Abstract
Phylogenetic and geographic evidence suggest that many parthenogenetic organisms have evolved recently and have spread rapidly. These patterns play a critical role in our understanding of the relative merits of sexual versus asexual reproductive modes, yet their interpretation is often hampered by a lack of detail. Here we present a detailed phylogeographic study of a vertebrate parthenogen, the Australian gecko Heteronotia binoei, in combination with statistical and biophysical modeling of its distribution during the last glacial maximum. Parthenogenetic H. binoei occur in the Australian arid zone and have the widest range of any known vertebrate parthenogen. They are broadly sympatric with their sexual counterparts, from which they arose via hybridization. We have applied nested clade phylogeographic, effective migration, and mismatch distribution analyses to mitochondrial DNA (mtDNA) sequences obtained for 319 individuals sampled throughout the known geographic ranges of two parthenogenetic mitochondrial lineages. These analyses provide strong evidence for past range expansion events from west to east across the arid zone, and for continuing eastward range expansion. Parthenogen formation and range expansion events date to the late Pleistocene, with one lineage expanding from the northwest of its present range around 240,000 years ago and the second lineage expanding from the far west around 70,000 years ago. Statistical and biophysical distribution models support these inferences of recent range expansion, with suitable climatic conditions during the last glacial maximum most likely limited to parts of the arid zone north and west of much of the current ranges of these lineages. Combination of phylogeographic analyses and distribution modeling allowed considerably stronger inferences of the history of this complex than either would in isolation, illustrating the power of combining complementary analytical approaches.
Introduction
All vertebrate parthenogenetic lineages examined in any detail have been found to be quite young in evolutionary terms, typically being no more than one million years old and often much younger [1]. Recent origins are also suggested by the ‘twiggy’ taxonomic distribution of parthenogenetic organisms [2]–[4], which are taxonomically widespread but extremely ‘species’ poor within any given lineage [with very few exceptions–see 5]. Despite the apparently limited life-spans of most parthenogenetic lineages, they can potentially be very successful in the short term, as evidenced by their often broad geographic distributions and by molecular signatures of rapid range expansions [6]–[8]. Considerable effort has gone into explaining these patterns and their implications for the importance of sexual reproduction in evolution [4], [9]–[12], but interpretations have often been hampered by a lack of detailed phylogeographic data.
To properly understand the evolutionary dynamics of parthenogenesis, it is necessary to compare the amount and distribution of genetic variation in parthenogenetic lineages with that in closely related sexual lineages [1]. This can allow the identification of parental taxa [13] as well as provide information on the number of clonal origins [14], the ages of clonal lineages [15], and the proportion of genetic variation in parthenogens due to post-formation mutation [16]. Recently developed molecular markers and analytical techniques have allowed for more detailed and informative genetic and phylogeographic comparisons between sexual and asexual taxa [7], [17]–[19]. In addition, combination of phylogeographic approaches with analyses of ecological tolerances and interactions can permit cross-validation of phylogeographic inferences [20] and lead to considerably more insight into the underlying processes that generate the observed patterns of geographic distributions, amounts and distributions of genetic variation, and ecological and climatic correlates [e.g. 21], [22], [23].
Here we present a detailed phylogeographic analysis of parthenogenesis in a vertebrate, the Australian gecko Heteronotia binoei. We then combine this with high-resolution statistical [24] and biophysical [25] distribution models to make inferences of their likely distributions during the last glacial maximum (LGM). Parthenogenetic H. binoei have the largest range of any known vertebrate parthenogen, including extensive areas where they overlap with the ranges of their sexual counterparts. These attributes make them an appealing subject for the study of adaptation and evolutionary success in parthenogens, and of interactions between parthenogens and their parental taxa. Heteronotia binoei is a complex of several diploid sexual chromosome races and two mitochondrially distinct lineages of triploid parthenogenetic clones that formed via hybridization between two of the sexual chromosome races [26]. The CA6 and SM6 sexual chromosome races were involved in reciprocal hybridization events giving rise to the 3N1 and 3N2 (so named because they are triploid) parthenogenetic mtDNA lineages [27]. A third sexual chromosome race, EA6, was not involved in the hybridization events but is geographically widespread and sympatric with 3N1 parthenogens in part of its range. Numerous other sexual chromosome races are much more geographically localized and not as well characterized [26]. Parthenogenetic H. binoei exhibit substantial nuclear genetic diversity within each mtDNA lineage, mostly as a result of repeated backcrossing events between putative diploid female hybrids and sexual males [16].
Considerable work has been done characterizing the sexual and parthenogenetic taxa using cytology [28], allozymes [16], and mtDNA restriction profiles [6]. Our recent detailed phylogeographic study of the three widespread sexual races, including the two involved in the hybridization events [29], indicates that they diversified approximately 6 million years ago and expanded into the Australian arid zone during an extended period of gradual aridification throughout much of continental Australia. We have also presented molecular and distributional evidence that H. binoei and an invertebrate from the Australian arid zone, the grasshopper Warramaba virgo, have evolved hybrid parthenogenesis in parallel and in a strikingly similar fashion, both geographically and temporally [30]. Here we extend this work with an analysis of the origin, spread, and current population structure of parthenogenetic H. binoei using more powerful molecular markers and coalescent-based population genetic techniques. We consider the formation and expansion of the parthenogenetic lineages within the context of the last few glacial cycles, in which glacial intervals in much of continental Australia have been associated with increased aridity [31]. In addition, we compare our results to statistical [24] and biophysical [25] distribution analyses of the H. binoei complex, and extend these analyses to consider climatic conditions during the LGM. Our combined analyses allow for robust descriptions of the formation and expansion of H. binoei parthenogenetic lineages during the last two glacial cycles, and they suggest further avenues of research into the evolutionary dynamics of this complex.
Results
DNA sequences for all parthenogens ranged from 1283 to 1286 bases. Sequences were aligned manually, and at eight places gaps of one to two bases were inserted to keep all sequences in alignment. All indels occurred within or between adjacent tRNA genes. The aligned DNA sequences consisted of 1289 characters. Summary sequence diversity data for each lineage and for regions within lineages are shown in Table 1.
Table 1. Summary sequence information for each mtDNA lineage.
| Lineage/Region | N | # Haplotypes | % Nuc Div (SE) | Nesting Clades |
| 3N1 Total | 230 | 43 | 0.170 (0.065) | |
| 3N1 East Central | 48 | 12 | 0.085 (0.040) | 1-1, 1-2, 1-4 (2-1, 2-2) |
| 3N1 Far West | 17 | 2 | 0.034 (0.025) | 1-4, 1-5 (2-2) |
| 3N1 Northeast | 26 | 4 | 0.029 (0.018) | 1-1, 1-2 (2-1) |
| 3N1 Northwest | 10 | 2 | 0.047 (0.027) | 1-2, 1-4 (2-1, 2-2) |
| 3N1 Southeast | 21 | 3 | 0.104 (0.057) | 1-1, 1-2, 1-4 (2-1, 2-2) |
| 3N1 Southwest | 55 | 16 | 0.078 (0.029) | 1-3, 1-4, 1-6, 1-8, 1-9 (2-2) |
| 3N1 West Central | 52 | 13 | 0.062 (0.027) | 1-4, 1-7 (2-2) |
| 3N2 Total | 89 | 16 | 0.135 (0.050) | |
| 3N2 Central | 27 | 4 | 0.090 (0.051) | 1-2, 1-3, 1-6 (2-1, 2-3) |
| 3N2 Far West | 22 | 6 | 0.078 (0.037) | 1-2, 1-3 (2-1) |
| 3N2 Northeast | 15 | 7 | 0.203 (0.067) | 1-4, 1-5, 1-6, 1-7 (2-2, 2-3) |
| 3N2 Southeast | 24 | 4 | 0.110 (0.055) | 1-1, 1-2, 1-3, 1-6 (2-1, 2-3) |
Nested Clade Phylogeographic Analyses
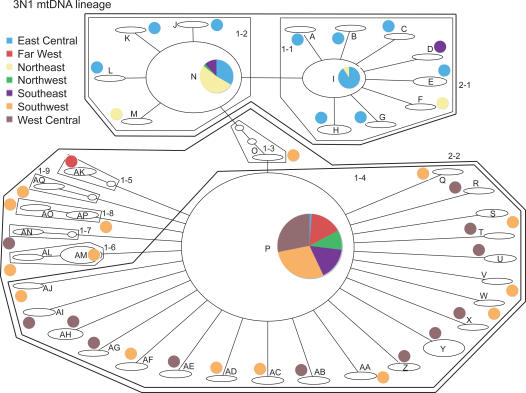
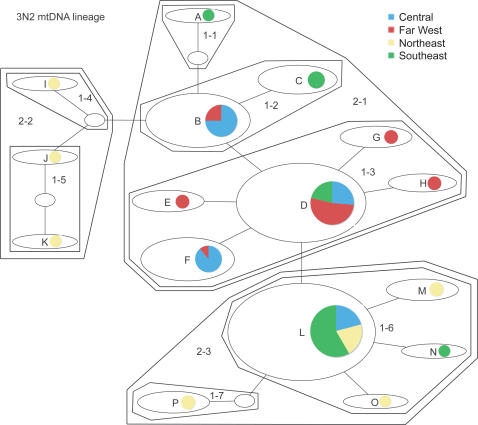
Haplotype networks for the 3N1 and 3N2 mtDNA lineages are shown in Figures 1 and 2, respectively. Geographic distributions of the nesting clades are given in Table 1. In the 3N1 lineage, clade 2–1 is almost exclusively (81 of 82 individuals) restricted to eastern populations, and clade 2–2 is mostly (133 of 148 individuals) restricted to western populations. In the 3N2 lineage, clade 2–2 is restricted to the northeastern part of the range, and clade 2–1 is restricted to the rest of the range; clade 2–3 occurs in all but the Far West region. Significant clade and nested clade distances and NCPA inferences for the two lineages are given in Table 2. Clades with nonsignificant distance values or for which the interpretation was ambiguous are not included.
