Summary
Progression through meiosis in yeast is governed by the cyclin-dependent kinase, Cdk1, in concert with a related kinase called Ime2. It remains unclear how these kinases collaborate to meet the unique demands of meiotic progression. We demonstrate that Ime2 and Cdk1 phosphorylate an overlapping substrate set, and that the two kinases overlap functionally as inhibitors of the ubiquitin ligase APCCdh1 and replication origin licensing. Surprisingly, Ime2 phosphorylates Cdk1 substrates at distinct phosphorylation sites that are highly resistant to dephosphorylation by the phosphatase Cdc14. We propose that Ime2-dependent phosphorylation of a subset of cell-cycle proteins limits the effects of Cdc14 in meiosis.
Introduction
Modulation of the phosphorylation state of proteins is a major mechanism of cellular control. The predominant focus of studies of phosphoregulation is the substrate specificity of protein kinases. Less emphasis is placed on the ability of phosphatases to selectively remove phosphates, partially due to the difficulty of studying these enzymes but also due to a widely held view that phosphatase specificity is limited. Nevertheless, phosphatase specificity can have profound effects: more complex information processing is possible in a regulatory system if phosphatases evolve the ability to distinguish between the phosphates placed on substrates by distinct kinases.
Control of the eukaryotic cell cycle provides the best understood example of a complex phosphoregulatory system. The mitotic cell cycle is driven by oscillations in cyclin-dependent kinase (Cdk) activity, leading to large numbers of phosphorylation events that trigger chromosome duplication in S phase and segregation in M phase (Morgan, 2007). Cdks are also the master regulators of the two meiotic divisions (Benjamin et al., 2003; Marston and Amon, 2004; Petronczki et al., 2003). An understanding of how Cdk function is modulated to transform the single mitotic division into the two meiotic divisions could provide insight into the evolution of phosphoregulation.
In a mitotic cell cycle, the chromosomes are duplicated once and only once. To prevent over-replication and genome instability, replication origins must fire only a single time per cell cycle. Robust control of origin firing is achieved by dividing the DNA replication process into two steps. First, in late mitosis and G1, pre-replicative complexes (pre-RCs) are loaded onto origins, which are thereby licensed to initiate. Second, at the onset of S phase, Cdk activity initiates replication by phosphorylating initiator proteins at the origin. Cdks also trigger disassembly of the pre-RC by phosphorylating its components, thereby ensuring that origins cannot be re-licensed until Cdks are inactivated at the end of the cell cycle (Bell and Dutta, 2002; Blow and Dutta, 2005; Diffley, 2004; Nguyen et al., 2001).
After S phase, the duplicated sister chromatids are held together by a protein complex called cohesin (Nasmyth, 2002). When the sister chromatids are properly bi-oriented on the spindle in metaphase, a ubiquitin ligase called the anaphase-promoting complex (APC) is activated by its Cdc20 regulatory subunit (Peters, 2006). APCCdc20 ubiquitinates the chaperone securin to signal its destruction, releasing separase, a protease that cleaves a cohesin subunit, allowing the two sister chromatids to be pulled to opposite poles of the spindle (Nasmyth, 2002; Uhlmann et al., 2000). APCCdc20 also decreases Cdk1 activity by promoting partial destruction of the cyclins (Wäsch and Cross, 2002; Yeong et al., 2000).
The APC is also regulated by a second activator subunit, Cdh1, whose activity is suppressed from S phase until the end of mitosis by inhibitory phosphorylation by Cdk1 (Jaspersen et al., 1999; Zachariae et al., 1998). In budding yeast, separase activation at anaphase, in addition to separating the sister chromatids, activates the phosphatase, Cdc14, which dephosphorylates and thereby activates Cdh1 (Stegmeier et al., 2002). Ubiquitination by APCCdh1 then targets cyclins for destruction. Cdc14 also dephosphorylates and activates the Cdk1 inhibitor, Sic1. The combined activation of Cdh1 and Sic1 leads to a complete loss of Cdk1 activity. Cdc14 and other phosphatases then catalyze the dephosphorylation of Cdk1 substrates (D′Amours and Amon, 2004), thereby resetting the cell to a G1 biochemical state and allowing the re-licensing of replication origins.
Meiosis is a specialized form of nuclear division that involves a single round of DNA replication followed by two rounds of chromosome segregation. The meiotic divisions exact some unique demands on the balance of activities of Cdk1 and counteracting phosphatases. Most notably, it is necessary to uncouple the chromosome and spindle cycles between the first (MI) and second (MII) meiotic divisions, such that after MI the spindle reduplicates but DNA replication remains completely inhibited. In budding yeast, it is known that activation of the Cdc14 phosphatase at anaphase I is required for the reduplication of the spindle (Marston et al., 2003), but it is unclear why Cdc14 does not dephosphorylate pre-RC components or reset the meiotic cell to a G1 state.
Two models can explain the uncoupling of events between the meiotic divisions. One possibility is that the role of Cdc14 in driving mitotic exit is specifically restricted between MI and MII, preventing Cdh1 and Sic1 activation and thereby allowing Cdk1 to remain partially active. The partial destruction of cyclins by APCCdc20 might reduce Cdk1 activity to levels that allow spindle disassembly but still prevent the licensing of DNA replication origins. This model is supported by studies in the fission yeast Schizosaccharomyces pombe, where the APC inhibitor, Mes1, prevents complete cyclin destruction after MI (Izawa et al., 2005), and in the frog Xenopus laevis, where Cdks must remain partially active between the meiotic divisions to prevent re-initiation of DNA synthesis (Iwabuchi et al., 2000).
An alternative, but not mutually exclusive, model is that an auxiliary kinase activity, present during the meiotic divisions, inhibits origin licensing even in the absence of Cdk1 activity and in the presence of Cdc14 activity. Such auxiliary mechanisms probably exist in mammalian meiosis, where Cdk1 is completely inactivated between MI and MII (Choi et al., 1991). This auxiliary activity should be capable of phosphorylating pre-RC components while permitting the spindle cycle to continue.
In the budding yeast Saccharomyces cerevisiae, Ime2 is a good candidate for an auxiliary factor that allows the unique aspects of the meiotic divisions to occur robustly. Ime2 is a Cdk-related kinase, expressed only in meiosis, that is required for Sic1 destruction and the meiotic G1/S transition, fulfilling a role played by G1 cyclin-Cdk1 complexes in a mitotic cycle (Dirick et al., 1998; Stuart and Wittenberg, 1998). Ime2 is also required for the meiotic divisions (Benjamin et al., 2003; Schindler and Winter, 2006). An extra round of DNA replication is observed in cells lacking Ime2 after prolonged incubation in sporulation media, perhaps indicating that Ime2 helps prevent DNA re-replication during meiosis (Foiani et al., 1996; Guttmann-Raviv et al., 2001). A small number of potential Ime2 substrates have been described, the majority of which (Cdh1, Sic1 and the meiotic transcription factor Ndt80) are also Cdk1 substrates (Honigberg, 2004). It therefore seems likely that Ime2 is acting as an auxiliary kinase to Cdk1 in meiosis.
Here, we demonstrate that Ime2 is capable of phosphorylating a large number of known Cdk1 substrates in vitro, including Sic1, Cdh1 and multiple pre-RC components. We define the consensus phosphorylation site for Ime2 and demonstrate that Ime2 and Cdk1 phosphorylate distinct sites both in vitro and in vivo. Although the two kinases phosphorylate different sites, Ime2 and Cdk1 phosphates can have the same effect on substrate function: both kinases inactivate Cdh1 and inhibit licensing of chromosome replication. Most importantly, Ime2-dependent phosphates are highly resistant to Cdc14. Ime2 therefore fulfills all of the requirements for a factor that modifies the overall kinase/phosphatase balance to allow the events of the meiotic divisions to occur faithfully.
Results
Ime2 phosphorylates many Cdk1 substrates
To investigate the potential for Ime2 to act as an auxiliary to Cdk1, we compared the ability of Ime2 and Cdk1 to phosphorylate 109 proteins, including 71 Cdk1 substrates identified in our previous work (Ubersax et al., 2003), as well as 38 additional proteins potentially important for meiotic progression.
