Abstract
LamB-LacZ fusion proteins have classically been used in studies of the general secretion pathway of Escherichia coli. Here we describe how increasing signal sequence hydrophobicity routes LamB-LacZ Hyb42-1 to the signal recognition particle (SRP) pathway. Secretion of this hydrophobic fusion variant (H*LamB-LacZ) was reduced in the absence of fully functional Ffh and Ffs, and the translocator jamming caused by Hyb42-1 was prevented by efficient delivery of the fusion to the periplasm. Finally, we found that in the absence of the ribosome-associated chaperone, trigger factor (Tig), LamB-LacZ localized to the periplasm in a SecA-dependent, SRP-independent fashion. Collectively, our results provide compelling in vivo evidence that there is an SRP-dependent cotranslational targeting mechanism in E. coli and argue against a role for trigger factor in pathway discrimination.
All cells face a common mechanistic challenge: how to target proteins synthesized in the cytosol to and across phospholipid bilayers. In eukaryotic cells, presecretory and integral membrane proteins are targeted to the endoplasmic reticulum via interactions with the signal recognition particle (SRP). Eukaryotic SRP is a complex comprised of the 7SL RNA and six proteins, including SRP54, which promotes signal sequence binding. This complex interacts at the ribosome-nascent preprotein interface to promote arrest of translation elongation, targeting, and finally cotranslational secretion into the endoplasmic reticulum lumen via the Sec61p translocation channel (reviewed in references 7, 15, and 16).
SRP homologues are found in all of the domains of life. In bacteria, the SRP is formed by Ffh protein (for “fifty-four homologue”) complexed with 4.5S RNA (homologous to the stem-loop of the 7SL RNA recognized by SRP54). Homologues of specific SRP components known to impart cotranslational targeting functions in eukaryotes, however, have not been found in bacteria (7). Despite this observation, the assumption that secretion and translation occur coordinately pervades discussions of the bacterial SRP. Experimental data that support this assumption are generally lacking.
An additional disputed question in the bacterial SRP field concerns the mechanism of secretion pathway discrimination. Unlike the eukaryotic system, in which all targeted proteins utilize the SRP, secreted Escherichia coli proteins reach the translocator via multiple targeting pathways.
Prevailing models argue that in E. coli, integral membrane proteins are preferentially targeted by the SRP, while proteins with N-terminal signal sequences reach the translocator via the SecB pathway. Despite these apparent preferences, structural studies show that the SRP interacts efficiently with N-terminal signal sequences (3). Precisely how the bacterial cell discriminates secreted substrates is not yet clear. Two distinct models have been put forth to explain pathway preference. One model holds that the ribosome-associated chaperone, trigger factor (Tig), binds near N-terminal signal sequences, thereby blocking SRP interaction (4). The other model holds that the hydrophobic character of the signal sequence alone can dictate pathway preference (17).
In order to address whether SRP-dependent secretion in E. coli can occur cotranslationally and to address basic questions of pathway discrimination, we adapted LamB and a LamB-LacZ fusion protein, which are known classically for their role in the identification and elucidation of the components and mechanism of the general secretion pathway and translocation channel, to use as genetic tools for characterization of the SRP pathway. By studying the secretion of a LamB-LacZ translational fusion, routed to the SRP pathway, we confirmed that hydrophobicity alone can dictate secretory pathway preference and eliminate trigger factor as a component of the selection mechanism. LamB-LacZ, secreted by the SecB pathway, jams the translocation machinery. Jamming is relieved and the fusion localizes to the periplasm when the protein is routed via SRP. These observations provide strong evidence that in vivo SRP-dependent targeting in E. coli is cotranslational. Finally, we demonstrated the utility and sensitivity of this genetic system for probing the SRP pathway to a level of allele specificity.
MATERIALS AND METHODS
Media and chemicals.
Standard microbiological growth media and chemicals were used (28). For pulse-labeling experiments, M63 glycerol liquid minimal medium was supplemented with maltose at a final concentration of 0.4% (wt/vol) for nonfusion strains and 0.8% for fusion strains, to induce lamB expression. [35S]methionine (specific activity, 1,175 Ci/mmol) was purchased from ICN Biomedicals, Inc. Protein A-Sepharose CL4B and ECL Western blotting reagents were obtained from Amersham Biosciences. Iodoacetamide was purchased from Sigma, St. Louis, Mo. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was purchased from Angus. Anti-LamB and anti-MalE antibodies were obtained from laboratory stocks.
Bacterial strains and plasmids.
The strains and plasmids used are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or relevant gene | Reference or source |
|---|---|---|
| BZR60 | MC4100 Φ(lamBΔ60′-′lacZ) Hyb42-1 [λp1(209)] | 10 |
| CWB96 | DY329 tig<>kan | This study |
| CWB168 | RO182 lamB (A16L,G17V) | This study |
| CWB170 | RO182 H*lamB (A13I,A15L,A16L,G17V) | This study |
| CWB172 | RO192 lamB (A16L,G17V) | This study |
| CWB174 | RO192 H*lamB (A13I,A15L,A16L,G17V) | This study |
| CWB178 | H*lamB pCWB36 | This study |
| CWB193 | Φ(lamB′-′lacZ) Hyb42-1 [λp1(209)] lamBΔ60 | This study |
| CWB197 | Φ(H*lamB′-′lacZ) Hyb42-1 [λp1(209)] lamBΔ60 | This study |
| CWB210 | MC4100 H*lamB | This study |
| CWB213 | MC4100 secA51(Ts) leuB::Tn10 | This study |
