Abstract
RNAi is a powerful method for suppressing gene expression that has tremendous potential for therapeutic applications. However, because endogenous RNAi plays a role in normal cellular functions, delivery and expression of siRNAs must be balanced with safety. Here we report successful stable expression in primates of siRNAs directed to chemokine (c-c motif) receptor 5 (CCR5) introduced through CD34+ hematopoietic stem/progenitor cell transplant. After hematopoietic reconstitution, to date 14 months after transplant, we observe stably marked lymphocytes expressing siRNAs and consistent down-regulation of chemokine (c-c motif) receptor 5 expression. The marked cells are less susceptible to simian immunodeficiency virus infection ex vivo. These studies provide a successful demonstration that siRNAs can be used together with hematopoietic stem cell transplant to stably modulate gene expression in primates and potentially treat blood diseases such as HIV-1.
Keywords: short-hairpin RNA, siRNA, rhesus macaque, gene therapy
siRNAs recognize cognate mRNAs and induce sequence specific RNA degradation through a highly conserved cellular mechanism (1). Because siRNAs have the potential for therapeutic application, a number of vector systems have been developed to express short-hairpin RNAs (shRNAs) to produce siRNAs within mammalian cells in tissue culture and in animal model systems (2–7). The results of these studies indicate that expression of siRNAs can potentially be used to effectively down-regulate gene expression in vivo for therapeutic purposes; however, it is important to control for the negative effects of expressing siRNAs in mammalian cells. Several recent studies have reported adverse effects of expressing siRNAs in mammalian cells, including induction of IFN responsive genes (8), global changes of mRNA expression profiles caused by off-target effects (9), and cytotoxic effects induced by microRNA (miRNA) dysregulation (5). We reported cytotoxic effects of shRNAs in human T lymphocytes as a result of overexpression (10). Any adverse effects of siRNAs would be more problematic in situations where siRNA expression is to be maintained in a living organism for a long period, such as for intracellular immunization against HIV-1 (11).
We selected the HIV-1 coreceptor, (c-c motif) chemokine receptor 5 (CCR5), as a model to examine the feasibility of long-term and stable down-regulation of cellular target gene by siRNA. CCR5, is also an ideal target because it is essential for infection by most strains of HIV-1, yet it is apparently dispensable in normal humans. Indeed, individuals who are homozygous for the CCR5 delta 32 allele that prevents CCR5 cell surface expression are resistant to HIV-1 infection but otherwise apparently normal (12–15), and heterozygous individuals with ≈50% decrease in CCR5 surface expression have lower plasma viral load and a substantially prolonged course of disease (14). Recently, several CCR5 antagonists were examined in clinical trials and resulted in reduction in plasma viral loads by 1.0–1.6 log10 copies/ml during treatment (16, 17). Although, CCR5 antagonists hold a great promise, there are drawbacks, such as transient antiviral effects and drug toxicity. Because HIV-1 infects predominately T lymphocytes and macrophages, hematopoietic stem cell transplant could in theory be used to stably express siRNA and down-regulate CCR5 in progeny cells that are targets for HIV-1 infection.
We previously demonstrated that down-regulation of CCR5 by siRNA can protect primary human T lymphocytes in culture from CCR5 tropic HIV-1 infection (4). Although effective inhibition of HIV-1 infection was observed, we observed cytotoxicities in T lymphocytes that correlated with CCR5 siRNA expression levels (10). By using the weaker H1 promoter, rather than the U6 promoter, to express shRNA reduced toxicities, however, the potency of the siRNAs was also attenuated. We report here the identification of a potent and noncytotoxic shRNA directed to CCR5 that stably down-regulates CCR5 when introduced via hematopoietic stem cell transplant.
Results
Given the cytotoxicities observed in primary human T lymphocytes with siRNAs expressed using the U6 promoter (10), we screened a random library of shRNA directed to human CCR5 (huCCR5) sequences expressed using the H1 promoter within a lentiviral vector. One shRNA sequence (CCR5 shRNA 1005) was identified that had no obvious toxicities in human peripheral blood (PB) T lymphocytes and was the most potent at inhibiting CCR5 among shRNAs characterized to date (Fig. 1). Unlike potent shRNAs expressed from the U6 promoter (10), expression of this shRNA from the H1 promoter did not alter the growth kinetics of transduced T lymphocytes over a 12-day period of culture (data not shown).
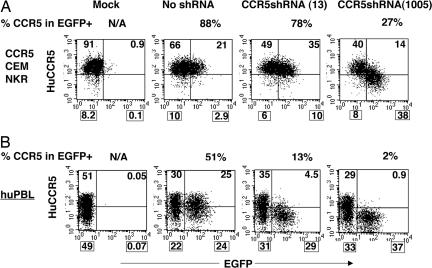
Fig. 1.