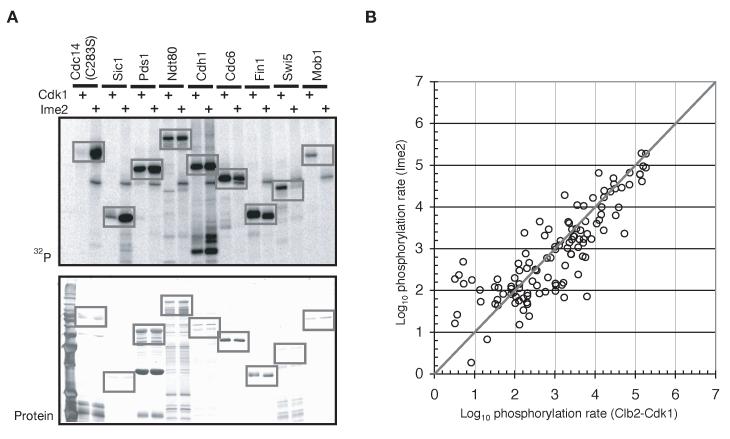
To obtain high specific-activity Ime2, we used an N-terminal fragment (amino acids 1-404) that lacks a C-terminal inhibitory domain necessary for kinase inactivation in response to glucose and nitrogen (Donzeau and Bandlow, 1999; Purnapatre et al., 2005). Native substrates were either purified from yeast using a TAP-tag or were expressed in baculovirus-infected cells or bacteria. Substrates were incubated with purified Clb2-Cdk1 or Ime2, whose activities were normalized to Ndt80, a known dual-specificity substrate that we find exhibits a similar KM for Cdk1 and Ime2 (data not shown). The rate of substrate phosphorylation by Cdk1 and Ime2 was calculated by determining the ratio of the amount of phosphate incorporated to the amount of substrate protein. A representative group of substrates is shown in Figure 1A. In Figure 1B, the log10 of the rate of substrate phosphorylation by Clb2-Cdk1 is plotted against the value for Ime2 for all 109 substrates (the data are also provided in Supplemental Table 1).
Figure 1.
Comparison of Cdk1 and Ime2 substrate phosphorylation
(A) Various known Cdk1 substrates were purified and incubated with purified Cdk1 or Ime2 (an N-terminal fragment, residues 1-404) and 32P-γ-ATP, and the reaction products analyzed by SDS-PAGE, silver staining (bottom) and autoradiography (top). A catalytically-inactive mutant of Cdc14 was used. A known dual-specificity substrate (Ndt80) was used to normalize the amount of Cdk1 and Ime2 kinase activities used in the reactions.
(B) Relative rates of phosphorylation of 109 substrates by Clb2-Cdk1 and Ime2. The rate of phosphorylation for each kinase was defined as the amount of phosphate incorporated divided by the amount of substrate protein present. A list of the substrates tested in these experiments is provided in Supplemental Table 1.
In general, we observed a high degree of correlation between the substrate specificities of Ime2 and Cdk1, supporting a model in which Ime2 and Cdk1 have the potential to carry out overlapping functions in the meiotic divisions.
A small number of substrates were specific for one or the other kinase. Cdc14, for example, was more rapidly phosphorylated by Ime2 (Figure 1A; note that we used a catalytically-inactive version of Cdc14, thereby preventing auto-dephosphorylation). Swi5 and Mob1, on the other hand, were better substrates for Clb2-Cdk1. Although Sic1 appeared to be an Ime2-specific substrate, this apparent difference resulted from the ability of Sic1 to inhibit Cdk1: if a stoichiometric complex of Sic1 bound to inactive Clb2-Cdk1 was used as a substrate, Sic1 was phosphorylated equally well by Cdk1 and Ime2 (data not shown).
Ime2 and Cdk1 phosphorylate distinct consensus sites
A major determinant of kinase specificity is the amino acid sequence around the phosphorylated site (the consensus site). The simplest mechanism by which two kinases could achieve a high degree of correlation of substrate specificity would be for both kinases to recognize a similar consensus site. Clb2-Cdk1 specificity is directed primarily by recognition of the Cdk1 consensus site (S/T*-P-x-K/R). The extensive overlap of Cdk1 and Ime2 substrates (Figure 1B) suggested that Ime2 might phosphorylate a similar sequence.
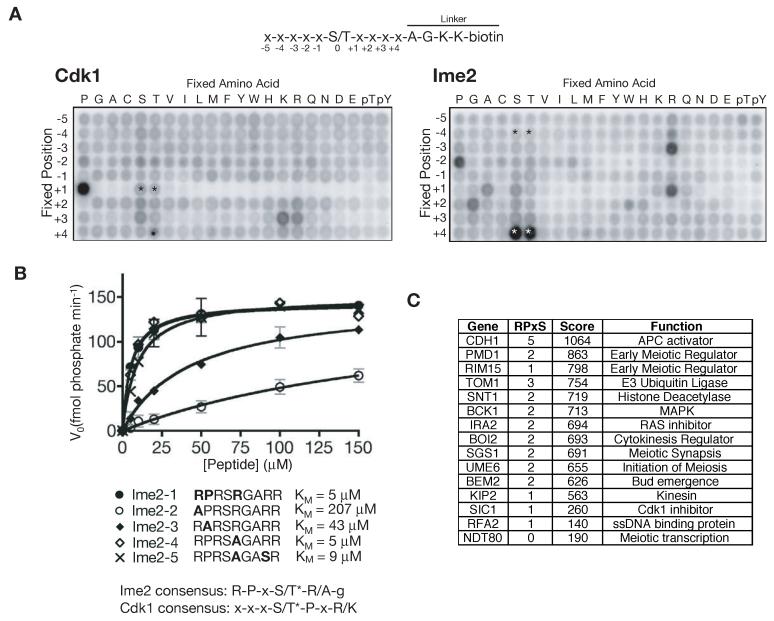
We tested this possibility using a positional scanning peptide library (Hutti et al., 2004) (Figure 2A; Supplemental Table 2). Each well of an array plate contains a degenerate library of peptides in which the central position is an equal mixture of serine and threonine. In each well, one position in the peptide is fixed to a single amino acid: each row in the array represents a different fixed position relative to the central serine/threonine, and in each column that position is fixed to a different amino acid. All other positions in the peptide are randomized to an equal mix of every amino acid except cysteine, serine or threonine (Figure 2A, top). Peptides are incubated in solution with purified kinase and 32P-γ-ATP, transferred to membranes, washed and dried for autoradiography. Any well corresponding to a preferred amino acid at a particular position will exhibit higher 32P incorporation than all other wells in the same row.
Figure 2.
Determination of the Ime2 consensus phosphorylation site
(A) A positional scanning peptide array was used to determine the consensus sites of Cdk1 (left) and Ime2 (right). Each well of a 22 × 9 plate contains a mixed pool of biotinylated peptides with a central S/T, one fixed position and every other position randomized to an equal mix of all amino acids except S/T/C (top). When a position is fixed to S or T, this can lead to artifacts (asterisks, see text). Each plate was incubated with purified kinase and 32P-γ-ATP, phosphorylated peptides were transferred to an avidin-coated membrane, excess 32P-γ-ATP was washed away, and phosphate incorporation in each peptide pool was quantified by autoradiography. The ratio of intensity of each well divided by the average intensity of the entire row gives the relative preference for a given amino acid at each position of the consensus site (quantification of the average of two repeats of each array is provided in Supplemental Table 2).
(B) The initial velocity of Ime2-dependent phosphorylation was determined at various concentrations of an idealized Ime2 substrate peptide, and derivative peptides were used to test the importance of each of the major consensus site determinants. Data are represented as mean +/- SD. The kcat with the ideal peptide was 266 min-1.
(C) The entire translated yeast genome was scanned for potential Ime2 phosphorylation sites, and each ORF was assigned a rank score based on the number and quality of Ime2 sites present. Shown is a list of selected proteins from the top hits, along with some previously described Ime2 substrates.
