Abstract
The U.S. National Cancer Institute has used a panel of 60 diverse human cancer cell lines (the NCI-60) to screen >100,000 chemical compounds for anticancer activity. However, not all important cancer types are included in the panel, nor are drug responses of the panel predictive of clinical efficacy in patients. We asked, therefore, whether it would be possible to extrapolate from that rich database (or analogous ones from other drug screens) to predict activity in cell types not included or, for that matter, clinical responses in patients with tumors. We address that challenge by developing and applying an algorithm we term “coexpression extrapolation” (COXEN). COXEN uses expression microarray data as a Rosetta Stone for translating from drug activities in the NCI-60 to drug activities in any other cell panel or set of clinical tumors. Here, we show that COXEN can accurately predict drug sensitivity of bladder cancer cell lines and clinical responses of breast cancer patients treated with commonly used chemotherapeutic drugs. Furthermore, we used COXEN for in silico screening of 45,545 compounds and identify an agent with activity against human bladder cancer.
Keywords: bladder neoplasms, breast neoplasms, microarray expression profiling, NCI-60 anticancer compound screening, coexpression extrapolation
Tumors have traditionally been classified by descriptive characteristics, such as organ of origin, histology, aggressiveness, and extent of spread. That empirical rubric is being challenged, however, as molecular-level classifications, made possible by microarrays and other high-throughput profiling technologies, become increasingly common and persuasive (1–3). Several recent studies have predicted the clinical outcome of human cancer patients by using molecular signatures (4, 5). This experience suggests that, eventually, all differences among traditional tumor types will be reduced to statements about molecules in the tumors and about the interactions among those molecules. It might then be possible to study physiological processes in one type of cancer and extrapolate the results predictively to another type through commonalities in their molecular constitutions. More recently, several studies have demonstrated that genomic biomarkers can be used to predict chemotherapeutic responses of human cancer patients (6–8).
What if we want a more ambitious prediction at the pharmacological level to extrapolate and predict drug sensitivity from one type of cancer to another? The challenge is greater, but, we think, approachable. Toward that end, we present here a generic algorithm we term “coexpression extrapolation” (COXEN). The specific algorithm in essence uses specialized molecular profile signatures as a Rosetta Stone for translating the drug sensitivity signature of one set of cancers into that of another set.
The system that motivated development of COXEN was the NCI-60 cell line screen, composed of cell lines from diverse human cancers. The NCI-60 panel has been used by the Developmental Therapeutics Program (DTP) of the U.S. National Cancer Institute (NCI) to screen >100,000 chemically defined compounds plus a large number of natural product extracts for anticancer activity since 1990 (9–11). It has been controversial, however, whether tumor cell activities in such in vitro assays can predict human patient chemotherapeutic responses (12). Furthermore, it was not feasible to include all important tumor types in the NCI-60. For example, there are no lymphomas, sarcomas, head and neck tumors, squamous cell carcinomas, small cell lung cancers, pancreatic cancers, or urothelial bladder cancers. Even if cancer cells of the additional histological types were added to the panel now, all compounds screened in the past 16 years would have to be tested again in the updated panel to gain the full predictive power of the database for the legacy compounds.
Those limitations raise three practical questions: Can drug sensitivity data on the NCI-60 (or analogous screening panels) be extrapolated to predict the sensitivity of cell lines and cell line types not included? More ambitiously, can such in vitro screening data be used to obtain predictive power for clinical responses of human cancers? Finally, can such extrapolation be useful in the drug discovery process? Here we address those questions in three different applications.
Results
Detailed descriptions of the COXEN algorithm and its slightly different implementations for those three applications are in Materials and Methods and supporting information (SI) Materials and Methods. We focus first on a generic description of the algorithm and then on the results for the three applications.
COXEN Algorithm.
The COXEN algorithm is composed of six distinct steps. The end result is what we term the “COXEN score,” which reflects the predicted sensitivity of a particular cell line or human tumor to the specific drug being evaluated by the algorithm. Generically, the steps for prediction of a drug's activity in cells belonging to some set 2 on the basis of its activity pattern in different cells of some set 1 are as follows:
Step 1. Experimentally determine the drug's pattern of activity in cells of set 1.
Step 2. Experimentally measure molecular characteristics of the cells in set 1.
Step 3. Select a subset of those molecular characteristics that most accurately predicts the drug's activity in cell set 1 (“chemosensitivity signature” selection).
Step 4. Experimentally measure the same molecular characteristics of the cells in set 2.
Step 5. Among the molecular characteristics selected in step 3, identify a subset that shows a strong pattern of coexpression extrapolation between cell sets 1 and 2.
