Abstract
Background
It is not clear which species of bacteria may be involved in inflammatory bowel disease (IBD). One way of determining which bacteria might be likely candidates is to use culture‐independent methods to identify microorganisms that are present in diseased tissues but not in controls.
Aims
(1) To assess the diversity of microbial communities of biopsy tissue using culture‐independent methods; (2) to culture the bacteria found in the tissues of patients with IBD but not in the controls; (3) to identify potential virulence factors associated with cultured bacteria.
Methods
84 biopsy specimens were collected from 15 controls, 13 patients with Crohn's disease (CD) and 19 patients with ulcerative colitis (UC) from a population‐based case–control study. Ribosomal intergenic spacer analysis (RISA) was conducted to identify unique DNA bands in tissues from patients with CD and UC that did not appear in controls.
Results
RISA followed by DNA sequencing identified unique bands in biopsy specimens from patients with IBD that were classified as Escherichia coli. Targeted culture showed a significantly (p<0.05) higher number of Enterobacteriaceae in specimens from patients with IBD. The B2+D phylogenetic group, serine protease autotransporters (SPATE) and adherence factors were more likely to be associated with tissues from patients with UC and CD than with controls.
Conclusions
The abundance of Enterobacteriaceae is 3–4 logs higher in tissues of patients with IBD and the B2+D phylogenetic groups are more prevalent in patients with UC and CD. The B2+D phylogenetic groups are associated with SPATE and adherence factors and may have a significant role in disease aetiology.
Inflammatory bowel disease (IBD) is a collective term for UC and CD.1,2,3 These diseases are chronic inflammatory diseases of the digestive tract, potentially leading to severe inflammation, ulceration and obstruction, the end point of which may be surgical resection.1,2,3 The high incidence of IBD in the Western world is not explained by simple genetic drift of a susceptible human genome, prompting the hypothesis that environmental factors are important in disease aetiology.4,5
These diseases are thought to be a result of recognition of a microbial antigen(s) by a dysfunctional immune system in a genetically predisposed host.6,7 Many bacteria have been linked to IBD, but specific bacteria may have been missed because not more than 30% of the microbial diversity in the gut can be cultured.8,9,10,11,12 Cultivation‐independent methods now allow simultaneous surveys of microbial diversity.11,12 We maintain that culture‐independent surveys of microbial diversity should form a prelude to the targeted cultivation of bacteria. However, it is important to emphasise that, wherever possible, microorganisms identified using culture‐independent methods, as being associated with disease, should be cultured so that their virulence mechanisms can be evaluated.
Several bacteria have been implicated in the aetiology of IBD, the most prominent among these being Mycobacterium paratuberculosis. Other bacteria that have been associated are members of the Enterobacteriaceae, Helicobacter pylori and Bacteroides species.2,3 In this paper, we describe the use of ribosomal intergenic spacer analysis (RISA)13 of biopsy tissue to identify bands that were consistently associated with tissue from patients with IBD. This allowed us to use highly focused cultivation methods, including resuscitation methods, to specifically culture Enterobacteriaceae.
Materials and methods
Study subjects
We utilised 84 biopsy specimens from 15 controls, 13 patients with Crohn's disease (CD) (3 with ileal disease, 6 with ileocolonic disease and 4 with isolated colonic disease) and 19 patients with ulcerative colitis (UC) (3 with proctitis, 8 with left‐sided colitis and 8 with pancolitis) from a population‐based case–control study undertaken at the University of Manitoba, Winnipeg, Manitoba, Canada (table 1) as described previously.14
Table 1 Biopsy samples used in this study*.
| Controls (15 subjects, 28 tissues) | CD (13 patients, 27 tissues) | UC (19 patients, 29 tissues) | IBD (32 patients, 56 tissues) | |||||
|---|---|---|---|---|---|---|---|---|
| Site | End | Hist | End | Hist | End | Hist | End | Hist |
| Rectum | 0(13) | 0(13) | 5(13) | 5(13) | 9(10) | 9(10) | 14(23) | 14(23) |
| Caecum | 0(15) | 0(15) | 4(9) | 6(9) | 2(15) | 4(15) | 6(24) | 10(24) |
| Colon | 0(0) | 0(0) | 3(5) | 3(5) | 2(4) | 2(4) | 5(9) | 5(9) |
CD, Crohn's disease; End, endoscopic examination; Hist, histological examination; UC, ulcerative colitis.