We found that Cdk1 has a strict requirement for proline at the +1 position (Figure 2A, left), as indicated by the dark well in the proline column of the +1 row. In all other columns of this row, the +1 position is fixed to a non-proline amino acid and almost no phosphorylation was observed. The exceptions are the serine and threonine columns in the +1 row (indicated by asterisks in Figure 2A, left), in which phosphorylation presumably occurred on the subset of peptides with a proline at +2 (e.g. the peptide x-S(0)-S*(+1)-P(+2)-x can be phosphorylated at S(+1)). As demonstrated previously, Cdk1 also displayed a minor preference for lysine or arginine at the +3 position.
A comparison of the left (Cdk1) and right (Ime2) panels of Figure 2A reveals that Ime2 and Cdk1 have distinct consensus sites. The Ime2 consensus site is relatively complex, with at least three strong determinants and numerous minor preferences. In particular, there is a preference for arginine at -3, proline at -2 and arginine at +1.
The intense labeling of the +4 position in the serine and threonine rows (white asterisks in Figure 2A, right) is likely to be an artifact that arises from the nature of the linker sequence that follows the +4 position (x(+4)-A-G-K-K-biotin; Figure 2A, top). When serine or threonine is present at +4, this linker contains two residues (+1 A and +2 G relative to the +4 S/T) that are fixed to preferred amino acids. In these wells, it is almost certainly S/T(+4) rather than the central S/T that is being phosphorylated. Indeed, we found no evidence of a preference for S/T at +4 in our subsequent peptide analyses (see below).
To better characterize the Ime2 consensus sequence, we analyzed the activity of Ime2 toward peptides designed to test the relative importance of the different determinants (Figure 2B). The optimal Ime2 peptide displayed an excellent KM of 5 μM, ten-fold better than the KM of Clb2-Cdk1 for a model histone peptide (Loog and Morgan, 2005). The most important Ime2 consensus determinant is the -3 arginine (substitution increases the KM by two orders of magnitude), followed by the -2 proline (substitution increases the KM by one order of magnitude). Substitution of the +1 arginine with alanine had no effect. We also designed a peptide to confirm that the apparent preference for S/T at the +4 position was an artifact: changing the +4 position to serine did not improve the KM. An ideal consensus site for Ime2 is therefore R-P-x-S/T*, while minimal consensus sites are R-x-x-S/T* and x-P-x-S/T*. Because Cdk1 and Ime2 sites can be either independent or overlapping (e.g. R-P-x-S/T*-P-x-K/R), there is clear potential for Cdk1 and Ime2 phosphoregulation to synergize.
The lack of consensus site similarity between the two kinases suggests that the correlation between Cdk1 and Ime2 substrate phosphorylation derives from the evolution of a functional overlap between the two kinases. It seems likely, therefore, that Ime2 phosphorylation sites are easily evolved: a minimal independent Ime2 site requires only two amino acids, while evolution of a Cdk1 site into a dual-specificity Ime2/Cdk1 site simply requires acquisition of a -3 arginine or -2 proline. Indeed, a statistical analysis of the residues surrounding the 1885 S/T*-P motifs of 180 Cdk1 substrates (Ubersax et al., 2003) revealed a minor enrichment of proline at -2, suggesting that there has been evolutionary pressure to generate overlapping Cdk1/Ime2 sites (e.g. x-P-x-S/T*-P, Supplemental Figure 1).
Phosphorylation of distinct sites by Ime2 and Cdk1 can have similar functional consequences
Knowledge of the Ime2 consensus site allows prediction of potential Ime2 substrates based on primary sequence data. We assigned a score to each yeast ORF based on the number of occurrences of potential Ime2 phosphorylation sites, weighted according to how closely each site conforms to the ideal Ime2 consensus defined in Figures 2A and 2B. A selected list of potential substrates is shown in Figure 2C. The best predicted substrate is the APC activator Cdh1, which contains five R-P-x-S/T* motifs.
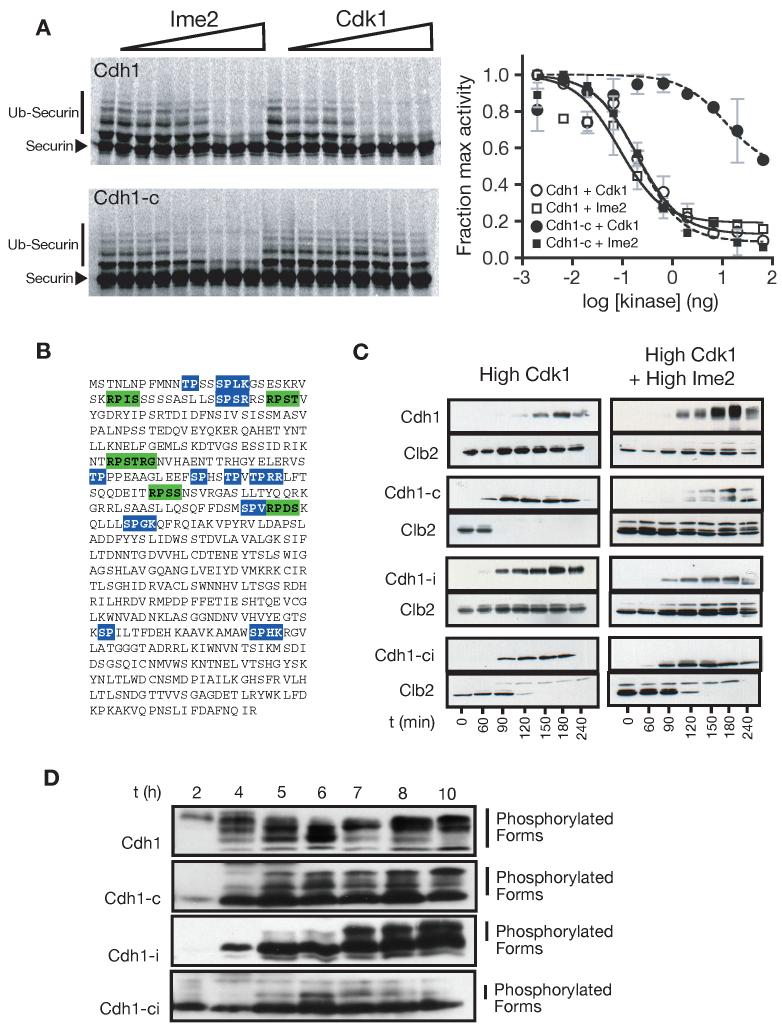
Previous work showed that exogenous expression of Ime2 leads to inhibition of Cdh1 in vivo (Bolte et al., 2002). We tested the ability of Ime2 to directly phosphorylate and inhibit Cdh1 in a reconstituted APCCdh1 assay in vitro. Ime2 and Cdk1 inhibited APCCdh1 with similar potency (Figure 3A, top). We also tested the Cdh1-c protein, in which all 11 Cdk1 consensus sites are mutated to alanine (Figure 3A, bottom). Clb2-Cdk1 did not inhibit Cdh1-c except at very high kinase concentrations. In contrast, Ime2 inhibited Cdh1-c just as well as wild-type Cdh1, suggesting that Ime2 phosphorylates and inhibits Cdh1 at non-Cdk1 sites.
Figure 3.
Functional comparison of Ime2- and Cdk1-dependent phosphates
(A) Ime2 inhibits Cdh1 by phosphorylating non-Cdk1 sites. The ability of Cdk1 and Ime2 to inhibit Cdh1 was tested by monitoring 35S-securin ubiquitination in a reconstituted APCCdh1 assay (SDS-PAGE and autoradiography on the left, quantitation on the right. Slower migrating bands correspond to ubiquitinated securin. Data are represented as mean +/- SD). Reactions were performed with wild-type Cdh1 (top left) or Cdh1-c, in which the eleven Cdk1 sites are changed to alanine (bottom left).
(B) The primary sequence of Cdh1, showing the 11 consensus Cdk1 sites (blue) and 5 consensus Ime2 sites (green).