Step 6. Use a multivariate algorithm to predict the drug's activity in set 2 cells on the basis of the drug's activity pattern in set 1 and the molecular characteristics of set 2 selected in step 5. The output of the multivariate analysis is a COXEN score.
In our first application of the COXEN algorithm, for example, cell sets 1 and 2 were the NCI-60 and BLA-40 cell panels, respectively; the step 1 drug activities were those assessed by the DTP in the NCI-60; the “molecular characteristics” in steps 2 and 4 were transcript expression levels, as assessed using Affymetrix HG-U133A microarrays (13); the algorithm in step 3 was significance analysis of microarrays (14) or similar statistical testing for differential expression; and step 5 was a coexpression extrapolation algorithm we developed. This coexpression extrapolation procedure is conceptually illustrated in SI Fig. 4 and detailed in SI Materials and Methods; step 6 was a refined classification algorithm, “misclassification-penalized posterior” (MiPP) (15), which we recently introduced for selection of the best mathematical “models” for such predictions. MiPP generates the final COXEN score. As will be discussed below, COXEN predictions for both cell line and clinical trial drug responses were prospectively and independently validated.
Although it may not be intuitively obvious, steps 3 and 5 are key and cannot be omitted; the algorithm does not use the entire molecular signature but only those optimal aspects of the signature that most strongly predict the drug's activity and that also reflect a pattern of concordant coexpression between the two sets of cancer cells. As will be shown below, simply using the entire molecular signature (or even the entire chemosensitivity signature portion of it) does not provide very much predictive power. Note that step 5 is based only on microarray coexpression patterns between set 1 and set 2 and does not use any drug activity information on set 2 in the gene selection. Likewise for step 6, prediction model discovery and training are performed strictly within set 1 to maintain statistical rigor for the prediction on any independent sets, avoiding the pitfalls of overfitting when such information from the test sets is used (15). In fact, in step 5 set 2 can be completely replaced by another (historical) set of the same type of tumor, not using any molecular information directly from set 2.
Predicting Drug Activity in Bladder Cancer Cells.
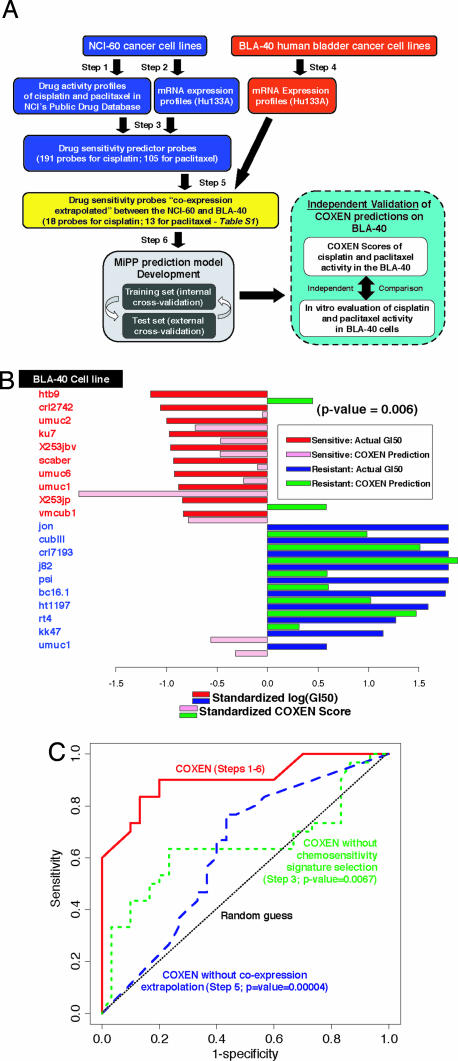
Applying the particular implementation of COXEN shown in Fig. 1A and described in Materials and Methods, we used the NCI-60 data to predict drug activities in the BLA-40, a panel of 40 human urothelial bladder carcinomas, profiled at the mRNA level. We first developed the MiPP prediction models for two drugs (cisplatin and paclitaxel, which are used clinically against bladder cancer) on the basis of their NCI-60 cell line screening data; as expected, the models performed extremely well on that training set with mean prediction accuracies of 93–96% in leave-one-out cross-validation (data not shown). We then exposed bladder cell lines in the BLA-40 panel to cisplatin and paclitaxel and thus generated the data to test the predictions prospectively. For that test, we focused first on prediction for the 10 most sensitive and 10 most resistant BLA-40 lines. For those cells, prediction accuracies for the top three MiPP models averaged 85% for cisplatin and 78% for paclitaxel (SI Table 1 and SI Fig. 4 B and C). As expected, those prediction accuracies were lower than the ones obtained for the training NCI-60 set but, nonetheless, were highly statistically significant (binomial test P = 0.002 for cisplatin and 0.012–0.042 for paclitaxel, against random coin tossing). For cisplatin, nine sensitive cell lines (all except umuc9) and eight resistant cell lines (all except crl7197 and kk47) were consistently correctly classified by the three prediction models. For paclitaxel, one sensitive (X235jp) and one resistant (umuc1) cell line were consistently misclassified by the top three models.
