated with stress resistance. Interestingly, several genes identified as σB dependent represent putative or known virulence genes (e.g., inlE and bsh, respectively) or stress response genes that have been shown to contribute to the ability of L. monocytogenes to survive within an infected host (e.g., opuC [60, 66]) or under conditions similar to those encountered in an infected host (e.g., gadB [11]). These results suggest that σB, in addition to enhancing bacterial survival under stress conditions, also contributes to virulence in L. monocytogenes.
Role of σB in L. monocytogenes stress resistance.
Phenotypic characterization of L. monocytogenes strains lacking sigB has shown that σB plays an important role in resistance to osmotic, oxidative, and acid stresses (4, 18, 19, 22, 67). Similar studies with B. subtilis ΔsigB mutants also provided evidence for the importance of a functional σB for survival under ethanol, oxidative, osmotic, and acid stresses (2, 25, 65). The σB-dependent L. monocytogenes genes identified here provide additional insight into the functional bases of the reduced stress resistance in L. monocytogenes ΔsigB mutants. For example, gadB, which encodes a glutamate decarboxylase important for acid stress survival in L. monocytogenes (11), was shown in this work to be part of the L. monocytogenes σB regulon.
Interestingly, both the L. monocytogenes and the B. subtilis σB regulons include multiple genes that have been shown experimentally to contribute to protection against osmotic stress, including genes encoding several solute transporters. The ABC transporter OpuC contributes to osmotic stress resistance by facilitating uptake of the osmoprotectants choline and glycine betaine in both L. monocytogenes and B. subtilis (1, 35, 36, 60), and expression of the encoding operon is σB dependent in L. monocytogenes. Similarly, lmo1421 and the B. subtilis homologue opuB, each of which is a member of the σB regulon in its species, both encode a putative choline transporter which may also contribute to osmotic stress resistance (22, 36). In addition, ctc, which shows σB-dependent expression in both L. monocytogenes (this study) and B. subtilis (29, 33), was recently shown to contribute to osmotolerance in L. monocytogenes (26).
Finally, the σB regulon in both L. monocytogenes and B. subtilis also includes genes that appear to contribute to oxidative stress resistance, consistent with the observation that an L. monocytogenes ΔsigB mutant shows increased susceptibility to oxidative stress (18). From our data, we propose that σB-dependent resistance to oxidative damage in L. monocytogenes is likely due, at least in part, to glutathione reductase, which is encoded by lmo1433. Glutathione reductase is an enzyme that provides protection from oxidative stress by reducing glutathione disulfide to glutathione (9). L. monocytogenes has been shown to accumulate glutathione, supporting the possible activity of this enzyme during oxidative stress (47). While no glutathione reductase homologue was found in B. subtilis, oxidative stress protection in B. subtilis is dependent on the DNA-binding protein Dps, as well as on the trxA-encoded thioredoxin, both of which are σB dependent (59). Our HMM searches did not identify L. monocytogenes trxA as bearing a σB-dependent promoter.
Role of σB in virulence gene expression.
Characterization of L. monocytogenes ΔsigB mutants in both tissue culture and murine models of infection has provided initial evidence that σB contributes to the ability of L. monocytogenes to cause infection. While it has been shown that transcription from one of three prfA promoters (specifically the P2 promoter) is abolished in a ΔsigB mutant, no additional evidence for other functional contributions of σB to virulence have been reported. Our results provide clear evidence that σB in L. monocytogenes contributes to transcriptional activation of genes directly associated with virulence as well as those that likely contribute indirectly to virulence (58).
Virulence genes defined as being part of the σB regulon include bsh (encoding a bile salt hydrolase) as well as two genes from the internalin family (inlA and inlB). Although σB-dependent transcription of inlA and bsh has also been independently verified by reverse transcription-PCR (D. Sue and M. Wiedmann, unpublished data), and while RACE-PCR confirmed the presence of functional σB promoters for these genes, it is important to note that both genes also appear to be transcribed from σB-independent promoters (14, 15).
