Abstract
The Escherichia coli cytoplasmic protein thioredoxin 1 can be efficiently exported to the periplasmic space by the signal sequence of the DsbA protein (DsbAss) but not by the signal sequence of alkaline phosphatase (PhoA) or maltose binding protein (MBP). Using mutations of the signal recognition particle (SRP) pathway, we found that DsbAss directs thioredoxin 1 to the SRP export pathway. When DsbAss is fused to MBP, MBP also is directed to the SRP pathway. We show directly that the DsbAss-promoted export of MBP is largely cotranslational, in contrast to the mode of MBP export when the native signal sequence is utilized. However, both the export of thioredoxin 1 by DsbAss and the export of DsbA itself are quite sensitive to even the slight inhibition of SecA. These results suggest that SecA may be essential for both the slow posttranslational pathway and the SRP-dependent cotranslational pathway. Finally, probably because of its rapid folding in the cytoplasm, thioredoxin provides, along with gene fusion approaches, a sensitive assay system for signal sequences that utilize the SRP pathway.
Proteins that are exported from the cytosol by the Sec pathways of either prokaryotes or eukaryotes have special features that promote efficient translocation across membranes. These features include the amino-terminal hydrophobic signal sequences that are cleaved during the translocation process. Furthermore, some feature of the protein or of the export process must ensure that the protein does not fold into its stable three-dimensional structure in the cytoplasm, which would prevent export. A protein may be maintained in an export-competent state in several different ways: (i) the protein may be translocated across a membrane simultaneously with translation of the protein, thus ensuring that not even its secondary structures are formed in the cytoplasm; (ii) for proteins that are not translocated simultaneously with translation, chaperones or antifolding factors may prevent folding in the cytoplasm (25); (iii) in some cases, signal sequences act as intrapolypeptide chaperones to prevent rapid folding (19); and (iv) proteins that are exported posttranslationally may have features in their final structures (e.g., disulfide bonds) that do not form in the environment of the cytoplasm so that the proteins cannot attain their final folded conformations in the cytoplasm (8).
In eukaryotes, the signal recognition particle (SRP), made up of six proteins and an RNA molecule, directs rapid cotranslational translocation of many proteins (20). Protein synthesis itself is thought to provide the energy to push the proteins through the endoplasmic reticulum membrane-embedded translocase. The prokaryotic SRP is composed of one protein (Ffh) and a 4.5S RNA and is probably mainly involved in the cotranslational assembly of integral membrane proteins (20). In eubacteria, most, if not all, proteins with cleavable signal sequences are thought to be exported by a relatively slow, partially posttranslational pathway (13); that is, they do not begin to pass through the SecYEG translocase in the cytoplasmic membrane until substantial amounts of the polypeptide chain have been synthesized in the cytoplasm. The energy for this translocation is thought to come from the ATPase activity of the SecA component of the translocase.
Many, but not all, slowly exported Escherichia coli proteins bind the chaperone SecB, which prevents the large nascent polypeptide from folding into a conformation that would interfere with translocation (19). Among such proteins is the maltose binding protein (MBP) encoded by the malE gene; in a secB mutant, MBP folds in the cytoplasm and is inefficiently exported (4, 16). Mutations in the mature portion of MBP that interfere with folding of the protein restore substantial amounts of export of the protein in a secB background (5). Similar folding mutations were obtained with ribose binding protein and MBP containing signal sequence alterations, although ribose binding protein export is not SecB dependent (18, 27, 32).
Workers in numerous laboratories have encountered difficulties in translocating cytoplasmic proteins across the membrane into the periplasm. In many cases, the inefficient export may be explained by the more rapid folding of these proteins in the cytoplasm than of proteins destined to be secreted. It can be imagined that cytoplasmic proteins, in contrast to, for example, periplasmic proteins, have evolved to fold more rapidly and to avoid antifolding factors. For example, it has been shown that when the signal sequence of the periplasmic enzyme alkaline phosphatase (PhoAss) is fused directly to the cytoplasmic protein thioredoxin 1 (referred to below as thioredoxin), very little (perhaps a few percent) of the thioredoxin reaches the periplasm (6). This result was explained by the proposal that thioredoxin, which is known to have a rapid protein folding pathway, assembles into its stable three-dimensional structure before it has a chance to be exported by the Sec pathway. However, Jonda et al. (11) reported that with an analogous fusion in which the signal sequence of DsbA (a periplasmic disulfide bond oxidoreductase) (DsbAss) is fused directly to thioredoxin, the thioredoxin is efficiently translocated into the periplasm.