Figure 1. Haplotype network for the 3N1 mtDNA lineage, showing nesting levels.
Clades correspond to those listed in Table 2. Small ovals without letter names are haplotypes not sampled but which are necessary to connect sampled haplotypes. Pie charts next to each haplotype indicate the proportion of individuals with that haplotype sampled from the various regions described in the analytical methods and Figure 7.
Figure 2. Haplotype network for the 3N2 mtDNA lineage, showing nesting levels.
Clades correspond to those listed in Table 2. Small ovals without letter names are haplotypes not sampled but which are necessary to connect sampled haplotypes. Pie charts next to each haplotype indicate the proportion of individuals with that haplotype sampled from the various regions described in the analytical methods and Figure 7.
Table 2. Results of NCPA for each mtDNA lineage.
| Clone | Clade | Chain of Inference | Result | Age (MYA) | C.I. (MYA) |
| 3N1 | 1-1 | 1-2-3-5-6-TOO FEW-7-YES | DRD in eastern part of range, range expansion to southeast | 0.06 | 0.001–0.23 |
| 1-2 | 1-2-3-4-NO | DRD in east/central part of range | |||
| 1-4 | 1-2-3-4-NO | DRD in western and central parts of range | |||
| 2-1 | 1-2-3-4-NO | DRD in eastern part of range | |||
| 2-2 | 1-2-3-4-NO | DRD in western and central parts of range | |||
| Total | 1-19-20-2-11-12-13-YES | Range expansion from west to east, possibly followed by some fragmentation between east and west | 0.24 | 0.025–0.73 | |
| - | - | Age based on divergence from CA6 mtDNA | 2.65 | 0.54–6.67 | |
| 3N2 | 1-2 | 1-2-3-4-NO | DRD in west and central | ||
| 1-3 | 1-2-11-12-NO | Range expansion from west/central to north/west parts of range | 0.06 | 0.001–0.23 | |
| 1-6 | 1-2-11-12-NO | Range expansion from south/central to north/east parts of range | 0.06 | 0.001–0.23 | |
| 2-1 | 1-2-3-4-NO | DRD throughout most of range | |||
| 2-3 | 1-2-3-4-NO | DRD in central, northern, and eastern parts of range | |||
| Total | 1-2-11-12-NO | Range expansion from west/central to north/east parts of range | 0.07 | 0.006-0.22 | |
| - | - | Age based on divergence from SM6 mtDNA | 1.21 | 0.21-3.20 |
Only clades with significant values are shown. DRD = dispersal restricted by distance. For dates and confidence intervals, point estimates are based on an estimate of 1.3% sequence divergence per million years for this portion of the mtDNA genome. 95% Confidence intervals are based on a gamma probability distribution for coalescence time and a range of 1.22-1.4% divergence per million years (see methods). MYA = million years ago.
For the 3N1 lineage, most inferences at low and intermediate nesting levels are dispersal restricted by distance. There is evidence for recent range expansion into the narrow southeastern portion of its range, where this lineage coexists with EA6 sexuals. The oldest inference is a range expansion from west to east, corresponding to the initial expansion following their formation in the west via hybridization between the CA6 and SM6 sexuals. This event is dated at 0.24 MYA (range 0.025–0.73 MYA). The fact that lower and intermediate nesting levels have signatures of dispersal restricted by distance, including in the central and eastern parts of the range, suggests that any continuing range expansion is relatively slow. However, it does still appear to be occurring at the eastern edge of the range, as evidenced by the inference of range expansion to the southeast in clade 1–1 (Table 2). There is also evidence at the highest nesting level for fragmentation between eastern and western 3N1 populations.
In the 3N2 lineage, there are inferences of dispersal restricted by distance at both lower and intermediate nesting levels. However, there are also several inferences of range expansion at multiple nesting levels. These inferences suggest that 3N2 parthenogens were formed in roughly the southern or western portion of their current range approximately 0.07 MYA (range 0.006–0.22 MYA) and have been spreading, and are continuing to spread, to the north and east.
Dates for origins of the 3N1 and 3N2 lineages based solely on their mtDNA divergence from the mostly closely related sampled CA6 and SM6 mtDNA haplotypes, respectively, are 2.65 MYA for 3N1's (range 0.54–6.67 MYA) and 1.21 MYA for 3N2's (range 0.21–3.20 MYA), suggesting that they are older than their respective earliest NCPA inferences. Dates for NCPA-inferred initial range expansions are lower bounds for the ages of each event, because dating is based on the youngest monophyletic clade of the haplotype network for which the inference of range expansion applies [20]. However, the limited mtDNA diversity within each lineage makes it very unlikely that they are as old as their divergence from related sexual haplotypes would suggest. A selective sweep within each group is an unlikely explanation for this limited diversity because nuclear backcross clonal diversity is much higher (16, and Strasburg and Kearney in prep), and since the cytoplasmic and nuclear genomes are in complete linkage in these parthenogens, a sweep in one would affect the other as well. The most likely explanation is that more closely related sexual haplotypes were not sampled. This is a plausible explanation because regional divergence among CA6 and SM6 mtDNA populations can be as much as 4–5% and 7–8%, respectively [29]. Minimum divergence between 3N1 and CA6 haplotypes is 4–5%, and between 3N2 and SM6 haplotypes it is 2–3%.
Effective Migration
The validity of NCPA for inferring population structure and historical events has been questioned [32], [33]. Although many of these criticisms have been rebutted [34], the inherent uncertainty in any such analysis warrants multiple alternative methods of inference. Consequently, we have also implemented coalescent-based analyses of effective migration in addition to more traditional distance-based analyses.
Results from coalescent-based migration analyses are shown in Table 3. Effective migration rate estimates generally had very large confidence intervals, with lower ends of those intervals often far below 0.1, suggesting that a high degree of subdivision among these particular regions cannot be rejected. Only effective migration rate estimates with confidence intervals completely above 0.1 are considered significant.
Table 3. Effective migration rates (average number of effective migrants per generation) among regions within each mtDNA lineage.
| Clone/Source Region | Receiving Region | ||||||
| 3N1 | East Central | Far West | Northeast | Northwest | Southeast | Southwest | West Central |
| East Central | - | 0–0.14 | 0.42–2.13 | 0–0.07 | 0.05–4.06 | 0–0.65 | 0–0.55 |
| Far West | 0–0.17 | - | 0–0.34 | 0.05–0.21 | 0.69–7.17 | 4.99–11.36 | 0–0.55 |
| Northeast | 0–0.17 | 0–0.14 | - | 0–0.03 | 1.15–8.57 | 0–0.65 | 0–0.55 |
| Northwest | 0–0.17 | 1.64–3.30 | 0–0.34 | - | 5.93–18.60 | 0–0.65 | 3.55–8.57 |
| Southeast | 0–0.17 | 0–0.14 | 0–0.34 | 0.03–0.17 | - | 0–0.65 | 0–0.55 |
| Southwest | 0.01–0.39 | 0–0.14 | 0–0.34 | 0–0.03 | 0–1.77 | - | 0–0.55 |
| West Central | 0–0.17 | 0–0.14 | 0–0.34 | 0–0.03 | 0.05–4.06 | 0–0.65 | - |
| 3N2 | Central | Far West | Northeast | Southeast | |||
| Central | - | 1.27–4.19 | 0–0.68 | 0–0.22 | |||
| Far West | 0–0.19 | - | 0–0.68 | 0.14–1.06 | |||
| Northeast | 0 | 0 | - | 0 | |||
| Southeast | 0.03–0.51 | 0–0.43 | 1.06–4.79 | - | |||
Values shown are 95% confidence intervals for Nefm = effective migration rate = inbreeding effective population size times proportion of individuals migrating. Direction of migration is from the region listed at left to the region listed across the top. Significant migration events (defined as those estimates whose confidence intervals are completely above 0.1) are shown in bold italics.
Effective migration results for 3N1 strongly support NCPA inferences of formation in the western portion of the range and spread to the east and southeast. All significant migration occurs within the western portion of the range or from west to east; no migration was inferred out of the southeast or from east to west. This highly asymmetric migration includes significant migration inferred from most other regions, and from the Northwest region in particular, to the Southeast region, where NCPA inferred a recent and possibly continuing range expansion event.
While it is clear from NCPA and from phylogenetic relationships between 3N1 and CA6 haplotypes that 3N1 parthenogens originated in the western portion of their range, neither analysis offers a more precise estimate of location. These migration analyses suggest that the most likely location of origin is the northern part of the western portion of the range (the Northwest region). There has been asymmetric migration from this region to the far western part of the range, and to the east and southeast, with no evidence of significant migration into the Northwest region.