(C) Cdk1 and Ime2 inhibit Cdh1 using distinct phosphorylation sites in vivo. Cells were arrested in mitosis with nocodazole, and different forms of CDH1 were expressed under the control of the GAL-L promoter, in the presence or absence of Ime2. Cdh1 phosphorylation is indicated by slower migrating forms on HA3-Cdh1 western blots. APC activity was assessed by the degradation of Clb2. In addition to wild-type Cdh1, we tested mutant Cdh1 proteins lacking the 11 Cdk1 sites (Cdh1-c), lacking the five Ime2 sites (Cdh1-i), or lacking all Cdk1 and Ime2 sites (Cdh1-ic).
(D) Cdh1 is phosphorylated on both Cdk1 and Ime2 sites during meiosis. Two hours after transfer to sporulation media, wild-type and mutant forms of Cdh1 were exogenously expressed from the GAL-L promoter using the Gal4-ER system (Benjamin et al., 2003). Cdh1 mobility was monitored during meiosis by western blotting against an N-terminal HA3 tag. Time in sporulation media is indicated.
Figure 3B shows the primary sequence of Cdh1 with the five Ime2 and eleven Cdk1 consensus sites highlighted. Mutant forms of Cdh1 were generated in which the five Ime2 sites (Cdh1-i), the eleven Cdk1 sites (Cdh1-c), or all sixteen Cdk1 and Ime2 sites (Cdh1-ci) are mutated to alanine. These mutants were tested for their ability to activate the APC in vivo in the presence of either high levels of active Cdk1 or high levels of both Cdk1 and Ime2. Cells were arrested in mitosis with nocodazole (a state of high Cdk1 activity), and expression of the CDH1 mutants was induced from the GAL-L promoter. Cells either contained no Ime2 (Figure 3C, left side) or expressed the high-activity N-terminal fragment of Ime2 from the GAL-L promoter (Figure 3C, right side). Activation of the APC was assessed by destruction of the cyclin Clb2. The Cdh1-c mutant activated the APC in the presence of high Cdk1 activity but was completely inhibited in the presence of Ime2, indicating that Ime2 inhibits Cdh1 at non-Cdk1 sites in vivo. The Cdh1-ci double mutant activated the APC in the presence of both Ime2 and Cdk1, indicating that the R-P-x-S/T* sites are the major Ime2 sites in vivo.
We also assessed the phosphoregulation of Cdh1 by Ime2 in meiosis. When expressed from the GAL-L promoter in meiotic cells, Cdh1 displayed a complex pattern of mobility shifts on polyacrylamide gels (Figure 3D). Removal of Cdk1 (Cdh1-c) or Ime2 (Cdh1-i) phosphorylation sites each partially abolished the low-mobility forms of Cdh1, and the removal of all sixteen sites (Cdh1-ci) caused Cdh1 to migrate almost entirely as a single high-mobility species. Cdh1 therefore appears to be extensively phosphorylated at Cdk1 and Ime2 sites during meiosis. Cdh1-c is known to be lethal in mitotic cells, but we found that it did not appear to be toxic when expressed in meiotic cells (data not shown), suggesting that endogenous Ime2 is capable of keeping Cdh1-c inactive during meiosis. However, because we were unable to achieve good exogenous expression of any of the three Cdh1 phosphomutants, interpretation of phenotypes was difficult. Replacement of endogenous Cdh1 with Cdh1-i did not lead to any detectable phenotype: spore viability and the timing of the meiotic divisions were normal (data not shown).
Ime2-dependent phosphates are resistant to Cdc14 but can be removed by PP2A and PP2C
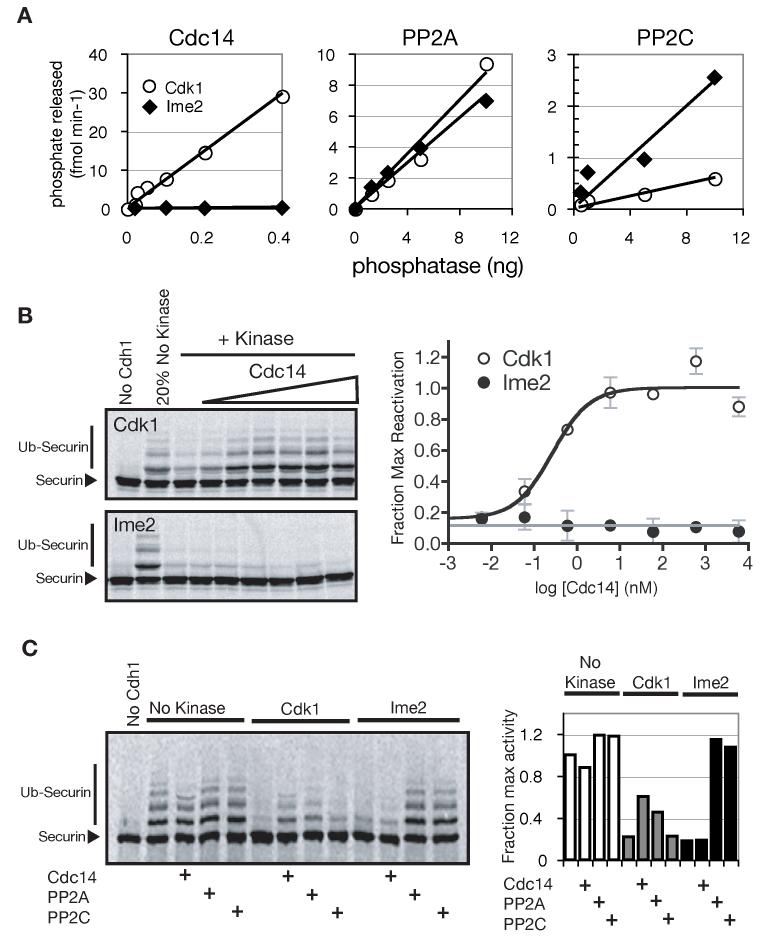
If two kinases phosphorylate the same substrate at different sites with similar functional consequences, a regulatory niche is created in which phosphatase selectivity can play a key role. Between the meiotic divisions, for example, Cdc14 is activated to allow the spindle to reduplicate. Cdc14 could also potentially dephosphorylate and activate other Cdk1 targets such as pre-RC components (leading to DNA re-replication) or Cdh1 (potentially leading to premature exit from the meiotic program). We investigated the possibility that phosphates placed on substrates by Ime2 are resistant to Cdc14, and that Ime2 might therefore limit the effects of Cdc14 activation after meiosis I.
We found that Cdc14 removed Cdk1-dependent phosphates from a model substrate (Fin1), but Ime2-dependent phosphates were remarkably resistant to Cdc14 (Figure 4A, left). Cdc14 was also unable to remove Ime2-dependent phosphates from Cdc6 (Supplemental Figure 2A), Cdh1 (Figure 4B, C), Ndt80, and securin (data not shown). Cdc14 efficiently removed Cdk1-dependent phosphates from all of these substrates. The degree of Cdc14 specificity for Cdk1 phosphates over Ime2 phosphates was greater than four orders of magnitude - far higher than the previously published ten-fold selectivity for a proline at the +1 position (Gray et al., 2003), suggesting that additional sequence determinants govern substrate recognition by Cdc14.
Figure 4.
Ime2 phosphates are completely resistant to Cdc14 activity
(A) A model substrate (Fin1) was phosphorylated by Cdk1 or Ime2 and the rate of phosphate release upon incubation with Cdc14, PP2A-Cdc55 or PP2C was determined by TCA precipitation and scintillation counting of soluble phosphate.
(B) APCCdh1 can be reactivated by Cdc14 after phosphorylation by Cdk1 but not Ime2. A reconstituted APC assay was performed using Cdh1 that was first phosphorylated by either Cdk1 or Ime2 and then treated with increasing amounts of Cdc14 (SDS-PAGE and autoradiography of 35S-securin on the left; quantitation on the right. Data are represented as mean +/- SD).
(C) APCCdh1 is controlled by a complex variety of kinases and phosphatases. The ability of different phosphatases to reactivate Cdh1 following Ime2 or Cdk1 phosphorylation was assessed as in panel B.
