Abstract
TolC is the outer-membrane component of several multidrug resistance (MDR) efflux pumps and plays an important role in the survival and virulence of many gram-negative bacterial animal pathogens. We have identified and characterized the outer-membrane protein-encoding gene tolC in the bacterial plant pathogen Erwinia chrysanthemi EC16. The gene was found to encode a 51-kDa protein with 70% identity to its Escherichia coli homologue. The E. chrysanthemi gene was able to functionally complement the E. coli tolC gene with respect to its role in MDR efflux pumps. A tolC mutant of E. chrysanthemi was found to be extremely sensitive to antimicrobial agents, including several plant-derived chemicals. This mutant was unable to grow in planta and its ability to cause plant tissue maceration was severely compromised. The tolC mutant was shown to be defective in the efflux of berberine, a model antimicrobial plant chemical. These results suggest that by conferring resistance to the antimicrobial compounds produced by plants, the E. chrysanthemi tolC plays an important role in the survival and colonization of the pathogen in plant tissue.
The bacterium Erwinia chrysanthemi is a gram-negative broad-host-range phytopathogen causing soft-rot disease. Production of plant cell wall-degrading enzymes and avirulence proteins by the phytopathogen contributes to the destruction of plant structural barriers and evasion of certain host defense responses, respectively (4, 9, 10, 21, 37). Nutrient acquisition in E. chrysanthemi from degraded plant cell-wall pectin is facilitated by transport systems for pectin-derived oligomers and monomers (18, 20, 22, 46). Secretion of pathogenesis-related proteins across the bacterial cytoplasmic and outer membranes requires several export systems that have been extensively studied. Pectin-degrading enzymes are secreted by a type II-dependent mechanism (encoded by the out genes), and secretion of avirulence proteins into the host is mediated by a type III-dependent process (encoded by the hrp genes) (9, 33, 35). Finally, secretion of metalloproteases by a sec-independent type I mechanism (prt genes) in E. chrysanthemi has also been demonstrated (11, 25, 26).
TolC is an important though low-abundance protein in the outer membrane of gram-negative bacteria (17). The crystal structure of this protein has recently been determined (24). The protein exists functionally as a trimer forming a β-barrel with a large internal diameter, facilitating movement of both large and small molecules through the outer membrane (2). TolC functions as a component of multidrug resistance (MDR) efflux systems in the removal of a broad range of toxic chemicals from the cell (16, 41). In Escherichia coli, type I-dependent secretion of certain virulence-associated proteins also requires TolC (48). The role of TolC in virulence and survival strategies of the pathogenic enteric bacteria E. coli (49), Vibrio cholerae (5), Salmonella enterica serovar Enteritidis (40), and Serratia marcescens (6) has been established. While the existence and role of TolC in many bacterial pathogens of mammals have been established, the occurrence and role of a TolC homologue in a plant pathogenic bacterium have not been conclusively demonstrated. Here we present the characterization of the E. chrysanthemi tolC and demonstrate its importance in resistance to antimicrobial plant chemicals as well as phytopathogenesis.
MATERIALS AND METHODS
Bacterial strains and plasmids, media, and growth conditions.
The bacterial strains, plasmids, and phages used in this study are listed in Table 1. E. chrysanthemi and E. coli cells were grown (while being shaken at 200 rpm for all liquid cultures) in Luria-Bertani (LB) medium at 30 and 37°C, respectively. Antibiotics were added where required at the following concentrations unless specified otherwise: kanamycin (Kan), 50 μg/ml; ampicillin (Amp), 50 μg/ml; tetracycline (Tet), 10 μg/ml; streptomycin (Str), 25 μg/ml; and novobiocin (NOV), 1 to 20 μg/ml. Deoxycholate (DOC) was used in LB growth medium at a final concentration of 0.1 to 10%. Plant-derived chemicals such as rhein (9,10-dihydro-4,5-dihydroxy-9,10-dioxo-2-anthracenecarboxylic acid), genistein (4′,5,7-trihydroxyisoflavone), plumbagin (5-hydroxy-2-methyl-1,4-naphthalenedione), p-coumaric acid and t-cinnamic acid (phenolic acids), pyrithione (2-mercaptopyridine-1-oxide), esculetin (6,7-dihydroxycoumarin), and berberine (5,6-dihydro-9,10-dimethoxybenzo-1,3-benzodioxoloquinolizimium) were purchased from the Sigma-Aldrich Chemical Co. Using appropriate solvents, stock solutions of all the plant-derived chemicals were made at 10 mg/ml. Esculetin was dissolved in 100% methanol, berberine and pyrithione were dissolved in water, and rhein and genistein were dissolved in 50 mM NaOH, while the rest of the plant-derived chemicals were dissolved using 95% ethanol. Appropriate solvent controls were incorporated into the experiments to account for the solvent effect.
TABLE 1.
Bacterial strains, phages, and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F φ80dlacZΔM15 Δ(lacZY-argR)U169 deoR recA endA1 hsdR17(rK− mK−) | Gibco-BRL |
| HB101 | supE44 hsdS20 recA13 ara-14 proA2 lacY1 GalK2 rpsL20 xyl-5 mtl-I | 34 |
| LM512 | Parent of LM1151 | B. Bachman |
| LM1151 | LM 512 tolC recA Strr Tetr | J.A. Fralick |
| LM1051 | LM1151 carrying pLAFR1051 | This study |
| Erwinia chrysanthemi | ||
| EC16 | Wild type | A. K. Chatterjee |
| SF1603 | EC16 carrying a nptI insertion in tolC gene, Kanr | This study |
| SF1603a | SF1603 carrying pBR2316, Ampr, Kanr | This study |
| SF1603b | SF1603 carrying pBR2353, Ampr, Kanr | This study |
| Plasmids | ||
| pLAFR3 | Tetrcos Tra− Mob+ IncP | 11 |
| pLAFR1051 | pLAFR3 with EC16 tolC genomic DNA fragment | This study |
| pBR2316 | pBR322 derivative carrying EC16 tolC | This study |
| pBR2353 | pBR322 derivative carrying E. coli tolC | This study |
DNA methods.