*Endoscopic and histological examination was made on biopsy specimens, and only histologically positive samples were considered inflamed. Total numbers of biopsies for each group are presented in parentheses, and the numbers not in parentheses represent inflamed biopsies.
In brief, a population‐based study refers to a process by which selection of study subjects proceeds by accounting for bases related to various factors such as lifestyle (eg, smoking), geographical location (eg, urban vs rural), age, sex or ethnicity. In IBD research, it is challenging to obtain untainted biopsy controls because endoscopy is normally performed on persons only when it is clinically required. The controls were true controls in the sense that the subjects voluntarily submitted to endoscopy and were drawn from the same population‐based study. No antibiotics were prescribed to any of the subjects during the 6 weeks before the colonoscopy.
Colonoscopy and biopsies
Following standard oral Fleet Phospho‐soda (CB Fleet Company, Lynchburg, Virginia, USA) treatment, biopsy specimens were taken from the caecum and the rectum. In subjects with a previous caecal resection, biopsies were obtained from the right colon distal to the ileocolonic anastomosis. All biopsy samples were snap frozen in liquid nitrogen and stored at −70°C. Biopsy specimens were subject to standard histological staining with H&E for evaluation of inflammation. A site was considered inflamed if it had histological evidence of inflammation and uninflamed if it was histologically normal.
DNA extraction for RISA analysis
Tissue samples were suspended in 150 μl lysis buffer (10 mM Tris‐HCl, pH 8.0; 5 mM EDTA, pH 8.0; 4 M guanidinium isothiocyanate, pH 7.5; 50 g sarcosyl/l, 2.5 g SDS/l, 5 g sodium citrate/l and 5 g Triton X‐100/l). 300 μl of chloroform and Tris‐saturated phenol (pH 6.9) were added to each tube. The samples were placed at −20°C for 1 h. Subsequently, samples were centrifuged in microfuge tubes at 4°C for 20 min at 10 000 g. Supernatants were transferred to fresh tubes. Isopropanol was added to 1/4 volume of the supernatants and the mixtures loaded on to silica‐cellulose membranes in columns. Samples were allowed to filter through the membrane by gravity. The membranes were washed twice with 300 μl of 95% ethanol (by gravity). DNA was eluted with 400 μl hot (about 75°C) Tris‐EDTA buffer (by gravity) and precipitated with two portions of 95% ethanol. The resulting pellets were suspended in 25 μl 0.5× Tris‐EDTA buffer (pH 8.0) and stored at −20°C until further analysis.
Table 2 lists the primer sequences used for RISA and the amplified intergenic transcribed spacers between the 16S and 23S rDNA.13 PCR products were subjected to electrophoresis using 2% agarose. DNA fragments found only in patients with UC and CD were purified from agarose gels, cloned into the pCR2.1‐TOPO TA vector (Invitrogen, Carlsbad, California, USA) and sequenced. Standard bioinformatics analysis was used to taxonomically classify the sequence fragment.