Identification of a potent shRNA against huCCR5. (A) CEM-NKR-CCR5 cells were transduced with lentiviral vectors expressing random shRNAs against huCCR5 in 96-well plates, cultured for 3 days, and analyzed by flow cytometry for CCR5 expression in EGFP-expressing populations. Among the 400 shRNAs screened, CCR5 shRNA(1005) reduced CCR5 more efficiently than our previously published CCR5-shRNA (13). The CCR5-shRNA (13) was selected based on the initial criteria by Elbashir et al. (19). (B) Efficient reduction of endogenous CCR5 expression in human primary lymphocytes (huPBLs). PHA/IL-2-activated huPBL were transduced with lentiviral vectors bearing shRNA(1005) and analyzed 8 days after infection by monoclonal antibody staining and flow cytometry for CCR5 expression in the EGFP+ population. Mock, no vector transduction; no shRNA, vector transduction without shRNA expression; N/A, not applicable. The percentage of CCR5-expressing cells within the EGFP+ population (% CCR5 in EGFP+) was calculated and is indicated on the top of each image. The percentage number in each quadrant is also indicated.
The rhesus macaque hematopoietic stem/progenitor cell transplant model is arguably the closest model to that of humans (18). We tested the function and safety of this shRNA sequence by lentiviral vector-mediated transduction of cytokine mobilized PB rhesus CD34+ cells followed by autologous transplant into myeloablated rhesus macaques. We mutated a single mismatched nucleotide in the sequence of the huCCR5 shRNA so that it would be 100% homologous to the corresponding rhesus CCR5 target sequence. This rhesus CCR5 (rhCCR5) shRNA sequence inhibited rhCCR5 expression but not huCCR5 (Fig. 2A). Similar to the homologous huCCR5 shRNA, there was no apparent cytotoxicity (data not shown). CCR5 expression was also inhibited by the expression of the rhCCR5 shRNA in primary rhesus T lymphocytes (Fig. 2B).
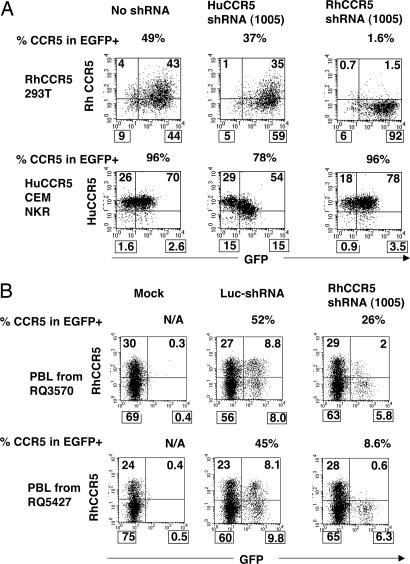
Fig. 2.
Reduction of rhCCR5 by an shRNA(1005). An shRNA against rhCCR5(1005) was tested in rhCCR5-expressing 293T cells. Cells were transduced with an SIV-based lentiviral vector bearing shRNA(1005) against rhCCR5 and analyzed for CCR5 and EGFP expression by monoclonal antibody staining and flow cytometry at 4 days after transduction. The rhCCR5 shRNA reduced rhCCR5 expression in rhCCR5–293T cells but did not reduce huCCR5 in huCCR5-expressing CCR5NKRCEM cells because of a single-nucleotide mismatch in target sequence. Similarly, huCCR5 shRNA reduced huCCR5 but not rhCCR5. (B) Reduction of CCR5 in primary rhesus macaque lymphocytes. Before a study of lentiviral vector transduction and transplant of CD34+ cells, PBLs from two rhesus macaques (animal identifications, RQ3570 and RQ5427) were isolated, PHA/IL-2-activated, and transduced with an SIV vector expressing shRNA against rhCCR5(1005) in vitro. CCR5 expression was analyzed by flow cytometry in EGFP+ cells. A vector expressing an shRNA against firefly luciferase was used as a control. Mock, no vector transduction; N/A, not applicable. The percentage of CCR5-expressing cells within EGFP+ population (%CCR5 in EGFP+) was calculated and is indicated on the top of each image. The percentage number in each quadrant is also indicated.
Two rhesus macaques were subsequently transplanted with autologous PB CD34+ cells transduced with the rhCCR5–shRNA lentiviral vector. The vector also expresses the EGFP marker for tracking transduced cells. Recovery from transplant was consistent with our previous studies (20, 21). Marking as monitored by EGFP expressed from the vector increased over time in PB granulocyte, monocyte, and lymphocyte lineages, stabilizing at ≈6 months after transplant and monitored for 14 months (Fig. 3A).