| CWB249 | CWB210 secA51(Ts) leuB::Tn10 recA::kan | This study |
| CWB277 | CWB210 ffs69 zba-3054::Tn10 | This study |
| CWB278 | CWB210 pheA3141::Tn10Kan ffh87 | This study |
| CWB281 | Φ(lamB′-′lacZ) Hyb42-1 [λp1(209)] lamB | This study |
| CWB288 | CWB281 pCLC11(degP) | This study |
| CWB289 | CWB281 tig<>kan | This study |
| CWB294 | CWB281 secA51(Ts) leuB::Tn10 | This study |
| CWB295 | CWB299 secY39(Cs) zhc::Tn10 | This study |
| CWB296 | CWB299 secY40(Cs) zhc::Tn10 | This study |
| CWB299 | Φ(H*lamB′-′lacZ) Hyb42-1 [λp1(209)] H*lamB | This study |
| CWB304 | CWB299 pCLC11 (degP) | This study |
| CWB305 | CWB299 tig<>kan | This study |
| CWB309 | CWB299 secA51(Ts) leuB::Tn10 | This study |
| CWB312 | CWB289 pCLC11 (degP) | This study |
| CWB313 | CWB305 pCLC11 (degP) | This study |
| CWB316 | CWB299 ffs69 zba-3054::Tn10 | This study |
| CWB318 | MC4100 tig<>kan | This study |
| CWB341 | CWB289 secA51(Ts) leuB::Tn10 | This study |
| CWB342 | CWB305 secA51(Ts) leuB::Tn10 | This study |
| CWB350 | CWB213 tig<>kan | This study |
| CWB351 | HPT244 tig<>kan | This study |
| DY329 | W3110 ΔlacU169 nadA::Tn10 gal490 λc1857 Δ(cro-bioA) | 33 |
| HPT 242 | MC4100 Δara714 zba-3054::Tn10 | 30 |
| HPT244 | MC4100 Δara714 ffs69 zba-3054::Tn10 | 30 |
| HPT404 | MC4100 Δara714 pheA3141::Tn10Kan | 31 |
| HPT406 | MC4100 Δara714 pheA3141::Tn10Kan ffh87 | 31 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 ptsF25 deoC1 thi | Lab collection |
| NT1001 | MC4100 malBΔ1 | Lab collection |
| Pop3186 | MC4100Φ(lamB′-′lacZ) Hyb42-1 [λp1(209)] | 27 |
| RO104 | MC4100 secB::Tn5 | Lab collection |
| RO182 | MC4100 spc[R] malBΔ1-linked Tn10 | Lab collection |
| RO192 | MC4100 spc[R] malBΔ1-linked Tn10 secB::Tn5 | Lab collection |
| pCLC11 | degP in pACYC184 (Cmr) | 8 |
| pCWB9 | tig gene cloned in pBluescript KS(+) | This study |
| pCWB10 | kan gene from pACYC177 inserted into tig of pCWB9 | This study |
| pCWB27 | malK5′ Δ160-lamB in pBR322 | This study |
| pCWB35 | H*lamB (A13I,A15L,A16L,G17V) quadruple mutant | This study |
| pCWB37 | lamB (A16L, G17V) double mutant | This study |
Cloning and site-directed mutagenesis.
In order to efficiently introduce site-specific changes into the chromosomal region encoding the lamB signal sequence, a 2.2-kb malK-lamB fragment was PCR amplified to incorporate 5′ HindIII and 3′ BamHI sites and cloned into pBR322 to generate plasmid pCWB27. This construction encodes a 3′ fragment of the malK gene with the first 160 nucleotides of malK deleted and the entire lamB coding region and is sufficient to repair the chromosomal malBΔ1 lesion via recombination, as described previously (8). With the Quickchange site-directed mutagenesis system (Stratagene), primer A16LG17Vu (5′-CCT CTG GCG GTT GCC GTC GCA CTG GTC GTA ATG TCT GCT CAG GCA ATG-3′) and its complement, A16LG17Vd, were used to generate the double mutant allele A16LG17V. Using the A16LG17V pCWB27 derivative as the starting template, we introduced two additional changes using primer A1315ILu (5′-CGC AAA CTT CCT CTG GCG GTT ATC GTC CTA CTG GTC GTA ATG TCT-3′) and its complement, A13IA15Ld, to generate the quadruple mutant allele A13IA15LA16LG17V (designated H*).
For recombination into the chromosome, lamB plasmid derivatives bearing signal sequence mutations were transformed into NT1001, a malBΔ1 strain. malBΔ1 encodes a large deletion spanning the 3′ sequences of malK and the N-terminal coding region of lamB DNA. Mal+ recombinants repair malBΔ1, resulting in transfer of the DNA with the signal sequence mutations to the chromosomal lamB locus. Final signal sequence changes were confirmed by DNA sequence analysis.
Construction of the fusion strains.
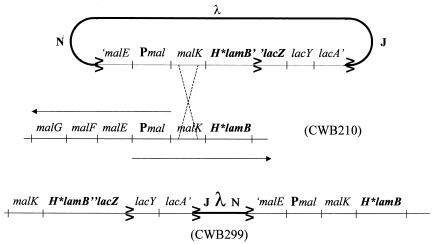
The goal of the following strain manipulations was to introduce the H*LamB quadruple amino acid signal sequence mutations into a LamB-LacZ translational fusion. This was accomplished by a modification of a strategy used by Emr et al. (11). A specialized λ transducing phage carrying the lamB′Δ60-′lacZ fusion was isolated from strain BZR60. The Δ60 deletion removes 10 codons internal to the LamB signal sequence. Because this deletion blocks hybrid protein secretion, lysogens carrying the phage were phenotypcially Lac+. A lysate of the specialized phage was used to infect both MC4100 (wild-type LamB) and CWB210 (H*LamB), and lysogens were picked from Luria-Bertani (LB) agar containing X-Gal. Lac+ and Lac− lysogens were screened for maltose sensitivity and other LamB-LacZ fusion localization-associated phenotypes (Table 2). Lysogens with the following genotypes were identified: lamB′-′lacZ/Δ60lamB (CWB192) and H*lamB′-′lacZ/Δ60lamB (CWB197). Transducing phage isolated from these strains were then used to infect MC4100 (lamB+) and CWB210 (H*lamB), respectively. The resulting strains were CWB281 (lamB′-′lacZ/lamB+) and CWB299 (H*lamB′-′lacZ/H*lamB) (Fig. 1). Because CWB281 and CWB299 were homogenotes, the fusion phenotypes could not be altered by homologous recombination.
TABLE 2.
Summary of phenotypes conferred by lamB′-′lacZ fusions
| Fusion | Location | Maltose sensitive | Toxicity | Suppressed by increased DegP | Lac phenotypea |
|---|---|---|---|---|---|
| LamBΔ60-LacZ | Cytoplasm | No | None | No | + |
| LamB-LacZ 42-1 | Inner membraneb | Yes | Folding based | No | + |
| LamB-LacZX90 | Periplasm | Yes | Periplasmic | Yes | − |
| H*LamB-LacZ | Periplasm | Yes | Periplasmic | Yes | − |
Lac phenotypes were assessed in the absence of the inducer maltose.
The exact cellular location of LamB-LacZ 42-1 is not known.
FIG. 1.
Structure of the diploid fusion strains. A λ specialized transducing phage carrying the H*lamB′-′lacZ gene fusion integrated at the chromosomal malB locus of strain CWB210 to produce a lysogen (CWB299) that carried both H*lamB and the H*lamB′-′lacZ fusion.