We decided to explore the strikingly different efficiencies of export of thioredoxin by PhoAss and DsbAss. The results reported here indicate that DsbAss directs thioredoxin to the SRP pathway, where cotranslational translocation obviates the inhibitory effect of protein folding on export. Analogously, when DsbAss is fused to MBP, it directs MBP to the SRP pathway, thus bypassing the usual posttranslational export of MBP and obviating the requirement for the SecB antifolding factor. Our results show that even when export is proceeding via the SRP pathway, reductions in SecA activity interfere with this export. These findings indicate that SecA plays a role in cotranslational export. They also suggest that, via gene fusion analysis, thioredoxin can provide a sensitive assay system for signal sequences that direct proteins to the SRP pathway.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used are listed in Table 1. Cells were generally grown at 37°C in NZ medium, which has been described previously (26). For pulse-labeling, cells were grown in minimal glucose medium (M63 salts with 0.2% glucose, 1 mg of vitamin B1per ml, 1 mM MgSO4, and a solution containing 18 amino acids, each at a concentration of 50 μg/ml [methionine and cysteine were not included in the amino acid solution]). Strains containing secB mutant alleles were grown in minimal glycerol medium (M63 salts with 0.2% glycerol, 1 mg of vitamin B1 per ml, and 1 mM MgSO4). Temperature-sensitive secA mutant strains were grown at 30°C for permissive growth and were shifted to 37°C for nonpermissive conditions. Antibiotic selection was maintained for selected markers at the following concentrations: ampicillin, 200 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 40 μg/ml; tetracycline, 15 μg/ml; and rifampin, 100 μg/ml. Induction of lac promoter constructs was accomplished by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or features | Reference or source |
|---|---|---|
| Strains | ||
| MC1000 | F−araD139 Δ(ara-leu)7697 rpsL150 galU galK thi | 7 |
| MC4100 | F−araD139 ΔlacU169 rpsL150 thi rbsR | 34 |
| DHB3 | MC1000 ΔmalF3 Δ(phoA[PvuII]) phoR | 10 |
| DHB4 | DHB3/F′ lac-pro lacIq | 10 |
| DRH223 | JAH143 ΔtrxA | This study |
| DRH226 | JAH143 ΔtrxA secA51 | This study |
| JAH143 | MC1061 ΔmalE444 | This study |
| WP570 | DHB4 ΔtrxA | Lab stock |
| CFS330 | JAH143 secA51 | This study |
| CFS401 | MC4100 Δara714 ffh87 ΔmalE444 | This study |
| CFS456 | CFS403 ffh+ | This study |
| CFS403 | MC4100 Δara714 ffh87 ΔtrxA | This study |
| CFS406 | MC4100 secB::Tn5 ΔmalE444 | This study |
| CFS469 | MC4100 Δara714 ffh87 ΔmalE444 secB::Tn5 | This study |
| Plasmids | ||
| pTRC99a | IPTG inducible, high expression | Promega |
| pDSW204 | pTRC99a, attenuated promoter | 36 |
| pDSW206 | pTRC99a, attenuated promoter | 36 |
| pCFS117 | pDSW206 + malEss | This study |
| pCFS118 | pCFS117 + trxA | This study |
| pCFS119 | pDSW204 + dsbAss | This study |
| pCFS122 | pDSW206 + phoAss | This study |
| pCFS123 | pCFS119 + trxA | This study |
| pCFS126 | pCFS122 + trxA | This study |
| pCFS127 | pDSW206 + malE | This study |
| pCFS128 | pCFS119 + malE(ΔmalEss) | This study |
| pCH3 | PBAD18 dsbA+ | 9 |
Strain and plasmid construction.
Standard genetic and molecular techniques were used for strain and plasmid construction (21, 29). Plasmids containing only signal sequences were generated by digesting the parent plasmid with NcoI and ligating in a double-stranded oligonucleotide with compatible cohesive ends. This resulted in elimination of the NcoI site at the 5′ end of the signal sequence and retention of an in-frame NcoI site at the 3′ end of the signal sequence, which served as an acceptor for coding regions of proteins to be exported. As we found that the DsbAss-TrxA and MBP signal sequence (MBPss)-TrxA constructs were expressed at different levels when they were fused to the trc promoter of pTRC99a, we incorporated the two constructs into different versions of pTRC99a carrying trc promoters with different transcription efficiencies (plasmids pDSW204 and pDSW206). This was done to ensure that the two thioredoxin constructs were expressed at comparable levels.
Fractionation.
Cells were grown as described above to saturation and then subcultured 1:100 and grown to an optical density at 600 nm of 0.5. Cultures were centrifuged, and pellets were resuspended in 50 mM Tris (pH 8)-18% sucrose-1 mM CaCl2. EDTA (0.5 mM) and lysozyme (0.5 μg/ml) were added, and samples were left on ice for 30 min before centrifugation at 3,140 × g in a bench top centrifuge for 5 min. The supernatants and pellets were used as periplasmic and spheroplast fractions, respectively.
Western blotting.
Cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes by using a semidry apparatus obtained from Bio-Rad. The membranes were probed by using laboratory stocks of antiserum. Rabbit anti-thioredoxin and anti-β-lactamase antibodies for probing membranes were obtained from Sigma and 5′-3′, respectively. All other antisera used for probing membranes were obtained from laboratory stocks. Immunodetection was done by using the ECL protocol (Amersham) and streptavidin-horseradish peroxidase.
Labeling with [35S]methionine and immunoprecipitation.
Cells were grown at 37°C in M63 salts supplemented with all the amino acids except methionine and cysteine, 0.2% glucose, and 1 mM IPTG. When the culture reached an optical density at 600 nm of approximately 0.2, aliquots for the control sample were removed, and 3 mM azide was added for 5 min. The cells were labeled with 60 μCi of [35S]methionine per ml and 10 μg of methionine per ml to give a final methionine concentration of 90 mM for 15 s. For identification of polypeptides with or without their signal peptides, standards were prepared by labeling for 5 min in the presence or absence of azide, which allowed full incorporation of [35S]methionine. Labeling was terminated by addition of trichloroacetic acid to a concentration of 5%, and the resultant precipitate was washed once with acetone to remove the excess trichloroacetic acid. Samples were resuspended in 3× loading dye (Sigma B7709S) and were immunoprecipitated by using anti-MBP antibodies and protein A as described previously (15). Three immunoprecipitated samples were pooled and resolved in an SDS-PAGE gel. This gel was designated the first-dimension gel.
Estimating signal sequence hydrophobicity.