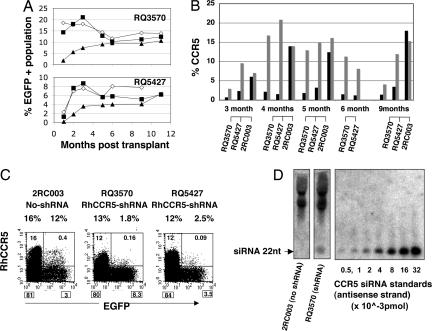
Fig. 3.
In vivo vector marking, CCR5 down-regulation, and siRNA expression. (A) Stable EGFP marking in PB cells in transplanted animals. After CD34+ cell transplant, the percentage of EGFP expression was monitored in granulocyte (■), monocyte (◇), and lymphocyte (▲) populations by flow cytometric analysis. (B) Stable CCR5 reduction in EGFP+ lymphocytes. The percentage of CCR5 expression in EGFP+ (black bar) and in EGFP− (gray bar) cells was monitored by flow cytometry. Control animal 2RC003 was previously transplanted with a lentiviral vector bearing EGFP (20) but no shRNA expression unit. (C) A representative CCR5/EGFP plot at 5 months after transplant. PB from transplanted macaques was stained for CCR5 and CCR5 and EGFP expression in lymphocyte population and analyzed by flow cytometry. Based on the percentage of events in each quadrant (shown in each quadrant), the percentage of CCR5 expression in EGFP+ and EGFP− lymphocyte populations was calculated and is shown on the top of each image. (D) Detection of siRNA in rhesus macaque lymphocytes. The 22-nt antisense-strand siRNA was detected by micro-Northern blot analysis in the small RNA fraction of PHA/IL-2-stimulated lymphocytes from an shRNA-transduced animal (animal identification, RQ3570) but not in cells from control animal (animal identification, 2RC003).
Cell-surface expression of CCR5 was reduced in the EGFP+ population relative to the EGFP− population at all time points assayed and ranged from 3- to 10-fold (Fig. 3 B and C). In contrast, a control animal (2RC003) transplanted with vector expressing EGFP but without shRNA (20) showed no evidence of CCR5 down-regulation. Measurement of CCR5 mRNA levels by RT-PCR showed 5- to 10-fold reduction of mRNA levels in EGFP+ lymphocytes, consistent with the flow cytometric analysis of CCR5 cell-surface expression (data not shown).
Expression of siRNA was demonstrated by micro-Northern blot analysis of lymphocytes from one transplanted rhesus (RQ3570). The presence of a 22-nt band corresponding to the antisense strand of CCR5 siRNA demonstrated processing of the shRNA into the proper effector siRNA (Fig. 3D). More accurate quantitation of the levels of siRNA in both animals by real-time RT-PCR indicated expression of ≈3 × 104 siRNA molecules per EGFP+ cell in phytohemagglutinin (PHA)/IL-2-stimulated PB T lymphocytes and ≈5-fold lower levels in nonstimulated PB mononuclear cells. The level of siRNA in stimulated T lymphocytes is similar to the level of siRNA expressed from stimulated rhesus-T lymphocytes transduced in vitro (data not shown).
Importantly, no apparent toxicity was observed despite expression of siRNA during hematopoietic cell differentiation over the period of this study. The level of EGFP-marked cells increased after transplant with normal kinetics and has remained stable over the course of the study, 14 months to date. These results are in accordance with our previous studies demonstrating stable EGFP expression and marking in differentiated hematopoietic cells in rhesus macaques transplanted with hematopoietic stem cells transduced with lentiviral vectors (20–22). The flow cytometric profiles of CD4, CD8, chemokine (c-x-c motif) receptor 4 (CXCR4), CD45RA, and CD95(fas) are nearly identical for EGFP+ and EGFP− subpopulations [supporting information (SI) Fig. 5]. The EGFP-marked lymphocytes respond normally in culture to PHA/IL-2 stimulation with the same kinetics as nontransduced cells and are maintained at the same frequency for up to 12 days (SI Fig. 6). Over the same period of ex vivo culture, the reduction of CCR5 surface expression in the EGFP+ population persists even though the overall expression of CCR5 in the EGFP− population increases from 5% to 35% of the cell population, because of IL-2-induced CCR5 up-regulation on activated T cells, as described (23, 24) (SI Fig. 7).
We tested the susceptibility of the PB lymphocytes (PBLs) to simian immunodeficiency virus (SIVmac239) infection. Lymphocytes from animal ID RQ3570 were sorted for EGFP+ and EGFP− populations, activated with PHA/IL-2 for 2 days and IL-2 for 2 days (total of 4 days), and infected with SIVmac239 at a multiplicity of infection of 0.04. Both subpopulations proliferated at comparable efficiencies. At the time of infection (d0), the mean fluorescent intensity and percentage of CCR5 expression were 8 and 16.2 (Fig. 4A) and 4% and 9% (Fig. 4B) in the EGFP+ and the EGFP− population, respectively. The CCR5 cell surface expression increased overtime because of PHA/IL-2 stimulation. The EGFP+ population of cells produced ≈3-fold lower levels of p27 SIV gag capsid antigen in the culture supernatant than did the EGFP− population at every time point over the 9-day period of culture (Fig. 4C).