Table 2 Primers used.
| Primer sequences | Target sequences | References |
|---|---|---|
| Autotransporters | ||
| SPATE1 5′ GAGGTCAACAACCTGAACAAACGTATGGG | The genes encoding SPATE | This study |
| SPATE2 5′ CCGGCACGGGCTGTCACTTTCCAG | ||
| E coli phylogenetic groups | ||
| ChuAf 5′ CGGACGAACCAACGGTCAGGAT | The chuA gene is required for haemtransport in E coli O157:H7 | Modified from Clermont et al15 |
| ChuAr 5′ TGCCGCCAGTACCAAAGACACG | ||
| Yjaf 5′ CGTGAAGTGTCAGGAGACGCTGC | The yjaA gene coding for protein of unknown function | Modified from Clermont et al15 |
| Yjar 5′ TGCGTTCCTCAACCTGTGACAAACC | ||
| Tsp1 5′ GGGAGTAATGTCGGGGCATTCAG Tsp2 5′ CATCGCGCCAACAAAGTATTACGCAG | Tsp encodes for a putative DNA fragment (TSPE4.C2) in E coli | Modified from Clermont et al15 |
| E coli toxins | ||
| Cnff 5′ AGTACTGACACTCACTCAAGCCGC | Cytotoxic necrotising factors (Cnf1 and Cnf2) | This study |
| Cnfr 5′ GCAGAACGACGTTCTTCATAAGTATCACC | ||
| IpgDf 5′ CGACTTCTCTTCTGACGCCGAC IpgDr 5′ CAACATTCCTCCAGCCTAAGCCC | ipgD gene modulates entry of bacteria into epithelial cells | This study |
| Vt1f 5′ CGCATAGTGGAACCTCACTGACGC | Verocytotoxin 1 | This study |
| Vt1r 5′ CATCCCCGTACGACTGATCCC | ||
| Vt2f 5′ CGGAATGCAAATCAGTCGTCACTCAC | Verocytotoxin 2 | This study |
| Vt2r 5′ TCCCCGATACTCCGGAAGCAC | ||
| HlyAf 5′ TGCAGCCTCCAGTGCATCCCTC | The hlyA gene encoding α haemolysin | |
| HlyAr 5′ CTTACCACTCTGACTGCGATCAGC | ||
| STaf 5′ GTGAAACAACATGACGGGAGG | Heat‐stable enterotoxin 1 | This study |
| STar 5′ ATAACATCCAGCACAGGCAGG | ||
| STbf 5′ GGGGTTAGAGATGGTACTGCTGGAG | Heat‐stable enterotoxin 2 | This study |
| STbr 5′ GACAATGTCCGTCTTGCGTTAGGAC | ||
| LTf 5′ CCGTGCTGACTCTAGACCCCCA | Heat‐labile enterotoxin LT | This study |
| LTr 5′ CCTGCTAATCTGTAACCATCCTCTGC | ||
| Eaef 5′ CCAGGCTTCGTCACAGTTGCAGGC Eaer 5′ CGCCAGTATTCGCCACCAATACC | The eae gene coding for intimin present in AEEC strains | This study |
| Enterotoxigenic Bacteroides fragilis (ETBF) | ||
| Bf1F 5′ GTTAGTGCCCAGATGCAGGATGCGG | The genes coding for Bacteroides fragilis enterotoxins (Bft1, Bft2 and Bft3) | This study |
| Bf2F 5′ GAACTCGGTTTATGCAGTTCATGGACTG | ||
| Bf3R 5′ TGGGTTGTAGACATCCCACTGGCTT | ||
| Bf4R 5′ GGATACATCAGCTGGGTTGTAGACATCCC | ||
| E coli adhesins | ||
| PapF 5′ CCGGCGTTCAGGCTGTAGCTG PapR 5′ GCTACAGTGGCAGTATGAGTAATGACCGTTA | The genes coding for pathogenicity islands (PAI I, PAI II)16 and sfp gene cluster | This study |
| PAI1 5′ TAGCTCAGACGCCAGGATTTTCCCTG | ||
| pai2 5′ CCTGGCGCCTGCGGGCTGACTATCAGGG | ||
| BmaEf 5′ CTAACTTGCCATGCTGTGACAGTA | The bmaE gene for M‐agglutinin subunit; Afa‐8 gene cluster | Martin et al17 |
| BmaEr 5′ TTATCCCCTGCGTAGTTGTGAATC | ||
| Sfaf 5′ CGGAGGAGTAATTACAAACCTGGCA | S‐fimbrial adhesins encoded by sfaD to sfaE | Martin et al17 |
| Sfar 5′ CTCCGGAGAACTGGGTGCATCTTAC | ||
| Afaf 5′ TATGGTGAGTTGGCGGGGATGTACAGTTACA | AfaE‐3 gene cluster | This study |
| Afar 5′ CCGGGAAAGTTGTCGGATCCAGTGT | ||