The hydrophobicity of the signal sequences was assessed by using an algorithm modified from the algorithm of Boyd et al. (3). The software scanned a given sequence by using various window lengths (in amino acids) and recorded the average hydrophobicity of the most hydrophobic window. Using three different previously published hydrophobicity scales (3) and various window lengths (ranging from 6 to 13 amino acids), we found that DsbAss consistently appeared to be more hydrophobic than MBPss and PhoAss.
Two-dimensional limited proteolysis.
The method used to analyze processing of nascent chains by limited proteolysis has been described previously (14). The same principles were applied for analysis of the processing of MBPss-MBP and DsbAss-MBP. The entire lane from the first-dimension SDS-PAGE gel containing the polypeptides to be digested was excised and laid lengthwise on top of a second SDS-PAGE gel. The samples were subjected to limited proteolysis by Staphylococcus aureus V8 protease in the stacking gel. The gel was dried and subjected to fluorography by using sodium salicylate as a fluor, as described previously (2).
RESULTS
Comparison of efficiencies of thioredoxin export by different signal sequences.
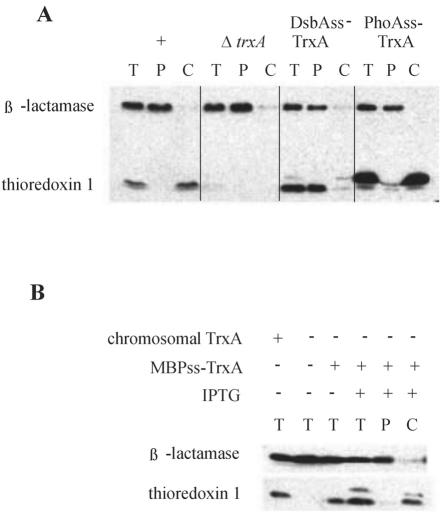
Previous experiments examining the export of thioredoxin by PhoAss and experiments of Jonda et al. in which DsbAss was used were performed with different plasmids and promoters (6, 7, 11). Therefore, we first verified the previously reported differences in export efficiency by cloning both constructs into derivatives of the same plasmid in which the constructs were under the control of a version of the lac promoter (trc) (see Materials and Methods). The expression and secretion of thioredoxin were studied in a strain in which the chromosomal gene (trxA) encoding thioredoxin was deleted. Western blotting with thioredoxin antibody resulted in findings that replicated the differences reported by Jonda and coworkers: PhoAss allowed export to the periplasm of only a tiny fraction (a few percent) of thioredoxin, while DsbAss efficiently exported the same protein (in some experiments up to 100% of the protein was exported) (Fig. 1). We also examined the ability of a third signal sequence, that of MBP, to promote export of thioredoxin. MBPss was fused directly to the amino terminus of thioredoxin. In this case, we found that there was inefficient export of thioredoxin, comparable to that seen with PhoAss (Fig. 1).
FIG. 1.
Export of thioredoxin fused to various signal sequences. (A) Western analysis of fractionated extracts from cells expressing wild-type thioredoxin (+) (DHB4), cells with the trxA gene deleted (ΔtrxA) (WP570), cells expressing thioredoxin fused to DsbAss (DsbAss-TrxA) (WP570/pCFS120), and cells expressing thioredoxin fused to PhoAss (PhoAss-TrxA) (WP570/pCFS122). β-Lactamase was included as a periplasmic control. (B) Western analysis of fractionated extracts from cells expressing either chromosomal thioredoxin or thioredoxin fused to MBPss (MBPss-TrxA) (WP570/pCFS118). IPTG was used to induce expression of the MBPss-TrxA construct, as indicated. β-Lactamase was included as a periplasmic control. T, total extract; P, periplasmic fraction; C, cytoplasmic fraction plus cytoplasmic membrane fraction.
These results led us to consider two general explanations for the different efficiencies of the three signal sequences: (i) DsbAss acted as an inhibitor of the folding of thioredoxin, thus allowing effective posttranslational export; or (ii) DsbAss was able to direct more rapid engagement of the protein with the secretory machinery.
DsbAss promotes export of proteins by the SRP pathway.
We next attempted to determine differences between the signal sequence that efficiently exported thioredoxin (DsbAss) and the two signal sequences that did not (PhoAss and MBPss). The most obvious feature to examine was the levels of hydrophobicity of the three signal sequences. Using different methods for estimating hydropathy, we consistently found that DsbAss was significantly more hydrophobic than the other two signal sequences (data not shown) (see Materials and Methods). Since at about this time Lee and Bernstein (17) reported that they had engineered cleavable signal sequences with higher hydrophobicity than normal and that proteins attached to such signal sequences were exported by the SRP pathway, we asked whether the DsbAss-thioredoxin fusion protein was dependent on the SRP pathway for export. This possibility appeared to be a satisfying explanation for our results, as the rapid folding of thioredoxin might require a truly cotranslational mode of export for the protein to be translocated into the periplasm.
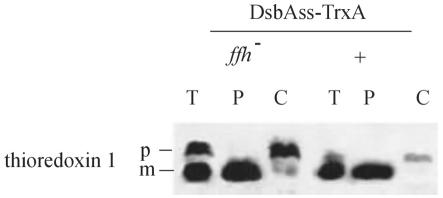
Strains carrying mutations in the genes for SRP components have been described previously (33, 34). These strains are viable, as the mutations have only weak effects on the pathway. Such mutants are useful for critically analyzing the dependence of proteins on the SRP pathway, as mutations in the depletion strains ordinarily used in such studies have more profound consequences for cell physiology and the observed effects may be indirect. We constructed strains expressing the DsbAss-thioredoxin fusion from a plasmid in a background containing a trxA deletion and an ffh (viable) mutation. In this strain thioredoxin export was reduced by 40% compared to the export in an Ffh+ strain (Fig. 2); that is, the wild-type strain contained very little thioredoxin in the cytoplasm, while nearly one-half of the thioredoxin was localized to the cytoplasm in the ffh background.