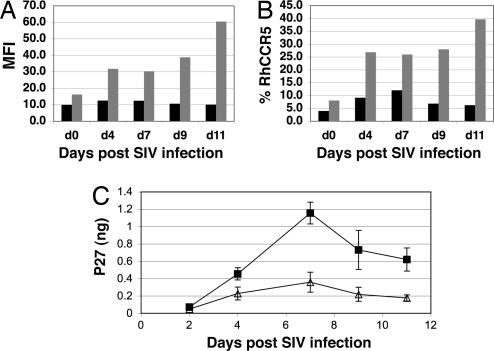
Fig. 4.
Inhibition of SIV replication ex vivo. PBLs from RQ3570 at 13 months after transplant were sorted for EGFP+ and EGFP− population, stimulated with PHA/IL-2 for 2 days and IL-2 for 2 days, and infected with SIVmac239. (A) Mean fluorescent intensity (MFI) of CCR5 expression in EGFP+ sorted (black bar) and EGFP− sorted (gray bar) PHA/IL-2 activated lymphocyte. (B) The percentage of CCR5 expression in EGFP+ and EGFP− sorted lymphocytes. CCR5 expression was monitored during ex vivo culture by flow cytometric analysis and compared between EGFP+ (black bar) and EGFP− sorted (gray bar) lymphocyte populations. (C) SIV p27 production in EGFP+ and EGFP− sorted lymphocytes. After stimulation, 1 × 105 EGFP+ (△) or EGFP− (■) cells were infected with 100 μl of SIVmac239 at a multiplicity of infection of 0.04 (the infectious unit of the virus stock was 4 × 104 ml by titrating on MAGI-CCR5 cells) for 1 h and monitored for p27 production for 11 days in culture supernatant. The infection experiment was done in triplicate. The average p27 production in culture supernatant and error bar (standard deviation) are shown.
The degree of resistance to SIVmac239 was relatively modest; however, the CCR5 levels significantly elevated ex vivo because of PHA/IL-2 stimulation resulting in only a 2-fold difference in CCR5 surface expression at the time of infection. In addition, although CCR5 has been reported as the primary coreceptor for SIVmac239 infection, SIVmac239 has also been reported to use other coreceptors with lower efficiency (25). The use of other coreceptors may contribute to SIV production in EGFP+ cells. It is noteworthy that even 2-fold reductions in CCR5 surface expression can slow disease progression, as in the case of CCR5 delta 32 heterozygote individuals (14).
Discussion
One advantage of stem cell gene therapy over lifetime antiretroviral drug therapy is that a successful gene therapy protocol would be designed to be effective after a single treatment. When applied therapeutically for HIV-1 disease, like gene therapy for X-linked and adenosine deaminase deficiency SCIDs (26, 27), we anticipate that the intense HIV-1-driven selection pressures will result in selection over time of relatively HIV-1-resistant cells. However, given the high rates of HIV production in infected individuals and the ability of HIV to mutate to use other coreceptors, effective therapeutic application of siRNAs for HIV disease will likely involve combination with other “genetic immunization” reagents directed against HIV. Given further optimization, stable RNAi, introduced via hematopoietic stem cell transplant has a potential to treat HIV-1 disease.
Our studies demonstrate that, given appropriate care in selecting shRNAs, it is possible to produce siRNAs in vivo that are both potent and safe for long-term therapy. Importantly, we did not observe apparent adverse effects of siRNA on hematopoietic stem cell differentiation, T cell levels, subset phenotypes, or by gross measures of lymphocyte activation. Continued long-term followup of hematopoietic cell function and gene expression profiles will be informative. These studies demonstrate successful down-regulation of an endogenous gene using RNAi through hematopoietic stem cell transplant in a primate. Not only do these results provide the basis for a potential therapeutic avenue for HIV-1 disease, but they also provide evidence that RNAi could be used as a therapeutic modality for other diseases.
Methods
Construction of CCR5 shRNA Library.