Plasmid or genomic DNA preparation and DNA purification from agarose gels were carried out using respective Qiagen kits (QIAGEN Inc). Restriction digestion, ligation, DNA electrophoresis, and E. coli transformations were performed following manufacturer recommendations or as described by Sambrook et al. (34). For transformation of E. chrysanthemi, a single colony of EC16 culture was inoculated in 2 ml of LB broth and grown while being shaken overnight at 30°C. The culture was diluted 1:50 in 50 ml of LB broth and grown while being shaken at 40°C for 6 h. The culture was centrifuged and washed three times with 20 ml of ice-cold 10% glycerol. The cell pellet was resuspended in 0.2 ml of 10% glycerol and maintained on ice. The cell suspension (0.02 ml) was mixed with 0.005 ml of plasmid DNA-0.1 to 1 μg DNA in water, chilled on ice for 5 min, and pulsed (using a Cell-Porator with a voltage booster [BRL Life Technologies Inc.]) at 2 kV and 4 kΩ. Cells were transferred immediately to 1 ml of LB medium and grown while being shaken at 30°C for 1 h. The culture was then centrifuged, and the pellet was resuspended in 0.1 ml of LB medium. Aliquots of resuspended cells were spread on LB agar plates carrying appropriate antibiotics.
Isolation of E. chrysanthemi tolC.
An E. chrysanthemi EC16 cosmid library in pLAFR3 vector (provided by Noel T. Keen [11]) was mated with LM1151 in the presence of a helper strain (E. coli HB101) containing a self-transmissible plasmid, pRK2013 (14). Following triparental mating, transconjugants were screened for restoration of DOC resistance by direct selection on LB agar plates containing 5% DOC. The cosmid clone (pLAFR1051) that conferred DOC resistance was isolated, and its functional complementation ability was confirmed by repeating the triparental mating. DOC- and Tet-resistant colonies were restreaked on selective media before further analysis.
Subcloning and DNA sequencing.
A 5.4-kb ClaI and PstI functionally complementing fragment of the cosmid pLAFR1051 was cloned into pBluescript, subcloned into E. coli DH5α, and sequenced. The initial partial sequence obtained was used to design internal primers to completely sequence both strands of the E. chrysanthemi tolC gene from pLAFR1051. The sequence information was utilized to design nucleotide primers upstream of the E. chrysanthemi tolC transcription start site and downstream of the translational stop codon. Using these primers, a PCR product of about 2.0 kb (full-length tolC gene) was obtained using E. chrysanthemi EC16 genomic DNA. The PCR product was cloned into pCR2.1-TOPO cloning vector (Invitogen) and sequenced. The functional complementing ability of the PCR product was confirmed by transforming this construct into LM1151 and direct selection on LB plates containing 5% DOC. Using an ABI Prism 310 automated DNA sequencer (Perkin Elmer), all DNA sequencing was carried out at the Texas Tech University Center for Biotechnology and Genomics core facility.
Complementation analysis.
Cosmid pLAFR1051 carrying E. chrysanthemi tolC was transformed into LM1151 for complementation analysis (23). Transformants were screened on appropriate antibiotics and then tested for resistance to MDR efflux pump substrates (NOV, DOC, and carbonyl cyanide m-chlorophenylhydrazone [CCCP]) and sensitivity to colicin E1 and bacteriophage U3. Drug sensitivity was tested using a filter disk zone inhibition assay (16). Phage or colicin sensitivity was determined by cross-streaking or spotting the phage or colicin with bacterial cells on LB agar plates and incubating overnight. The occurrence of clearing at the intersection of cross-streaks or plaques on bacterial lawns was used as an indication of phage or colicin sensitivity.
Immunoblot analysis of TolC.
E. chrysanthemi outer membranes were prepared as previously described (23, 18). Following protein separation on SDS-10% polyacrylamide gels and transfer to polyvinylidene difluoride (PVDF) membranes, immunological analysis was performed (using E. coli anti-TolC rabbit polyclonal antiserum) on outer-membrane preparations and detected with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G and 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium (Sigma Co.). Coomassie blue staining was performed according to standard procedures (34).
Marker-exchange mutagenesis.
Marker-exchange mutagenesis was carried out as described previously (32) with few modifications. E. chrysanthemi EC16 carrying the tolC::nptI (Kanr) construct in pBR322 (Ampr) vector was grown overnight at 30°C in 2 ml of LB medium containing 25 μg of Kan/ml. The culture was centrifuged and washed in basal A-P medium (containing, per liter, 12.1 g of Tris, 0.1 g of MgSO4 • 7H2O, 1 g of NH4SO4, 0.5 g of sodium citrate, and 2 ml of glycerol, with the pH adjusted to 7.4) and resuspended in 1 ml of the same medium. Cells were diluted 1:1,000 into 1 ml of the basal A-P medium containing 1 mM potassium phosphate buffer (pH 7.4) and 15 μg of Kan/ml and grown for 12 h at 30°C. Following this, the culture was again diluted 1:1,000 into 1 ml of the basal A-P medium containing 1 mM potassium phosphate buffer (pH 7.4) and grown for 12 h at 30°C. Appropriate dilutions (10−6 to 10−7) of the culture were then plated on LB agar (agar at 1.2% concentration) containing 15 μg of Kan/ml and then screened for Kanr Amps colonies to obtain the EC16 tolC mutant.