FIG. 2.
Export of thioredoxin fused to DsbAss is impaired by a mutation in the ffh gene: Western analysis of fractionated cell extracts from either Ffh+ cells (+) (CFS456) or ffh mutant cells (ffh−) (CFS403) expressing thioredoxin fused to DsbAss (pCFS123). T, total extract; P, periplasmic fraction; C, cytoplasmic fraction plus cytoplasmic membrane fraction; p, precursor containing the signal sequence; m, mature protein.
These results lend themselves to a straightforward interpretation of the differences between the efficiency of export of thioredoxin when it is fused to PhoAss or MBPss on the one had and the efficiency of export of thioredoxin when it is fused to DsbAss on the other. The first two signal sequences are comparatively weakly hydrophobic and are capable of directing export only by the slow (non-SRP) pathway. In this pathway, thioredoxin, which folds rapidly in the cytoplasm, has ample time to assemble into its three-dimensional structure, thus preventing export. In contrast, DsbAss directs thioredoxin to export through the rapid cotranslational SRP pathway. In this case, the protein is exported before it has a chance to fold.
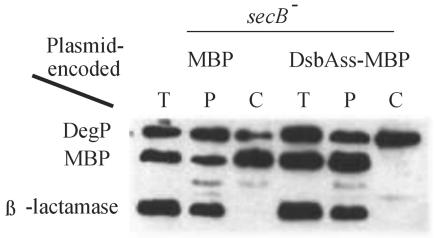
If this interpretation is correct, then we would predict that DsbAss could promote export of other proteins via the SRP pathway. MBP appeared to be an excellent candidate to test this prediction as export of this protein has been well characterized. Wild-type MBP is exported via the slow pathway and thus requires the SecB chaperone to maintain it in an export-competent state. When we fused DsbAss to the mature sequence of MBP, we found that the protein was exported efficiently, even in the absence of SecB, in contrast to MBP containing its native MBPss (Fig. 3).
FIG. 3.
DsbAss alleviates the SecB dependence of MBP export: Western analysis of fractionated extracts from secB mutant cells expressing wild-type MBP (MBP) (CFS406/pCFS127) or MBP with its signal sequence replaced by that of DsbA (DsbAss-MBP) (CFS406/pCFS128). DegP and β-lactamase were included as examples of SecB-dependent and SecB-independent protein export, respectively. T, total extract; P, periplasmic fraction; C, cytoplasmic fraction plus cytoplasmic membrane fraction.
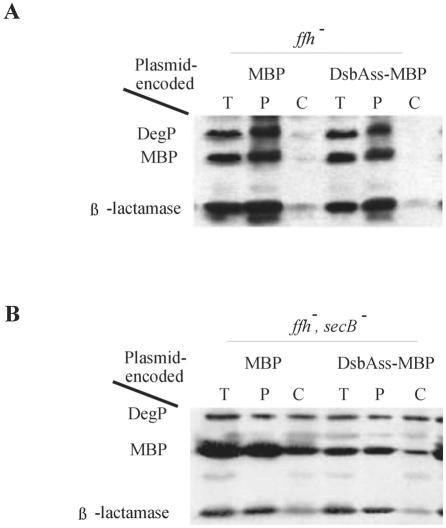
We also asked whether an SRP mutation would interfere with DsbAss-MBP export in a strain carrying an ffh mutant which was otherwise wild type (SecB+). In contrast to the results obtained with DsbAss-thioredoxin, the export of DsbAss-MBP was not affected by the SRP mutation (Fig. 4). This is not surprising and even was expected, as (i) the effects of the ffh mutation are weak and (ii) in the somewhat defective SRP background DsbAss-MBP should still be perfectly capable of utilizing the posttranslational pathway especially because of the presence of SecB. However, this interpretation predicted that in an ffh secB background, in which both co- and posttranslational pathways should be partially disabled, defects in MBP export would be seen. This was the case (Fig. 4).
FIG. 4.
Mutation in ffh restores the SecB dependence of export of MBP fused to DsbAss. (A) Western analysis of fractionated extracts of ffh mutant cells expressing wild-type MBP (MBP) (CFS401/pCFS127) or MBP with its signal sequence replaced by that of DsbA (DsbAss-MBP) (CFS401/pCFS128). (B) Western analysis of fractionated extracts of ffh secB double mutant cells expressing wild-type MBP (MBP) (CFS469 and CFS127) or MBP with its signal sequence replaced by that of DsbA (DsbAss-MBP) (CFS469 and CFS128). DegP and β-lactamase were included as examples of SecB-dependent and SecB-independent protein export, respectively. T, total extract; P, periplasmic fraction; C, cytoplasmic fraction plus cytoplasmic membrane fraction.
We wished to ask directly whether the export of DsbA itself was affected by ffh mutations. We were able to detect the effects of an ffh mutation on MBP export because of the reliance of MBP on SecB when it is exported posttranslationally. However, DsbA, like many cell envelope proteins, is not SecB dependent (data not shown). This is either because DsbA is one of the proteins that use the posttranslational pathway without a SecB requirement or because it is uses the SRP pathway and therefore does not need SecB. At any rate, we found that DsbA export was not affected in the ffh secB mutant strain (data not shown). Thus, we were not able to establish whether DsbA is normally exported by the SRP pathway by this means. We might have used strains from which SRP components could be depleted to assess DsbA export, but we chose not to take this approach as (i) depletion experiments can result in broader effects on cell physiology, making interpretation difficult, and (ii) given the results obtained with MBP, it is likely that if SRP were disabled, DsbA could still be exported efficiently by the Sec pathway.