We generated a random shRNA library directed to huCCR5 sequences expressed using an H1 promoter within a lentiviral vector by adapting the method of enzymatic production of RNAi libraries from cDNAs (EPRIL) (28). huCCR5 cDNA was PCR-amplified from pBABE.CCR5 plasmid DNA (National Institutes of Health AIDS research and reference reagent program) by using primer pairs (5′-GATGGATTATCAAGTGTCAAGTCCA-3′) (5′-GTCACAAGCCCACAGATATTTCC-3′) and KOD hot-start DNA polymerase (Novagen, Madison, WI). The CCR5 cDNA was partially digested by DNaseI (Qiagen, Chatsworth, CA) to generate 100- to 200-bp DNA fragments. DNA fragments were ligated to a hairpin adopter1 (Ad1) DNA linker containing an MmeI restriction enzyme site (5′-GTCGGACAATTGCGACCCGCATGCTGCGGGTCGCAATTGTCCGAC-3′). The ligated DNA fragments were then treated with T4 DNA polynucleotide kinase (NEB, Beverly, MA) followed by Escherichia coli DNA ligase treatment (NEB) to fill in a nick between the 5′ end of Ad1 and the 3′ end of CCR5 DNA fragments. The Ad1-ligated DNAs (≈40 nucleotides) were digested with MmeI (NEB) and purified from a native PAGE gel. The purified DNA fragments were ligated to an Ad2 linker (+strand, 5′-GGGGATCCCTTCGGTACTCCAGACCGTGAGTC-3′) (−strand, 5′-TACCGAAGGGATCCCCNN-3′) and purified from a native PAGE gel. The purified DNA fragments were treated with T4 polynucleotide kinase (NEB) and T4 DNA ligase to fill a nick. The Ad2-ligated by DNA fragments were subjected to a primer extension reaction using primer (5′-GACTCACGGTCTGGAGTACCGAAG-3′) and BstDNA polymerase large fragment (NEB), and the resulting products were purified from a PAGE gel. The purified DNA fragments were digested with BpmI, blunt-ended with Klenow fragment (NEB), digested with BamHI, and ligated to pBShH1–5 plasmid DNA, which contains a human H1 RNA polymerase III promoter and 4T termination signal. The ligation mixture was introduced into E. coli (XL1 blue) by electroporation and plated on 2×YT agar plate with 10 μg/ml carbenicillin overnight. Approximately 8,000 colonies were combined, and plasmid DNAs were prepared. The plasmid DNAs were digested with BcgI, blunt-ended with T4 DNA polymerase (NEB), and religated to remove excess DNA sequences. The religated plasmid DNAs were further treated with MfeI to eliminate contamination of BcgI incompletely digested plasmids and used to transform E. coli. Twenty clones of the plasmid DNA were sequenced to confirm the randomness of shRNA sequences targeting CCR5. After sequence confirmation, shRNA expression units consisting of an H1 promoter, shRNA sequence, and 4Ts termination signal were excised from the pBShH1–5 plasmid DNAs by XbaI and XhoI digestion and inserted into XbaI/XhoI sites of the FG12 lentiviral vector (4).
Lentiviral Vector Production and shRNA Library Screening.
Four hundred clones of vesicular stomatitis virus (VSV)-G pseudotyped lentiviral vector were individually produced in 293T cells in 96-well plates. Vector supernatant from each well was harvested at 48 h after transfection and used to infect CCR5NKRCEM cells in 96-well plates. Reduction of CCR5 expression in EGFP+ cells was analyzed at 3 days after infection by monoclonal staining against huCCR5 (2D7 APC; BD Biosciences, San Jose, CA) and flow cytometric analysis.
shRNAs Against huCCR5 and rhCCR5.
The target sequence of huCCR5-shRNA(1005) identified from our shRNA library screening consisted of 5′-GAGCAAGCUCAGUUUACACC-3′. The corresponding rhesus shRNA CCR5(1005) target sequence in rhCCR5 mRNA is 5′-GAGCAAGUUCAGUUUACACC-3′. A rhCCR5 shRNA expression unit was generated by inserting hybridized oligo DNAs (sense 5′-gatccccGAGCAAGTTCAGTTTACACC-TTGTCCGAC-GGTGTAA A CTGAACTTGCTC-TTTTTc-3′, antisense 3′-gggCTCGTTCAAGTCAAATGTGG-AACAGGCTG-CCACATTTGACTTGAACGAG-AAAAAGAGCT-5′) into pBShH1–3 plasmid DNA. The rhCCR5 shRNA sequence was confirmed by sequencing reaction. The target sequence of CCR5-shRNA (13) has been described (10).
SIVmac251-Based Lentiviral Vector Construction.
To insert a rhCCR5 shRNA expression unit into an SIV-based vector for in vitro transduction studies, XbaI and XhoI sites were created by an oligo DNA insertion into a ClaI site in pSIV-R4SAW10, which is derived from pSIV-games (29, 30). The rhCCR5 shRNA expression unit was excised from the pBShH1–3 by XbaI and XhoI digestion and inserted into the XbaI and XhoI site of pSIV-R4SAW10.