Interestingly, both bsh and inlA are regulated by PrfA (14, 15), a positive transcriptional regulator of virulence gene expression. While PrfA binding has been shown to activate transcription by binding to the −35 promoter region, none of the PrfA binding boxes upstream of the σB-dependent virulence genes described here overlap the σB consensus promoter −35 region. However, the σB-dependent P2 promoter of prfA contains a sequence resembling a PrfA box (23), and Milohanic et al. (42) reported a gene, lmo0596, with a putative PrfA box located at the −35 region of a predicted σB-dependent promoter. We conclude that transcription of a subset of L. monocytogenes virulence genes is regulated by a complex regulatory network that can include activation by both PrfA and σB. We hypothesize that this PrfA/σB regulatory mechanism coordinates transcriptional activation of subsets of L. monocytogenes virulence genes in specific host environments, e.g., bsh and inlA have been shown to be critical for listerial pathogenesis in the intestine (15, 39).
While bsh, inlA, and inlB represent experimentally verified virulence genes which were identified as being σB dependent, we also defined as members of the σB regulon additional putative L. monocytogenes virulence genes, including additional internalins. The L. monocytogenes internalins represent a diverse group of surface proteins with confirmed or putative virulence functions (39, 49, 57). While internalin B does not display an LPXTG motif, the other internalins include this cell wall anchor domain. Proteins with LPXTG motifs are common among gram-positive bacteria, and their functions are broad in range and frequently affect virulence (reviewed in reference 46). In addition to inlA, we identified four other L. monocytogenes genes encoding putative cell wall-anchored proteins displaying an LPXTG motif (including the internalin genes inlC2 and inlE) as being σB dependent. While inlC2 and inlE, which are part of the inlC2DE operon in L. monocytogenes 10403S, have not been shown experimentally to contribute to virulence, members of the homologous inlGHE operon present in other L. monocytogenes strains (e.g., EGD-e) have been shown to contribute to host cell internalization (5, 57). In addition to two other σB-dependent genes encoding proteins with LPXTG motifs (lmo2085 and lmo0880), we also identified one σB-dependent gene (lmo0994) that is unique to L. monocytogenes (i.e., absent from Listeria innocua and B. subtilis). Further studies with appropriate null mutants will be necessary to determine the specific functions of these putative σB-dependent virulence genes.
In addition to the established or putative virulence genes discussed above, we also identified σB-dependent stress response genes that had been shown previously to contribute to L. monocytogenes virulence, infection, and intrahost survival. Examples include the σB-dependent opuC operon; an L. monocytogenes LO28 opuC mutant showed reduced colonization of the mouse upper small intestine following peroral inoculation (60) and reduced numbers of bacteria in the spleens and livers of infected mice (66), although the phenotype appears to be strain specific (60). The σB-dependent gadB was previously shown to contribute to L. monocytogenes acid survival, including survival in synthetic gastric and ex vivo porcine stomach fluids (11). In conjunction with our findings described above, these data support a broad role for σB-dependent genes in L. monocytogenes virulence and intrahost survival. The majority of L. monocytogenes virulence studies thus far have used the mouse model of infection, but the mouse lacks the appropriate E-cadherin receptor, which is particularly critical for gastrointestinal invasion by L. monocytogenes (67). Use of more appropriate animal models to study L. monocytogenes pathogenesis (e.g., guinea pigs [39]) may help us to more clearly define the in vivo contributions of σB to L. monocytogenes virulence.
Studies in other gram-positive pathogens have provided further evidence that σB contributes to directing gene expression during host infection. In S. aureus, the virulence determinant SarA is expressed from a promoter that is strictly σB dependent (6, 41). SarA specifically activates transcription of agr, which encodes a positive regulator of extracellular virulence gene expression (43, 48). In both S. aureus and S. epidermidis, σB is also required for biofilm formation, a probable prerequisite for establishing infections (37, 54, 55). σB contributes to virulence in Bacillus anthracis; a sigB mutation in B. anthracis severely attenuates virulence in a murine model of infection (21). In addition to σB, stress-responsive sigma factors may contribute broadly to bacterial virulence. For example, the gram-negative stress-responsive alternative sigma factor RpoS has also been shown to contribute to virulence in a variety of pathogens, including Yersinia spp., Salmonella spp., Pseudomonas aeruginosa, and Legionella pneumophila (3, 17, 34, 61).