| AIDA1 5′ TATGCCACCTGGTATGCCGATGAC AIDA2 5′ ACGCCCACATTCCCCCAGAC | The aidA gene coding E coli AIDA‐I adhesin in DAEC strains | This study |
| AggRf 5′ GAGTTAGGTCACTCTAACGCAGAGTTG | The aggR gene for adhesin of aggregative adherence fimbria I | This study |
| AggRr 5′ GACCAATTCGGACAACTGCAAGCATCTAC | ||
| Ag43F 5′TGACACAGGCAATGGACTATGACCG Ag43R 5′GGCATCATCCCGGACCGTGC | The agn43 gene coding for antigen involved in E coli autoaggregation | This study |
| Flagella and type 1 fimbrae typing | ||
| Primer1 5′CAAGTCATTAATAC(A/C)AACAGCC | The fliC genes coding for flagellin proteins | Machado et al18 |
| Primer2 5′GACAT(A/G)TT(A/G)GA(G/A/C)ACTTC(G/C)GT | ||
| FimHf 5′ CTGGTCATTCGCCTGTAAAACCGCCA FimHr 5′ GTCACGCCAATAATCGATTGCACATTCCCT | The fimH gene encoding FimH subunit of type 1 pili | This study |
| Ribosomal DNA‐based primers | ||
| ITSF 5′ GTCGTAACAAGGTAGCCGTA | 16S‐23S rRNA intergenic transcribed spacers | Cardinale et al13 |
| ITSReub 5′ GCCAAGGCATCCACC | ||
| 27f 5′ AGAGTTTGATCMTGGCTCAG | Conserved 16S rDNA used to amplify ribosomal genes | Lane19 |
| 342r 5′ CTGCTGCSYCCCGTAG |
SPATE, serine protease autotransporter.
Bacterial cultures
Once RISA analysis had determined that the bacteria that appeared in patients with UC and CD and not in controls were E coli, targeted bacterial cultivation was carried out. All cultivation of the bacteria was done with untreated biopsy tissue, and no procedures were used to wash the tissues or to remove the mucus. To ensure that as many E coli cells as possible were cultured, resuscitation in buffered peptone water was performed, followed by decimal dilution and culturing on chromogenic E coli/coliform medium (Oxoid CM0956; Oxoid, Nepean, Ontario, Canada). Resuscitation was done by incubation of the biopsy specimen in 1 ml of 100 mM buffered peptone water for 16 h at 37°C. Ten μl droplets were pipetted (Maxipettor, Eppendorf; Eppendorf AG, Hamburg, Germany) onto media, allowed to dry, and then inverted and incubated at 37°C. After 18 h, E coli (purple colonies) and non‐E coli coliforms (blue, pink and white colonies) were counted separately.
Colour differentiation on chromogenic agar is a good first approximation of E coli identity. Five putative E coli (purple colonies) were picked from each positive tissue and subcultured in Luria‐Bertani broth, then recultured on E coli/coliform medium, and tested for reactivity to indole, methyl red, Vogues Proskauer and citrate utilisation to differentiate E coli from non‐E coli. The 16S rDNA gene sequences for all the non‐E coli were determined using standard primers (table 2).
DNA extraction from bacterial cultures
For DNA extraction from cultures, 1 ml suspensions of each culture were centrifuged and pellets were suspended in lysis buffer, and subsequently mixed with chloroform and Tris‐saturated phenol. All other steps were the same as for the extraction of DNA from tissue samples.