To construct an SIV vector for animal transplant studies, we inserted a SacII/XhoI DNA fragment containing the RhMLV RNA polymerase II promoter sequence excised from pCS-Rh-MLV-E plasmid DNA (31) into the ClaI/SalI sites of the SIV vector (pSIV-RMES GAE) using an oligo DNA linker (+strand 5′-CGATACCCTAGGACGGCTGACGC-3′, −strand 5′-GTCGACCGTCCTAGGGTAT-3′) (29). This resulted in vector pSIV GAE RhMLV-E. EGFP expression has been shown to be efficiently expressed in non-human primate lymphocytes by using the RhMLV promoter (31). An XbaI/XhoI-digested DNA fragment containing an H1 promoter-shRNA expression unit was inserted into the AvrII and SalI sites in front of the RhMLV promoter of the pSIV-GAE RhMLV-E vector, resulting in the final pSIV GAE rhCCR5 shRNA RhMLV-E vector.
Lentiviral Vector Transduction.
VSV-G pseudotyped retroviral/lentiviral vector stocks were produced by calcium phosphate-mediated transfection of HEK-293 T cells, as previously described. Briefly, HEK-293 T cells were cultured in Iscove's modified Eagle's medium containing 10% FCS, 100 units of penicillin, and 100 μg/ml streptomycin. The pBabe-rhCCR5 retroviral vector was produced by cotransfecting vector plasmid, the MLV packaging plasmid (pSV-psi-env-MLV) (32), and the VSV-G expression plasmid. HIV vectors were produced by cotransfecting vector plasmid, the HIV-1 lentiviral packaging plasmids pRSVREV and pMDLg/pRRE, and pHCMVG. SIV-based vector was produced by cotransfecting SIV vector, SIV packaging vector pSIV15, pGREV, pSI-k-VPX plasmid DNAs into HEK-293T cells by calcium phosphate transfection, as described (29). Virus culture supernatants were harvested at day 2 after transfection and concentrated 300-fold by ultracentrifugation. The concentrated virus stocks were titrated on HEK-293 T cells based on EGFP expression.
Cells.
rhCCR5–293T was created by infecting HEK-293T cells with a VSV-G pseudotyped pBabe-rhCCR5 retroviral vector followed by puromycin selection (1 μg/ml). CCR5-NKR-CEM was obtained from the National Institutes of Health AIDS reagents program. Human primary PB mononuclear cells were isolated from leukopack by Ficoll-Paque PLUS (GE Healthcare Life Sciences, Piscataway, NJ). Rhesus primary PB mononuclear cells were isolated from PB by Ficoll-Paque PLUS.
Rhesus Macaque Cell Transduction and Transplant.
Animals used in this study were colony-bred rhesus macaques (Macaca mulatta) maintained and used in accordance with guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Department of Health and Human Services publication no. NIH85-23). The protocol was approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services. These animals were free of specific pathogens (serologically negative for simian T lymphotropic virus, SIV, simian retrovirus, and herpesvirus B). CD34+ hematopoietic stem/progenitor cells were mobilized into the PB and purified as described (20). The PB CD34+ cells from the two animals (7 × 107 and 1 × 108 for animals RQ3570 and RQ5427, respectively) were transduced ex vivo with the VSV-G pseudotyped SIV rhCCR5 shRNA RhMLV-E vector at a multiplicity of infection of 2.4 once per day for 2 days in the presence of stem cell factor (50 ng/ml) and IL-6 (50 ng/ml) in X-Vivo 10 Serum Free Medium with Gentamicin (50 μg/ml; Cambrex Bio Science, Walkersville, MD). Transduction efficiency was analyzed by quantitating EGFP expression by flow cytometry 64 h after first transduction. Eighteen percent and 7.4% of the cells were EGFP+ for animals RQ3570 and RQ5427, respectively. The two animals received autologous transplants with 1.6 × 108 transduced PB CD34+ cells. All animals received 10 Gy of total-body gamma-irradiation as a 5-Gy fractionated dose given on 2 consecutive days before transplantation (days −1 and 0, with day 0 being the date of reinfusion) and supported with antibiotic, blood, and fluid support accordingly.
Monoclonal Antibodies, Staining Procedures, and Flow Cytometric Analysis.
We used a whole-blood staining method to detect CCR5 on cell surface by monoclonal antibodies in PBLs (33). Briefly, EDTA-treated whole blood was centrifuged, and plasma was removed. Fifty microliters of packed blood was mixed with monoclonal antibodies and stained at room temperature for 30 min, followed by ammonium chloride-mediated red blood cell lysis, and fixed with 2% formaldehyde in PBS. For the staining of ex vivo PHA/IL-2-stimulated cells, the cells (1 × 105) were mixed with 5 μl of anti-CCR5 monoclonal antibodies (2D7 for human; 3A9 for rhesus) in 100 μl of PBS in 2% FCS, incubated at room temperature for 30 min, washed with PBS with 2% FCS, and fixed with 2% formaldehyde in PBS. The monoclonal antibodies used for this study included: anti-huCCR5 (2D7 APC, 556903; BD Biosciences), rhCCR5 (3A9 APC, 550586; BD Biosciences), CD4 PerCP (550631; BD Biosciences), CD8PE (555367; BD Biosciences), CXCR4 PE (555974; BD Biosciences), CD45RO PE-Cy7 (337167; BD Biosciences), and CD95 PE (556641; BD Biosciences). The stained cells were analyzed by a FACScalibur (BD Biosciences) or a Cytomics FC500 (Beckman Coulter, Fullerton, CA).