Fig. 1.
Application and performance characteristics of the COXEN algorithm for prediction of drug sensitivity in the BLA-40 human urothelial cancer cell lines. (A) Summary schematic diagram of the development and validation of chemosensitivity predictions. Step numbers relate to those of the COXEN algorithm, as described in the text. (B) Direct comparison between COXEN prediction scores and experimentally measured paclitaxel activities in the BLA-40 cell lines. The activity here and elsewhere is expressed as −log(GI50), where GI50 is the drug concentration leading to 50% growth inhibition of cells compared with control. The cell lines are ordered on the basis of their −log(GI50) values. COXEN scores and GI50 values were standardized by subtracting the overall mean and dividing by the SD across the BLA-40. The statistical significance of the set of predictions (two-tailed P = 0.006) on all 40 cells of the BLA-40 was assessed by Spearman correlation. (C) ROC analysis. ROC curves were computed for COXEN scores generated for cisplatin sensitivity from the full COXEN algorithm (steps 1–6) and for variations in which either the drug chemosensitivity signature selection step (step 3; χ2 statistic P = 0.0067) or the coexpression extrapolation step (step 5; χ2 statistic P = 4.0 × 10−5) was omitted.
Because classification of the sensitive and resistant cells does not provide predictive results for cells of intermediate sensitivity, we next analyzed the quantitative relationship between COXEN-predicted and actual activity values for all 40 cell lines. The results for the top MiPP model were highly significant (Spearman correlation coefficient P = 0.016 for cisplatin and 0.006 for paclitaxel); their standardized COXEN prediction scores, in fact, predicted the standardized log(GI50) values well, as shown in Fig. 1B and SI Fig. 4 B and D for cisplatin and paclitaxel, respectively.
The predictive power of the COXEN algorithm can also be expressed in a receiver–operator characteristic (ROC) analysis. As is often useful in biomarker studies, the ROC formulation permits free choice of a set-point to use in balancing the costs of false-positive and false-negative predictions. Fig. 1C contrasts the ROC curves obtained for cisplatin from the full COXEN algorithm with those obtained by leaving out either the drug chemosensitivity signature step (step 3; χ2 statistic P = 0.0067) or the coexpression step (step 5; P = 4.0 × 10−5) (16). The overall predictive power of an algorithm is indicated by the area between its ROC curve and the straight dashed line representing classification at random. Clearly, the predictions were far superior when the entire algorithm was used. Again, note that no chemosensitivity data on the BLA-40 cells were used to “tune” any part of the COXEN algorithm to obtain the results described here.
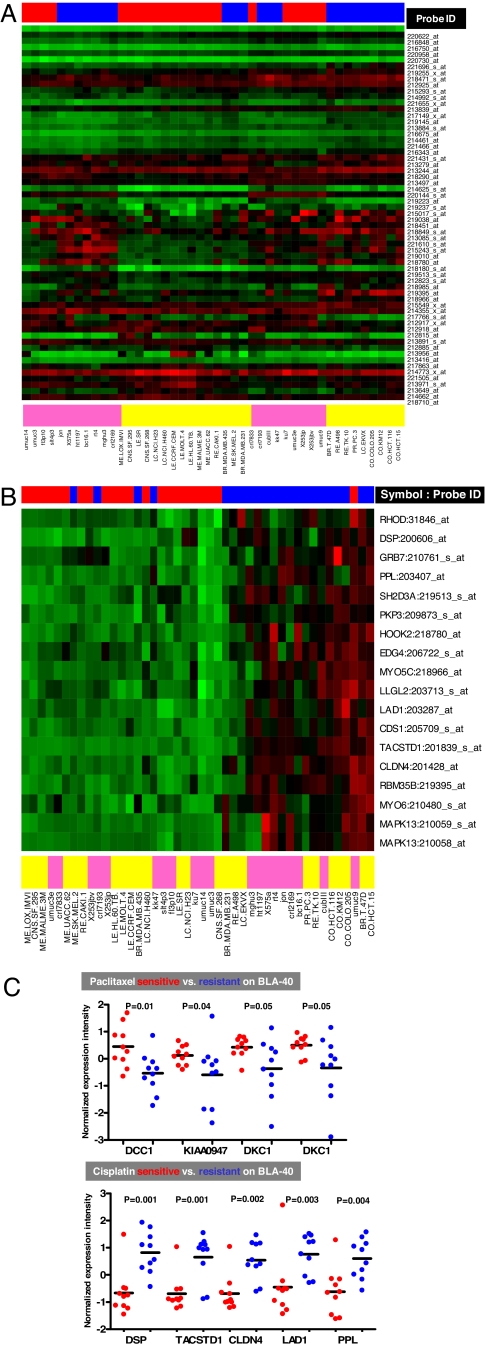
The clustered image maps (heat maps) (17) in Fig. 2 further illustrate in graphical terms the raison d'etre for the coexpression extrapolation step (step 5) in COXEN. Without that step (Fig. 2A), the cell types tend to sort themselves out according to whether they are NCI-60 or BLA-40; with that step (Fig. 2B), the cells of the two panels tend to intermingle and (as one would wish) cluster according to their sensitivity to cisplatin. SI Fig. 5 A and B shows similar results for paclitaxel. In essence, step 5 transforms clustering by cell panel or histological type into clustering by sensitivity to the drug. Of 18 and 13 COXEN prediction biomarkers for cisplatin and paclitaxel, individual