Construction of a trigger factor null strain.
A tig<>kan null allele was generated in MC4100 by using the method of Yu et al. (33). First, the tig gene was amplified from MC4100 by PCR by using primers tigKPNup (5′-CGT ATA ATG CCG GGT ACC ATT TGA ATG C-3′) and tigSCAdn (5′-GTG CGA ATT TAG CGA GCT CTG CTG CGT A-3′), which incorporated 5′ KpnI and 3′ SacI sites at the ends of the resulting fragment. PCR products were digested with KpnI and SacI and cloned into pBluescript KS(+) to create pCWB9, a high-copy-number expression plasmid for tig. Next, a second cloning step was performed to introduce the kan gene, amplifed by PCR from pACYC177, by using oligonucleotides kanHNCup (5′-CCA CGT TGT GTC AAC AAA TCT CTG-3′) and kanECOdn (5′-GTT ACA ACG AAT TCA CCA ATT C-3′), which incorporated 5′ HincII and 3′ EcoRI sites into the kan gene. The resulting kan PCR DNA was digested and cloned into pCWB9, inactivating tig, to create plasmid pCWB10 tig<>kan. tig<>kan was next PCR amplified from the plasmid with the original tig primers to generate linear DNA for electorporation into strain DY329 (33). The resulting construction had deleted 785 nucleotides from the tig locus, including 34 nucleotides upstream of the tig coding sequence. Kanamycin-resistant transformants were selected and sequenced to confirm that tig inactivation occurred. Finally, tig<>kan was moved by P1 transduction into MC4100 to create CWB318.
For additional strain construction we used standard microbiological techniques with P1 transduction and DNA transformation, as described elsewhere (28).
Pulse-chase and immunoprecipitation.
Overnight cultures grown in M63 minimal medium containing 0.4% glycerol were subcultured with 1/100 inocula in fresh glycerol minimal medium and grown to an optical density at 600 nm of 0.3 to 0.5. For nonfusion strains, lamB expression was induced for 1 h at 37°C by addition of maltose to a final concentration of 0.4% (wt/vol). Fusion strains were induced for 2 h with maltose at a final concentration of 0.8% (wt/vol). Samples were pulsed for 30 s and chased for 10 s to 2 min, as indicated below. Reaction mixtures were precipitated with trichloroacetic acid on ice for 30 min, frozen in an ethanol-dry ice bath, and boiled for 3 min. Extracts were centrifuged, and the supernatants were added to immunoprecipitation reaction mixtures. The reaction mixtures were incubated with a 50% protein A-Sepharose bead slurry for at least 1 h prior to washing and resuspension in sodium dodecyl sulfate (SDS) protein sample buffer. Samples were resolved on 8% SDS protein gels and visualized by autoradiography.
SDS-PAGE.
LacZ localization was assessed by performing an SDS-polyacrylamide gel electrophoresis (PAGE) analysis under reducing conditions (in the presence of 360 mM β-mercaptoethanol) or nonreducing conditions (in the presence of 2 mM iodoacetiamide), as described previously (29).
RESULTS
Increasing signal sequence hydrophobicity routes LamB through the SRP pathway.
In order to develop a genetic system for probing SRP function, we sought to redirect LamB to the SRP pathway. Lee and Bernstein (17) demonstrated that signal sequence hydrophobicity is sufficient to direct MalE to the SRP pathway. By analogy, we increased LamB signal sequence hydrophobicity incrementally by introducing double and quadruple amino acid substitutions. Since our goal was to perform genetic analysis of SRP-dependent secretion, a thorough assessment of the secretion mode of the hydrophobic LamB variants was our first objective.
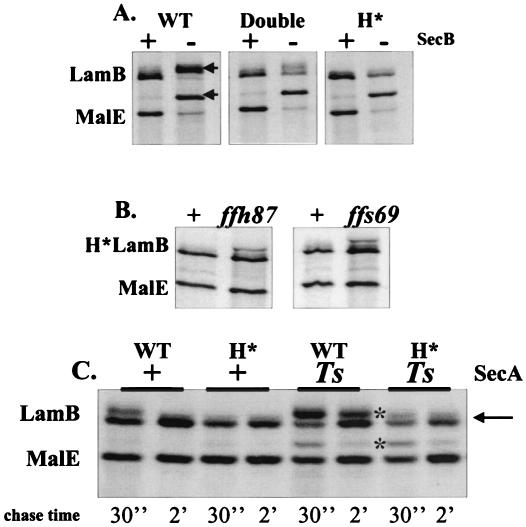
Secretion of LamB in its native state is predominantly dependent on the general secretion pathway components SecA and SecB. SecB, the export dedicated chaperone, binds to the mature sequences of LamB in order to maintain a conformation competent for translocation. SecA, a molecular motor, provides the energy for driving insertion of the preprotein at the translocation channel (reviewed in reference 9). Ffh, the protein component of the signal recognition particle, is not required for LamB secretion. Guided by these facts, we assessed the secretion profiles of the double (A16LG17V) and quadruple (A13IA15LA16LG17V, designated H*) hydrophobicity change variants in SecB+ and SecB− backgrounds. Pulse-chase analysis of LamB with the wild-type signal sequence clearly revealed precursor accumulation in the absence of SecB. The double mutant exhibited decreased precursor accumulation, while SecB dependence was completely eliminated in the quadruple H* mutant (Fig. 2 A). As expected, MalE secretion remained SecB dependent in all of the LamB backgrounds. In subsequent analyses we focused on the quadruple H* mutant, which exhibited complete SecB independence.
FIG. 2.
Increasing signal sequence hydrophobicity reroutes LamB to the SRP pathway. (A) SecB dependence. E. coli cells were pulse-labeled and chased for 30 s, and this was followed by immunoprecipitation. The precursor LamB and MalE bands are indicated by arrows. (B) SRP dependence. E. coli with H*lamB and either wild-type or mutant SRP alleles (ffh87 or ffs69) was pulse-labeled for 30 s and chased for 10 s. (C) Reduced SecA requirement. E. coli with lamB+ or H*lamB in the presence or absence of secA51(Ts) was grown overnight at 30°C, subcultured, and then induced with 0.4% maltose. A pulse-chase analysis was performed at 30°C. The precursor band is indicated by an asterisk. The arrow indicates the position of the presumed H*LamB precursor. WT, wild type.