Migration analysis of 3N2 is also concordant with NCPA inferences, which suggested an origin in the southern or western portion of their range and subsequent spread to the north and east. There has been significant migration from western regions to the southeast, and migration from the southeast to the northeast.
Mismatch Distributions, Analyses of Molecular Variance
Other evidence for population growth can be obtained from an examination of the distributions of pairwise differences among haplotypes, or mismatch distributions [35], [36]. Populations that have undergone or are undergoing periods of growth tend to have a unimodal distribution of pairwise differences, with the mode shifting to the right with time following growth. Conversely, stable populations tend to show multimodal “ragged” mismatch distributions [37].
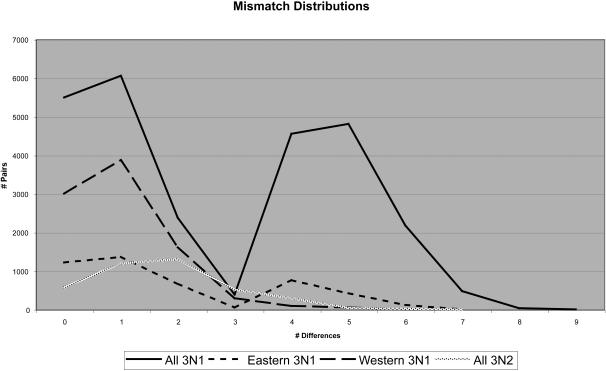
Graphs of mismatch distributions for each mtDNA lineage, and for eastern and western portions of the 3N1 range, are shown in Figure 3. In each case, the distribution is clearly unimodal or bimodal. The 3N2 clone has a strongly unimodal mismatch distribution; the estimate of τ, time since expansion measured in units of 1/(2u) generations [where u is total substitution rate over all sites–38], based on the least-squares method implemented in Arlequin is 1.82, and a sudden expansion model cannot be rejected for this data. Based on our substitution rate estimate of 0.65% per lineage per million years, this corresponds to a timing of approximately 0.11 MYA for the initial expansion of 3N2's following formation. This time is very consistent with the estimate made based on the NCPA inference of northward and eastward expansion (0.07 MYA, range 0.006–0.22 MYA). In addition, estimates of Tajima's [39] D and Fu's [40] Fs were significantly negative, indicating population expansion.
Figure 3. Mismatch distributions for 3N1 and 3N2 lineages.
Distributions are shown for all 3N1 or 3N2 populations together and for eastern and western 3N1 populations.
The overall 3N1 distribution shows two peaks, at one and five differences. The peak at five differences corresponds to a timing of approximately 0.30 MYA, which coincides well with the timing of the initial expansion event inferred by NCPA (0.24 MYA, range 0.025–0.73 MYA). The peak at one difference corresponds to a timing of approximately 0.06 MYA, which is the same time as the estimate for range expansion to the southeast portion of the range inferred by NCPA (0.06 MYA, range 0.001–0.23 MYA). Further examination of Figure 3 reveals that this bimodality is due to eastern 3N1 individuals, while western 3N1 individuals show a unimodal mismatch distribution. This bimodality is likely the result of contraction/fragmentation during the LGM and subsequent continued expansion to the east and south.
The estimate of τ for western 3N1 is 1.04, corresponding to 0.06 MYA for this expansion-considerably more recent than estimates for eastern expansion. Based on a variety of evidence (NCPA, effective migration, affinity of 3N1 mtDNA with western CA6 mtDNA, and affinity of many 3N1 chromosomal and allozyme variants with western CA6 and SM6 variants–16), it is clear that the 3N1 mtDNA clone originated in western Australia. Therefore, this expansion in western 3N1 may also reflect Holocene expansion following contraction during the LGM. Eastern, western, and overall 3N1 fit a sudden expansion model of population growth. Tajima's D is significantly negative for western and overall 3N1 (western D = −2.38, p<0.0001; overall D = −1.81, p = 0.006), and Fu's Fs is significantly negative for all three groups (eastern Fs = −6.57, p = 0.009; western Fs<−100, p<0.0001; overall Fs = −10.65, p = 0.002).
Results from AMOVA of mtDNA for each lineage are shown in Table 4. Groups for AMOVAs are the same regions that were used for effective migration analyses. For both mtDNA lineages, among region, within region, and within population comparisons all explain a significant portion of genetic variation. However, the distribution of variation is quite different between the two lineages. Relatively little variation is distributed among regions in the 3N2 lineage, and most of the remaining variation is found within populations; this is consistent with the more recent origin of the 3N2's and their comparatively small range. In the 3N1 lineage, more than two thirds of the variation is distributed among regions. However, in an AMOVA with eastern and western populations as the groups, an almost identical amount of variation (66.5%) is distributed between groups, suggesting that almost all of this regional variation is distributed between eastern and western populations. This is consistent with the NCPA inference of a possible relatively old fragmentation event between eastern and western populations. Within regions, almost all variation is found within rather than among populations.
Table 4. AMOVA results for mtDNA sequence.
| Race | Source | d. f. | Sum Sq. | % Variation | P |
| 3N1 | Among Regions | 6 | 195.471 | 66.75 | <0.00001 |
| Among Pops | 34 | 24.751 | 3.30 | 0.00131 | |
| Within Pops | 188 | 85.909 | 29.95 | <0.00001 | |
| Total | 228 | 306.131 | |||
| 3N2 | Among Regions | 3 | 13.844 | 13.80 | 0.00875 |
| Among Pops | 14 | 24.250 | 28.42 | <0.00001 | |
| Within Pops | 70 | 36.474 | 57.78 | <0.00001 | |
| Total | 87 | 74.568 |
Mantel tests [41] of correlation between geographic distance and genetic distance were performed on each mtDNA lineage as a whole and within the 3N1 mtDNA lineage for eastern and western populations separately. For all tests, there is a significant correlation between geographic and genetic distance. In the 3N1's, the correlation was lowest (but still significant) among western populations (western 3N1 r = 0.19, p = 0.049; eastern 3N1 r = 0.47, p = 0.002; overall 3N1 r = 0.53, p<0.0001; 3N2 r = 0.30, p = 0.003). Mantel tests were also run on the same regions used in previous analyses, but almost all results were not significant even if correlation coefficients were high, most likely due to small sample sizes.
Distribution Modeling
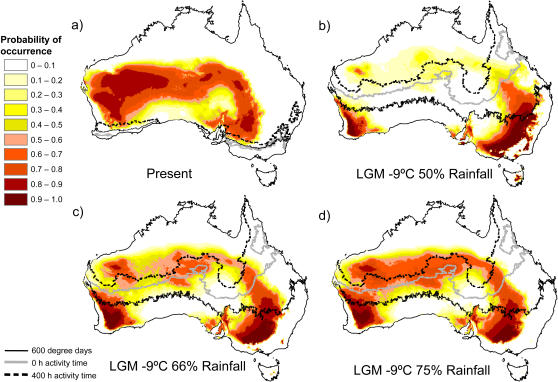
Kearney et al. [24] found that the current distribution of parthenogenetic H. binoei coincides fairly closely with their expected distribution based on correlations with six temperature and rainfall variables in western Australia, while considerable similar but unoccupied habitat exists in central and eastern Australia. Taking a more mechanistic approach, Kearney and Porter [25] found that the current southern distribution of the H. binoei complex is partially limited by temperature requirements for successful egg development and foraging activity. Here we have applied these approaches using estimates of climatic conditions during the LGM. Average air temperatures in the interior of Australia were around 9°C cooler 16–45 KYA than at present [42]. The arid zone was also considerably drier during the LGM, although estimates of the degree of aridification vary [31]. Predicted correlative distribution models for parthenogenetic H. binoei and biophysical predictions for the temperature limits for successful egg developments and minimal foraging activity are shown in Figure 4. Three scenarios are presented, with mean annual rainfall reductions of 1/2, 1/3, and 1/4 (all with a 9°C average temperature reduction).
Figure 4. Statistical distribution models for parthenogenetic Heteronotia binoei.
Models are for (a) present conditions and (b–d) last glacial maximum conditions with a 9°C reduction in mean air temperature and three different rainfall reduction scenarios. All statistical models are based on the AICc model reported in Kearney et al (2003). Overlayed on the predicted distributions are the contours for biophysical predictions of the limit for successful development of eggs (600 degree days), of the limit for potential activity time (0 hours) and of the number of hours of potential activity at the current distribution limit of the Heteronotia complex (400 hours). Any regions roughly south of the contours are outside the fundamental niche of Heteronotia. The biophysical predictions use either (a) current climatic conditions or (b–d) a 9°C decrease in monthly maximum and minimum temperatures. Methods for biophysical predictions are described in Kearney and Porter (2004).
Probability of occurrence based on correlations with temperature and rainfall variables decreases dramatically throughout much of the interior of Australia under all three scenarios; probability density is shifted to southeastern and southwestern Australia, where rainfall amounts are similar to current levels in the interior. However, the 9°C temperature decrease shifts the contours for biophysical predictions of minimal temperatures for successful egg development and foraging far to the north. Under our assumptions of temperature and rainfall conditions during the LGM, and assuming that climatic correlates and biophysical requirements of current H. binoei are comparable to those of the LGM, the regions where they were most likely to persist during the LGM were the northwest and north-central parts of the arid zone.