Comparative genomics of the σB-dependent stress response.
The definition of 54 σB-dependent genes in L. monocytogenes provides a unique opportunity for a comparative evaluation of the predicted functions of the σB-dependent stress response among low-GC-content, gram-positive bacteria. Overall, the range of protein functions associated with the L. monocytogenes σB-dependent genes defined in this work appears to be similar to the range of functions encoded by the genes described for the B. subtilis σB regulon (51, 53). Through similarity searches, we determined that 31 of the 54 σB-dependent genes in L. monocytogenes have homologous gene sequences in B. subtilis and 12 of these 31 genes show σB-dependent expression in both L. monocytogenes (this work) and B. subtilis (51, 53). These 12 genes include the stress response genes ctc, ltrC, and lmo1602, the rsbV operon, and metabolic genes (see Table 2). These genome-scale comparisons of the L. monocytogenes and the B. subtilis σB regulons thus suggest that the σB-dependent stress response system has adapted in L. monocytogenes to facilitate pathogen-host interactions.
The analysis of the S. aureus σB regulon performed by Gertz et al. (27) identified predominantly uncharacterized proteins. While 20 of the 27 σB-dependent S. aureus proteins identified had homologues in B. subtilis, only 7 of those homologues were σB dependent in B. subtilis. This finding parallels our own in that only a portion of the B. subtilis genes that are homologous to the σB-dependent genes identified in L. monocytogenes are also σB dependent in B. subtilis. Taken together, these observations suggest that the σB regulon has evolved to serve different roles among these bacteria.
Towards defining the complete L. monocytogenes σB regulon.
Stress-responsive alternative sigma factors, including RpoS in gram-negative bacteria (32) and σB in low-GC-content, gram-positive bacteria, contribute to regulation of large sets of genes, both directly and indirectly (51). As commercial, full-genome microarrays become available, more genes than those found here can be identified as σB dependent, including those not identified by the original HMM search. Still, defining all genes regulated by an alternative sigma factor represents a significant challenge, even with the availability of technologies such as two-dimensional gel electrophoresis and full-genome microarrays.
While our identification of 54 σB-dependent genes likely represents about one-third of the L. monocytogenes σB regulon, which is estimated to contain around 150 genes, the functional diversity represented by the proteins encoded by these genes provides valuable new insight into the specific functions of the L. monocytogenes σB regulon. In addition to the 54 genes reported here, many additional members of the L. monocytogenes σB regulon may have been correctly predicted by our HMM search, but their detection may have been masked by redundant regulation of transcription. Additional approaches such as in vitro transcription analyses with a purified L. monocytogenes σB-RNAP complex (8) or transcriptional profiling of an L. monocytogenes strain with an inducible sigB (53) will likely be necessary to identify and confirm additional members of the σB regulon. We have demonstrated, however, that a combination of HMM-based similarity searches and construction of a microarray as described here represents an efficient and economical approach for defining genes regulated by a specific mechanism, such as an alternative sigma factor.
Acknowledgments
We thank Paul Debbie and Nai-Chun Lin for technical assistance in creating and working with microarrays and John Helmann for helpful discussions about the project and manuscript.
This research was supported in part by the Cornell University Agricultural Experiment Station federal formula funds, project no. NYC-143422, received from the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Agriculture.