A screen was undertaken to find phosphatases that can remove Ime2 phosphates. Candidate serine/threonine phosphatase complexes were purified from yeast and tested for their ability to remove Ime2-dependent phosphates from Cdh1. PP2A (Cdc55-TAP; Figure 4A, center panel) and PP2C (Ptc2-TAP; Figure 4A, right panel) both removed Ime2-dependent phosphates efficiently. That PP2A and PP2C can dephosphorylate Ime2 sites on Fin1 argues against the possibility that the inability of Cdc14 to remove Ime2 phosphates is due to phosphate inaccessibility or protein aggregation. Furthermore, the data in Figure 4A reveal a wide range of phosphatase specificities: Cdc14 is highly specific for Cdk1-dependent phosphates, PP2C is somewhat specific for Ime2 phosphates, and PP2A is a dual-specificity phosphatase. These differences suggest that phosphatase specificity could play a decisive role in meiotic phosphoregulation.
Cdh1 that was phosphorylated and inactivated by Cdk1 was reactivated by incubation with Cdc14 (Figure 4B, top; Jaspersen et al., 1999). In contrast, when Cdh1 was phosphorylated by Ime2, even high levels of Cdc14 could not reactivate Cdh1 and restore APC activity. However, PP2A and PP2C completely restored Cdh1 activity after phosphorylation and inactivation by Ime2 (Figure 4C), demonstrating that Ime2 does not inactivate Cdh1 in a non-specific manner. Thus, the presence of Ime2 phosphates on Cdh1 during meiosis may help limit the effects of Cdc14 between the meiotic divisions and allow rapid progression from MI to MII.
Ime2 inhibits DNA re-replication
A key feature of the transition from meiosis I to meiosis II is that the spindle must reduplicate but the chromosomes must not. The activation of Cdc14 is required for spindle cycle progression (Marston et al., 2003), but it is not clear how Cdc14 is prevented from dephosphorylating pre-RC components and thereby re-licensing DNA replication. The ability of Ime2 to place Cdc14-resistant phosphates on Cdk1 substrates provides a possible mechanism for the robust inhibition of origin licensing between the meiotic divisions: Ime2 could help prevent pre-RC assembly during periods of high Cdc14 and low Cdk1 activities. Ime2 is capable of phosphorylating a number of pre-RC components, including Cdc6, Mcm6, and Orc6 (Figure 1, Supplemental Table 1), and Cdc6 has previously been reported to interact with Ime2 during meiosis (Ofir et al., 2004). Furthermore, we found that Ime2 phosphates cannot be removed from Cdc6 by Cdc14 (Supplemental Figure 2A).
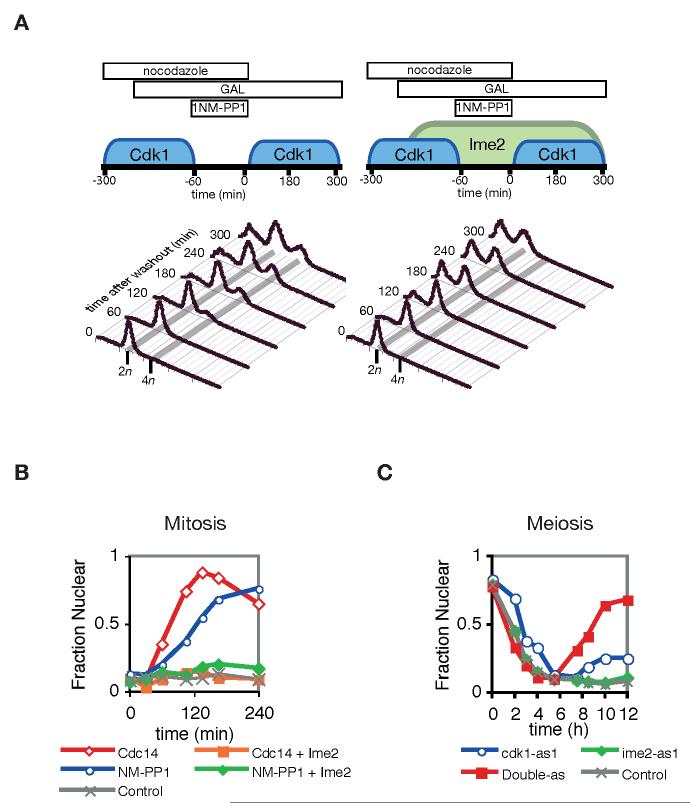
To test whether Ime2 can inhibit origin licensing, we used a chemically-inhibitable Cdk1 mutant to artificially oscillate Cdk1 activity in mitotic cells. In the analog-sensitive Cdk1 mutant (Cdk1-as1), a bulky phenylalanine residue in the ATP-binding pocket is mutated to glycine, thereby expanding the active site and rendering the kinase uniquely sensitive to a bulky inhibitor, 1-NM-PP1 (Bishop et al., 2000). The experimental scheme is illustrated in Figure 5A. Cells were arrested after S phase in nocodazole, and galactose was added to the media of either control cells or cells expressing IME2 (1-404) from the GAL-L promoter. 1-NM-PP1 was then added to the cultures for one hour, after which the inhibitor was washed out to restore Cdk1 activity. Samples were taken to monitor DNA content by flow cytometry (Figure 5A). 30% of control cells re-licensed their origins and re-duplicated their DNA, yielding a 4n peak in the flow-cytometry histogram (Figure 5A, left). In contrast, the 4n peak was completely suppressed when Ime2 was exogenously expressed (Figure 5A, right). Similar results were obtained in cells progressing through S phase from a G1 arrest (Supplemental Figure 2C). These data demonstrate that Ime2 is sufficient to block DNA reduplication in vivo.
Figure 5.
Ime2 inhibits DNA re-replication and nuclear accumulation of the Mcm2-7 complex
(A) Ime2 prevents licensing of DNA origins and re-replication. Mitotic cdk1-as1 cultures were arrested in nocodazole with a 2n DNA content, and Cdk1-as1 activity was artificially oscillated by a one-hour treatment with 1-NM-PP1 and subsequent removal of inhibitor, either in a control strain (left) or in a strain expressing IME2 (1-404) under the control of the GAL-L promoter (right). DNA content was measured by flow cytometry in the cell populations after removal of 1-NM-PP1.
(B) The localization of the Mcm2-7 complex was monitored using Mcm7-GFP, expressed at the MCM7 locus under the control of its own promoter. Cells were arrested in nocodazole and Cdk1-as1 was inhibited with 20 μM 1-NM-PP1, or CDC14 was expressed from PGAL1, in the presence or absence of exogeneous PGAL-L-IME2 (1-404) expression. The fraction of cells with nuclear Mcm2-7 was determined.
(C) Cells were sporulated and allowed to progress through the meiotic program until most cells had excluded Mcm7-GFP from the nucleus (4-5 h). At this time point, most cells have completed meiotic S phase but have not yet entered the first meiotic division. cdk1-as1 cells were then treated with 20 μM 1-NM-PP1, ime2-as1 cells were treated with 20 μM 1-Bn-PP1, or double mutants were treated with both inhibitors. Control double-mutant cells were treated with solvent (DMSO). Reaccumulation of Mcm7-GFP in the nucleus was measured.
We also analyzed spindle pole reduplication by in situ fluorescence of Spc42-GFP, and found no evidence that Ime2 blocks spindle pole body licensing or duplication (Supplemental Figure 2B).