In order to determine whether the SecB independence was due to the rerouting of LamB, we examined the targeting of H*LamB in the presence of two mutant SRP alleles, ffs69, which encodes two cytosines changed to thymines at the 3′ end of the 4.5S RNA, and ffh87, which encodes a serine-to-threonine change at Ffh position 382 (30). Both of these alleles, which were isolated in the Beckwith laboratory, confer mild but measurable defects on SRP-dependent secretion (30, 31). These mutations were introduced into our strains by transduction with selection for a linked marker. Because colonies bearing the mutant SRP alleles were phenotypically indistinguishable from wild-type colonies, transductants were screened by pulse-chase analysis, followed by DNA sequence analysis. Of 10 transductants for ffh87 and ffs69, 4 and 5 isolates, respectively, were found to have measurable accumulations of the H*LamB precursor. All of these isolates were confirmed by DNA sequence analysis to possess the mutant alleles, while all isolates that exhibited normal LamB secretion profiles were found to be wild type at the SRP loci. The accumulation of the precursor H*LamB in the presence of both ffs69 and ffh87 demonstrated that H*LamB secretion was SRP dependent. Neither lesion caused the precursor MalE to accumulate, which was what was expected since MalE is a SecB pathway substrate (Fig. 2B). Similarly, neither ffs69 nor ffh87 conferred detectable secretion defects on wild-type LamB (data not shown).
It is important to note that the mutation encoding the H* signal sequence preceded the signal sequence processing site. Once the H* signal sequence was removed, the LamB protein was indistinguishable from LamB secreted via the SecB pathway. Not surprisingly, therefore, the stability of H*LamB was comparable to that of wild-type LamB, and H*LamB was correctly processed and appropriately targeted (data not shown).
Secretion of H*LamB is insensitive to SecA51(Ts) at the permissive growth temperature.
A role for SecA in SRP-dependent secretion when large periplasmic loops are present in a translocating polypeptide has been suggested (22, 26). We examined H*LamB secretion in the presence of wild-type SecA and SecA51(Ts) at the permissive growth temperature, since the secA51(Ts) allele confers measurable defects on LamB secretion even at the permissive temperature. Both MalE and LamB showed marked precursor accumulation in the presence of secA51(Ts) (Fig. 2C). In contrast, secA51(Ts) had little effect on H*LamB secretion. This observation was supported by phenotypes of a lamB′-′lacZ fusion strain bearing the H* signal sequence (see below).
H* signal sequence relieves hybrid jamming of a LamB-LacZ fusion.
LamB-LacZ fusion proteins provided critical insight into the general secretion pathway of E. coli. Since H*LamB secretion is convincingly SRP dependent, we constructed an H*lamB′-lacZ gene fusion to establish a genetic system for the SRP pathway.
The utility of LamB-LacZ for genetic analysis of secretion hinges on the phenotypes that result from fusion protein localization (Table 2). The classical LamB-LacZ Hyb42-1 fusion protein includes the signal sequence and first 173 amino acids of LamB fused to a nearly complete LacZ protein (5). When the LamB signal sequence is functional, the fusion is targeted to the SecYEG translocator, where rapid folding of LacZ in the cytoplasm jams the secretion channel, which leads to accumulation of precursors of all secreted substrates in the cytoplasm. This condition, known as inducer sensitivity, ultimately leads to cell death. Inducer sensitivity is relieved by mutations which prevent fusion targeting, such as lesions in the signal sequence (lamBΔ60′-′lacZ). When the fusion remains cytoplasmic, the strain is resistant to maltose and exhibits a strongly Lac+ phenotype. The targeting of the fusion to any extracytoplasmic location results in a Lac− phenotype and under all known conditions also leads to inducer sensitivity. The lamB′-′lacZX90 allele encodes a mutant LacZ protein that does not fold properly, thereby preventing LacZ-mediated jamming and allowing fusion protein to exit the translocation channel and enter the periplasm. Extracytoplasmic toxicity is believed to result from the formation of toxic disulfide-bonded aggregates. In contrast to folding-based cytoplasmic toxicity, extracytoplasmic toxicity is suppressible by increased production of the periplasmic protease and folding factor, DegP (29).
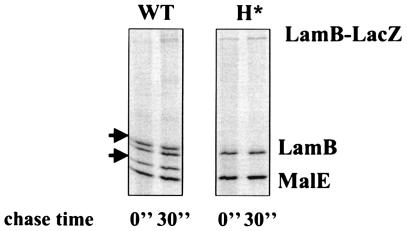
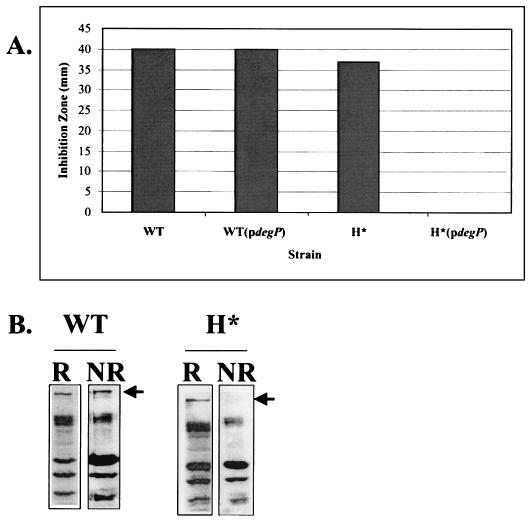
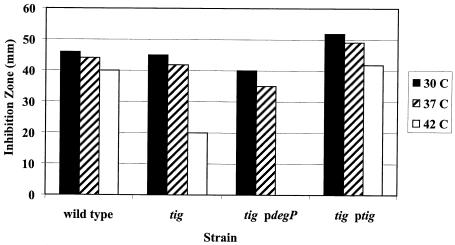
Targeting of H*LamB-LacZ (Hyb42-1 containing the quadruple hydrophobic signal sequence variant) led to a Lac−, maltose-sensitive phenotype, a profile similar to that seen for the periplasmically localized LamB-LacZX90 fusion (Table 2). H*LamB-LacZ was made at levels comparable to the levels of LamB-LacZ, but it did not jam the secretion apparatus (Fig. 3). Furthermore, genetic and biochemical analyses supported a periplasmic localization for H*LamB-LacZ. First, DegP overexpression suppressed the maltose-sensitive phenotype of strains carrying H*lamB′-′lacZ (Fig. 4A). Previous studies have shown that LacZ fusion proteins localized to the periplasm of E. coli form toxic, disulfide-bonded aggregates that are too large to enter SDS gels under nonreducing conditions (29). Consistent with periplasmic localization, H*LamB-LacZ was not detectable in SDS gels under nonreducing conditions. In contrast, wild-type LamB-LacZ, which remained at the cytoplasmic interface, did not form disulfide-bonded aggregates and was readily visible under nonreducing conditions (Fig. 4B). Thus, the results of a combination of both genetic and biochemical approaches suggest that the H*LamB-LacZ fusion is in the periplasm.