Zone inhibition assays.
Cultures of respective strains were grown overnight. Approximately 106 cells were mixed with 3 ml of 0.8% agar, overlaid on LB agar plates containing appropriate selection antibiotics, and allowed to solidify. Sterile paper disks (Whatman filters) 6 mm in diameter were placed on the surface of the agar. A total of 0.03 ml of NOV (20 mg/ml), SDS (10% [wt/vol]), DOC (5% [wt/vol]), or CCCP (8 mM) was pipetted onto separate disks. The plates were incubated for 36 h, and the diameters (in millimeters) of the zones of inhibition were measured.
Determination of MIC.
MIC assays were performed in a final volume of 0.2 ml in 96-well microtiter plates. Appropriate hydrophobic agents were serially diluted in LB broth containing the appropriate selection antibiotics for the different strains. Appropriate controls for the solvents used to dissolve the hydrophobic chemicals were also included. Bacteria from an overnight culture were inoculated at a final concentration of 106 cells/ml. The microtiter plates were shaken at 200 rpm at 30°C for 36 h. The MIC was determined as the concentration of the substance which prevented growth after 36 h.
Plant pathogenicity assay.
Heads of witloof chicory (Cichorium intybus L. [Belgian endive]) were purchased from a local supermarket. Cultures of appropriate strains grown overnight were centrifuged and washed twice with 5 mM MES (morpholineethanesulfonic acid), pH 6.5. Cells were resuspended in the same buffer at a density of approximately 108 cells/ml. Using a sterile toothpick, chicory leaves were inoculated by introducing 0.005 ml of inoculum into a small wound created on the inner surface of each leaf. Leaves were maintained in a moist chamber at 30°C for 72 h. Lesions were identified by browning and maceration around the site of infection. The area of macerated tissue was defined as the product of the length and width of the lesion. To estimate the total number of bacteria per infected leaf, the leaves were homogenized in 5 mM MES and serial dilutions were plated on appropriate antibiotic plates.
Determination of berberine accumulation.
Determination of berberine accumulation was performed as described previously (39) with a few modifications. Cultures of appropriate strains grown overnight were centrifuged and washed twice with 20 mM HEPES buffer (pH 7.0). Cells were resuspended to an optical density of 600 nm of 0.3 in HEPES buffer containing 0.2% glucose and incubated at 30°C for 90 min. The cells were washed and resuspended to an optical density of 600 nm of 0.15 in HEPES buffer. Assays were performed in 96-well flat-bottom black plates (Polyfiltronics) in a final volume of 0.2 ml. Berberine was added at 0.03 mg/ml, and fluorescence was measured using a Bio-Tek FL600 microplate fluorescence reader (Bio-Tek Instruments, Inc.), with a 360/40-nm excitation filter and a 530/25-nm emission filter.
RESULTS AND DISCUSSION
E. chrysanthemi tolC can functionally complement an E. coli tolC mutant.
TolC mutants of E. coli are hypersensitive to detergents such as SDS and a variety of hydrophobic agents such as DOC, NOV, and CCCP (31, 44). To screen for the presence of E. chrysanthemi EC16 tolC, the EC16 genomic library (cloned into pLAFR3) was conjugated into an E. coli tolC deletion strain (LM1151) by triparental mating and transconjugants were selected on LB agar plates containing DOC. This procedure allowed us to identify cosmid pLAFR1051, which conferred DOC resistance to the E. coli tolC mutant. To determine whether this DNA fragment complemented other tolC phenotypes, we examined its ability to rescue LM1151 to NOV, CCCP, and DOC resistance. Table 2 shows that the E. chrysanthemi tolC was able to complement the E. coli tolC mutant to drug resistance. Chemicals such as NOV and DOC are known to be substrates of the AcrAB/TolC efflux pump, while CCCP is known to be a substrate of the EmrAB/TolC efflux system (7, 29). Thus, it is likely that homologs of these efflux pumps in E. chrysanthemi can utilize TolC in conferring resistance to these substrates.
TABLE 2.
Functional analysis of E. chrysanthemi EC16 tolC in E. coli
| Strainb | Diam (mm) of zone of inhibitiona in the presence of:
|
||
|---|---|---|---|
| CCCP | NOV | DOC | |
| LM512 | 20 | 15 | 6 |
| LM1151 | 33 | 28 | 20 |
| LM1051 | 22 | 17 | 6 |
Disk diameter, 6 mm.
LM512, parent strain of LM1151; LM1151, E. coli tolC deletion strain; LM1051, LM1151 with pLAFR1051 and E. chrysanthemi tolC.
The E. chrysanthemi tolC gene product bears strong identity to its E. coli counterpart.
Southern dot blot analysis using the E. coli tolC gene as a probe for the pLAFR1051 cosmid DNA and genomic DNA from E. chrysanthemi indicated that there was nucleotide sequence homology for this gene between the two microorganisms (data not shown). Using primers generated from the cosmid-derived partial tolC sequence and E. chrysanthemi genomic DNA as the template, PCR amplification yielded a 2-kb fragment carrying the complete tolC gene, which was cloned and sequenced (GenBank AF421372).