Ime2 can block the ability of Cdc14 to drive nuclear import of the Mcm2-7 complex
Cdk1-dependent phosphorylation prevents pre-RC formation by multiple mechanisms, including degradation of Cdc6, inhibition of ORC function, and nuclear exclusion of Cdt1 and the Mcm2-7 complex (Diffley, 2004). To investigate the molecular mechanisms by which Ime2 inhibits DNA re-replication, we monitored the localization of the Mcm2-7 complex using an Mcm7-GFP strain (Nguyen et al., 2000). In control experiments with cdk1-as1 cells arrested in mitosis with nocodazole, we found that chemical inhibition of Cdk1-as1 caused the gradual accumulation of Mcm2-7 in the nucleus (Figure 5B). Because inhibition of Cdk1 alone does not trigger Cdc14 activation (Azzam et al., 2004), we also tested the effects of Cdc14 overexpression from the GAL-L promoter. Cdc14 promoted faster accumulation of Mcm2-7 in the nucleus (Figure 5B), and a combination of Cdc14 expression and Cdk1-as1 inactivation synergized to drive rapid nuclear Mcm2-7 accumulation (Supplemental Figure 2D).
We next determined if Ime2 could prevent nuclear accumulation of Mcm2-7. Expression of IME2 from the GAL-L promoter prevented accumulation of nuclear Mcm2-7 both when Cdk1-as1 was inhibited and when Cdc14 was overexpressed (Figure 5B). These data provide a direct in vivo correlate to our in vitro results suggesting that Ime2 can block a subset of Cdc14-catalyzed events.
Ime2 and Cdk1 collaborate to block nuclear accumulation of Mcm2-7 in meiotic cells
To test whether Ime2 contributes to the suppression of DNA re-replication in meiotic cells, we generated Mcm7-GFP SK1 strains containing a cdk1-as1 mutation, an ime2-as1 mutation (Benjamin et al., 2003), or both. In the absence of kinase inhibition, Mcm7-GFP was excluded from the nucleus as these cells progressed through meiotic S phase, and Mcm7-GFP did not re-enter the nucleus between the meiotic divisions (Figure 5C, control). We never observed cells with two DNA masses and nuclear Mcm7-GFP. To test the effects of specific kinase inhibition, we treated cdk1-as1 cells with 1-NM-PP1 or ime2-as1 cells with 1-Bn-PP1 near the end of meiotic S phase, when most Mcm2-7 had been excluded from the nucleus. As seen previously (Benjamin et al., 2003), inhibition of either Ime2-as1 or Cdk1-as1 blocked further meiotic progression, so that most cells arrested before or during the first division with a single DNA mass. Inhibition of Ime2-as1 alone did not cause nuclear accumulation of Mcm2-7, and inhibition of Cdk1-as1 caused only a minor percentage of cells to accumulate nuclear Mcm2-7. However, simultaneous inhibition of both kinases caused a significant fraction of cells to re-accumulate nuclear Mcm2-7 (Figure 5C).
It is possible to distinguish between Cdk1-as1 and Ime2-as1 pharmacologically: the bulkier drug 1-NM-PP1 efficiently inhibits Cdk1-as1 but does not affect Ime2-as1, while the smaller drug 1-Bn-PP1 inhibits both kinases. This allowed us to compare the effects of inhibition of Cdk1-as1 alone or together with Ime2-as1 in a single sporulating culture (Supplemental Figure 2E). Again, inhibition of both kinases caused more rapid and efficient re-accumulation of nuclear Mcm2-7 than inhibition of Cdk1-as1 alone.
We conclude that after cells initiate meiotic S phase, both Cdk1 and Ime2 contribute to the nuclear exclusion of Mcm2-7 and thereby help suppress DNA re-replication. In anaphase I, when Cdk1 activity decreases and Cdc14 is activated, we speculate that Ime2 assumes a critical role in keeping Mcm2-7 out of the nucleus.
Discussion
This study was motivated by the observation that Ime2, a Cdk-related kinase, shares several substrates with Cdk1. We investigated the extent to which Ime2 might act in parallel with Cdk1 in meiosis by comparing the ability of Cdk1 and Ime2 to phosphorylate a large substrate set. We found that Ime2 is capable of phosphorylating many Cdk1 substrates, supporting the idea that Ime2 and Cdk1 act in concert to define the phosphoregulatory background for meiotic development.
Based on the extensive overlap of their substrates, we expected Ime2 and Cdk1 to phosphorylate a similar consensus site. Surprisingly, however, our studies revealed that Cdk1 and Ime2 phosphorylate distinct sites: S/T*-P-x-K/R and R-P-x-S/T*, respectively. Our results are consistent with recent papers (Clifford et al., 2005; Sedgwick et al., 2006) that describe Ime2 phosphorylation sites consistent with the R-P-x-S/T* motif.
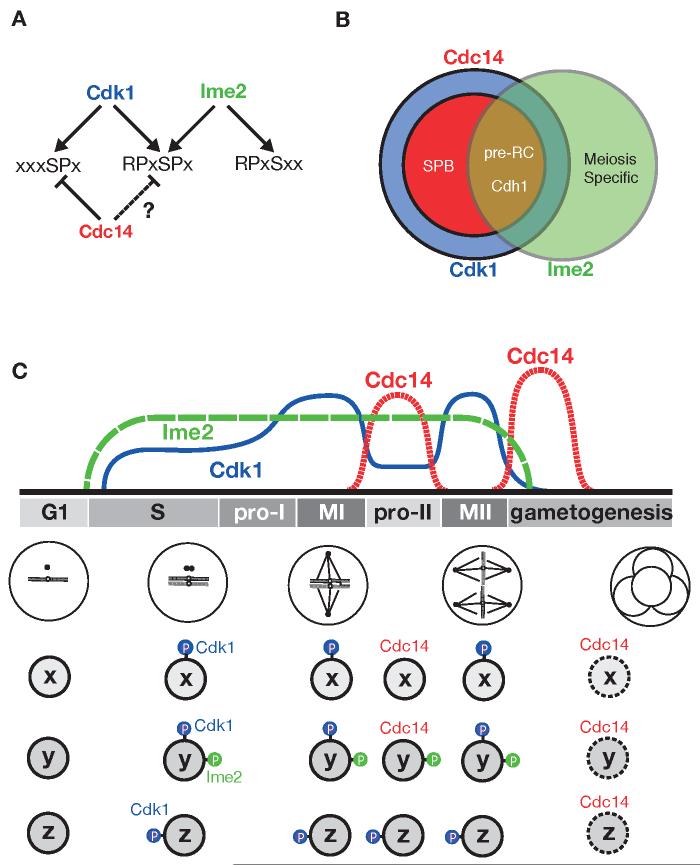
It is possible for Ime2 and Cdk1 sites to be either independent or overlapping (e.g. R-P-x-S/T*-P; Figure 6A). In the present study, we demonstrate that Cdh1 has independent Cdk1 and Ime2 sites, while Sedgwick et al. (2006) describe overlapping Cdk1/Ime2 sites in Sic1. Where phosphorylation sites overlap, the functional consequences of phosphorylation will be identical, but even in cases where Ime2 and Cdk1 phosphorylate distinct sites, the functional consequences of phosphorylation can be similar (as we saw in the case of Cdh1).
Figure 6.
A model for the phosphoregulation of the meiotic divisions
(A) Cdk1 sites (left) are sensitive to Cdc14 dephosphorylation; dual-specificity sites (middle) can be phosphorylated by both Cdk1 and Ime2 but their Cdc14 sensitivity is not known; and Ime2 sites (right) are resistant to dephosphorylation by Cdc14.
(B) The Cdk1 substrate set is represented by a blue circle, within which Cdc14 targets are represented as a red subset. Ime2 substrates are represented by a green circle. Ime2 may be specialized to regulate some processes unique to meiosis, such as recombination and gametogenesis, but also phosphorylates a subset of Cdk1 substrates. In the absence of Ime2, Cdc14 dephosphorylates a subset of Cdk1 substrates, including pre-RC components and Cdh1. In the presence of Ime2, however, we propose that Cdc14-resistant phosphates are placed on these and other substrates, helping to convert the single mitotic division into two consecutive meiotic divisions.