FIG. 3.
H*LamB-LacZ does not jam the secretion machinery. Strains were pulse-labeled and chased for 30 s. The arrows indicate the positions of the precursor LamB and MalE. WT, wild type.
FIG. 4.
H*LamB-LacZ localizes to the periplasm. (A) Maltose sensitivity disk assay. Overnight cultures of lamB′-′lacZ and H*lamB′-′lacZ strains, with and without DegP overexpression (pCLC11), were washed and concentrated 2.5-fold. Fifty microliters of cell suspension was added to 3 ml of molten F-top agar at 42°C and spread onto glycerol minimal plates. Fifteen microliters of 20% maltose was spotted in the center of a disk, and inhibition zones were measured after 15 h of incubation at 37°C. The results of one representative experiment are shown. (B) Disulfide bond formation in H*LamB-LacZ. Extracts of the lamB′-′lacZ and H*lamB′-′lacZ strains were prepared as described in Materials and Methods under reducing (R) and nonreducing (NR) conditions for SDS-PAGE and immunoblot analysis. Blots were probed with antibody against LamB. The position of the fusion band is indicated by an arrow. WT, wild type.
Secretion of H*LamB-LacZ is SRP dependent.
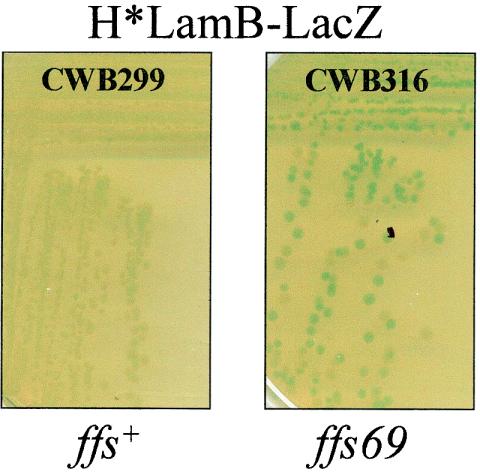
ffs69 was transduced into CWB299 (H*lamB′-′lacZ) via a 50%-linked Tn10 (30). Plates were incubated overnight at 37°C and then for 2 days at room temperature, as described by Tian and colleagues (30, 31). Two representative colonies which developed a Lac+ phenotype, as well as two Lac− colonies, were selected for DNA sequence analysis. The Lac+ transductants were found to have the ffs69 allele, while the Lac− colonies were wild type at the ffs locus, confirming that secretion of H*LamB-LacZ is SRP dependent (Fig. 5).
FIG. 5.
A 4.5S RNA lesion impairs H*LamB-LacZ secretion in vivo. H*lamB′-′lacZ strains with wild-type ffs or ffs69 were streaked onto LB agar containing X-Gal and incubated overnight at 37°C and then for 2 days at room temperature.
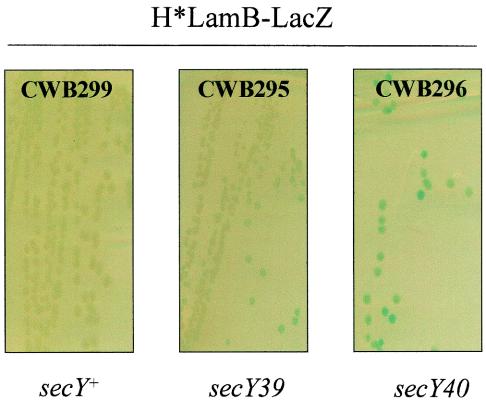
H*LamB-LacZ secretion is impaired in the presence of secY40 but not in the presence of secY39.
The secY39 and secY40 alleles both confer cold-sensitive phenotypes and specific defects on the secretion mechanism in E. coli. Newitt and Bernstein (23) showed that secY40 impairs inner membrane protein insertion and functions synergistically with the SRP pathway. They also suggested that the secY39 allele was synthetically lethal with genetic lesions in SRP. secY39 strains, however, exhibit strong defects in the export of at least one SecB pathway substrate, OmpA (2, 20). Strains bearing the H*lamB′-′lacZ fusion in combination with secY40 or secY39 were incubated overnight at 37°C and then for 2 days at room temperature to allow the cold-sensitive defects of the secY alleles to accumulate. CWB296 (H*lamB′-′lacZ secY40) exhibited approximately fivefold-higher Lac activity than CWB295 (H*lamB′-′lacZ secY39) exhibited. CWB295 exhibited a minimal increase in Lac activity compared to the activity of parent strain CWB299 (H*lamB′-′lacZ) (Fig. 6). Thus, H*LamB-LacZ secretion is strongly and specifically impaired in the presence of secY40 but is insensitive to secY39.
FIG. 6.
secY40 impairs H*LamB-LacZ secretion in vivo. H*lamB′-′lacZ strains with either secY+, secY39, or secY40 were streaked onto LB agar containing X-Gal and incubated overnight at 37°C and then for 2 days at room temperature.
H*LamB-LacZ secretion is insensitive to secA51(Ts).
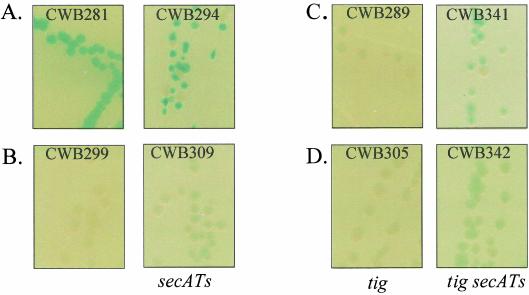
The secA51(Ts) mutation was identified as a conditional lethal mutation that increased the Lac activity of a malE′-′lacZ fusion strain at the permissive growth temperature. Under these conditions the secretion machinery was compromised, and a fraction of the hybrid protein remained in the cytoplasm, where LacZ folded properly. Although the wild-type lamB′-′lacZ fusion conferred a Lac+ phenotype on media containing X-Gal, secA51(Ts) discernibly made colonies smaller, and there was an increase in Lac activity (Fig. 7A, compare CWB281 and CWB294). Note that there was also an increase in the rate of fusion loss (white colonies) by homologous recombination (Fig. 1).
FIG. 7.