Direct evidence for enhanced cotranslational export directed by DsbAss.
Our evidence to this point was consistent with the proposal that DsbAss promoted rapid cotranslational export of proteins via the SRP pathway. To test this model directly, we employed a technique developed by Linda Randall and coworkers to analyze the processing of nascent polypeptides (14). In this experiment, the processing of signal sequences was used as an indication that the polypeptide had been translocated. Previous studies showed that the transmembrane translocation of several Sec-dependent exported proteins did not occur until substantial amounts of the polypeptide chain had been synthesized. In the case of MBP, it appeared that at least 50% of the mature MBP was synthesized before signal sequence cleavage took place (13).
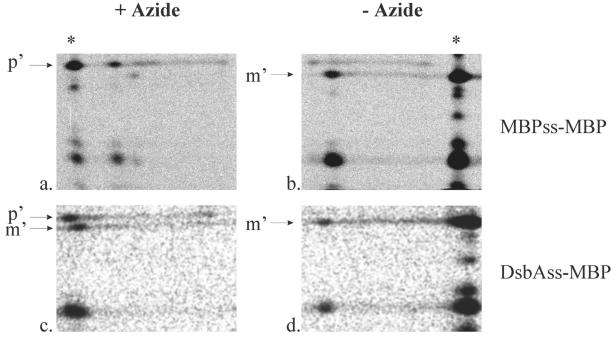
To analyze the timing of signal sequence processing relative to translation, immunoprecipitated MBPss-MBP and DsbAss-MBP polypeptides were fractionated by electrophoresis in a preparative SDS-PAGE gel, and the fractionated polypeptides were subjected to partial proteolysis in a second dimension. This treatment removed the heterogeneous C-terminal ends of nascent polypeptides, resulting in proteolytic products of defined length that either contained or lacked the signal sequence. To identify the characteristic signal sequence-bearing peptide (p′ in Fig. 5), the same manipulations were performed with immunoprecipitated precursor MBPss-MBP or DsbAss-MBP prepared by labeling cells in the presence of the SecA inhibitor sodium azide. This standard was applied to the second-dimension gel at the position indicated in Fig. 5. The characteristic processed peptide lacking its signal sequence (m′ in Fig. 5) was identified in an analogous manner by using mature MBP prepared by long-term labeling.
FIG. 5.
Replacement of MBPss with DsbAss results in cotranslational processing: two-dimensional analysis of the processing of MBP containing its native signal peptide (MBPss-MBP) (a and b) or DsbAss (DsbAss-MBP) (c and d). Exponentially growing cultures expressing either MBPss-MBP (pCFS127) or DsbAss-MBP (pCFS128) were radiolabeled for 15 s in the presence (a and c) or in the absence (b and d) of sodium azide, and extracts were subjected to immunoprecipitation with anti-MBP antibodies. The MBP immunoprecipitates were separated on SDS-12% PAGE gels (first dimension). The fractionated peptides were subjected to partial proteolysis by V8 protease in a second dimension and analyzed by using SDS-15% PAGE gels and fluorography. The arrows indicate the positions of the amino-terminal fragments derived from full-length unprocessed precursors (p′) and from mature processed MBP (m′). The positions of the standards used for identification of processed and unprocessed nascent polypeptide are indicated by asterisks. Only the relevant portions of the gels are shown.
To analyze the processing of nascent polypeptides, strains expressing MBPss-MBP or DsbAss-MBP were pulse-labeled for 15 s, and radiolabeled nascent polypeptides, as well as fully elongated radiolabeled proteins, were recovered from extracts by immunoprecipitation with anti-MBP antiserum. The nascent polypeptides were separated from the fully elongated polypeptides by preparative SDS-PAGE. Second-dimension proteolysis of these samples demonstrated that horizontal streaks comigrating at the positions of p′ or m′ could be detected. The streaks arose due to proteolytic cleavage of nascent polypeptides whose first-dimension mobility was greater than that of fully elongated proteins.
When MBPss-MBP was analyzed in this manner, both signal sequence-bearing and processed nascent polypeptides were observed (Fig. 5), which is consistent with previous observations (12). In contrast, when DsbAss-MBP was analyzed, only processed nascent polypeptides were observed (Fig. 5). Therefore, cotranslational processing of MBP was strongly enhanced by DsbAss, and posttranslational processing was not observed. For both proteins, treatment of the cells with sodium azide prior to and during labeling reduced the amount of cotranslational processing and increased the amount of posttranslational processing (Fig. 5), indicating that SecA was involved in the export of both proteins.
The results are consistent with a model in which DsbAss allows more rapid export of MBP via the SRP pathway. Whether DsbAss-MBP was translocated absolutely simultaneously with its synthesis could not be determined by these experiments due to the large size of the fragment that was generated by partial proteolysis.
We noted that export of DsbAss-MBP was significantly less sensitive to azide than export of normal MBP was. This difference and the finding that inhibition with SecA had an effect are discussed below.
Role of SecA in export promoted by DsbAss.
A favored explanation for the difference between the posttranslational and cotranslational pathways of export in E. coli is that the former uses the ATPase activity of SecA to push proteins through the membrane-embedded translocase and the latter uses the energy of protein synthesis for the same purpose. If this explanation for the role of SecA described its entire function in protein export, then SecA would not be required for cotranslational export via the SRP pathway. We studied this question by examining the effect of inhibition or partial inactivation of SecA on the export of proteins carrying DsbAss. We showed previously that in the case of DsbAss-MBP, the requirement for SecB, a protein whose function is to interfere with protein folding of some secretory proteins, disappears.