Discussion
Phylogeographic History of H. binoei Parthenogens
NCPA of the 3N1 and 3N2 parthenogens reveal a recent origin of each lineage and subsequent spread to the east and south (3N1) and east and north (3N2). Dating estimates of the oldest NCPA inferences, which correspond to initial expansion following formation, put the 3N1 expansion at 0.24 MYA (range 0.025–0.73 MYA), and the 3N2 expansion at 0.07 MYA (range 0.006–0.22 MYA). As mentioned above, these dates for NCPA inferences of initial range expansion are based on the youngest monophyletic clade of the haplotype network for which the inference of range expansion applies [20], which in each case corresponds to one or more of the highest level nesting clades; thus these range expansion dates set lower bounds for the ages of each lineage. There is no evidence suggesting that there would have been any substantial delay between formation and range expansion in either lineage, and we expect dates for formation to be close to dates for initial range expansion. Analyses of effective migration support northwestern and southwestern or central-western origins for the 3N1 and 3N2 lineages, respectively. NCPA and effective migration also indicate recent and possibly ongoing expansions to the southeast in 3N1's and to the east in 3N2's, and mismatch distributions also suggest rapid population growth in each lineage. Coalescent analyses of effective population growth show that overall, and in most regions, populations of both parthenogens are growing very quickly, as would be expected under a scenario of recent and rapid range expansion (data not shown).
There is also evidence at the highest NCPA nesting level for fragmentation between eastern and western 3N1 populations. In addition, AMOVA using the eastern and western areas as groups reveals a large amount (66%) of variation distributed between groups, and eastern and western haplotypes are mostly segregated at the highest levels of nesting in the haplotype network (Figure 1). However, analyses of effective migration (Table 3) provide no evidence of east/west fragmentation; in fact, there is strong signal of west to east migration. Sampling in the middle portion of the 3N1 range is somewhat sparse in comparison to more eastern and western areas, and this may be partially responsible for an inference of fragmentation; more sampling in the region may reveal intermediate haplotypes and more continuity between east and west. While this fragmentation inference may be considered slightly tentative, it is interesting that the predicted distribution of parthenogenetic H. binoei during the LGM under 33% and 50% rainfall reduction scenarios is somewhat discontinuous in this region (Figure 4), with an area of low probability of occurrence, corresponding roughly with the fragmentation event, separating two areas of higher probability of occurrence (see “Distribution Modeling” below).
Based on the mtDNA restriction profiles showing an affinity of 3N2 mtDNA with a clade of SM6 haplotypes from the extreme western part of their range along the northwest coast of Australia, Moritz and Heideman [27] concluded that the 3N2 mtDNA lineage had likely originated in the northwestern part of its range (see Figure 2 in 26). Under this scenario, 3N2 parthenogens then spread to the east and south to occupy their current range. However, based on our mtDNA sequence data [29] this SM6 clade also includes a haplotype sampled from near Shark Bay at the west-central edge of the 3N2 range. No other SM6 individuals were sampled within 400 km of this population (see Figure 2, 29), so it could well be a remnant population from a more southern historical distribution of this race. It seems likely that the CA6 and/or SM6 ranges in this area have changed substantially due to Pleistocene climatic changes (see below), and these range shifts facilitated hybridization between ecologically and genetically distinct races in this group [29], [43]. This, combined with NCPA and effective migration analyses showing range expansion and movement to the east and north in the 3N2 lineage, make it most likely that the 3N2 parthenogens were formed in the west-central or southwest part of their current range.
Distribution Modeling
Modeling of the climatic correlates of H. binoei distributions and biophysical modeling of limits for successful egg development and minimal foraging time strengthen our phylogeographic scenario. Kearney et al. [24] analyzed the bioclimatic envelopes of each asexual lineage and found that six climatic variables related to temperature and rainfall fairly accurately describe the distributions of each lineage in the western parts of their ranges, but that in each case large areas of climatically similar habitat exist to the east of their current ranges (see Figure 9 in 24). This result is in agreement with our inference of recent and continuing eastward expansion within each lineage. Concordance between the predicted 3N1 range based on these climatic variables and our inferred range expansion is especially striking–an uninhabitable area in the Lake Eyre basin around northeast South Australia, southeast Northern Territory, and southwest Queensland is mostly surrounded by more suitable habitat (see Figure 9 in 24), and the southern part of this circle corresponds to the recent southeastern range expansion and our predicted continuing expansions (Figure 5).
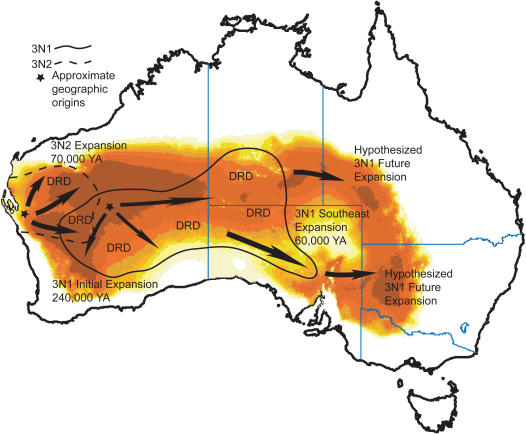
Figure 5. Proposed origin and spread of 3N1 and 3N2 parthenogens.
Also shown are timing estimates for expansions and hypothesized future expansions in 3N1 parthenogens. Phylogeographic events are overlaid on the predicted distribution for parthenogenetic Heteronotia binoei based on a statistical distribution model for present climatic conditions [24]. Times given here are point estimates; confidence intervals are given in Table 2. DRD = dispersal restricted by distance.
It is significant that the 3N1 lineage has not expanded further into the southwestern part of Australia, an area where no H. binoei exist. Kearney and Porter [25] showed that in many places the southern limit of the range of the EA6 sexual chromosome race (the most southerly distributed chromosome race) coincides very closely with the thermal limit for successful egg development; similar climatic constraints on the 3N1 southern distribution are likely to be in place.
We repeated these correlative analyses for both parthenogenetic lineages combined, under current climatic conditions as well as under three different scenarios for the LGM–a uniform 9°C decrease in average air temperature along with rainfall reductions of 25%, 33%, and 50% (Figure 4). During the LGM, rainfall conditions similar to those in present-day parthenogenetic H. binoei ranges would mostly have been restricted to extreme southwestern and southeastern Australia. Rainfall was strongly weighted in the correlative distribution model for parthenogenetic Heteronotia (Kearney et al. 2003) hence the prediction for a significant southward shift in the distribution.
We have also extended the biophysical modeling of Kearney and Porter [25] to include these LGM scenarios (overlaid contour lines on Figure 4b–d for the 600 degree days necessary for successful egg development and the zero and 400 hours potential activity time contours). Correlative distribution model predictions are discordant with those of the biophysical model, which shows that most of the areas of highest probability density in the correlative model are well south of the 600 degree day and zero hours potential activity contour lines, and so would likely have been outside the fundamental niche of H. binoei based on these biophysical requirements [25]. Regions of most similar habitat north of the contour lines are found in the northwest and north-central parts of the arid zone, and for the 33% and 50% rainfall reduction scenarios they are separated by an area of somewhat less similar habitat. The absence of extremely cold and arid environments in Australia at present is presumably why extrapolation of the regression model results in a biologically unrealistic prediction.
It is particularly interesting to note that the biophysical model predicts potential activity time to be more limiting than potential egg development time during glacial maxima, whereas the reverse is true under current climatic conditions. This occurs because egg development rate in the soil is affected by solar radiation and air temperature, while potential activity time in this nocturnal lizard is solely affected by air temperature. Potential activity time is more severely affected because our modeling assumes that the air temperature changes between glacial cycles but solar radiation does not. In this respect, it may be significant that parthenogenetic H. binoei have evolved greater aerobic endurance at low temperature when compared with their sexual relatives [44].
Concordance between phylogeographic analyses and distribution modeling
While our modeling for the LGM is somewhat crude in that it assumes geographically uniform changes in temperature and rainfall (probably not a realistic assumption–31), it is in substantial agreement with our phylogeographic results, summarized in Figure 5. We have inferred an origin of the 3N1 mitochondrial lineage approximately 240,000 years ago, likely during the previous glacial cycle, in the northwest part of its range. This would have been near the southern limit of the fundamental niche of the Heteronotia complex (assuming roughly similar conditions during the glacial maximum previous to the LGM), and it is reasonable to expect that the CA6 and SM6 sexual races would have come into contact in this region as the range of each was contracted northward. Following some, mostly eastward, expansion, the 3N1 range contracted to the northwest and north-central arid zone during the LGM, possibly into two disjunct regions (see Figure 4b–d). This is a likely cause of the fragmentation event inferred at higher levels of nesting in the 3N1 NCPA. Also during the LGM, the 3N2 parthenogens were formed via a second period of contact and hybridization between the CA6 and SM6 races in Western Australia. Under this scenario, the range of the SM6 sexuals during the LGM extended further to the south in this area, and the population from Shark Bay is a remnant of this southern range.
Results for both 3N1 and 3N2 lineages suggest that abiotic factors may play the most important role in determining their geographic distributions. However, it is worth pointing out that both lineages appear to still be expanding their ranges, and so are likely in a non-equilibrium state. In addition, Moritz et al. [45] found much higher rates of infection by parasitic mites for parthenogenetic H. binoei sampled throughout their range relative to their sexual counterparts. Studies of the environmental and physiological tolerances of different parthenogenetic clones are underway (Kearney and Strasburg in prep), and further studies involving direct competition and transplant experiments will help strengthen inferences of limiting factors in parthenogen distributions.