Molecular analysis of bacterial cultures
Table 2 lists all primer sequences used for molecular analysis of bacterial cultures. Primers were either modified from published primers or newly designed for the purposes of our studies. PCR was done by standard methods after optimisation for specificity of primer pairs by adjusting annealing temperature and salt concentration. Amplified products were run on agarose gels as described above.
Statistical analysis
For statistical analysis of data, we applied a χ2 test based on the Mantel–Haenszel method (Epi Info V.6.04).
Results
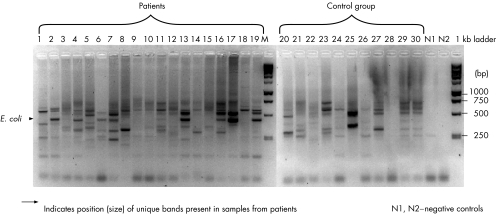
A total of 84 biopsies were collected from 15 controls, 13 patients with CD and 19 patients with UC (table 1). In most cases, more than one biopsy specimen was obtained from multiple sites, or from adjacent sites from the same subject. DNA was extracted from each biopsy sample and subjected to RISA analysis (fig 1). We were able to identify bands (∼450 bp) that were consistently present in approximately 70% of patients but in <30% of controls (fig 1). The bands in the controls were also of a much lower density than those from tissues of patients with IBD. Five bands from tissues of patients with IBD were retrieved from the gel, sequenced, aligned with Genbank sequence and found to be E coli.
Figure 1 Ribosomal intergenic spacer analysis of biopsy samples from tissues of patients with inflammatory bowel disease and control tissues.
Resuscitation of biopsies by resuspension in buffered peptone water, incubation for 16 h at 37°C, and subsequent plating on chromogenic agar allowed growth of coliform bacteria in over 90% of biopsy tissues. Chromogenic agar enriches for predominantly coliform bacteria, but only lactose‐fermenting bacteria (E coli) turn purple. We could culture purple colonies in only 46.7% of control subjects, 69.2% of patients with CD and 63.2% of patients with UC, even though non‐E coli coliforms (white and pink colonies) could be cultured from most biopsies (table 3).
Table 3 Microbial tissue phenotypes encountered from control tissues and tissues of patients with inflammatory bowel disease*.
| Disease | Patient | E coli† group | Non‐E coli‡ | SPATE | E coli adhesins | E coli H‐type | E coli type 1 fimbriae |
|---|---|---|---|---|---|---|---|
| Control | 41A | ND | Enterobacter sp | ND | – | – | – |
| Bacteroides fragilis Bft1§ | |||||||
| Control | 43A | B2 | Vat | Ag43, PAI I | H7 | Bovine | |
| Control | 48B | B2 | Vat, Pic‐like | Sfp¶ | H4 | APEC | |
| Control | 50A | B2 | Vat, Sat, Pic | Ag43 | H1 | APEC | |
| Control | 58 | ND | Enterobacter flavescens | ND | – | – | – |
| Control | 59B | ND | Klebsiella oxytoca | ND | – | – | – |
| Control | 73A | A | Klebsiella sp | ND | AIDA‐I | H12 | Bovine |
| Control | 69A | ND | Enterococcus sp | ND | – | – | – |
| Bacteroides fragilis Bft1 | |||||||
| Control | 76 | ND | Escherichia fergusonii | SigA, Sat††, SepA | – | – | – |
| Staphylococcus epidermidis | |||||||