Secretion of LamB-LacZ in a tig strain is SecA dependent. Strains were streaked onto LB agar plates containing X-Gal and incubated for 15 h at 30°C. (A) lamB′-′lacZ (wild-type) strains CWB281 and CWB294 [secA(Ts)]. (B) H*lamB′-′lacZ strains CWB299 and CWB309 [secA(Ts)]. (C) lamB′-′lacZ strains CWB289 (tig<>kan) and CWB341 [tig<>kan secA(Ts)]. (D) H*lamB′-′lacZ strains CWB305 (tig<>kan) and CWB 342 [tig<>kan secA(Ts)].
H*LamB-LacZ, like its nonfusion counterpart, H*LamB, is secreted in an SRP-dependent fashion. The large size of the LacZ sequences added to LamB in the fusion context potentially imposes additional constraints on the translocation machinery. Indeed, proteins bearing large periplasmic domains may require SecA for efficient periplasmic delivery (22, 26). H*LamB-LacZ secretion, however, appeared to be insensitive to secA51(Ts) at the permissive growth temperature (Fig. 7B, compare CWB299 and CWB309).
Trigger factor does not promote targeting pathway discrimination.
Trigger factor (Tig) has been evoked as a key component in secretory pathway discrimination (4, 21). If association with Tig prevents LamB-LacZ, a SecB pathway substrate, from interacting with SRP, then deletion of tig should allow LamB-LacZ to engage in SRP-dependent secretion. A lamB′-′lacZ strain null at the tig locus should exhibit all of the phenotypes associated with H*LamB-LacZ, which is secreted via the SRP pathway. Indeed, the lamB′-′lacZ strain CWB289, in the absence of Tig, exhibited a Lac− (Fig. 7C), maltose-sensitive phenotype that was partially suppressible by DegP overexpression (Fig. 8). Secretion of LamB-LacZ into the periplasm in the absence of Tig was confirmed by nonreducing SDS-PAGE analysis (data not shown). tig inactivation had no discernible impact on the Lac phenotype of the H*lamB′-′lacZ strain CWB305 (Fig. 7D).
FIG. 8.
LamB-LacZ localizes to the periplasm in a tig strain. Strains were processed as described in the legend to Fig. 3. In the presence of pCLC11 at 42°C, the maltose sensitivity of a lamB′-′lacZ strain in the tig strain background was partially suppressed.
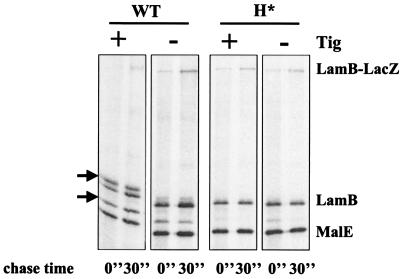
At first glance, it appeared that the predictions that Tig is a discrimination factor were true. Further scrutiny, however, revealed that although LamB-LacZ reached the periplasm in a Tig− background, it did so via a mechanism different than that for H*LamB-LacZ. First, in contrast to H*LamB-LacZ (Fig. 7B), LamB-LacZ targeting in the absence of Tig was strongly sensitive to SecA function (Fig. 7C, CWB341). Even in the absence of trigger factor, H*LamB-LacZ secretion was only mildly affected by secA51(Ts) (Fig. 7D, CWB342). Fusion-induced jamming, although reduced in the lamB′-′lacZ tig strain, was not eliminated completely, as it was in the H*lamB′-′lacZ strain with or without Tig (Fig. 9).
FIG. 9.
Fusion-induced jamming is reduced but not eliminated in tig fusion strains. Strains were pulse-labeled and chased (each for 30 s), immunoprecipitated with anti-LamB and anti-MalE antisera, and prepared for SDS-PAGE as described in Materials and Methods. The position of the precursor is indicated by arrows. WT, wild type.
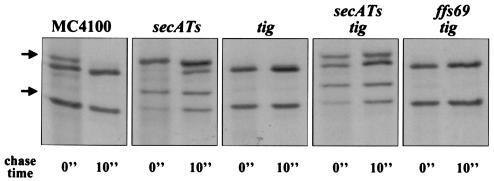
To explore the basis of the variations in LamB targeting in the absence of Tig, we performed a pulse-chase analysis and immunoprecipitation with the nonfusion variants of the tig strains. The secretion of LamB (wild type) was strongly SecA dependent (Fig. 10). Deletion of tig improved the efficiency of LamB secretion (no precursor was seen) but did not relieve the SecA dependence. LamB secretion in the tig background was completely unaffected by a lesion in the SRP pathway (ffs69) (Fig. 10), in contrast to H*LamB secretion, which was detectably sensitive to the presence of ffs69 (Fig. 2B).
FIG. 10.
Secretion of LamB is SecA dependent and SRP independent in tig strains. Pulse-labeling was performed at 30°C for 30 s with a 10-s chase. Samples were immunoprecipitated with anti-MalE and anti-LamB antisera.
Taken together, these observations show that LamB and the LamB-LacZ fusion, bearing wild-type signal sequences, are still targeted by the SecB pathway in the absence of Tig. As described below, we suggest that deletion of tig facilitates SecB- and SecA-dependent LamB-LacZ secretion, allowing the fusion to more effectively traverse the translocation channel. This improved translocation efficiency is not due to an increase in the levels of the cytoplasmic chaperones, GroEL. The steady-state levels of GroEL in the fusion strain were the same whether Tig was present or not (data not shown).
DISCUSSION
The H*lamB′-′lacZ fusion produces a hybrid protein that is routed to the SecYEG translocator by SRP, and this construct provides a useful genetic tool for analysis of this targeting mechanism.
H*LamB-LacZ secretion is cotranslational.
The presence of the rough endoplasmic reticulum in eukaryotic cells prompted the idea of a cotranslational targeting mode, and a vast body of evidence supports the hypothesis that there is a cotranslational mechanism for SRP-dependent secretion in eukaryotes (reviewed in references 15 and 16). Eukaryotic SRP binds to signal sequences of nascent polypeptides as they emerge from the ribosome and arrests translation. Translation resumes when the ribosome-nascent chain complex docks to form a tight seal at the translocation channel on the endoplasmic reticulum membrane. Presumably this translation arrest and the coupling between translation and secretion prevent secreted proteins from premature or incorrect folding in the cytosol. Several lines of evidence suggest that a cotranslational mechanism is conserved in prokaryotes. First, Powers and Walter (25) showed that in vitro the prokaryotic SRP components Ffh and FtsY could partially replace the function of the eukaryotic components SRP54 and SRα, respectively. Moreover, it was observed long ago that the 4.5S RNA of E. coli could associate with both the targeting factor, Ffh, and the translation elongation factor, EF-G (6). Finally, a cotranslational mechanism makes the most sense for the targeting of inner membrane proteins, the preferred substrate of prokaryotic SRP (32). Although these observations are consistent with a potential mechanism for coupling between secretion and translation, such an association has never been experimentally demonstrated.