Nucleotide sequence analysis of the 2,004-base PCR fragment indicates that the tolC homologue from E. chrysanthemi encodes an open reading frame of 1,410 nucleotides. A putative promoter sequence with −10 (TATATC) and −35 (TTGGCC) regions lies 50 nucleotides upstream of the start codon (ATG). The presumed initiator methionine is preceded by a canonical ribosome-binding site (AGGA) at six nucleotides upstream (36). The deduced amino acid sequence bears approximately 70% identity to that of the E. coli TolC. Multiple sequence alignment (CLUSTAL W [45]) of EC16 TolC with homologues from other bacteria (E. coli, S. marcescens, S. enterica serovar Enteritidis, and V. cholerae) revealed approximately 35% amino acid identity across all five sequences. The deduced amino acid sequence of the E. chrysanthemi tolC gene suggests that the protein possesses a signal peptide at its amino terminus. The 24-amino-acid sequence has the characteristics of a signal peptide (30) followed by a peptidase cleavage site between ala24 and glu25. This signal peptide is identical in sequence to the E. coli TolC signal peptide (17).
E. chrysanthemi carries an outer-membrane polypeptide with an apparent mass of 51 kDa that reacts with the anti-E. coli TolC rabbit antiserum (Fig. 1). This molecular mass was predicted from the deduced amino acid sequence and is slightly lower than that of the 52-kDa polypeptide observed in E. coli.
FIG. 1.
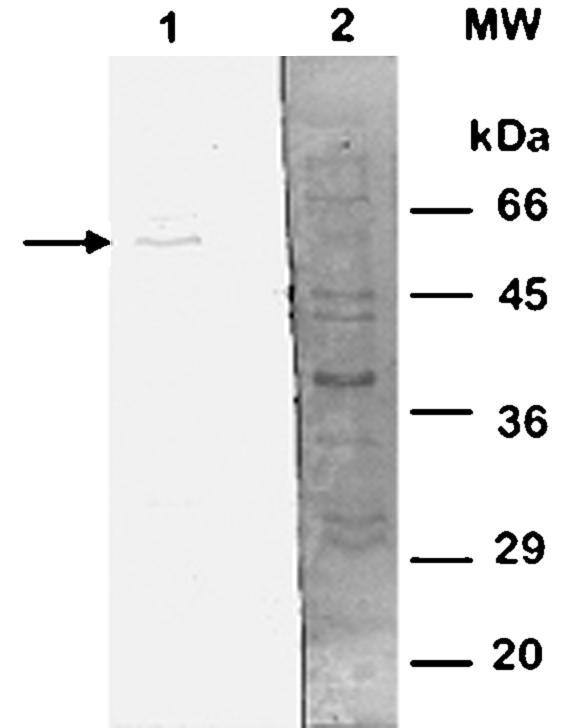
Immunoblot analysis of E. chrysanthemi outer-membrane proteins. Lane 1, results of EC16 outer-membrane protein immunoblot experiments using anti-E. coli TolC rabbit antiserum; lane 2, Coomassie-stained EC16 outer-membrane proteins. The arrow indicates a band representing an outer-membrane polypeptide with an apparent mass of 51 kDa. MW, molecular mass standards.
Construction and characterization of the E. chrysanthemi tolC mutation.
Using a Kan resistance (nptI) cassette, the tolC gene of E. chrysanthemi EC16 was insertionally inactivated by marker-exchange mutagenesis (32). Confirmation of the chromosomal gene disruption was made by Southern blotting and PCR analysis using gene-specific probes and primers, respectively (data not shown). To characterize the phenotype of the EC16 tolC mutant (SF1603), we examined its sensitivity to a variety of agents that are substrates for E. coli MDR efflux systems: SDS, DOC, and NOV. The results are presented in Table 3 and indicate that the tolC mutation sensitizes this strain to all of these agents, suggesting that the role of TolC in resistance to antimicrobial and hydrophobic chemicals in E. chrysanthemi appears to be similar to that found in E. coli. Replacement of the tolC gene in trans on a plasmid (strain SF1603a) in SF1603 rescued the bacterium to drug resistance (Table 3). Strain SF1603 harboring the E. coli tolC gene on a plasmid (strain SF1603b) was also resistant to these antimicrobial chemicals. These results, together with those of the complementation studies performed using the cloned EC16 tolC in strain LM1151, suggest that the tolC genes from the two genera are interchangeable for drug resistance to certain antimicrobial chemicals. The MICs of SDS, DOC, and NOV for E. chrysanthemi EC16 (Table 4) are similar to those observed for E. coli (16). The MICs of these chemicals for the EC16 tolC mutant SF1603 were dramatically reduced and were approximately 500-fold lower than those for the wild type (Table 4).
TABLE 3.
Sensitivity to antimicrobial agentsa
| Strainc | Diam (mm) of zone of inhibitionb in the presence of:
|
||
|---|---|---|---|
| NOV | SDS | DOC | |
| EC16 | 20.00 | 16.33 | 6.00 |
| SF1603 | 49.67 | 46.33 | 31.00 |
| SF1603a | 16.00 | 22.00 | 9.00 |
| SF1603b | 13.00 | 18.00 | 9.00 |
Results represent averages from three separate experiments.
Disk diameter, 6 mm.