genes have shown their significant differential expression patterns between the sensitive and resistant BLA-40 cell lines (Wilcoxon two-sample P = 0.001–0.05) (Fig. 2C). Of importance, none of those genes was selected on the basis of differences in expression pattern between the sensitive and resistant BLA-40 groups. Thus, the genes' expression patterns confirm the ability of COXEN to identify real biological chemosensitivity biomarkers on a completely independent set (and type) of tumor without any drug activity information on the independent set. Many of the genes have been reported to be relevant to cancer (SI Table 2).
Fig. 2.
Coclustering of NCI-60 and BLA-40 cells with and without the COXEN coexpression extrapolation step (step 5). (A) Clustered image map for the NCI-60 and BLA-40 cell lines using the first 50 chemosensitivity probe sets for cisplatin, omitting step 5. (Only the first 50 were used simply for readability of the figure.) Red, black, and green indicate high, intermediate, and low expression, respectively. Red and blue in the upper bar indicate sensitive and resistant cell types, respectively. Yellow and cyan in the lower bar indicate NCI-60 and BLA-40 cells, respectively. Most cell lines clustered on the basis of cell panel (NCI-60 vs. BLA-40) not sensitivity or resistance. Probe IDs were those provided by the commercial microarray manufacturer (Affymetrix). (B) Clustered image map for the NCI-60 and BLA-40 using the 18 COXEN probes obtained for cisplatin after step 5; cells clustered primarily on the basis of sensitivity and resistance rather than on the basis of the cell panel. (C) Normalized expression intensities of COXEN-identified genes shown for BLA-40 cells sensitive and resistant to cisplatin. The genes were selected on the basis of only NCI-60 chemosensitivity information yet showed significant differential expression between the sensitive and resistant cell lines of BLA-40. Many of the genes have been reported to be relevant to cancer (SI Table 2).
Prediction of Clinical Response to Chemotherapeutic Drugs in Breast Cancer Patients.
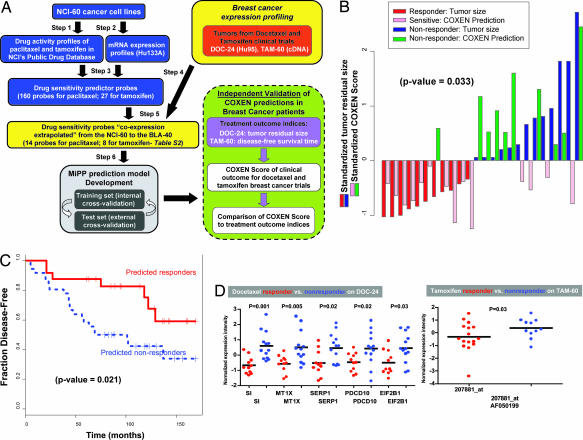
Given the finding that COXEN could predict drug sensitivity, even in cell lines of histological types not included in the NCI-60 panel, we wondered whether an analogous algorithm would also have predictive power for chemotherapeutic responses in human patients. Historically, it has proven difficult to predict drug activity in mouse xenografts from cell line data or clinical responses from mouse xenograft data. So our hope and our hypothesis was that by bypassing the intermediate animal model, we might be able to achieve predictiveness for the clinic on the basis of human cancer cell line in vitro assays. Hence, we developed a modification of COXEN that aligns the NCI-60 gene expression data with expression data from patients' tumors, rather than cell lines. Fig. 3A shows the algorithm in schematic form. To demonstrate that application, we chose two cohort-based breast cancer clinical trials, DOC-24 (24 patients treated with docetaxel) and TAM-60 (60 patients treated with tamoxifen) (4, 18). Those trials satisfied several criteria for our analysis, most important among them: (i) the clinical response data were publicly available; (ii) the patients' tumors had been transcript-profiled; and (iii) the treatment was a single agent, mirroring the single-agent treatments of the NCI-60 panel. The latter criterion was the hardest to satisfy, because most clinical efficacy trials are carried out with drug combinations.