The Australian arid zone is home to a diverse array of hybrid parthenogens [reviewed in 46], and those that have been studied in detail also appear to have late Pleistocene origins [30], [47]. Many explanations have been put forth for the persistence of parthenogens in the arid zone and elsewhere [9], [10], [48]–[50], and the role of climatic cycling in hybridization is well-documented [51]. It may be the case that similar climatic conditions have driven the hybridization events resulting in other arid zone parthenogens, and that similar factors constrain their distributions. We were able to make robust inferences about the history of the H. binoei complex in relation to climatic cycles by combining population genetic approaches with climatic and biophysical distribution modeling. This methodology should also be very valuable for understanding the prevalence of hybrid parthenogenesis in the Australian arid zone, and for addressing the role of abiotic factors in the formation, spread, and persistence of parthenogenetic lineages more generally.
Materials and Methods
Field
Our analyses are based on 319 specimens of parthenogenetic H. binoei, encompassing the ranges of the two mtDNA lineages known as 3N1 and 3N2. Of these samples, 127 were collected in the 1980's and early 1990's [26] and 192 were collected in 2000–2001 (Table 5 and Figure 6). In some cases, nearby populations with small population sizes were combined for analyses. For the 2001 collections, representative individuals from each population were euthanized for voucher specimens, and for the rest tail tips were taken and the individuals were released. Voucher specimens are deposited in the South Australian Museum, Australian National Wildlife Collection, Queensland Museum, and University of Michigan Museum of Zoology (for individuals collected by C. Moritz), and in the Western Australian Museum (for individuals collected in 2001). Museum catalog numbers for voucher specimens are given in Table 5.
Table 5. Sampling localities, sample sizes, and mtDNA haplotypes sampled at each locality.
| mtDNA Lineage | |||||||
| Locality | Latitude | Longitude | 3N1 | 3N2 | Museum Catalog Numbers | GenBank Accession Numbers | Haplotypes |
| 17kms S Leonora WA | −29.033 | 121.317 | 1 | ABTC32512 | DQ287414 | 3N1-P | |
| 22km NE Bonnie Rock WA | −30.383 | 118.500 | 1 | ABTC31222 | DQ287372 | 3N1-P | |
| 4km W Johnston Rocks WA | −29.800 | 119.833 | 1 | ABTC31220 | DQ287371 | 3N1-P | |
| Aileron NT | −22.648 | 133.352 | 5 | DQ287444, DQ287463, DQ287470, DQ287473, DQ287475 | 3N1-N (5) | ||
| Alice Springs NT | −23.700 | 133.867 | 2 | ABTC32565, ABTC32566 | DQ287419, DQ287420 | 3N1-N (2) | |
| Avoca Downs Station | −30.950 | 122.318 | 4 | R144987 | DQ287495, DQ287544, DQ287550, DQ287589 | 3N1-AD (2), 3N1-AM (2) | |
| Bandya WA | −27.550 | 121.828 | 7 | R146801 | DQ287496, DQ287498, DQ287516, DQ287521, DQ287531, DQ287545, DQ287571 | 3N1-P (2), 3N1-T, 3N1-Y, 3N1-Z (2), 3N1-AN | |
| Belele WA | −26.367 | 118.017 | 5 | ABTC32361, ABTC32373, ABTC32445, ABTC32457, ABTC32461 | DQ287604, DQ287608, DQ287626, DQ287632, DQ287633 | 3N2-B (2), 3N2-D (3) | |
| Billabalong WA | −27.417 | 115.833 | 3 | ABTC32371, ABTC32374, ABTC32385 | DQ287607, DQ287609, DQ287613 | 3N2-D (2), 3N2-E | |
| Billabong WA | −26.817 | 114.617 | 4 | ABTC32444, ABTC32526, ABTC32530, ABTC32542 | DQ287625, DQ287641, DQ287643, DQ287647 | 3N2-D (2), 3N2-E (2) | |
| Boologooroo WA | −24.333 | 114.033 | 2 | ABTC32378, ABTC32379 | DQ287610, DQ287611 | 3N2-H (2) | |
| Brickhouse WA | −24.817 | 113.783 | 5 | ABTC32796, ABTC32800, ABTC32916, ABTC33076, ABTC33078 | DQ287655, DQ287656, DQ287667, DQ287673, DQ287674 | 3N2-B (2), 3N2-G (3) | |
| Bulga Downs WA | −28.494 | 119.739 | 5 | R146770 | DQ287560, DQ287563, DQ287570, DQ287575, DQ287576 | 3N1-P (5) | |
| Bullabulling WA | −31.017 | 120.867 | 3 | ABTC32364, ABTC32470, ABTC32490 | DQ287385, DQ287403, DQ287411 | 3N1-P, 3N1-AC (2) | |
| Bullardoo Stn WA | −27.850 | 115.667 | 2 | 2 | ABTC32458, ABTC32471, ABTC32386, ABTC32447 | DQ287400, DQ287404, DQ287614, DQ287628 | 3N1-P (2), 3N2-D (2) |
| Bundarra Stn WA | −28.317 | 121.167 | 2 | 2 | ABTC32504, ABTC32534, ABTC32507, ABTC32537 | DQ287413, DQ287416, DQ287637, DQ287645 | 3N1-AB (2), 3N2-L (2) |
| Coondambo SA | −31.060 | 135.865 | 3 | ABTC32440 | DQ287398, DQ287456, DQ287488 | 3N1-P (3) | |
| Copper Hill SA | −27.950 | 134.313 | 1 | DQ287446 | 3N1-A | ||
| Cundeelee WA | −30.720 | 123.422 | 11 | R146757, R146792 | DQ287494, DQ287517, DQ287526, DQ287553, DQ287557, DQ287558, DQ287569, DQ287572, DQ287586, DQ287587, DQ287597 | 3N1-P (5), 3N1-AA (2), 3N1-AM (4) | |
| Cunyu WA | −26.017 | 120.117 | 1 | ABTC32404 | DQ287387 | 3N1-P | |
| Curbur WA | −26.467 | 115.933 | 4 | ABTC32354, ABTC32446, ABTC32450, ABTC32528 | DQ287602, DQ287627, DQ287630, DQ287642 | 3N2-D (3), 3N2-F | |
| Dalgetty Downs WA | −25.283 | 116.200 | 5 | ABTC32370, ABTC32387, ABTC33128, ABTC33130, ABTC33131 | DQ287606, DQ287615, DQ287675, DQ287676, DQ287677 | 3N2-D (2), 3N2-F (3) | |
| De Rose Hill SA | −26.417 | 133.250 | 2 | ABTC32431, ABTC32522 | DQ287394, DQ287415 | 3N1-I, 3N1-L | |
| Deep Well NT | −24.298 | 134.133 | 4 | DQ287465, DQ287468, DQ287471, DQ287600 | 3N1-I, 3N1-N (3) | ||
| Earaheedy WA | −25.550 | 121.583 | 1 | ABTC32397 | DQ287386 | 3N1-P | |
| East edge of Yeo Lake WA | −28.236 | 124.673 | 1 | DQ287542 | 3N1-P | ||
| East of Yeo WA | −28.136 | 124.465 | 7 | R146769, R146781, R146803, R146804, R146808 | DQ287504, DQ287512, DQ287537, DQ287549, DQ287564, DQ287581, DQ287596 | 3N1-P (3), 3N1-Y (4) | |
| Glenayle WA | −25.267 | 122.033 | 1 | ABTC31426 | DQ287381 | 3N1-P | |
| Goongarrie WA | −29.984 | 121.044 | 6 | R145049 | DQ287497, DQ287503, DQ287515, DQ287533, DQ287574, DQ287585 | 3N1-P (3), 3N1-AF (2), 3N1-AP | |
| Granite Downs SA | −26.937 | 133.495 | 5 | ABTC32435, ABTC32486 | DQ287396, DQ287409, DQ287452, DQ287486, DQ287490 | 3N1-E, 3N1-I, 3N1-N (3) | |
| Granite Peak WA | −25.633 | 121.350 | 1 | ABTC32541 | DQ287646 | 3N2-N | |
| Hampton Hill WA | −30.762 | 121.737 | 4 | R146787 | DQ287519, DQ287556, DQ287598, DQ287599 | 3N1-S (2), 3N1-AO, 3N1-AP | |
| Jimba Jimba WA | −25.033 | 115.133 | 1 | ABTC32930 | DQ287670 | 3N2-B | |
| Juna Downs WA | −22.883 | 118.483 | 4 | ABTC32773, ABTC32819, ABTC32820, ABTC32834 | DQ287652, DQ287659, DQ287660, DQ287662 | 3N2-L (3), 3N2-O | |
| Kathleen Valley WA | −27.400 | 120.650 | 1 | ABTC32509 | DQ287638 | 3N2-L | |
| Kingoonya SA | −30.912 | 135.315 | 2 | DQ287437, DQ287482 | 3N1-P (2) | ||
| Kirkalocka WA | −28.555 | 117.777 | 1 | 2 | ABTC32408, ABTC32417, ABTC32525 | DQ287389, DQ287622, DQ287640 | 3N1-P, 3N2-L (2) |