(C) The activities of Cdk1 (blue), Ime2 (green) and Cdc14 (red) are thought to rise and fall as shown during the meiotic program. Three hypothetical phosphoproteins are illustrated: protein x, a Cdk1 substrate that is dephosphorylated by Cdc14 between the meiotic divisions; protein y, a substrate for both Cdk1 and Ime2 whose Cdk1 sites, but not Ime2 sites, are dephosphorylated by Cdc14; and protein z, a Cdk1 substrate that remains phosphorylated between the meiotic divisions. Clb1-Cdk1, together with Ime2, drives the cell into meiosis I. Cdc14 is activated at the first metaphase-to-anaphase transition and removes Cdk1 phosphates (blue) from protein x and protein y. Protein x is thus dephosphorylated and activated (e.g. proteins involved in spindle cycle progression). However, the presence of an Ime2 phosphate (green) on protein y or a stable Cdk1 phosphate (blue) on protein z prevents them from being activated. We speculate that maintenance of phosphorylation on these proteins prevents origin licensing and resetting of the cell to a G1 state.
The fact that Ime2 and Cdk1 phosphorylate different sites raises questions about how large groups of phosphoproteins can evolve to be co-regulated by different kinases acting at distinct sites. We suggest a model of evolution of phosphocontrol in which the pattern of kinase expression is defined first, and substrates subsequently evolve phosphorylation sites that confer useful regulation in this background of kinases and phosphatases. This model of evolution is most effective if a highly evolvable mechanism of phosphoregulation is at play. Much emphasis has been placed on the notion that phosphate groups drive precise conformational changes. However, it is difficult to evolve such precise modes of regulation for multiple distinct phosphorylation sites. An alternative model of phosphoregulation is exemplified by Cdh1, whose inhibition by Cdk1 and Ime2 is not likely to depend on a precise conformational change but rather upon the disruption of protein-protein interactions by bulky and electrostatically-negative phosphates. The disruption of interactions in protein complexes is an easily evolvable mode of phosphoregulation and is facilitated by the composition of most cellular machinery from multiple subunits.
Cdk1 and Ime2 collaborate by phosphorylating an overlapping set of proteins on distinct sites, but what new opportunities does this provide for the regulation of the meiotic divisions? We investigated the ability of various phosphatases to counteract Cdk1 and Ime2 and discovered that Ime2-dependent phosphates are highly resistant to Cdc14. Cdc14 removes Ime2 phosphates at a rate four orders of magnitude slower than Cdk1 phosphates. This is a far greater level of Cdc14 specificity than previously described in peptide studies (Gray et al., 2003).
The extreme Cdc14-resistance of Ime2 phosphates provides a potential explanation for the distinct consequences of Cdc14 activation in mitosis and meiosis (Figures 6B and 6C). Ime2 phosphorylates several important regulators of mitotic exit, including Cdh1 and Sic1, and components of the pre-RC, including Cdc6. In late mitotic cells lacking Ime2, activation of these proteins by Cdc14 helps promote Cdk1 inactivation and licensing of origins of replication. In cells completing the first meiotic division, Ime2-dependent phosphorylation could prevent activation of Cdh1, Sic1, and Cdc6 even in the presence of active Cdc14. Ime2-dependent phosphorylation of pre-RC subunits could help block origin licensing, while phosphorylation of Cdh1 and Sic1 could help prevent complete Cdk1 inactivation and facilitate re-accumulation of Cdk1 activity to allow rapid entry into meiosis II. Consistent with this model, the Cdh1-specific substrate Cdc5 is not degraded between the meiotic divisions (Clyne et al., 2003; M. Sullivan and D.O.M., unpublished observations).
The suppression of origin licensing and other events between the meiotic divisions could also depend on the continued phosphorylation of some Cdk1-dependent sites (protein z in Figure 6C). Although Clb5-Cdk1 is thought to be inactivated in anaphase I (Oelschlaegel et al., 2005), other cyclin-Cdk1 complexes may retain activity. In addition, mechanisms may exist to prevent Cdc14 from removing some Cdk1-dependent phosphates.
In support of a role for Ime2 in the suppression of DNA re-replication in meiosis, we demonstrated that Ime2 helps to exclude the Mcm2-7 complex from the nucleus following meiotic S phase (Figure 5C). However, transient inhibition of Ime2-as1 during the meiotic divisions did not cause re-replication that was detectable by flow cytometry (data not shown; note that these experiments were performed in GAL-NDT80 strains that do not depend on Ime2 for progression into the meiotic divisions; Benjamin et al., 2003). We also tested the effects of inhibiting Cdk1, either alone or in combination with Ime2 inhibition, using cdk1-as1 and ime2-as1 cdk1-as1 strains. Unfortunately, transient inhibition of Cdk1-as1 at any stage caused a complete failure of meiosis, suggesting that Cdk1 activity must be maintained at some minimal level throughout meiotic development. Other approaches, including comprehensive analysis of Cdk1 and Ime2 phosphorylation site mutants, will be necessary to evaluate the importance of Ime2 and Cdk1 as suppressors of DNA replication between the meiotic divisions. It seems likely that multiple overlapping mechanisms are required to provide robust inhibition of origin licensing at this stage.
What are the implications of our work for the phosphoregulation of meiosis in mammals? In mouse oocytes, Cdk1 activity appears to decline to near basal levels between the meiotic divisions (Choi et al., 1991), suggesting that auxiliary kinases could be important. The closest mammalian relatives of Ime2 are Cdk2 and a meiosis-specific kinase called Mak (Male germ-cell associated kinase). In mice, deletion of the gene encoding Mak leads to a decrease in fertility (Shinkai et al., 2002). Mak is therefore not essential for meiosis but is required for its robust execution. The minor phenotype of Mak-/- mice may result from redundancy of Mak with the closely related kinases, Mok and Ick. Alternatively, the reduced importance of Mak relative to Ime2 may be due to differences between meiosis in yeast and mammals. There is an absolute requirement to reduplicate the spindle poles between the meiotic divisions in yeast. Higher eukaryotes, in contrast, sometimes employ a second pathway for spindle assembly that is independent of the centrosome and depends instead on spindle self-organization around the chromosomes. The meiotic divisions of oogenesis, for example, occur in the absence of centrosomes in many species (Schatten, 1994). The lack of centrosomes in female meiosis may reduce the need for mechanisms to uncouple the spindle and chromosome cycles. It is interesting to note that Mak is required for efficient spermatogenesis - a centrosomal process - but is dispensable for oogenesis.
The extreme selectivity of Cdc14 for Cdk1-dependent phosphates emphasizes the potential for phosphatases to play decisive roles in phosphoregulation. Recent work has highlighted specific roles for PP2A in the meiotic divisions (Kitajima et al., 2006; Riedel et al., 2006) and in the release of Cdc14 from the nucleolus (Queralt et al., 2006). Interestingly, we identified PP2A as a dual-specificity phosphatase capable of counteracting both Ime2 and Cdk1. What is becoming increasingly clear is that knowledge of both kinase and phosphatase specificities is necessary to understand phosphoregulation in the eukaryotic cell. Every phosphoprotein can be seen as a node for the integration of information from the balance of kinases and phosphatases. In many cases, evolution of new layers of phosphocontrol may simply involve the mutation of an amino acid at the interface between subunits of a multi-protein complex, either to create a new phosphorylation site or to subtly modulate the sensitivity of an existing site to the kinases and phosphatases already present in the cell.
Experimental Procedures
General Methods
All strains were derivatives of W303 or the fast-sporulating SK1 strain and were grown at 30°C. Epitope- and GFP-tagged strains were constructed as described (Longtine et al., 1998). Sporulation was carried out as previously described (Padmore et al., 1991). CDH1 was induced from the GAL-L promoter in meiosis by addition of 5μM β-estradiol to strains containing GAL4-ER (Benjamin et al., 2003). Mutagenesis of Cdh1 was achieved by single-stranded mutagenesis. The 3HA-tag was detected with the 16B12 antibody (Covance), and endogenous Clb2 was detected with polyclonal anti-Clb2 antibody (a generous gift of Doug Kellogg). Flow cytometry was by standard methods using cytox-green (Molecular Probes) as a DNA stain. Micrographs were acquired with a Zeiss Axiovert-200M microscope and a Hamamatsu Orca-ER CCD camera controlled by μManager acquisition software (Vale lab, UCSF; www.micro-manager.org). Cells were either viewed live or rapidly fixed in 4% formaldehyde for 5 min on ice (mitotic cultures) or in 1% formaldehyde for 1 min on ice (meiotic cultures).