Cell Sorting.
PB-derived mononuclear cells were isolated by Ficoll-Paque PLUS (GE Healthcare Life Sciences), and EGFP+ and EGFP− lymphocyte populations were isolated by FACS Aria cell sorter (BD Biosciences). The sorting purities of EGFP+ and EGFP− population were 92% and 96%, respectively, as determined by flow cytometric analysis of the sorted cells.
Small RNA Isolation.
Small RNA fraction was isolated from PHA/IL-2 stimulated rhesus monkey PBLs (≈4 × 108 cells) at day 9 after stimulation by using the PureLink microRNA isolation kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Northern Blot Analysis of siRNA.
Twenty-five micrograms of fractionated small RNA was resolved on a 15% urea-acrylamide-TBE gel (SequaGel; National Diagnostics, Atlanta, GA) and electrotransferred to nylon membrane (GeneScreen Plus; NEN, Waltham, MA) at 80 V for 1 h in 0.5× TBE. The membrane was dried, UV cross-linked, and baked at 80°C for 1 h. Oligonucleotide probes (sense: GAG CAA GTT CAG TTT ACA CC; antisense: GGT GTA AAC TGA ACT TGC TC) were labeled by using Starfire oligos kit (Integrated DNA Technologies, Coralville, CA) and [α-32P] dATP (6,000 Ci/mmol; PerkinElmer, Waltham, MA) according to the manufacturer's instructions. The probes were hybridized to the membranes at 37°C in ULTRAhyb-Oligo (Ambion, Austin, TX) overnight. The membrane was washed three times for 15 min at 37°C in 2× SSC/0.1% SDS. Signal detection and analysis of Northern blots were performed by exposing the blots to phosphorimaging plates followed by analysis on a phosphorimager (Storm System; Molecular Dynamics, Sunnyvale, CA) by using synthetic rhCCR5 siRNA (GGU GUA AAC UGA ACU UGC UC; Sigma–Proligo, St. Louis, MO) as a standard.
Real-Time RT-PCR Analysis.
We used published primer pairs and probes to detect rhCCR5 mRNA and β2 microglobulin (34). Total RNA (100 ng) was isolated from sorted EGFP+ or EGFP− rhesus PBLs by TRIzol Reagent (Invitrogen) and subjected to RT-PCR by using the Qiagen one-step RT-PCR kit and the following conditions: 50°C, 30 min, and 55°C, 10 min for reverse transcription reaction; 95°C, 15 min for reverse transcription inactivation and activation of HotStarTaq DNA Polymerase; 50 cycles of 95°C, 15 sec, 55°C, 30 sec, and 60°C, 90 sec for PCR. RNA standards for CCR5 and β2 microglobulin mRNA quantitation were made by serial dilution of in vitro transcribed rhCCR5 and β2 microglobulin RNAs by using T7 RNA polymerase (MEGAscript T7; Ambion).
RT-PCR Analysis of siRNA.
We used a real-time stem–loop RT-PCR method to quantitate the levels of the antisense strand of siRNA against rhCCR5 (35). Total RNA (500 ng) was isolated from rhesus PBLs by TRIzol reagent (Invitrogen) and subjected to reverse transcription reaction (16°C, 30 min; 42°C, 30 min; 85°C, 5 min), followed by PCR (95°C, 10 min, 1 cycle; 95°C, 15 sec and 58°C, 1 min, 50 cycles). Primer and probe sequences are as follows: reverse transcription stem–loop primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACA A G AGCAA-3′; forward primer, 5′-GCGCGGTGTAAACTGAAC-3′; reverse primer, 5′-GTGCAGGGTCCGAGGT; probe, 6-FAM-TGGATACGACAAGAGCAA-MGB. A set of serial diluted synthetic 22-nt antisense strand of siRNA against rhCCR5 (GGU GUA AAC UGA ACU UGC UC; Sigma–Proligo) was used as standard for quantitation.
SIV Production and Infection.
The 5′ and 3′ halves of SIVmac239 plasmid DNAs linearized by SphI digestion were transfected into HEK-293T cells. The virus produced was propagated in CEMX174 cells. Infectious titer of virus stocks (4 × 104 infectious units/ml) (p27 value = 72 ng/ml) was determined on MAGI CCR5 cells as described (36).
SIV Infection.