Fig. 3.
COXEN prediction of chemotherapeutic response in patients with breast cancer. (A) Schematic diagram of the prediction and validation processes. (B) Direct comparison between the COXEN predictive scores and the patients' residual tumor sizes. The scores and tumor sizes were standardized for comparison by subtracting the overall mean and dividing by the SD of each of the COXEN scores and residual tumor sizes. The statistical significance of the set of predictions (two-tailed test; P = 0.022) was assessed by Spearman correlation. (C) Kaplan–Meier survival curves for the 36 COXEN-predicted responders and the 24 COXEN-predicted nonresponders in the tamoxifen trial. The predicted responder group showed a significantly longer disease-free survival time than did the predicted nonresponder group (log-rank test; P = 0.021). (D) Normalized expression intensities of COXEN-identified genes between responder and nonresponder DOC-24 patients treated with docetaxel. The genes were selected based only on NCI-60 chemosensitivity information yet showed significant differential expression between responder and nonresponder DOC-24 patients. Many of them were found to be relevant to cancer (SI Table 3).
By analogy with our algorithm for bladder cancer cell lines, we first identified the drug signature probe sets with high degrees of coexpression between the NCI-60 and each of the clinical microarray data sets (i.e., those for the docetaxel and tamoxifen trials). We then derived the corresponding MiPP classification models on the basis of the NCI-60 drug responses and microarray data. COXEN response predictions were generated for the 11 responder and 13 nonresponder DOC-24 patients after four cycles of neoadjuvant therapy reported in the original study (4). Because complete docetaxel drug activity data were not available in the NCI-60 database, we used the data from another, very similar, taxane, paclitaxel, to make the predictions. A previous analysis had shown that essentially all of the taxane drugs have very similar activity profiles in the NCI-60 (10). We then compared the predictions with the actual clinical data on tumor response and, as summarized in SI Table 3, the classification prediction accuracies across the top three MiPP models were uniformly 75% (SI Fig. 6). As anticipated, the accuracy of clinical response prediction was lower than that for the BLA-40, but nevertheless statistically significant (binomial test P = 0.022 against random coin tossing). We next compared our COXEN scores with the patients' residual tumor sizes (Fig. 3B). The rank-based Spearman correlation of those results showed statistical significance (P = 0.033). The clustered image maps in SI Fig. 7 A and B (like those in the Fig. 2 A and B pair and the SI Fig. 5 A and B pair) show the importance of the coexpression extrapolation step for coclustering of data sets (and, therefore, for COXEN prediction).
In the tamoxifen trial (TAM-60), 60 postmenopausal breast cancer patients with estrogen receptor-positive tumors were uniformly treated with adjuvant tamoxifen alone and followed for up to 180 months (18). Genome-wide expression profiling was performed on the primary tumors using a customized cDNA microarray platform. The study data did not include measures of short-term tumor response but did include long-term disease-free survival and disease recurrence times. Within this cohort, 28 (46%) women developed distant metastases with a median time to recurrence of 4 years (“tamoxifen recurrences”), and 32 (54%) women remained disease-free with median follow-up of 10 years (“tamoxifen nonrecurrences”). Hence, we made the assumption that tamoxifen recurrences constituted tamoxifen nonresponders and tamoxifen nonrecurrences constituted responders (SI Fig. 9A). From those observations, we identified 11 responders and 16 nonresponders before, and independent of, making the COXEN predictions. Without knowing other confounding factors, e.g., other treatments after the failure of tamoxifen, disease recurrence is a more indirect measure of tumor response to a therapeutic agent than pathological responses determined shortly after treatment (as in the DOC-24 chemotherapy trial described above), so we would expect less, rather than more, power from the algorithm for prediction of disease-free survival.