However, as mentioned above, inhibition of SecA by sodium azide does cause some accumulation of the precursor of DsbAss-MBP, although the amount is considerably less than the amount seen with MBPss-MBP. This difference in the sensitivities of the two constructs was seen repeatedly under different sodium azide treatment conditions. In some experiments, no effect was seen on the DsbAss version of MBP (data not shown). In addition, we observed a difference in the effects of a secA(Ts) mutation on DsbAss-MBP and MBPss-MBP; the former was always less sensitive to inactivation of SecA. It may be that the more rapid delivery of the DsbAss-MBP protein to the translocation machinery allows it to compete more effectively for the available functional SecA.
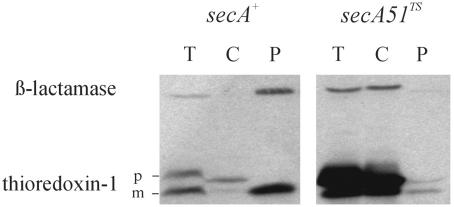
Therefore, we examined the SecA dependence of thioredoxin export by DsbAss. Figure 6 shows the results of experiments in which the secA(Ts) mutant was grown at the permissive temperature (30°C), at which the secretion defect can be quite weak depending on the protein examined. Despite the permissive nature of the mutation, we observed a very strong defect in export of the DsbAss-thioredoxin fusion protein. These results indicate that export of passenger proteins fused to DsbAss, thus directing them to the SRP pathway, still requires SecA.
FIG.6.
Export of thioredoxin by DsbAss is SecA dependent. DRH223 (secA+) and DRH226 [secA(Ts)] containing plasmid pCFS123 (DsbAss-TrxA) were grown at 30°C and subjected to fractionation. Western blot analysis with thioredoxin and β-lactamase was performed by using samples electrophoresed on an SDS-15% PAGE gel. The positions of the precursor of mature thioredoxin (DsbAss-TrxA) (p) and the mature thioredoxin (m) are indicated. T, total extract; P, periplasmic fraction; C, cytoplasmic fraction plus cytoplasmic membrane fraction.
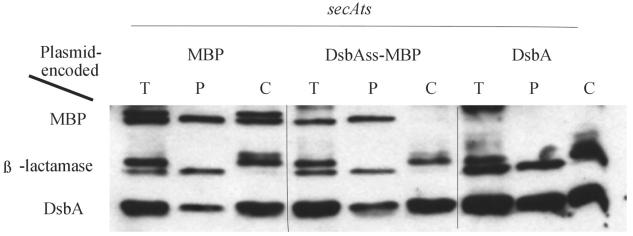
Finally, we examined the intact DsbA (DsbAss-DsbA) protein itself to determine its dependence on SecA. The export of DsbA was strongly inhibited in a secA(Ts) culture that was grown at 30°C and switched to 37°C for 2 h (Fig. 7). Under these conditions, large amounts of DsbA were found in the cytoplasm, whether the DsbA was expressed from its chromosomal locus or from a plasmid. The cytoplasmic location of DsbA was further verified by demonstrating that its two cysteines were in the reduced form, in contrast to their existence in disulfide-bonded form when they are located in the periplasm (data not shown). (Note also that this was one of the experiments in which DsbAss-MBP was insensitive to the interference with SecA activity.) Inhibition of SecA by azide also resulted in inhibition of DsbA export (data not shown).
FIG. 7.
Export of DsbA in a secA(Ts) strain. Cells were grown at 30°C and shifted to 37°C for 2 h. Extracts from secA(Ts) (secA51) cells expressing MBP (CF330/pCFS127), DsbAss-MBP (CFS330/pCFS128), and DsbA (CFS330/pCH3) were subjected to Western blot analysis. All strains expressed wild-type chromosomal DsbA. β-Lactamase was included as a control. T, total extract; P, periplasmic fraction; C, cytoplasmic fraction plus cytoplasmic membrane fraction.
DISCUSSION
Cleavable bacterial signal sequences engineered to increase their hydrophobicities direct their passenger proteins to the proposed cotranslational SRP pathway of protein export (17). Previously, Ng et al. (22) showed that the signal sequences of yeast proteins that utilize the SRP pathway are more hydrophobic than those that use the alternative posttranslational pathway. Recently, a native cleavable signal sequence of an E. coli protein was shown to enter the SRP export pathway (31). Here we report another example in bacteria of a naturally occurring cleavable signal sequence (DsbAss) that promotes SRP-based protein export. We attribute this property of DsbAss to its apparent hydrophobicity that is greater than the hydrophobicities of two other signal sequences that apparently do not promote SRP-dependent export.
The proposal that bacterial SRP promotes cotranslational export is based on analogies with the eukaryotic SRP pathway of protein translocation. While Powers and Walter (23) have shown that the components of the bacterial SRP can replace their eukaryotic counterparts in an in vitro system promoting cotranslational membrane translocation of proteins, there has been no evidence for cotranslational protein export in bacteria. Our experiments showed that replacing the native signal sequence of MBP with that of DsbA dramatically alters the pattern of MBP translocation. MBP with its own signal sequence is translocated slowly in a largely posttranslational manner. In contrast, MBP exported by the DsbAss version of MBP is not detectably exported posttranslationally. A definitive conclusion concerning whether the SRP pathway shifted the export of the latter construct to a purely cotranslational mode was limited by the detection technique which we used. The low molecular weight of the thioredoxin molecule makes it a poor candidate for the same approach to demonstrate its cotranslational export by DsbAss.