REFERENCES
- 1.Angelidis, A. S., L. T. Smith, L. M. Hoffman, and G. M. Smith. 2002. Identification of opuC as a chill-activated and osmotically activated carnitine transporter in Listeria monocytogenes. Appl. Environ. Microbiol. 68:2644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., S. Engelmann, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 4.Becker, L. A., M. S. Çetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 8.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. W. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 9.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 10.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 12.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dramsi, S., P. Dehoux, M. Lebrun, P. L. Goossens, and P. Cossart. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 65:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dramsi, S., C. Kocks, C. Forestier, and P. Cossart. 1993. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator PrfA. Mol. Microbiol. 9:931-941. [DOI] [PubMed] [Google Scholar]
- 15.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 16.Epstein, C. B., W. t. Hale, and R. A. Butow. 2001. Numerical methods for handling uncertainty in microarray data: an example analyzing perturbed mitochondrial function in yeast. Methods Cell Biol. 65:439-452. [DOI] [PubMed] [Google Scholar]
- 17.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, K. R., D. Sue, M. Wiedmann, and K. J. Boor. 2003. The role of σB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is σB dependent. Appl. Environ. Microbiol. 69:2015-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes PrfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 24.Furrer, B., U. Candrian, C. Hoefelein, and J. Luethy. 1991. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J. Appl. Bacteriol. 70:372-379. [DOI] [PubMed] [Google Scholar]
- 25.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardan, R., O. Duche, S. Leroy-Setrin, and J. Labadie. 2003. Role of ctc from Listeria monocytogenes in osmotolerance. Appl. Environ. Microbiol. 69:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gertz, S., S. Engelmann, R. Schmid, A.-K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. d. Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. G.-D. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueño, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. d. Pablos, J.-C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 29.Haldenwang, W. G., C. D. Banner, J. F. Ollington, R. Losick, J. A. Hoch, M. B. O'Connor, and A. L. Sonenshein. 1980. Mapping a cloned gene under sporulation control by insertion of a drug resistance marker into the Bacillus subtilis chromosome. J. Bacteriol. 142:90-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 32.Ibanez-Ruiz, M., V. Robbe-Saule, D. Hermant, S. Labrude, and F. Norel. 2000. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:5749-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igo, M. M., and R. Losick. 1986. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J. Mol. Biol. 191:615-624. [DOI] [PubMed] [Google Scholar]
- 34.Iriarte, M., I. Stainier, and G. R. Cornelis. 1995. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 63:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 37.Knobloch, J. K.-M., K. Bartscht, A. Sabottke, H. Rohde, H.-H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 40.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 43.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 44.Moszer, I., P. Glaser, and A. Danchin. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 45.Nadon, C., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. σB contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parida, S. K., E. Domann, M. Rohde, S. Muller, A. Darji, T. Hain, J. Wehland, and T. Chakraborty. 1998. Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol. Microbiol. 28:81-93. [DOI] [PubMed] [Google Scholar]
- 50.Petersohn, A., J. Bernhardt, U. Gerth, D. Hoper, T. Koburger, U. Volker, and M. Hecker. 1999. Identification of σB-dependent genes in Bacillus subtilis with a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 53.Price, C. W., P. Fawcett, H. Cérémonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 54.Raad, I., A. Alrahwan, and K. Rolston. 1998. Staphylococcus epidermidis: emerging resistance and need for alternative agents. Clin. Infect. Dis. 26:1182-1187. [DOI] [PubMed] [Google Scholar]
- 55.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raddatz, G., M. Dehio, T. F. Meyer, and C. Dehio. 2001. PrimeArray: genome-scale primer design for DNA-microarray construction. Bioinformatics 17:98-99. [DOI] [PubMed] [Google Scholar]
- 57.Raffelsbauer, D., A. Bubert, F. Engelbrecht, J. Scheinpflug, A. Simm, J. Hess, S. H. Kaufmann, and W. Goebel. 1998. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol. Gen. Genet. 260:144-158. [DOI] [PubMed] [Google Scholar]
- 58.Roberts, A., and M. Wiedmann. 2003. Listeria monocytogenes as a model for the pathogenesis of human foodborne and animal infectious diseases. Cell. Mol. Life Sci. 60:904-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sleator, R. D., J. Wouters, C. G. Gahan, T. Abee, and C. Hill. 2001. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tseng, G. C., M. K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress response confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wemekamp-Kamphuis, H. H., J. A. Wouters, R. D. Sleator, C. G. Gahan, C. Hill, and T. Abee. 2002. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]