For the tamoxifen case, prediction accuracies across the top three MiPP prediction models averaged 71%. That was lower than the accuracy scores for the DOC-24 case, yet statistically significant (binomial test P = 0.019–0.052 against random coin tossing) for responders and nonresponders in the tamoxifen trial (SI Table 3 and SI Fig. 8B). To examine the robustness of COXEN predictions for all 60 patients, we examined the data by Kaplan–Meier survival analysis. In that analysis, the predicted responder group based on the top MiPP prediction model showed a significantly longer disease-free survival time (Fig. 3C) than did the predicted nonresponder group (P = 0.021); for example, the 5-year disease-free survival rate was 88% for the COXEN-predicted responders compared with 49% for the COXEN-predicted nonresponders (19). Overall, the prediction performance is impressive given that (i) only a small proportion (≈11%) of probe sets were matched in their annotation between the Affymetrix HG-U133A and customized cDNA microarray data and (ii) we had to use the surrogate of disease-free survival time instead of a more conventional short-term outcome measure (such as complete or partial tumor response), which would probably have related more closely to the in vitro chemosensitivity data. Finally, as for the bladder cancer studies, it is important to note that validations were done prospectively, without any “tuning” of the model on the basis of response data from the clinical trials.
Of 14 and 8 COXEN prediction biomarkers for the docetaxel and tamoxifen responses, several individual genes have shown significant differential expression patterns between the responder and nonresponder DOC-24 and TAM-60 patients (Wilcoxon two-sample test; P = 0.001–0.03) (Fig. 3D). As for the BLA-40 analysis above, none of those genes was selected on the basis of differential expression patterns between responder and nonresponder patient groups; rather, they were identified by COXEN, which could identify common chemosensitivity biomarkers on the two completely different cancer cell line and patient populations. Most of those genes were found to be important in cancer-related processes (SI Table 3).
Using the Ingenuity pathway analysis software, we found those COXEN-identified biomarkers to be related to the pathways and molecular functions of each of the compounds. For example, paclitaxel and docetaxel are known to bind to microtubules and inhibit their depolymerization into tubulin monomers, thereby blocking a cell's ability to break down the mitotic spindle during mitosis. Most of our COXEN biomarkers belong to pathways related to DNA replication, recombination, repair, and cell-to-cell signaling (SI Fig. 5 C and D). Cisplatin (BLA-40) is known to act by binding DNA in several different ways, making it impossible for rapidly dividing cells to duplicate their DNA for mitosis; most COXEN biomarkers for cisplatin were also found to be associated with cell-to-cell signaling and DNA replication (SI Fig. 5E).
Use of COXEN for in Silico Drug Discovery.
Given the encouraging predictive performance of COXEN, both in vitro (for BLA-40 bladder cancer lines) and in patients (with breast cancer), we next applied it to drug discovery. For each of the 45,545 compounds whose NCI-60 drug screening data are publicly available from the NCI DTP, we used COXEN to predict in silico chemosensitivity patterns for cells in the BLA-40 panel as we had done for cisplatin and paclitaxel (SI Fig. 9 Upper). For prediction of each drug's activity in the BLA-40, we averaged the classification probabilities of the top five MiPP models identified. The compounds selected were then ranked by the number of BLA-40 cell lines predicted to be sensitive.
In an initial screen we identified 139 compounds for which COXEN predicted at least 35% sensitive cells among the BLA-40. For eight of those compounds, >50% of the BLA-40 were predicted to be sensitive. Not all of the eight candidate compounds were available from the NCI DTP but, fortunately, our top hit, NSC637993 (6H-imidazo[4,5,1-de]acridin-6-one, 5-[2-(diethylamino) ethylamino]-8-methoxy-1-methyl-, dihydrochloride), was (SI Fig. 10E), and we were able to assay it for growth inhibition in the BLA-40 panel. For NSC637993, COXEN predicted 62% sensitive cells of the BLA-40 (SI Fig. 9 Lower). That prediction compared favorably with the experimentally measured GI50 values, which were <10−6 M for >60% of the cells. In comparison, the equivalent in vitro parameters for cisplatin, which is one of the most potent current chemotherapeutic agents used in bladder cancer treatment, were <10−6 M for only 22% of the BLA-40 cells.
Discussion
We have developed an algorithm, COXEN, for in silico prediction of chemosensitivity. Here we have shown illustrative studies in which it was used (i) to extrapolate from chemosensitivity data on the NCI-60 cancer cell panel to an analogous cell line panel of bladder cancers, (ii) to extrapolate from the NCI-60 to data on clinical breast cancers, and (iii) to predict sensitivity of the bladder cancers to 45,545 candidate agents on the basis of NCI-60 data. In each case, the algorithm was run independently of the validating experimental results and not further tuned thereafter. One of the conceptual principles that our COXEN approach is based on is identifying networks of chemosensitivity genes concordantly expressed/regulated between different cancer cell types or subtypes (20).