However, the very properties of thioredoxin export that led us into these studies are most easily explained by attributing to DsbAss a cotranslational mode of export. In particular, we believe that the fact that the rapidly folding thioredoxin cannot be exported by PhoAss and MBPss is due to utilization of the slow posttranslational export pathway by these signal sequences. In the case of these two signal sequences, we imagine that thioredoxin folds into a stable structure before it can be exported. With DsbAss, thioredoxin never has a chance to fold if its translocation across the membrane is initiated as soon as the polypeptide begins to emerge from the ribosome.
These properties of thioredoxin provide an exquisitely sensitive assay system for the functioning of the SRP pathway and for the signal sequences that utilize this pathway. An indication of the sensitivity was provided by the finding that mutants with only weak defects in the SRP pathway showed dramatic effects on the export of thioredoxin by DsbAss. Thus, according to our explanation, a slight slowing down of this pathway is sufficient to give thioredoxin time to fold and prevent its own export. We are currently using this sensitivity to examine other signal sequences for the ability to direct thioredoxin to the SRP pathway. An analysis of the properties of these signal sequences may allow us to refine the criteria for defining the properties of signal sequences that can utilize this pathway. The identification of such signal sequences may also be useful for engineering fusions to improve the efficiency of translocation of difficult-to-export proteins, such as those of biotechnological interest.
Our findings suggest that signal sequences that direct proteins to the SRP pathway are not restricted to the use of this pathway. They are not SRP dependent. In the case of the DsbAss-MBP fusion, in the strain background with the weakened SRP system, MBP is still efficiently exported, but it is dependent on SecB for export again. Thus, it appears that when efficient export by the SRP pathway is interfered with, DsbAss-MBP is perfectly capable of entering the slower posttranslational pathway. We believe that findings such as these raise significant problems for the utilization of strong SRP mutants (e.g., depletion strains) in studies of protein export. Such mutants make it likely that when membrane proteins are blocked from entering the SRP pathway, they may enter the posttranslational pathway, thus overwhelming that system and blocking export of the proteins normally exported by this pathway. Thus, the effects of such mutants on the latter class of proteins would be a second-order consequence of blockage of the SRP pathway. The use of weaker SRP mutants, such as the ffh mutant used here, which has little or no effect on the growth of the bacteria, would be preferable for such studies.
The unexpected result from our point of view was the finding that export of proteins with the SRP pathway is SecA dependent (when thioredoxin and MBP are fused to DsbAss). In addition, the export of DsbA itself is SecA dependent. Current models for protein export suggest that cotranslational export directly utilizes the energy of protein synthesis that occurs while ribosomes are bound to the translocase. They propose that posttranslational export utilizes the energy generated by the ATPase of SecA for the translocation process. According to this simple model, when a protein such as thioredoxin is exported cotranslationally, it should no longer require SecA for translocation.
The results of previous studies are contradictory in terms of the role of SecA in insertion of proteins into the cytoplasmic membrane and translocation of the hydrophilic domains (1, 10, 24, 28, 30, 35, 37). Thus, the role of SecA in proteins that utilize the SRP pathway is not completely understood. We can imagine several explanations for our finding of SecA dependence of proteins directed to the SRP pathway by DsbAss: (i) although we avoided using depletion conditions for SecA in our experiments, all the conditions which we did use might lead to a considerable backup of proteins at the membrane translocase, resulting in blockage of protein export for proteins that do not directly require SecA (e.g., SRP-dependent proteins); (ii) SecA, in addition to using its ATPase activity for the translocation process, may also be an integral part of the translocase itself and is therefore required for export of all proteins through this complex; and (iii) neither protein export pathway is truly cotranslational (i.e., entirely simultaneous with translation), and therefore SecA is required to provide energy for translocation of all proteins. SRP may cotranslationally direct proteins to the membrane, but the ribosome-membrane contact does not provide the energy for translocation, or such contact is not made. The latter explanation could mean that DsbAss-thioredoxin is not translocated simultaneously with translation. However, since the most compelling interpretation of our data on the export of thioredoxin by different signal sequences is that the folding of thioredoxin must be prevented in order for export to occur, an explanation other than cotranslational translocation must be offered for the maintenance of the protein in the unfolded state. One possibility is that the cotranslational delivery of thioredoxin to the membrane-imbedded translocase is sufficient to maintain the protein in the export-competent conformation, perhaps through interaction with SecA itself.
Finally, it is interesting to consider whether there is an advantage to having some proteins with cleavable signal sequences exported by the SRP pathway, while other proteins are exported by the cotranslational pathway. Although we have not directly demonstrated that the DsbA protein itself uses this pathway, the fact that its signal sequence promotes SRP pathway export of MBP and thioredoxin makes this a plausible assumption. Assuming that DsbA is exported via the SRP pathway, we can conceive of two possible explanations for why this might be the case. First, DsbA is a protein that is important for the folding of a number of other cell envelope proteins (those containing disulfide bonds). Thus, its cotranslational transfer across the membrane might ensure that it is exported rapidly, perhaps under conditions under which there is competition for export. Second, DsbA is a member of the thioredoxin family of proteins. If its export were to occur posttranslationally, its interaction with other proteins in the cytoplasm, such as thioredoxin reductase, or substrates of thioredoxins might interfere with its export.
Acknowledgments
This work was supported by grant GM41883 from the NIGMS to J.B. and by grant AI38591 from the NIAID to C.K.