Sorted EGFP+ or EGFP− rhesus PBLs were stimulated with PHA (5 μg/ml; Sigma) and IL-2 for 2 days, PHA was removed, and the cells were stimulated with IL-2 for another 2 days. The cells (1 × 105) were infected with 100 μl of SIVmac239 (multiplicity of infection of 0.04) for 1 h at 37°C, washed two times with PBS with 2% FCS, washed one time with medium, and resuspended in RPMI 20% FCS with IL-2 and cultured.
ELISA.
The levels of SIV p27 core antigen in SIV-infected culture supernatant were measured by using the COULTER SIV core antigen assay according to the manufacturer's instructions (catalog no. 6604395; Beckman Coulter).
Supplementary Material
Acknowledgments
We thank Drs. Kathie Grovit Ferbas (University of California, Los Angeles), François-Loïc Cosset (Institut National de la Santé et de la Recherche Médicale, ENSINSERM, France), and Didier Nègre (ENSINSERM, France) for providing reagents and the animal management staff of 5 Research Court (National Heart, Lung and Blood Institute) for assistance. This research was supported by the National Institutes of Health (Grants AI39975-05 and AI28697 to I.S.Y.C. and 1R01HL086409-01 to D.S.A.) and in part by the Intramural Research Program of the National Institutes of Health.
Abbreviations
- shRNA
short-hairpin RNA
- CCR5
(c-c motif) chemokine receptor 5
- PB
peripheral blood
- PBL
PB lymphocyte
- rhCCR5
rhesus CCR5
- huCCR5
human CCR5
- SIV
simian immunodeficiency virus
- PHA
phytohemagglutinin
- VSV
vesicular stomatitis virus.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705474104/DC1.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, et al. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 3.Tiscornia G, Singer O, Ikawa M, Verma IM. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin XF, An DS, Chen ISY, Baltimore D. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA, et al. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 6.Li MJ, Bauer G, Michienzi A, Yee JK, Lee NS, Kim J, Li S, Castanotto D, Zaia J, Rossi JJ. Mol Ther. 2003;8:196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 7.Anderson J, Li MJ, Palmer B, Remling L, Li S, Yam P, Yee JK, Rossi J, Zaia J, Akkina R. Mol Ther. 2007;6:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 8.Fish RJ, Kruithof EK. BMC Mol Biol. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, Chen ISY. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baltimore D. Nature. 1988;335:395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 13.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 14.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, et al. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 16.Tsibris AMN, Kuritzkes DR, Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD, et al. Annu Rev Med. 2007;58:445–459. doi: 10.1146/annurev.med.58.080105.102908. [DOI] [PubMed] [Google Scholar]
- 17.Van Der Ryst E, Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD, Rockstroh JK, et al. Nat Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 18.Donahue RE, Kuramoto K, Dunbar C. Current Protocols in Immunology. New York: Wiley; 2005. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;24:411. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 20.Sander WE, Metzger ME, Morizono K, Bonifacino A, Penzak SR, Xie YM, Chen ISY, Bacon J, Sestrich SG, Szajek LP, et al. J Nucl Med. 2006;47:1212–1219. [PubMed] [Google Scholar]
- 21.Kung SK, An DS, Bonifacino A, Metzger ME, Ringpis GE, Mao SH, Chen ISY, Donahue RE. J Virol. 2000;74:1286–1295. [Google Scholar]
- 22.An DS, Kung KPS, Bonifacino A, Wersto RP, Metzger ME, Agricola BA, Mao SH, Chen ISY, Donahue RE. J Virol. 2001;75:3547–3555. doi: 10.1128/JVI.75.8.3547-3555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, Choe H, Sodroski J, Newman W, Koup RA, et al. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Lou B, Lal RB, Gettie A, Marx PA, Moore JP. J Virol. 2000;74:6893–6910. doi: 10.1128/jvi.74.15.6893-6910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, et al. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 27.Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, Morecki S, Andolfi G, Tabucchi A, Carlucci F, et al. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 28.Shirane D, Sugao K, Namiki S, Tanabe M, Iino M, Hirose K. Nat Genet. 2004;36:190–196. doi: 10.1038/ng1290. [DOI] [PubMed] [Google Scholar]
- 29.Mangeot PE, Duperrier K, Negre D, Boson B, Rigal D, Cosset FL, Darlix JL. Mol Ther. 2002;5:283–290. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- 30.Mangeot PE, Negre D, Dubois B, Winter AJ, Leissner P, Mehtali M, Kaiserlian D, Cosset FL, Darlix JL. J Virol. 2000;74:8307–8315. doi: 10.1128/jvi.74.18.8307-8315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kung S, An DS, Chen ISY. J Virol. 2000;74:3668–3681. doi: 10.1128/jvi.74.8.3668-3681.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landau NR, Littman DR. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chackerian B, Long EM, Luciw PA, Overbaugh J. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.