EC16, E. chrysanthemi EC16 parent strain; SF1603, EC16 tolC mutant strain; SF1603a, EC16 tolC mutant complemented with the functional EC16 tolC gene on plasmid pBR322; SF1603b, EC16 tolC mutant complemented with the functional E. coli tolC gene on plasmid pBR322.
TABLE 4.
Minimum inhibitory concentration of antimicrobial agentsa
Average values of three separate experiments rounded off to nearest whole number are presented.
EC16, E. chrysanthemi EC16 wild type parent strain; SF1603, EC16 tolC mutant strain.
The wild-type parent strain grew in the presence of 10,000 μg of SDS/ml (highest concentration tested).
The tolC mutant strain did not grow in the presence of 2 μg of NOV/ml (lowest concentration tested).
The tolC mutant strain did not grow in the presence of 20 μg of SDS/ml (lowest concentration tested).
An E. chrysanthemi tolC mutant is defective in causing disease in plant tissue.
For many members of the Enterobacteriaceae, TolC has been implicated in a variety of functions (such as toxin secretion and MDR) that might play a role in pathogenicity. For S. enterica serovar Enteritidis, for example, TolC has been implicated in hemolysin secretion and virulence in BALB/c mice (40). In E. coli, hemolysin and enterotoxin secretion have been shown to require a functional TolC (27, 49); a tolC mutant of V. cholerae was found to be sensitive to bile salts, defective in cytotoxin secretion, and impaired in intestinal colonization (5). Since TolC is an important component in a variety of efflux pumps for the removal of many antimicrobial chemicals and in the secretion of certain virulence-associated proteins, we investigated the phytopathogenic potential of a tolC mutant of E. chrysanthemi.
The E. chrysanthemi tolC SF1603 mutant strain was compared with its wild-type parent in plant infections to assess the contribution of the protein to phytopathogenicity. Using witloof chicory leaves in a bioassay, we observed 95% (± 6.4% standard deviation) reduction in the area of tissue macerated with the tolC mutant (Fig. 2). This significant (P < 0.001) reduction in phytopathogenic potential might reflect the inability of the bacterium to survive the toxic plant environment due to impairment of TolC-dependent chemical efflux pumps. Additionally, the mutation may impair secretion of virulence-associated proteins. However, no qualitative or quantitative differences were seen in the abilities of the tolC mutant strain and the wild-type parent strain to secrete proteases by using milk-derived proteins or gelatin as the substrate (11) or to secrete pectate lyases by using semisolid pectin agar and spectrophotometric assays (38) (data not shown).
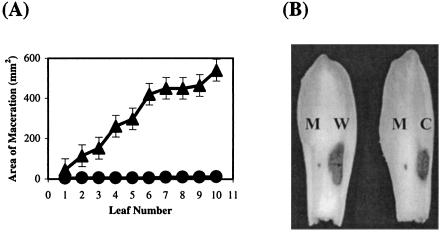
FIG. 2.
E. chrysanthemi EC16 tolC mutant has severely reduced ability to produce lesions in witloof chicory leaves. (A) A total of 10 witloof chicory leaves were infected with either the E. chrysanthemi EC16 wild-type parent strain (closed triangles) or the E. chrysanthemi EC16 tolC mutant strain (closed circles). To estimate disease severity after incubation at 30°C for 72 h, the area of maceration was measured. Percent reduction in maceration was calculated as follows: [(area of maceration by wild-type strain − area of maceration by mutant strain)/area of maceration by wild-type strain] × 100. The mean value (± standard deviation) of the percentage of reduction in maceration per 10 inoculations was used as a measure of the reduction in tissue maceration ability of the tolC mutant strain. (B) The E. chrysanthemi tolC mutant strain (M) was unable to macerate plant tissue but recovered almost wild-type (W) capacity to cause soft rot when the tolC gene was reintroduced into the mutant in trans (complemented strain) (C). Results representative of leaves after three independent experiments are shown. The relative positions (i.e., relative left-right orientations) of the inoculated strains did not have any effect on the extent of lesions produced (data not shown).
In planta survival and growth of the E. chrysanthemi tolC mutant.
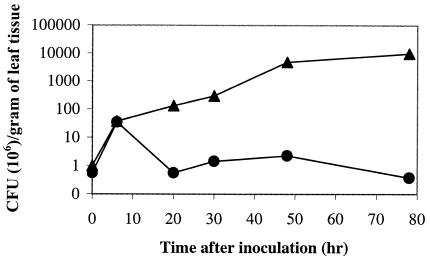
Because the E. chrysanthemi tolC mutant was sensitive to many antimicrobial chemicals, we hypothesized that the bacterium would be unable to tolerate the toxic plant environment. We therefore compared the abilities of the E. chrysanthemi tolC mutant strain and the wild-type parent strain to survive and multiply in plant tissue (Fig. 3). The E. chrysanthemi wild-type strain multiplied in plant tissue, with bacterial numbers increasing approximately 104-fold 78 h following inoculation. However, the total number of bacteria in the leaves inoculated with the tolC mutant was reduced by 40% 78 h following inoculation. The wild-type EC16 and the tolC mutant strains both multiplied to approximately 3.5 × 107 cells/gram of leaf tissue 6 h following inoculation, although no visible tissue maceration could be observed at this time. It was surprising that the tolC mutant was able to multiply in the plant tissue during the first 6 h. This could have been due to a lag time in the response of the leaves to infection.
FIG. 3.
Witloof chicory leaves inoculated with approximately 7 × 105 bacteria of either the wild-type (closed triangles) or the tolC mutant (closed circles) strain were harvested at different time intervals to assess the number of viable bacteria. Results for number of CFU per gram of leaf tissue are plotted in a logarithmic scale along the y axis. A graph representative of two independent experiments is shown.