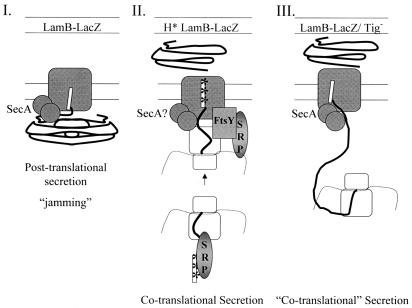
While a few studies have approached the question of a cotranslational secretion mechanism for prokaryotic SRP in vitro (14, 19, 22, 25), none has yet convincingly demonstrated that there is such a mechanism in vivo. Data presented here present for the first time strong support for the hypothesis that there is a cotranslational targeting mechanism in vivo. The targeting of H*LamB-LacZ to the periplasm by the SRP prevents fusion-dependent jamming of the translocation channel. It is difficult to imagine how a molecule as large as the LamB-LacZ fusion could get through the translocation channel without jamming unless secretion is closely coupled to translation (Fig. 11).
FIG. 11.
Possible secretion mechanisms: comparison of secretion modes for LamB-LacZ variants. In mechanism I SecA-dependent, posttranslational secretion of LamB-LacZ jams the translocation channel. In mechanism II cotranslational, SRP-dependent secretion of H*LamB-LacZ relieves jamming and results in localization of the fusion to the periplasm. The ribosome is shown in loose association with the translocation channel. SecA may or may not be required. In mechanism III cotranslational, SecA-dependent secretion of LamB-LacZ in the absence of Tig occurs. Secretion commences prior to completion of translation, reducing the potential for cytoplasmic, folding-based toxicity. SecA is clearly required.
Role of SecA.
Our finding that H*LamB-LacZ secretion was insensitive to the presence of secA51(Ts) was surprising because previous studies have shown that SRP substrates bearing large periplasmic domains require SecA for secretion (22, 26). LacZ effectively constitutes a large periplasmic domain, yet this substrate is efficiently secreted when SecA is impaired. This could indicate that SecA is not essential for the SRP pathway. Alternatively, it is possible that the increased hydrophobicity of the H*LamB signal sequence improves H*LamB affinity for SecA. Increased H*LamB affinity for SecA might attenuate the impact of the secA51(Ts) allele. The small amount of precursor H*LamB seen in the secA51(Ts) background and the slight effect of this mutation on H*LamB-LacZ-dependent Lac activity might reflect a true SecA requirement. Finally, we cannot exclude the possibility that translocation requirements may vary depending on the nature of the substrate received at SecYEG, irrespective of the mode of delivery to the channel (12). Studies are needed to clarify the role of SecA in this system. However, the observation that secA51(Ts) produces similar effects on H*LamB whether LacZ is fused or not is noteworthy by itself, given the hypothesis that SRP-dependent substrates require SecA for secretion of large periplasmic domains.
Trigger factor influences the secretion mode but not pathway discrimination.
Results presented here strongly favor the idea that signal sequence hydrophobicity is sufficient to influence targeting pathway preference. These results correlate with the observations of Lee and Bernstein (17), who showed that increasing signal sequence hydrophobicity rendered MalE secretion SRP dependent. While it is clear that hydrophobicity alone can route certain substrates to the SRP, there is evidence that additional factors, such as structural features of the signal sequence, may influence pathway discrimination (1). Moreover, there are several examples of proteins that bear signal sequences which appear to be sufficiently hydrophobic to promote SRP interaction but remain SecB and SecA dependent (24; J. Beckwith, personal communication). The basis for this discrepancy is not yet understood. It is noteworthy, however, that both LamB and the LamB-LacZ fusion protein, which are vastly different in terms of size and structural behavior, are efficiently secreted by the SRP with changes only to the signal sequence.
Some workers have proposed that an additional targeting factor precludes SRP interactions with certain substrates. In vitro cross-linking studies performed in the laboratory of Muller and colleagues showed that Tig cross-linked to SecB pathway substrates (like preOmpA) but not to SRP-dependent substrates, leading to the proposal that Tig promotes pathway discrimination (4; reviewed in references 13 and 21). The idea is that Tig association with presecretory proteins prevents SRP interaction, thereby delimitating pathway selection. From this model, it follows that deletion of Tig should remove the blocking activity that prevents secretory proteins from interacting with SRP. To the contrary, results presented here show that deletion of Tig influences the efficiency of secretion but not the pathway selected, which directly challenges this model.
Our observation that LamB-LacZ is secreted into the periplasm in the absence of Tig correlates instead with the findings of Lee and Bernstein (18), who demonstrated that Tig inactivation increased the overall efficiency of Sec secretion, suppressing the need for targeting factors such as SecB. Like Muller's group, Lee and Bernstein promoted the idea that Tig associates early in polypeptide synthesis with certain secreted substrates, thereby blocking association with export dedicated chaperone factors, such as SecB, until polypeptide synthesis is completed and Tig is released. Here the models diverge. According to Lee and Bernstein, when Tig is absent, translocation commences prior to completion of synthesis, obviating the need for chaperone activity and effectively rendering the Sec pathway cotranslational. Such a mechanism could explain how LamB-LacZ, which typically gets stuck in the secretion apparatus, can localize to the periplasm in the absence of Tig (Fig. 11). This mechanism differs from SRP-dependent cotranslational fusion targeting in that SecA is clearly required. Furthermore, the lack of SRP involvement highlights the possibility that in prokaryotes, translation arrest is not a necessary corollary to cotranslational targeting. An alternative explanation for the secretion of LamB-LacZ into the periplasm is that LacZ folding is retarded in the absence of Tig, which reduces jamming and allows more of the fusion to effectively exit the translocation channel. If this model is correct, then in the absence of Tig LamB-LacZ behaves like LamB-LacZX90 (Table 2).
Useful genetic tool.