In the drug discovery test case, the top hit, NSC637993 is an imidazoacridinone with structural similarities to drug classes such as the anthracyclines (e.g., doxorubicin), the anthracenediones (e.g., mitoxantrone), and the anthrapyrazoles (e.g., oxantrazole and biantrazole), which are known to intercalate in DNA and inhibit DNA topoisomerase II. In our validation studies, NSC637993 was a potent inhibitor of bladder cancer cell lines, as predicted. An almost identical compound, C1311, has shown significant cytotoxic activity in vitro and in vivo for a range of colon tumors (both murine and human) and is currently being prepared for clinical trials (21, 22). Interestingly, the selectivity of NSC637993 for bladder cancer compared with cancer cells from other tissues is indicated by its activity pattern in the NCI-60 panel (http://dtp.nci.nih.gov); only the leukemia call lines show inhibition. In contrast, as we show in this article, most bladder cell lines are inhibited. Although tissue selectivity in the NCI-60 screen does not necessarily translate into tumor type selectivity clinically, the results here suggest consideration of NSC637993 and/or C1311 for treatment of bladder cancer. By using human tumor tissue profiling information (such as that used here for breast cancer), the screening application of the COXEN algorithm could prove useful for discovery of candidate agents to treat other cancer types.
COXEN might also prove useful for determining subsets of patients or for “personalizing” their treatment. Currently, the hope is that gene expression profiles obtained from a patient's tumor can be compared with the expression profiles from other tumors of the same organ, grade, and stage to assist in prognosis and selection of therapy. The results described here for COXEN reinforce the idea that it is most advantageous to focus on the subset of probes that constitutes a signature of drug sensitivity. An additional possibility is the following: if, in the future, a drug has been found to be active in some patients with one type of cancer, its utility in at least some patients with a second type might be predicted by COXEN if both types have been molecularly profiled. In other words, the first type of cancer might provide a “training set” with at least some power to predict activity in the second, in line with the generic idea that drug response is influenced by signature molecular characteristics, not just by organ of origin. That strategy would be particularly useful with respect to orphan cancers for which clinical studies are insufficient and treatments are empirical.
The present study was conducted under at least two limitations that may not pertain to future applications. First, the available sample sizes were relatively small. Nevertheless, statistically significant results were achieved with 60 NCI-60 cell lines, 40 BLA-40 cell lines, and 84 breast cancer patients. Second, the microarray data from the breast cancers came from Affymetrix HG-U95 data in the docetaxel trial and custom cDNA arrays in the tamoxifen trial. Any lack of concordance introduced by differences in platform would presumably have confounded the predictions and made it harder, not easier, to obtain positive results. Similarly, any differences in factors such as cell culture conditions, cell heterogeneity, sample handling, purification of RNA, hybridization conditions, drug activity assay, or method of data analysis would be expected to have degraded our predictive results for the BLA-40. In the face of those limitations, the predictive results presented here demonstrate an important feature of the COXEN algorithm: generic algorithm steps 3 and 5 (which select probe sets related to drug activity and probe sets concordant between cell sets 1 and 2, respectively) tend to mitigate the influence of such confounding factors, making the overall algorithm and strategy quite robust. We also note that our COXEN approach needs to be further validated before its applications in clinical practice.
To share the COXEN algorithm with the scientific community, the development of a web-based COXEN system (www.coxen.org) is in progress, with which investigators with genomic profiling data from bladder and breast cancer cells or patient tumors can obtain chemosensitivity prediction results based on FDA-approved chemotherapeutic compounds.
Materials and Methods
Development of the COXEN Algorithm.
The COXEN algorithm consists of six distinct steps: input of drug activity and transcript expression profile data (steps 1, 2, and 4); identification of candidate “chemosensitivity biomarkers” in the NCI-60 panel (step 3); identification of coexpression extrapolation signatures (step 5); and development of chemosensitivity prediction models for the NCI-60 panel (step 6). These steps and the reagents used in them are discussed in detail in SI Materials and Methods.
Sensitivity of Human Bladder Cancer Cells to Cisplatin, Paclitaxel, and NSC637993 (Validation of COXEN Predictions).
To examine the performance of the COXEN prediction models objectively, we performed in vitro drug response experiments to determine the sensitivity of each bladder cell line to cisplatin and paclitaxel (Sigma, St. Louis, MO) and also to the compound NSC637993 (DTP; NCI; see SI Materials and Methods). The sensitivity to each agent (i.e., the GI50) was calculated from dose-response experiments carried out on the BLA-40 cells as described for the NCI-60 (http://dtp.nci.nih.gov). In each case, the cells were seeded in 96-well cell plates (Costar, Cambridge, MA) at a density of 1,000 cells per well on day 0, exposed to drug in triplicate from day 1 to 3 at 37°C, and then assayed fluorometrically using Alamar Blue aqueous dye (BioSource International, Camarillo, CA). Each experiment was repeated independently three to five times, and the results were expressed as the fractional difference between the initial cell count and the untreated control. Log10 (GI50) values were then estimated from the resulting dose–response curves (11, 23). Bladder cell lines were defined as sensitive or resistant, as described above for the NCI-60 panel.