Visible maceration of the plant tissue in the leaves infected with wild-type EC16 strain was seen beginning just 12 h after initial inoculation. The total number of wild-type EC16 bacteria corresponding to the 20-h time interval was approximately 1.3 × 108 cells/gram of leaf tissue. The bacteria multiplied exponentially, and the area of maceration grew correspondingly with time. In the case of the tolC mutant, after the initial increase in bacterial number the total number of bacteria decreased to the level of the inoculum after 20 h and to approximately 60% of the initial inoculum after 78 h. No visible tissue maceration was observed in the leaves inoculated with the tolC mutant at any time following inoculation. Hence, although the tolC strain survived, it was unable to grow and produce lesions in the leaf. This may have been due to the presence of some of the growth-inhibitory compounds produced by the leaves in response to infection. However, the amount of these compounds produced in the detached leaves (i.e., the ones used in this assay) apparently was not sufficient to kill all of the tolC mutant bacteria.
The E. chrysanthemi tolC mutant is sensitive to antimicrobial plant chemicals.
Plants produce a variety of chemicals with antimicrobial properties (15, 42). Many bacteria have evolved mechanisms to tolerate and rid themselves of plant-derived toxins (43). Since the growth of the E. chrysanthemi tolC mutant was suppressed in plant tissue, we used a MIC assay and a variety of plant-derived antimicrobial chemicals to compare the sensitivities of the wild-type and mutant strains (Table 5). The tolC mutant clearly showed increased sensitivity to most of the plant chemicals we tested. When compared to those for the wild-type parent strain, the MICs of rhein, genistein, berberine, and plumbagin for the tolC mutant strain were found to be at least 64-fold, 32-fold, 16-fold, and 8-fold lower, respectively. Rhein is a well-characterized antimicrobial of rhubarb and is known to be effective against bacteria and Saccharomyces cerevisiae (12, 19). Genistein is an isoflavonoid produced by both legumes and nonleguminous plants (47, 50). It is a common precursor in the biosynthesis of antimicrobial phytoalexins and phytoanticipins and is also antibacterial (1, 3). Plumbagin is a naphthoquinone with known antimicrobial properties (13). The phenolic compounds p-coumaric acid and t-cinnamic acid as well as pyrithione and esculetin were slightly more toxic to the tolC mutant than to the wild-type parent. Pyrithione was seen to inhibit growth even of wild-type cells at concentrations as low as 3.1 μg/ml. Wild-type strain EC16, as well as the tolC mutant strain, was resistant to relatively high levels (>2 mg/ml) of gossypol, a known antifungal phytoalexin of Gossypium species (8). This indicates that the tolC mutant is not nonspecifically sensitive to all chemicals. Berberine is an alkaloid with antibacterial properties. Interestingly, this compound has previously been observed to be toxic only to gram-positive bacteria. In gram-positive bacteria, the NorA pump has been implicated in resistance to this compound (39). Our data indicate that the loss of TolC sensitizes E. chrysanthemi to this compound because it accumulates inside the cells (Fig. 4).
TABLE 5.
MICs of model substrates and plant-derived chemicalsa
| Plant chemical | MIC (μg/ml) ofb:
|
||
|---|---|---|---|
| WT | ΔtolC | Several fold difference | |
| Berberine | >2,000 | 125 | >16 |
| t-Cinnamic acid | 500 | 250 | 2 |
| p-Coumaric acid | 500 | 125 | 4 |
| Esculetin | 500 | 125 | 4 |
| Genistein | >1,000 | 31.25 | 32 |
| Gossypol | >2,000 | >2,000 | |
| Plumbagin | 31.25 | 3.91 | 8 |
| Pyrithione | 3.13 | 1.56 | 2 |
| Rhein | >1,000 | 15.63 | 64 |
Results represent average values from three separate experiments.
WT, E. chrysanthemi parent strain; ΔtolC, E. chrysanthemi tolC mutant strain. Higher concentrations of some of the plant chemicals could not be tested due to limited solubility of these compounds.
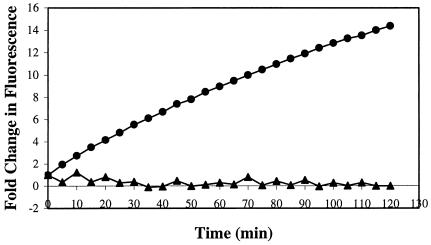
FIG. 4.
Berberine accumulation was studied with the addition of 0.03 mg of berberine/ml to wild-type (closed triangles) or tolC mutant (closed circles) cells energized in the presence of 100 mM formate. Fluorescence was measured using a 360/40-nm excitation filter and a 530/25-nm emission filter. A graph representative of three independent experiments is shown above. Severalfold changes in fluorescence were calculated using average values of triplicates within a single experiment.
Berberine has an increased fluorescence upon binding to DNA and thus represents a good model substrate for assessment of the efflux activity in the tolC mutant strain. The wild-type strain was found to be effective in keeping the intracellular concentrations of berberine low. However, the compound accumulated rapidly in the tolC mutant strain. These results suggest a role for TolC-mediated efflux of berberine in E. chrysanthemi and are compatible with those presented in Table 5.