| Lake Violet WA | −26.533 | 120.667 | 2 | ABTC32418, ABTC32421 | DQ287390, DQ287392 | 3N1-P (2) | |
| Leinster Downs WA | −27.850 | 120.600 | 3 | ABTC32349, ABTC32426, ABTC32505 | DQ287601, DQ287624, DQ287636 | 3N2-L (3) | |
| Lilla Ck. NT | −25.567 | 134.067 | 1 | ABTC31643 | DQ287383 | 3N1-I | |
| Mandilla WA | −31.376 | 121.537 | 1 | DQ287566 | 3N1-AJ | ||
| Mt Augusta WA | −24.300 | 116.917 | 4 | ABTC32847, ABTC32929, ABTC32932, ABTC32934 | DQ287665, DQ287669, DQ287671, DQ287672 | 3N2-F (4) | |
| Mt Cavenagh NT | −25.915 | 133.133 | 5 | DQ287432, DQ287433, DQ287443, DQ287462, DQ287476 | 3N1-I (5) | ||
| Mt Ebenezer NT | −25.100 | 132.567 | 1 | ABTC31384 | DQ287378 | 3N1-J | |
| Mt Gould WA | −25.800 | 117.383 | 4 | ABTC32358, ABTC32367, ABTC32390, ABTC32449 | DQ287603, DQ287605, DQ287617, DQ287629 | 3N2-B (3), 3N2-F | |
| Mt Willoughby SA | −27.958 | 134.145 | 7 | ABTC32436, ABTC32441, ABTC32469 | DQ287397, DQ287399, DQ287402, DQ287449, DQ287454, DQ287458, DQ287474 | 3N1-E, 3N1-H (3), 3N1-I, 3N1-N, 3N1-P | |
| Munarra WA | −26.283 | 118.700 | 2 | 4 | ABTC31373, ABTC32472, ABTC32409, ABTC32413, ABTC32416, ABTC32477 | DQ287376, DQ287405, DQ287618, DQ287620, DQ287621, DQ287635 | 3N1-N, 3N1-P, 3N2-B (4) |
| Nallan WA | −27.317 | 117.967 | 4 | ABTC32388, ABTC32412, ABTC32422, ABTC32467 | DQ287616, DQ287619, DQ287623, DQ287634 | 3N2-C, 3N2-D (3) | |
| NE of Yamarna WA | −28.127 | 123.699 | 4 | R146760 | DQ287508, DQ287530, DQ287538, DQ287562 | 3N1-P, 3N1-Y (3) | |
| Neale Junction WA | −28.304 | 125.816 | 1 | R146765 | DQ287541 | 3N1-U | |
| Neds Creek WA | −25.483 | 119.650 | 1 | ABTC32432 | DQ287395 | 3N1-P | |
| New Springs WA | −25.817 | 120.000 | 2 | ABTC32485, ABTC32544 | DQ287408, DQ287418 | 3N1-P (2) | |
| Ninghan WA | −29.431 | 117.287 | 2 | DQ287582, DQ287583 | 3N1-AK (2) | ||
| North Well SA | −30.843 | 135.310 | 6 | DQ287424, DQ287429, DQ287445, DQ287447, DQ287453, DQ287492 | 3N1-D, 3N1-N (2), 3N1-P (3) | ||
| Oakden Hills SA | −31.667 | 137.033 | 3 | ABTC32427, ABTC32473, ABTC32535 | DQ287393, DQ287406, DQ287417 | 3N1-P (3) | |
| Old Andado NT | −25.383 | 135.283 | 2 | ABTC31374, ABTC31417 | DQ287377, DQ287380 | 3N1-I, 3N1-N | |
| Old Bandya WA | −27.697 | 122.134 | 6 | R146761 | DQ287501, DQ287502, DQ287511, DQ287532, DQ287535, DQ287539 | 3N1-P (5), 3N1-AG | |
| Orange Ck. #1 NT | −24.450 | 133.450 | 1 | ABTC31179 | DQ287370 | 3N1-M | |
| Orange Ck. #2 NT | −23.997 | 133.592 | 5 | DQ287440, DQ287448, DQ287455, DQ287459, DQ287467 | 3N1-F, 3N1-N (4) | ||
| Pack Saddle Camp WA | −22.917 | 118.750 | 1 | ABTC32839 | DQ287663 | 3N2-L | |
| Parallel Road #2 WA | −26.998 | 125.545 | 2 | R145133, R146785 | DQ287559, DQ287584 | 3N1-AI (2) | |
| Pinjin WA | −30.080 | 122.732 | 6 | R145007, R145086, R145140 | DQ287518, DQ287523, DQ287524, DQ287529, DQ287543, DQ287548 | 3N1-P (4), 3N1-V, 3N1-AM | |
| Queen Victoria Springs WA | −30.790 | 123.363 | 1 | R146783 | DQ287578 | 3N1-AM | |
| Rocklea Stn WA | −22.883 | 117.450 | 4 | ABTC32771, ABTC32772, ABTC32774, ABTC32817 | DQ287650, DQ287651, DQ287653, DQ287657 | 3N2-L, 3N2-M (3) | |
| Ross River NT | −23.597 | 134.485 | 2 | DQ287421, DQ287426 | 3N1-I, 3N1-N | ||
| Sandstone WA | −27.992 | 119.292 | 3 | 1 | R146762, R146777 | DQ287500, DQ287520, DQ287536, DQ287678 | 3N1-P (3), 3N2-L |
| Sherwood WA | −26.567 | 118.533 | 4 | ABTC32381, ABTC32453, ABTC32511, ABTC32532 | DQ287612, DQ287631, DQ287639, DQ287644 | 3N2-L (4) | |
| Sylvania WA | −23.583 | 120.050 | 1 | ABTC32770 | DQ287649 | 3N2-I | |
| The Garden NT | −23.282 | 134.417 | 4 | DQ287477, DQ287479, DQ287481, DQ287485 | 3N1-N (4) | ||
| Thundelarra WA | −28.967 | 117.117 | 1 | ABTC31352 | DQ287375 | 3N1-P | |
| Ti Tree NT | −22.132 | 133.267 | 3 | DQ287425, DQ287480, DQ287489 | 3N1-N (3) | ||
| Tieyon SA | −26.208 | 133.855 | 4 | DQ287435, DQ287442, DQ287461, DQ287487 | 3N1-E, 3N1-I, 3N1-N (2) | ||
| Uluru NT | −25.417 | 131.967 | 1 | ABTC31233 | DQ287373 | 3N1-N | |
| Umbeara NT | −25.748 | 133.683 | 5 | DQ287430, DQ287450, DQ287469, DQ287472, DQ287483 | 3N1-G, 3N1-I (2), 3N1-K, 3N1-N | ||
| Uranerz Camp WA | −30.159 | 123.443 | 1 | R146782 | DQ287522 | 3N1-W | |
| Victory Downs SA | −25.988 | 132.970 | 5 | DQ287423, DQ287457, DQ287460, DQ287464, DQ287491 | 3N1-I (4), 3N1-N | ||
| Warakurna WA | −25.033 | 128.250 | 1 | ABTC31392 | DQ287379 | 3N1-N | |
| Welbourn Hill SA | −27.357 | 134.085 | 6 | ABTC31653 | DQ287384, DQ287422, DQ287427, DQ287438, DQ287441, DQ287451 | 3N1-I (2), 3N1-N (4) | |
| Wheelerrana Stn WA | −23.983 | 120.000 | 5 | ABTC32769, ABTC32778, ABTC32818, ABTC32821, ABTC32925 | DQ287648, DQ287654, DQ287658, DQ287661, DQ287668 | 3N2-J, 3N2-K, 3N2-P (3) | |
| White Cliffs WA | −28.479 | 122.803 | 10 | R145139, R146802 | DQ287510, DQ287513, DQ287525, DQ287540, DQ287546, DQ287552, DQ287555, DQ287567, DQ287591, DQ287595 | 3N1-P (5), 3N1-X, 3N1-AE (4) | |
| Windsor WA | −28.012 | 118.580 | 2 | 5 | R146775 | DQ287514, DQ287534, DQ287683, DQ287684, DQ287685, DQ287688, DQ287689 | 3N1-P (2), 3N2-L (5) |
| Wintinna SA | −27.712 | 134.115 | 3 | DQ287434, DQ287466, DQ287484 | 3N1-B, 3N1-C, 3N1-I | ||
| Wirraminna SA | −31.190 | 136.228 | 7 | ABTC31429, ABTC32495 | DQ287382, DQ287412, DQ287428, DQ287431, DQ287436, DQ287439, DQ287478 | 3N1-N (3), 3N1-P (4) | |
| Woomarel WA | −25.733 | 114.283 | 1 | ABTC32849 | DQ287666 | 3N2-D | |
| Yamarna WA | −28.161 | 123.668 | 4 | R145083, R146771, R146784, R146800 | DQ287493, DQ287547, DQ287551, DQ287565 | 3N1-P, 3N1-R, 3N1-U, 3N1-AH | |
| Yarri WA | −29.777 | 122.359 | 7 | R145005, R145061 | DQ287505, DQ287528, DQ287573, DQ287577, DQ287580, DQ287592, DQ287594 | 3N1-O, 3N1-P (5), 3N1-AL | |
| Yellowdine WA | −31.300 | 119.650 | 1 | ABTC32488 | DQ287410 | 3N1-P | |
| Yeo WA | −28.077 | 124.318 | 5 | R145047, R146766, R146772, R146791, R146796 | DQ287499, DQ287509, DQ287527, DQ287590, DQ287593 | 3N1-P (5) | |
| Yindi WA | −30.384 | 122.507 | 6 | R145006, R145059 | DQ287506, DQ287507, DQ287554, DQ287568, DQ287579, DQ287588 | 3N1-P (5), 3N1-Q | |
| Yoothapinna WA | −26.533 | 118.517 | 1 | ABTC32846 | DQ287664 | 3N2-L | |
| Yowergabbie WA | −28.242 | 117.661 | 1 | 6 | DQ287561, DQ287679, DQ287680, DQ287681, DQ287682, DQ287686, DQ287687 | 3N1-P, 3N2-A, 3N2-C (4), 3N2-D | |
| Yundamindra WA | −29.250 | 122.100 | 2 | ABTC32407, ABTC32480 | DQ287388, DQ287407 | 3N1-P (2) | |
| Yunndaga WA | −29.783 | 121.150 | 2 | ABTC32420, ABTC32466 | DQ287391, DQ287401 | 3N1-P, 3N1-AQ | |
| TOTALS | 230 | 89 | |||||
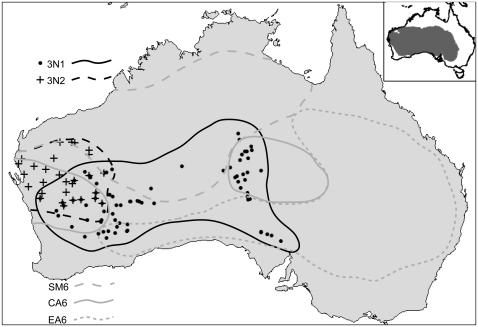
Figure 6. Sampling localities for the 3N1 and 3N2 mitochondrial clones.