By coupling the SRP pathway to Lac genetics we introduced a promising tool for probing SRP-dependent secretion. The utility of the system was demonstrated in our studies of H*LamB-LacZ secretion in the presence of secY40, a genetic lesion known to impair SRP-dependent secretion. Newitt and Bernstein (23) found that in rich media SecY40 specifically impaired inner membrane protein insertion and functioned synergistically with the SRP. Consistent with these findings, H*LamB-LacZ secretion is defective in the presence of secY40. Consistent with the studies of Baba et al. (2) and Mori and Ito (20), H*LamB-LacZ was insensitive to secY39. SecY39 confers a strong defect on secretion of precursor OmpA, a SecB pathway substrate (2, 20). With the H*lamB′-′lacZ fusion strain these allele-specific differences are reflected by clear changes in colony color. By analogy, the H*lamB′-′lacZ gene fusion should simplify the isolation and characterization of other mutations that specifically affect the SRP targeting pathway.
Acknowledgments
We thank the Beckwith laboratory for providing the SRP mutant strains and for sharing data prior to publication. We thank Daniel Isaac, Martin Braun, Natacha Ruiz, and other members of the Silhavy laboratory for insightful discussions and invaluable support during the course of this study.
C.W.B. gratefully acknowledges funding provided by the U.S. Department of Energy through a Life Sciences Research Foundation postdoctoral fellowship. T.J.S. was supported by NIGMS MERIT award GM34821.
REFERENCES
- 1.Adams, H., P. A. Scotti, H. de Cock, J. Luirink, and J. Tommassen. 2002. The presence of a helix breaker in the hydrophobic core of signal sequences of secretory proteins prevents recognition by the signal-recognition particle in Escherichia coli. Eur. J. Biochem. 269:5564-5571. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., A. Jacq, E. Brickman, J. Beckwith, T. Taura, C. Ueguchi, Y. Akiyama, and K. Ito. 1990. Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172:7005-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batey, R. T., R. P. Rambo, L. Lucast, B. Rha, and J. A. Doudna. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287:1232-1239. [DOI] [PubMed] [Google Scholar]
- 4.Beck, K., L.-F. Wu, J. Brunner, and M. Muller. 2000. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19:134-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, S. A., E. Bremer, and T. J. Silhavy. 1984. Intragenic regions required for LamB export. Proc. Natl. Acad. Sci. USA 81:3830-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S. 1987. Mutations in the gene for EF-G reduce the requirement for 4.5S RNA in the growth of E. coli. Cell 49:825-833. [DOI] [PubMed] [Google Scholar]
- 7.Bui, N., and K. Strub. 1999. New insights into signal recognition and elongation arrest activities of the signal recognition particle. J. Biol. Chem. 380:135-145. [DOI] [PubMed] [Google Scholar]
- 8.Cosma, C. L., M. D. Crotwell, S. Y Burrows, and T. J. Silhavy. 1998. Folding-based suppression of extracytoplasmic toxicity conferred by processing-defective LamB. J. Bacteriol. 180:3120-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 10.Emr, S. D., and T. J. Silhavy. 1980. Mutations affecting localization of an Escherichia coli outer membrane protein, the bacteriophage lambda receptor. J. Mol. Biol. 141:63-90. [DOI] [PubMed] [Google Scholar]
- 11.Emr, S. D., M. Schwartz, and T. J. Silhavy. 1978. Mutations altering the cellular localization of the phage lambda receptor, an Escherichia coli outer membrane protein. Proc. Natl. Acad. Sci. USA 75:5802-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froderberg, L., E. Houben, J. C. Samuelson, M. Chen, S.-K. Park, G. J. Phillips, R. Dalbey, J. Luirink, J., and J.-W. L. de Gier. 2003. Versatility of inner membrane protein biogenesis in Escherichia coli. Mol. Microbiol. 47:1015-1027. [DOI] [PubMed] [Google Scholar]
- 13.Herskovits, A. A., E. S. Bochkareva, and E. Bibi. 2001. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 38:927-939. [DOI] [PubMed] [Google Scholar]
- 14.Herskovits, A. A., A. Seluanov, R. Rajsbaum, C. M. ten Hagen-Jongman, T. Henrichs, E. S. Bochkareva, G. J. Phillips, F. J. Probst, T. Nakae, M. Ehrmann, J. Luirink, and E. Bibi. 2001. Evidence for coupling of membrane targeting and function of the signal recognition particle (SRP) receptor FtsY. EMBO J. 2:1040-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalies, K.-U., and E. Hartmann. 1998. Protein translocation into the endoplasmic reticulum (ER): two similar routes with different modes. Eur. J. Biochem. 245:1-5. [DOI] [PubMed] [Google Scholar]
- 16.Keenan, R. J., D. M. Freymann, R. M. Stroud, and P. Walter. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755-775. [DOI] [PubMed] [Google Scholar]
- 17.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, H. C., and H. D. Bernstein. 2002. Trigger factor retards protein export in Escherichia coli. J. Biol. Chem. 277:43527-43535. [DOI] [PubMed] [Google Scholar]
- 19.Macfarlane, J., and M. Muller. 1995. The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem. 233:766-771. [DOI] [PubMed] [Google Scholar]
- 20.Mori, H., and K. Ito. 2003. Biochemical characterization of a mutationally altered translocase: proton motive force stimulation of the initial phase of translocation. J. Bacteriol. 185:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller, M., H.-G. Koch, K. Beck, and U. Schafer. 2001. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog. Nucleic Acid Res. Mol. Biol. 66:107-157. [DOI] [PubMed] [Google Scholar]
- 22.Neumann-Haefelin, C., U. Schafer, M. Muller, and H.-G. Koch. 2000. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newitt, J. A., and H. D. Bernstein. 1998. A mutation in the Escherichia coli secY gene that produces distinct effects on inner membrane protein insertion and protein export. J. Biol. Chem. 273:12451-12456. [DOI] [PubMed] [Google Scholar]
- 24.Newitt, J. A., N. D. Ulbrandt, and H. D. Bernstein. 1999. The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of Escherichia coli inner membrane proteins. J. Bacteriol. 181:4561-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers, P., and T. Walter. 1997. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16:4880-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi, H-Y, and H. D. Bernstein. 1999. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J. Biol. Chem. 274:8993-8997. [DOI] [PubMed] [Google Scholar]
- 27.Silhavy, T. J., H. A. Shuman, J. Beckwith, and M. Schwartz. 1977. Use of gene fusions to study outer membrane protein localization in Escherichia coli. Proc. Natl. Acad. Sci. USA 74:5411-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Snyder, W. B., and T. J. Silhavy. 1995. β-Galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J. Bacteriol. 177:953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian, H. P., and J. Beckwith. 2002. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 184:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian, H. P., D. Boyd, and J. Beckwith. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. USA 97:4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 33.Yu, D., H. M. Ellis, E. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]