In this study we have shown that E. chrysanthemi harbors a functional homologue of the E. coli TolC protein. The protein was localized to the outer membrane and found to be immunologically similar to E. coli TolC. The E. chrysanthemi TolC was able to replace its E. coli counterpart in chemical efflux. We also were able to demonstrate that a tolC mutant of E. chrysanthemi could not effectively infect witloof chicory leaves. The plant environment is replete with antimicrobial enzymes and chemicals, many of which are induced following infection by microorganisms. Thus, the inducible plant cellular defensive response functions to block pathogen amplification and impair ingress into plant tissue. Plant-derived secondary chemicals such as phytoalexins and hydrophobic chemicals like phenolics are recognized for their antimicrobial properties. Survival and sustained growth of successful plant pathogens in the plant necessitates the functioning of mechanisms to detoxify and/or expel plant-derived toxins. In E. chrysanthemi EC16, the sapA-sapF locus conferring resistance to antimicrobial peptides from plants has been shown to be important in survival of the bacterium in planta (28).
However, although much is understood about the virulence capabilities of many bacterial plant pathogens, little is known about the mechanisms employed by phytopathogens to survive the onslaught of the plant chemical environment. The tolC mutant of E. chrysanthemi was severely impaired in plant tissue maceration and in causing soft rot of witloof chicory. This mutation did not impair secretion of extracellular proteases or pectinases. This suggested that because of its role in antimicrobial chemical resistance, TolC is essential for bacterial survival in planta. We found that the E. chrysanthemi tolC mutant was unable to multiply effectively in plant tissue and was therefore unable to initiate soft rot and disease. This indicates that the infected leaf tissue was able to suppress the growth of the tolC mutant strain. The E. chrysanthemi tolC mutant strain was found to be sensitive to several plant chemicals, as has been observed with an E. coli tolC mutant strain (47). Analysis of one of the model plant chemicals, berberine, showed that the tolC mutant was defective in the extrusion of this compound. These data suggest that TolC-mediated MDR efflux may be vital to E. chrysanthemi during plant infection. We are presently examining the role of MDR efflux systems and their substrates in the phytopathogenicity of E. chrysanthemi.
Acknowledgments
This work was supported in part by a seed grant from Texas Tech University to M.J.D.S.F. and J.A.F. and NIH grant A148696 to J.A.F.
The assistance of Jennifer Cole, Howard Hughes Undergraduate Research Fellow, is appreciated.
Footnotes
Dedicated to the memory of Noel Keen.
REFERENCES
- 1.Akeda, Y., K. Nagayama, K. Yamamoto, and T. Honda. 1997. Invasive phenotype of Vibrio parahaemolyticus. J. Infect. Dis. 176:822-824. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, C., E. Koronakis, C. Hughes, and V. Koronakis. 2002. An aspartate ring in the TolC tunnel entrance determines ion selectivity and presents a target for blocking by large cations. Mol. Microbiol. 44:1131-1139. [DOI] [PubMed] [Google Scholar]
- 3.Bae, E. A., M. J. Han, and D. H. Kim. 2001. In vitro anti-Helicobacter pylori activity of irisolidone isolated from the flowers and rhizomes of Pueraria thunbergiana. Planta Med. 67:161-163. [DOI] [PubMed] [Google Scholar]
- 4.Barras, F., F. van Gijsegem, and A. K. Chatterjee. 1994. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32:201-234. [Google Scholar]
- 5.Bina, J. E., and J. J. Mekalanos. 2001. Vibrio cholera tolC is required for bile resistance and colonization. Infect. Immun. 69:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binet, R., and C. Wandersman. 1996. Cloning of the Serratia marcescens hasF gene encoding the Has ABC exporter outer membrane component: a TolC analogue. Mol. Microbiol. 22:265-273. [DOI] [PubMed] [Google Scholar]
- 7.Borges-Walmsley, M. I., and A. R. Walmsley. 2001. The structure and function of drug pumps. Trends Microbiol. 9:71-79. [DOI] [PubMed] [Google Scholar]
- 8.Bugeja, V., G. Charles, D. Collier, and D. Wilkie. 1988. Primary mitochondrial activity of gossypol in yeast and mammalian cells. Biochem. Pharmacol. 37:4217-4224. [DOI] [PubMed] [Google Scholar]
- 9.Collmer, A. 1998. Determinants of pathogenicity and avirulence in plant pathogenic bacteria. Curr. Opin. Plant Biol. 1:329-335. [DOI] [PubMed] [Google Scholar]
- 10.Collmer, A., and N. T. Keen. 1986. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24:383-453. [Google Scholar]
- 11.Dahler, G. S., F. Barras, and N. T. Keen. 1990. Cloning of genes encoding extracellular metalloproteases from Erwinia chrysanthemi EC16. J. Bacteriol. 172:5803-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didry, N., L. Dubreuil, and M. Pinkas. 1994. Activity of anthraquinonic and naphthoquinonic compounds on oral bacteria. Pharmazie 49:681-683. [PubMed] [Google Scholar]
- 13.Didry, N., L. Dubreuil, F. Trotin, and M. Pinkas. 1998. Antimicrobial activity of aerial parts of Drosera peltata Smith on oral bacteria. J. Ethnopharmacol. 60:91-96. [DOI] [PubMed] [Google Scholar]
- 14.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 27:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon, R. A. 2001. Natural products and plant disease resistance. Nature 411:843-847. [DOI] [PubMed] [Google Scholar]
- 16.Fralick, J. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackett, J., R. Misra, and P. Reeves. 1983. The TolC protein of Escherichia coli K12 is synthesized in a precursor form. FEBS Lett. 156:307-310. [DOI] [PubMed] [Google Scholar]
- 18.Haseloff, B. J., T. L. Freeman, V. Valmeekam, M. W. Melkus, F. Oner, M. S. Valachovic, and M. J. D. San Francisco. 1998. The exuT gene of Erwinia chrysanthemi EC16: nucleotide sequence, expression, localization, and relevance of the gene product. Mol. Plant-Microbe Interact. 11:270-276. [DOI] [PubMed] [Google Scholar]
- 19.Hatano, T., H. Uebayashi, H. Ito, S. Shiota, T. Tsuchiya, and T. Yoshida. 1999. Phenolic constituents of Cassia seeds and antibacterial effect of some naphthalenes and anthraquinones on methicillin-resistant Staphylococcus aureus. Chem. Pharm. Bull. 47:1121-1127. [DOI] [PubMed] [Google Scholar]
- 20.Hugouvieux-Cotte-Pattat, N., and S. Reverchon. 2001. Two transporters, TogT and TogMNAB, are responsible for oligogalacturonide uptake in Erwinia chrysanthemi 3937. Mol. Microbiol. 41:1125-1132. [DOI] [PubMed] [Google Scholar]
- 21.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 22.Hugouvieux-Cotte-Pattat, N., N. Blot, and S. Reverchon. 2001. Identification of TogMNAB, an ABC transporter which mediates the uptake of pectic oligomers in Erwinia chrysanthemi 3937. Mol. Microbiol. 41:1113-1123. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, O. L. 1998. The TolC homologue of Erwinia chrysanthemi. Ph.D. dissertation. Texas Tech University, Lubbock, Tex.