Protein Purification
APC, Ubc4, Uba1 and Cdh1 were purified as described (Carroll and Morgan, 2005). Clb2 and the N-terminal Ime2 fragment (amino-acids 1-404) were expressed from a 2μ PGAL1-TAP plasmid and kinase complexes were purified using a C-terminal TAP tag as described (Puig et al., 2001). PP2A (Cdc55) and PP2C (Ptc2) were purified from mitotic cells using a C-terminal TAP tag. GST-Cdc14 was purified from bacteria as described (Jaspersen et al., 1999). The majority of substrate proteins were purified from a C-terminal TAP tag library (Ghaemmaghami et al., 2003) by immobilization on IgG-magnetic beads. Meiotic substrates were cloned by gap repair into a 2μ PGAL1-TAP plasmid and expressed exogenously in mitotic cells. Some substrates were expressed in bacteria or baculovirus-infected insect cells and purified by conventional affinity chromatography.
Kinase Assays
Kinase assays were carried out as previously described (Loog and Morgan, 2005) except that the reaction buffer was 25 mM Hepes pH 7.4, 150 mM NaCl, 10% glycerol, 0.1 mg/ml BSA, 0.1 mM ATP, 10 mM MgCl2, and 2 μCi 32P-γ-ATP in a 20 μl reaction volume. Positional scanning peptide arrays were performed as described (Hutti et al., 2004), and the consensus site was confirmed using peptides obtained from Elim-bio as described (Loog and Morgan, 2005). The rate of phosphorylation of substrates was determined as described (Loog and Morgan, 2005). Ime2 and Cdk1 activities were normalized for Ndt80. All comparative Cdk1 and Ime2 kinase assays were performed in parallel. If substrate phosphorylation rates by Cdk1 had been previously described (Ubersax et al., 2003), these rates were used in Figure 1B.
Phosphate release assays
Phosphate release assays were carried out as previously described (Jaspersen et al., 1999), except that 6HIS-Fin1 was phosphorylated with either Cdk1 or Ime2 (1-404). 32P-Fin1 was purified by metal affinity chromatography on a cobalt-IDA column (Talon metal affinity resin, Clontech) and incubated with Cdc14, PP2A or PP2C in a 20 μl reaction containing 25 mM Hepes pH 7.4, 150 mM NaCl, 10% glycerol, 0.1 mg ml-1 BSA, 10 mM MgCl2 and 10 mM MnCl2. Reactions were terminated after 5 min by addition of 10 μl 10 mg ml-1 BSA and 180 μl ice-cold 20% TCA, incubation for 5 min on ice and centrifugation to precipitate protein (16,000 g, 10 min 4°C). Phosphate released into the supernatant was quantified by scintillation counting.
Ubiquitination assays
All ubiquitination assays were performed as previously described (Carroll and Morgan, 2005), except that Cdh1 was pre-incubated with Ime2 (1-404) or Cdk1-Clb2 in a 10 μl reaction volume containing 25 mM Hepes pH 7.4, 150 mM NaCl, 10% glycerol, 0.1 mg ml-1 BSA, 0.1 mM ATP, 10 mM MgCl2 for 20 min at room temperature. Kinase reactions were terminated by 5 min incubation with 50 mM EDTA. For Figure 4B, phosphatase was then added to this mixture for a further 20 min at room temperature.
Supplementary Material
Acknowledgements
We thank K. Benjamin, M. Loog, P. O’Farrell, M. Sullivan, R. Morreale, J. Li, A. Moore, A.D. Johnson, B. Tuch and E. Woodbury for technical assistance and thoughtful discussions; W. Zachariae, D. Kellogg, C. Zhang and K. Shokat for reagents; and J. Feldman, M. Matyskiela, M. Sullivan, and E. Woodbury for critical reading of the manuscript. This work was supported by funding from the National Institute of General Medical Sciences (GM50684 to D.O.M. and GM56203 to L.C.C.) and a fellowship from the National Science Foundation (L.J.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte M, Steigemann P, Braus GH, Irniger S. Inhibition of APC-mediated proteolysis by the meiosis-specific protein kinase Ime2. Proc Natl Acad Sci U S A. 2002;99:4385–4390. doi: 10.1073/pnas.072385099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. Enzymology of the Anaphase-Promoting Complex. Meth Enzymol. 2005;398:219–230. doi: 10.1016/S0076-6879(05)98018-X. [DOI] [PubMed] [Google Scholar]
- Choi T, Aoki F, Mori M, Yamashita M, Nagahama Y, Kohmoto K. Activation of p34cdc2 protein kinase activity in meiotic and mitotic cell cycles in mouse oocytes and embryos. Development. 1991;113:789–795. doi: 10.1242/dev.113.3.789. [DOI] [PubMed] [Google Scholar]
- Clifford DM, Stark KE, Gardner KE, Hoffmann-Benning S, Brush GS. Mechanistic insight into the Cdc28-related protein kinase Ime2 through analysis of replication protein A phosphorylation. Cell Cycle. 2005;4:1826–1833. doi: 10.4161/cc.4.12.2214. [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- D′Amours D, Amon A. At the interface between signaling and executing anaphase--Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Dirick L, Goetsch L, Ammerer G, Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- Donzeau M, Bandlow W. The yeast trimeric guanine nucleotide-binding protein alpha subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol Cell Biol. 1999;19:6110–6119. doi: 10.1128/mcb.19.9.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O′Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmann-Raviv N, Boger-Nadjar E, Edri I, Kassir Y. Cdc28 and Ime2 possess redundant functions in promoting entry into premeiotic DNA replication in Saccharomyces cerevisiae. Genetics. 2001;159:1547–1558. doi: 10.1093/genetics/159.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM. Ime2p and Cdc28p: co-pilots driving meiotic development. J Cell Biochem. 2004;92:1025–1033. doi: 10.1002/jcb.20131. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- Iwabuchi M, Ohsumi K, Yamamoto TM, Sawada W, Kishimoto T. Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M-M transition in Xenopus oocyte extracts. Embo J. 2000;19:4513–4523. doi: 10.1093/emboj/19.17.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature. 2005;434:529–533. doi: 10.1038/nature03406. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- Marston AL, Lee BH, Amon A. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev Cell. 2003;4:711–726. doi: 10.1016/s1534-5807(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. New Science Press; London: 2007. [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Ofir Y, Sagee S, Guttmann-Raviv N, Pnueli L, Kassir Y. The role and regulation of the preRC component Cdc6 in the initiation of premeiotic DNA replication. Mol Biol Cell. 2004;15:2230–2242. doi: 10.1091/mbc.E03-08-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Purnapatre K, Gray M, Piccirillo S, Honigberg SM. Glucose inhibits meiotic DNA replication through SCFGrr1p-dependent destruction of Ime2p kinase. Mol Cell Biol. 2005;25:440–450. doi: 10.1128/MCB.25.1.440-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- Schindler K, Winter E. Phosphorylation of IME2 regulates meiotic progression in Saccharomyces cerevisiae. J Biol Chem. 2006;281:18307–16. doi: 10.1074/jbc.M602349200. [DOI] [PubMed] [Google Scholar]
- Sedgwick C, Rawluk M, Decesare J, Raithatha SA, Wohlschlegel J, Semchuk P, Ellison M, Yates Iii JR, Stuart DT. Saccharomyces cerevisiae Ime2 phosphorylates Sic1 at multiple PXS/T sites but is insufficient to trigger Sic1 degradation. Biochem J. 2006;399:151–160. doi: 10.1042/BJ20060363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Satoh H, Takeda N, Fukuda M, Chiba E, Kato T, Kuramochi T, Araki Y. A testicular germ cell-associated serine-threonine kinase, MAK, is dispensable for sperm formation. Mol Cell Biol. 2002;22:3276–3280. doi: 10.1128/MCB.22.10.3276-3280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Wäsch R, Cross F. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28- Clb2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the Anaphase Promoting Complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.