Discovery of Candidate Anticancer Compounds from the NCI-60 Screening Data.
To identify candidates in the NCI public database of 45,545 compounds that might be active against bladder cancer cells, we automated and applied our COXEN algorithm with several additional filtering criteria. First, compounds with flat activity profiles across the NCI-60 were eliminated. Mathematically that filter was defined by the slope coefficient estimate from a simple linear regression for each drug compound for detecting/excluding nonspecific toxic or placebo compounds from our drug screening. Second, we excluded compounds that did not provide >10 probe sets capable of differentiating sensitive and resistant cell groups with statistical significance (e.g., significance analysis of microarrays false discovery rate <0.1).
Supplementary Material
Acknowledgments
We thank Drs. Henry Frierson, Garret Hampton, and Martin Safo for constructive suggestions and comments on the manuscript and Ms. Jill Johnson at the National Cancer Institute for greatly facilitating the investigation of the NCI-60 drug database and obtaining compounds. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (J.N.W.).
Abbreviations
- COXEN
coexpression extrapolation
- DTP
Developmental Therapeutics Program
- NCI
National Cancer Institute
- MiPP
misclassification-penalized posterior
- GI50
50% growth inhibition
- ROC
receiver operator characteristic.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610292104/DC1.
References
- 1.Goodman MT, Hernandez BY, Hewitt S, Lynch CF, Cote TR, Frierson HF, Jr, Moskaluk CA, Killeen JL, Cozen W, Key CR, et al. Hum Pathol. 2005;36:812–820. doi: 10.1016/j.humpath.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, et al. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- 3.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 4.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, Mohsin S, Osborne CK, Chamness GC, Allred DC, et al. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 6.Ma XJ, Patel R, Wang X, Salunga R, Murage J, Desai R, Tuggle JT, Wang W, Chu S, Stecker K, et al. Arch Pathol Lab Med. 2006;130:465–473. doi: 10.5858/2006-130-465-MCOHCU. [DOI] [PubMed] [Google Scholar]
- 7.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 8.Dressman HK, Berchuck A, Chan G, Zhai J, Bild A, Sayer R, Cragun J, Clarke J, Whitaker RS, Li L, et al. J Clin Oncol. 2007;25:517–525. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- 9.Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, Boyd MR. J Natl Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 10.Shi LM, Fan Y, Lee JK, Waltham M, Andrews DT, Scherf U, Paull KD, Weinstein JN. J Chem Inf Comput Sci. 2000;40:367–379. doi: 10.1021/ci990087b. [DOI] [PubMed] [Google Scholar]
- 11.Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A, et al. J Natl Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 12.Hayon T, Dvilansky A, Shpilberg O, Nathan I. Leuk Lymphoma. 2003;44:1957–1962. doi: 10.1080/1042819031000116607. [DOI] [PubMed] [Google Scholar]
- 13.Shankavarum UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, Reimers MA, Scherf U, Kahn A, Dolginow D, et al. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soukup M, Cho H, Lee JK. Bioinformatics. 2005;21(Suppl 1):i423–i430. doi: 10.1093/bioinformatics/bti1020. [DOI] [PubMed] [Google Scholar]
- 16.DeLong ER, DeLong DM, Clarke-Pearson DL. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 17.Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 18.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Fleming TR, Green SJ, Harrington DP. Exp Suppl. 1982;41:469–484. [PubMed] [Google Scholar]
- 20.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Qi S, et al. Proc Natl Acad Sci USA. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Den Brok MW, Nuijen B, Kettenes-Van Den Bosch JJ, Van Steenbergen MJ, Buluran JN, Harvey MD, Grieshaber CK, Beijnen JH. PDA J Pharm Sci Technol. 2005;59:285–297. [PubMed] [Google Scholar]
- 22.Hyzy M, Bozko P, Konopa J, Skladanowski A. Biochem Pharmacol. 2005;69:801–809. doi: 10.1016/j.bcp.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Shoemaker RH, Monks A, Alley MC, Scudiero DA, Fine DL, McLemore TL, Abbott BJ, Paull KD, Mayo JG, Boyd MR. Prog Clin Biol Res. 1988;276:265–286. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.