REFERENCES
- 1.Andersson, H., and G. von Heijne. 1993. sec dependent and sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J. 12:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonner, W. M., and R. A. Laskey. 1974. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur. J. Biochem. 46:83-88. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D., C. Schierle, and J. Beckwith. 1998. How many membrane proteins are there. Protein Sci. 7:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier, D. N., V. A. Bankaitis, J. B. Weiss, and P. J. Bassford, Jr. 1988. The antifolding activity of secB promotes the export of the E. coli maltose-binding protein. Cell 53:273-283. [DOI] [PubMed] [Google Scholar]
- 5.Collier, D. N., and P. J. Bassford, Jr. 1989. Mutations that improve export of maltose-binding protein in SecB− cells of Escherichia coli. J. Bacteriol. 171:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debarbieux, L., and J. Beckwith. 1998. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc. Natl. Acad. Sci. USA. 95:10751-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debarbieux, L., and J. Beckwith. 2000. On the functional interchangeability, oxidant versus reductant, of members of the thioredoxin superfamily. J. Bacteriol. 182:723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derman, A. I., and J. Beckwith. 1991. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J. Bacteriol. 173:7719-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guilhot, C., G. Jander, N. L. Martin, and J. Beckwith. 1995. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc. Natl. Acad. Sci. USA. 92:9895-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jander, G., J. E. Cronan, Jr., and J. Beckwith. 1996. Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J. Bacteriol. 178:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonda, S., M. Huber-Wunderlich, R. Glockshuber, and E. Mossner. 1999. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 18:3271-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josefsson, L.-G., and L. L. Randall. 1981. Processing in vivo of precursor maltose-binding protein in Escherichia coli occurs post-translationally as well as co-translationally. J. Biol. Chem. 256:2504-2507. [PubMed] [Google Scholar]
- 13.Josefsson, L.-G., and L. L. Randall. 1981. Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell 25:151-157. [DOI] [PubMed] [Google Scholar]
- 14.Josefsson, L.-G., and L. L. Randall. 1983. Analysis of cotranslational proteolytic processing of nascent chains using two-dimensional gel electrophoresis. Methods Enzymol. 97:77-85. [DOI] [PubMed] [Google Scholar]
- 15.Katzen, F., and J. Beckwith. 2002. Disulfide bond formation in the periplasm of Escherichia coli. Methods Enzymol. 348:54-66. [DOI] [PubMed] [Google Scholar]
- 16.Kumamoto, C. A., and P. M. Gannon. 1988. Effects of Escherichia coli secB mutations on pre-maltose binding protein conformation and export kinetics. J. Biol. Chem. 263:11554-11558. [PubMed] [Google Scholar]
- 17.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, G., T. B. Topping, W. H. Cover, and L. L. Randall. 1988. Retardation of folding as a possible means of suppression of a mutation in the leader sequence of an exported protein. J. Biol. Chem. 263:14790-14793. [PubMed] [Google Scholar]
- 19.Liu, G., T. B. Topping, and L. L. Randall. 1989. Physiological role during export for the retardation of folding by the leader peptide of maltose-binding protein. Proc. Natl. Acad. Sci. USA. 86:9213-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luirink, J., and B. Dobberstein. 1994. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 11:9-13. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor N.Y.
- 22.Ng, D. T. W., J. D. Brown, and P. Walter. 1996. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers, T., and P. Walter. 1997. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16:4880-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi, H. Y., and H. D. Bernstein. 1999. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J. Biol. Chem. 274:8993-8997. [DOI] [PubMed] [Google Scholar]
- 25.Randall, L. L., T. B. Topping, V. F. Smith, D. L. Diamond, and S. J. S. Hardy. 1998. SecB: a chaperone from Escherichia coli. Methods Enzymol. 290:444-459. [DOI] [PubMed] [Google Scholar]
- 26.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan, J. P., M. C. Duncan, V. A. Bankaitis, and P. J. Bassford, Jr. 1986. Intragenic reversion mutations that improve export of maltose-binding protein in Escherichia coli malE signal sequence mutants. J. Biol. Chem. 261:3389-3395. [PubMed] [Google Scholar]
- 28.Sääf, A., H. Andersson, G. Gafvelin, and G. von Heijne. 1995. SecA dependence of the translocation of a large periplasmic loop in the Escherichia coli MalF inner membrane protein is a function of sequence context. Mol. Membr. Biol. 12:209-215. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Scotti, P. A., Q. A. Valent, E. H. Manting, M. L. Urbanus, A. J. M. Driessen, B. Oudega, and J. Luirink. 1999. SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274:29883-29888. [DOI] [PubMed] [Google Scholar]
- 31.Sijbrandt, R., M. L. Urbanus, C. M. Ten Hagen-Jongman, H. D. Bernstein, B. Oudega, B. R. Otto, and J. Luirink. 2003. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J. Biol. Chem. 278:4654-4659. [DOI] [PubMed] [Google Scholar]
- 32.Teschke, C. M., J. Kim, T. Song, S. Park, C. Park, and L. L. Randall. 1991. Mutations that affect the folding of ribose-binding protein selected as suppressors of a defect in export in Escherichia coli. J. Biol. Chem. 266:11789-11796. [PubMed] [Google Scholar]
- 33.Tian, H., and J. Beckwith. 2002. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 184:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian, H. P., D. Boyd, and J. Beckwith. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. USA 97:4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traxler, B., and C. Murphy. 1996. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J. Biol. Chem. 271:12394-12400. [DOI] [PubMed] [Google Scholar]
- 36.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner, P. K., M. H. Saier, Jr., and M. Müller. 1992. Membrane insertion of the mannitol permease of Escherichia coli occurs under conditions of impaired secA function. J. Biol. Chem. 267:24523-24532. [PubMed] [Google Scholar]