| Staphylococcus capitis | |||||||
| Bacteroides fragilis Bft2§ | |||||||
| Control | 80 | ND | E fergusonii | SigA, Sat††, SepA | – | – | – |
| Klebsiella sp | |||||||
| Control | 81 | ND | Enterococcus durans | ND | – | – | – |
| Control | 87 | ND | Klebsiella sp | ND | – | – | – |
| Control | 90 | A | ND | AIDA‐I, Ag43 | H11 | Bovine | |
| Control | 17B | D | ND | Ag43 | H4 | UPEC | |
| Control | 88 | A | Klebsiella sp | ND | AIDA‐I | H21 | Bovine |
| CD | 15B | B2 | Vat, Pic‐like | Ag43 | H4 | APEC | |
| CD | 79A | B2 | Vat, Sat, Pic | Ag43 | H1 | APEC | |
| CD | 91A | B1 | Acidovorax sp | ND | AIDA‐I | H21 | Bovine |
| CD | 118 | ND | Staphylococcus auricularis | ND | – | – | – |
| CD | 120A | D | EspI | PAI I, AIDA‐I, Ag43 | H52 | Bovine | |
| CD | 124A | ND | E fergusonii Enterococcus faecium | SigA, Sat††, SepA | – | – | – |
| Bacteroides fragilis Bft1 | |||||||
| CD | 125B | ND | Klebsiella oxytoca | ND | – | – | – |
| CD | 126C | B2 | ND | AIDA‐I, Ag43 | H39 | Bovine | |
| CD | 132 | B2 | Sat | AfaE‐3, Ag43, AIDA‐I | H4 | Bovine | |
| CD | 137 | ND | Klebsiella sp | ND | – | – | – |
| CD | 138 | ND | Staphylococcus aureus | ND | – | – | – |
| Pseudomonas putida | |||||||
| Klebsiella sp | |||||||
| CD | 146 | B2 | Vat | Sfa**, Ag43, PAI I | H7 | APEC | |
| CD | 149 | B2 | Vat, Pic | Sfa, Ag43, PAI I | H5 | APEC | |
| CD | 128 | B2 | E fergusonii | Vat, SigA, Sat††, SepA | Sfa | H7 | UPEC |
| UC | 117A | D | ND | AIDA‐I, Ag43 | H18 | Bovine | |
| UC | 119A | ND | Enterococcus faecium | ND | – | – | – |
| UC | 121 | B1 | ND | ND | H10 | Bovine | |
| UC | 122 | ND | Enterococcus faecalis | ND | – | – | – |
| Bacteroides fragilis Bft1 | |||||||
| UC | 127A | B2 | Enterobacter cloacae | Vat, Sat, Pic | F1C | H7 | Bovine |
| UC | 130 | ND | Bacillus licheniformis | ND | – | – | – |
| Bacillus pumilus | |||||||
| UC | 131A | B2 | E fergusonii | Vat, Sat††, Pic, SigA, Sat, SepA | Ag43, PAI I | H10 | APEC |
| Bacteroides fragilis Bft2 | |||||||
| UC | 133A | ND | Klebsiella sp | ND | – | – | – |
| UC | 135 | B2 | Klebsiella oxytoca | Pic | AIDA‐I, Ag43 | H31 | Bovine |
| UC | 136 | B2 | ND | Vat | Ag43, PAI I | H7 | Bovine |
| UC | 139 | D | ND | ND | AIDA‐I | H6 | APEC |
| UC | 140A | ND | Enterococcus sp | ND | – | – | – |
| UC | 141 | B2 | ND | ND | Sfa, Ag43, PAI I | H7 | APEC |
| UC | 142 | B2 | ND | Vat | AfaE‐3, Ag43 | H5 | APEC |
| UC | 143 | B2 | ND | Sat, Pic‐like | AfaE‐3, Ag43 | H4 | APEC |
| UC | 145B | B2 | ND | Vat, Pic‐like | Ag43 | H4 | Bovine |
| UC | 147 | B2 | Klebsiella sp | Vat | Sfa, Ag43 | H5 | APEC |
| UC | 134A | ND | Klebsiella sp | ND | – | – | – |
APEC, avian pathogenic E coli; ND, E coli not detected; PAI, pathogenicity island; SPATE, serine protease autotransporters; UPEC, uropathogenic E coli.
*A total of 84 biopsy specimens from 47 subjects were used. Data are not shown by site, and were pooled because the correlation between bacterial species and site, within and between subjects, was low (r = 0.22).