Latitude/longitude data and sample sizes are given in Table 5. Ranges of the CA6, EA6, and SM6 sexual chromosome races are shown in light gray. The inset in the upper right shows the extent of the arid zone in dark gray.
Molecular
Techniques for DNA extraction, amplification and sequencing are described in Strasburg and Kearney [29]. We sequenced the ND2 (NADH dehydrogenase subunit two) gene and flanking tRNA genes, a region particularly useful for intraspecific and intrageneric studies because of its relatively high rate of evolution [52], [53]. All sequences have been submitted to GenBank, and accession numbers are given in Table 5.
Analytical
AMOVAs were performed for mtDNA sequence for both lineages using the computer program Arlequin 2.001 [54]. Uncorrected pairwise differences were used as the distance measure, and significance was assessed with 16,000 permutations. Mantel tests and mismatch analyses were also performed in Arlequin, with 10,000 permutations for Mantel tests and 1000 bootstrap replicates for mismatch analyses. Nucleotide diversities were calculated using Mega 2.1 [55], with standard errors calculated using the bootstrap method with 1,000 resamples.
Nested clade phylogeographic analysis (NCPA–34) was performed separately on the 3N1 and 3N2 lineages. Haplotype networks were inferred under the criterion of statistical parsimony [56] using TCS 1.16 [57], and permutations and significance tests were performed using Geodis 2.0 [58] with 10,000 permutations. Single-locus phylogeographic studies are typically limited by the fact that they cannot account for inter-locus variability due to both mutational and coalescent stochasticity. While we acknowledge the former limitation with this study, the latter is not an issue here because these geckos reproduce clonally.
Dating of NCPA inferences was performed using the method of Templeton [20]. This method allows for calculation of a point estimate for the age of a given event, and a confidence interval around that estimate that accounts for evolutionary stochasticity by modeling the distribution of time to coalescence as a gamma function [59]. Point estimates were obtained by comparing sequence diversity in the youngest monophyletic clade of the haplotype network for which the inference applies and sequence divergence from the nearest clade: divergence time t = (Dxy-0.5*(Dx+Dy))*substitution rate, where Dxy is average divergence between the focal clade and its neighboring clade, and Dx and Dy are average diversity within each clade [60]. For the section of mtDNA sequenced here, Macey et al. [61] estimated the rate of evolution in Agamid lizards to be 0.65% per lineage per million years (range based on geological dating estimates 0.61–0.70%), corresponding to a divergence rate of 1.3% per million years. Other studies have found highly concordant rates in other reptile, amphibian, and fish taxa [62]. In our 95% confidence intervals, we used a range of 0.61–0.70% per lineage per million years (corresponding to 1.22–1.4% divergence per million years) to account for some error in the estimate of evolutionary rate. In order to verify our assumption of equal rates of evolution along lineages for NCPA dating, a likelihood ratio test of a molecular clock [63] was performed on a tree of all sexual and parthenogenetic H. binoei haplotypes (including the EA6 sexual chromosome race) rooted with a single H. planiceps haplotype. We were unable to reject a molecular clock (2δ = 285.234, df = 255, p = 0.094; for details on maximum likelihood analysis conditions see ref. 29).
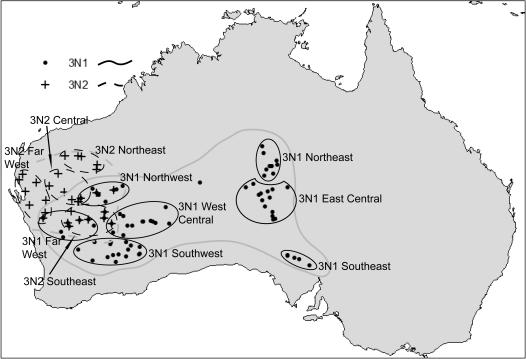
Effective migration rates among populations and regions within each race were measured using the computer program Migrate 1.7.6 [64]. Migrate uses a Markov chain Monte Carlo approach with importance sampling [65] to estimate Nefm, where Nef is the long-term inbreeding effective size and m is the average proportion of individuals migrating per generation. Analyses were run with 20 short chains with 1,000,000 genealogies sampled and 10,000 genealogies recorded, and 2 long chains with 10,000,000 genealogies sampled and 100,000 genealogies recorded. Analyses in Migrate were run both with individual populations and with nearby populations combined into regions to increase sample sizes and for ease of interpretation. 3N1 populations were grouped into Far West, Northwest, Southwest, West Central, East Central, Northeast, and Southeast regions, and 3N2 populations were grouped into Far West, Central, Northeast, and Southeast regions (Figure 7). Populations were grouped by eye, and in a few cases populations that were distant from any others were not included in a region. Combining populations that may show some genetic structure violates an assumption of the models underlying the coalescent techniques used in these programs; however, this is often a reasonable step to facilitate computation and interpretation of analyses [66]. Summed results from individual populations were very consistent with results from regions, suggesting that the analyses are in fact quite robust to violations of this assumption.
Figure 7. Regions used for various analyses.
Ranges for each mtDNA lineage as a whole are shown in light gray.
Distribution modeling
We used two contrasting approaches to predict the distribution of parthenogenetic H. binoei during current and LGM conditions: a correlative approach and a mechanistic approach. The correlative approach was based on a previously generated logistic regression model using six climatic predictor variables including mean annual temperature rainfall and humidity, as well as temperature and rainfall variability [24]. Predictions were made using current climatic conditions, as reported in Kearney et al. [24], as well as estimated conditions during the LGM. These estimates involve a 9°C reduction in mean annual air temperature [42] and three scenarios of reduced mean annual rainfall (3/4, 2/3 and 1/2 of current rainfall levels), with the other four climatic variables held constant. We used a range of rainfall reduction scenarios because there is considerable uncertainty in this respect (31 and P. Hope pers. comm.).
The mechanistic approach involved applying biophysical models to predict regions where egg development and above-ground activity are possible. This approach provides a means to map the fundamental niche of an organism (see [25] and [67] for details). Previous research has shown that H. binoei require approximately 600 degree days above 20°C for successful egg development, and that these lizards rarely forage at air temperatures below 15°C. Biophysical predictions were made using current climatic conditions, as reported in Kearney and Porter (2004), as well as an inferred 9°C reduction in monthly maximum and minimum air temperature during the LGM [42].
We assume here that habitat preferences for parthenogenetic H. binoei have not changed significantly since the LGM. While there are physiological differences between parthenogenetic and sexual H. binoei [44] which may have been a consequence of hybridization or evolved post-hybridization, there are no obvious differences in how they use microhabitats–both shelter and lay their eggs under a wide variety of surface debris as well as in burrows.
Acknowledgments
Paul Doughty, Geordie Torr, Jane Melville, Dale Roberts, Dave O'Conner, and Ben Phillips provided essential expert help with field collecting. We would like to thank Jane Melville for her generous financial and logistical assistance during field work, and the many station owners throughout Australia who gave us permission to collect on their properties. MK specimens were collected under permits S24357 1 (South Australia) and 8758 (Northern Territory) and exported from Australia under permit WT 2002-1168.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a Howard Hughes Medical Institute predoctoral fellowship to JLS and by grants from the Ethel Mary Read Fund, Peter Rankin Trust Fund for Herpetology, and Australian Research Council to MK.