- 24.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 25.Letoffe, S., J. M. Ghigo, and C. Wandersman. 1994. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J. Bacteriol. 176:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letoffe, S., P. Delepelaire, and C. Wandersman. 1990. Protease secretion by Erwinia chrysanthemi: the specific functions are analogous to those of Escherichia coli alpha-hemolysin. EMBO J. 9:1375-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, K. 1999. Multidrug resistance efflux, p. 15-40. In J. K. Broome-Smith, S. C. Baumberg, J. Stirling, and F. B. Ward (ed.), Transport of molecules across microbial membranes. Cambridge University Press, Cambridge, United Kingdom.
- 28.Lopez-Solanilla, E., F. Garcia-Olmedo, and P. Rodriguez-Palenzuela. 1998. Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and bacterial pathogenesis. Plant Cell 10:917-924. [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Otsuji, N., T. Soejima, S. Maki, and H. Shinagawa. 1982. Cloning of colicin E1 tolerant tolC (mtcB) gene of Escherichia coli K12 and identification of its gene product. Mol. Gen. Genet. 187:30-36. [DOI] [PubMed] [Google Scholar]
- 32.Roeder, D. L., and A. Collmer. 1985. Marker exchange mutagenesis of pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmond, G. P., and P. J. Reeves. 1993. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem. Sci. 18:7-12. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
- 35.Shevchik, V. E., J. Robert-Baudouy, and G. Condemine. 1993. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shine, J., and L. Dalgarno. 1974. Identical 3′-terminal octanucleotide sequence in 18S ribosomal ribonucleic acid from different eukaryotes. A proposed role for this sequence in the recognition of terminator codons. Biochem. J. 141:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starr, M. P., and A. K. Chatterjee. 1972. The genus Erwinia: enterobacteria pathogenic to plants and animals. Annu. Rev. Microbiol. 26:389-426. [DOI] [PubMed] [Google Scholar]
- 38.Starr, M. P., A. K. Chatterjee, P. B. Starr, and G. E. Buchanan. 1977. Enzymatic degradation of polygalacturonic acid by Yersinia and Klebsiella species in relation to clinical laboratory procedures. J. Clin. Microbiol. 6:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stermitz, F. R., P. Lorenz, J. N. Tawara, L. A. Zenewicz, and K. Lewis. 2000. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by a 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA 97:1433-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone, B. J., and V. L. Miller. 1995. Salmonella enteritidis has a homolog of tolC that is required for virulence in BALB/c mice. Mol. Microbiol. 17:701-712. [DOI] [PubMed] [Google Scholar]
- 41.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swain, T. 1974. Biochemical evolution in plants, p. 125-302. In A. M. Florkin and E. H. Stotz (ed.), Comparative biochemistry, molecular evolution, vol. 29, part I. Elsevier Scientific Publishing Company, New York, N.Y.
- 43.Tegos, G., F. R. Stermitz, O. Lomovskaya, and K. Lewis. 2002. Multidrug pump inhibitors uncover remarkable activity of plant antimicrobials. Antimicrob. Agents Chemother. 46:3133-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Gijsegem, F., and A. Toussaint. 1983. In vivo cloning of Erwinia carotovora genes involved in the catabolism of hexuronates. J. Bacteriol. 154:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Rhijn, R., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamanaka, H., T. Nomura, Y. Fujii, and K. Okamoto. 1998. Need for TolC, an Escherichia coli outer membrane protein, in the secretion of heat-stable enterotoxin I across the outer membrane. Microb. Pathog. 25:111-120. [DOI] [PubMed] [Google Scholar]
- 50.Yu, O., J. Woosuk, J. Shi, R. A. Croes, G. M. Fader, B. McGonigle, and J. T. Odell. 2000. Production of the isoflavones genistein and daidzein in non-legume and monocot tissues. Plant Physiol. 124:781-793. [DOI] [PMC free article] [PubMed] [Google Scholar]