References
- 1.Dawley RM, Bogart JP. Albany): (New York State Museum Press; 1989. [Google Scholar]
- 2.White MJD. Modes of Speciation. San Francisco): (W. H. Freeman; 1978. [Google Scholar]
- 3.Vrijenhoek RC, Dawley RM, Cole CC, Bogart JP. In: Evolution and Ecology of Unisexual Vertebrates, Dawley RM, Bogart JP, editors. Albany): (New York State Museum; 1989. pp. 19–23. [Google Scholar]
- 4.Bell G. The Masterpiece of Nature: the Evolution and Genetics of Sexuality. Berkeley): (University of California Press; 1982. [Google Scholar]
- 5.Butlin R. Nature Reviews Genetics. 2002;3:311–317. doi: 10.1038/nrg749. [DOI] [PubMed] [Google Scholar]
- 6.Moritz C. Genetics. 1991;129:211–219. doi: 10.1093/genetics/129.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Law JH, Crespi BJ. Molecular Ecology. 2002;11:1471–1489. doi: 10.1046/j.1365-294x.2002.01547.x. [DOI] [PubMed] [Google Scholar]
- 8.Chaplin JA, Ayre DJ. Heredity. 1997;78:57–67. doi: 10.1038/sj.hdy.6881110. [DOI] [PubMed] [Google Scholar]
- 9.Glesener RR, Tilman D. The American Naturalist. 1978;112:659–673. [Google Scholar]
- 10.Kearney M. Trends in Ecology&Evolution. 2005;20:495–502. doi: 10.1016/j.tree.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Templeton AR. In: Evolution and the Genetics of Life Histories, Dingle H, Hegmann JP, editors. Berlin): (Springer-Verlag; 1982. pp. 75–101. [Google Scholar]
- 12.Vandel A. Bull. Biol. France Belgique. 1928;62:164–281. [Google Scholar]
- 13.Cole CJ, Dessauer HC, Barrowclough GF. American Museum Novitates, 1988:1–38. [Google Scholar]
- 14.Fu JZ, Murphy RW, Darevsky IS. Copeia. 2000;2000:432–440. [Google Scholar]
- 15.Brown WM, Wright JW. Science. 1979;203:1247–1249. doi: 10.1126/science.424751. [DOI] [PubMed] [Google Scholar]
- 16.Moritz C, Donnellan S, Adams M, Baverstock PR. Evolution. 1989;43:994–1003. doi: 10.1111/j.1558-5646.1989.tb02545.x. [DOI] [PubMed] [Google Scholar]
- 17.Pongratz N, Storhas M, Carranza S, Michiels NK. BMC Evolutionary Biology. 2003;3:23. doi: 10.1186/1471-2148-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paland S, Colbourne JK, Lynch M. Evolution. 2005;59:800–813. [PubMed] [Google Scholar]
- 19.De Gelas K, De Meester L. Molecular Ecology. 2005;14:753–764. doi: 10.1111/j.1365-294X.2004.02434.x. [DOI] [PubMed] [Google Scholar]
- 20.Templeton AR. In: Evolutionary Theory and Processes: Modern Horizons, Wasser SP, editor. Netherlands): (Kluwer Academic Publishers; 2003. pp. 1–22. [Google Scholar]
- 21.Adamowicz SJ, Gregory TR, Marinone MC, Hebert PDN. Molecular Ecology. 2002;11:1209–1217. doi: 10.1046/j.1365-294x.2002.01517.x. [DOI] [PubMed] [Google Scholar]
- 22.Brisson JA, De Toni DC, Duncan I, Templeton AR. Evolution. 2005;59:1046–1059. [PubMed] [Google Scholar]
- 23.Hugall A, Moritz C, Moussalli A, Stanisic J. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6112–6117. doi: 10.1073/pnas.092538699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearney M, Moussalli A, Strasburg J, Lindenmayer D, Moritz C. Evolutionary Ecology Research. 2003;5:953–976. [Google Scholar]
- 25.Kearney M, Porter WP. Ecology. 2004;85:3119–3131. [Google Scholar]
- 26.Moritz C. Genetica. 1993;90:269–280. [Google Scholar]
- 27.Moritz C, Heideman A. Systematic Biology. 1993;42:293–306. [Google Scholar]
- 28.Moritz C. Chromosoma. 1984;89:151–162. [Google Scholar]
- 29.Strasburg JL, Kearney M. Molecular Ecology. 2005;14:2755–2772. doi: 10.1111/j.1365-294X.2005.02627.x. [DOI] [PubMed] [Google Scholar]
- 30.Kearney M, Blacket M, Strasburg JL, Moritz C. Molecular Ecology. 2006;15:1743–1748. doi: 10.1111/j.1365-294X.2006.02898.x. [DOI] [PubMed] [Google Scholar]
- 31.Hesse PP, Magee JW, van der Kaars S. Quaternary International. 2004;118–119:87–102. [Google Scholar]
- 32.Knowles LL, Maddison WP. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 33.Panchal M, Beaumont MA. Evolution. 2007;61:1466–1480. doi: 10.1111/j.1558-5646.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- 34.Templeton AR. Molecular Ecology. 2004;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- 35.Slatkin M, Hudson RR. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers AR, Harpending H. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 37.Harpending HC, Sherry ST, Rogers AR, Stoneking M. Current Anthropology. 1993;34:483–496. [Google Scholar]
- 38.Rogers AR, Jorde LB. Human Biology. 1995;67:1–36. [PubMed] [Google Scholar]
- 39.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu YX. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mantel N. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- 42.Miller GH, Magee JW, Jull AJT. Nature. 1997;385:241–244. [Google Scholar]
- 43.Moritz C, Brown WM, Densmore LD, Wright JW, Vyas D, Donnellan S, Adams M, Baverstock PR. In: Evolution and Ecology of Unisexual Vertebrates, Dawley RM, Bogart JP, editors. Albany): (New York State Museum Press; 1989. pp. 87–112. [Google Scholar]
- 44.Kearney M, Wahl R, Autumn K. Physiological and Biochemical Zoology. 2005;78:316–324. doi: 10.1086/430033. [DOI] [PubMed] [Google Scholar]
- 45.Moritz C, McCallum H, Donnellan S, Roberts JD. Proceedings of the Royal Society of London Series B-Biological Sciences. 1991;244:145–149. [Google Scholar]
- 46.Kearney MR. Trends in Ecology&Evolution. 2003;18:605–607. [Google Scholar]
- 47.Adams M, Foster R, Hutchinson MN, Hutchinson RG, Donnellan SC. Evolution. 2003;57:2619–2627. doi: 10.1111/j.0014-3820.2003.tb01504.x. [DOI] [PubMed] [Google Scholar]
- 48.Lynch M. The Quarterly Review of Biology. 1984;59:257–290. [Google Scholar]
- 49.Bierzychudek P. Experientia. 1985;41:1255–1264. [Google Scholar]
- 50.Peck JR, Yearsley JM, Waxman D. Nature. 1998;391:889–892. [Google Scholar]
- 51.Hewitt GM. Evolutionary Patterns and Processes. London): (Linnean Society of London; 1993. pp. 95–123. [Google Scholar]
- 52.Janzen FJ, Krenz JG, Haselkorn TS, Brodie ED. Molecular Ecology. 2002;11:1739–1751. doi: 10.1046/j.1365-294x.2002.01571.x. [DOI] [PubMed] [Google Scholar]
- 53.Macey JR, Strasburg JL, Brisson JA, Vredenburg VT, Jennings M, Larson A. Molecular Phylogenetics and Evolution. 2001;19:131–143. doi: 10.1006/mpev.2000.0908. [DOI] [PubMed] [Google Scholar]
- 54.Schneider S, Roessli D, Excoffier L. Switzerland): (Genetics and Biometry Laboratory, University of Geneva; 2001. [Google Scholar]
- 55.Kumar S, Tamura K, Jakobsen IB, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 56.Templeton AR, Crandall KA, Sing CF. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clement M, Posada D, Crandall KA. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 58.Posada D, Crandall KA, Templeton AR. Molecular Ecology. 2000;9:487–488. doi: 10.1046/j.1365-294x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- 59.Kimura M. Genetical Research. 1970;15:131–133. doi: 10.1017/s0016672300001439. [DOI] [PubMed] [Google Scholar]
- 60.Nei M. Molecular Evolutionary Genetics. New York): (Columbia University Press; 1987. [Google Scholar]
- 61.Macey JR, Schulte JA, Ananjeva NB, Larson A, Rastegar-Pouyani N, Shammakov SM, Papenfuss TJ. Molecular Phylogenetics and Evolution. 1998;10:118–131. doi: 10.1006/mpev.1997.0478. [DOI] [PubMed] [Google Scholar]
- 62.Weisrock DW, Macey JR, Ugurtas IH, Larson A, Papenfuss TJ. Molecular Phylogenetics and Evolution. 2001;18:434–448. doi: 10.1006/mpev.2000.0905. [DOI] [PubMed] [Google Scholar]
- 63.Felsenstein J. Journal of Molecular Evolution. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 64.Beerli P, Felsenstein J. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Iorio M, Griffiths RC. Advances in Applied Probability. 2004;36:417–433. [Google Scholar]
- 66.Pfenninger M, Posada D, Magnin F. BMC Evolutionary Biology. 2003;3:8. doi: 10.1186/1471-2148-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kearney M. Oikos. 2006;115:186–191. [Google Scholar]