†A total of 28 E coli strains and 34 non‐E coli were picked and assessed.
‡Identity was determined by sequencing the 16S rDNA between with 27f and 342r.
§Bft1 and Bft2 are enterotoxins produced by Bacteroides fragilis detected from tissue by the nested‐PCR method.
¶sfp is a gene cluster of sorbitol‐fermenting enterohaemorrhagic E coli O157:H‐.
**sfa gene coding for S‐fimbriae minor subunit of E coli UTI89.
††Sat gene coding for autotransporter toxin Sat identified in Escherichia fergusonii.
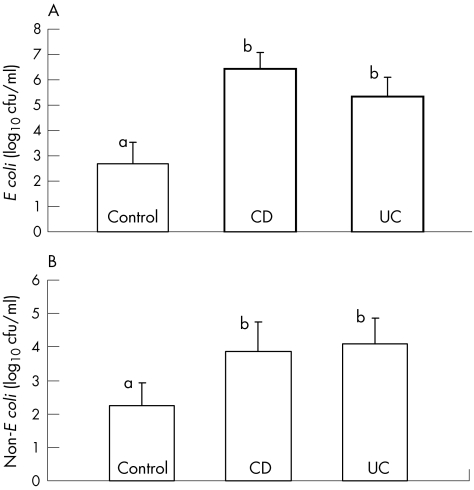
Serial dilution and plating on chromogenic agar allowed for quantification of total coliforms and E coli (fig 2). The numbers of E coli (4×102/ml) and non‐E coli coliforms (6.3×105/ml) cultured were higher (p<0.05) in CD and UC than in controls. There was a poor correlation (r = 0.22) between the site of inflammation and the presence of E coli, and data were pooled by subject.
Figure 2 Proportions of (A) E coli and (B) non‐E coli in biopsy tissues from controls, and patients with ulcerative colitis (UC) and Crohn's disease (CD).
Five purple colonies were picked from all agar plates (total 135 colonies) that showed E coli growth. Taxonomic identity of the E coli was confirmed by reactivity to indole, methyl red, Vogues Proskauer and citrate utilisation. There were also few polymorphisms in the RISA product, confirming that isolates were predominantly clonal by subject, irrespective of site. We selected 150 colonies of non‐E coli (five per biopsy) of pink, blue and white colonies from plates and were able to confirm that none of these colonies were E coli by checking for reactivity to indole, methyl red, Vogues Proskauer and citrate utilisation. The identities of these colonies were determined by 16S‐rDNA sequence analysis (table 3).
E coli comprises four phylogenetic groups (A, B1, B2 and D) with virulent types typically belonging to groups B2 and D.15 These groups can be identified by a simple PCR procedure of the chuA and yjaA genes and a cryptic DNA fragment (table 2). Clermont et al15 indicated that these groups could be identified in a single multiplex PCR for all DNA targets, but we did not get reproducible results. We modified the primers (table 2) and the PCR conditions and were able to generate highly reproducible results from a suite of pathogenic and non‐pathogenic E coli in our laboratory (data not shown). The abundance of the pathogenic B2+D groups was significantly (p = 0.04) greater in IBD than in controls (table 4).
Table 4 Distribution of virulence features of E coli isolated from patients with inflammatory bowel disease and controls.
| Item* | Control (15 subjects, 28 tissues) | UC (19 patients, 27 tissues) | CD (13 patients, 29 tissues) |
|---|---|---|---|
| Group | |||
| A | 3 (20)† | 0 | 0 |
| B1 | 0 | 1 (5.3) | 1 (7.7) |
| B2 | 3 (20) | 9 (47.4) | 7 (53.8) |
| D | 1 (6.7) | 2 (10.5) | 1 (7.7) |
| B2+D | 4 (26.7) | 11 (57.9) | 8 (61.5) |
| SPATE | |||
| Vat | 3 (20) | 7 (36.8) | 4 (30.8) |
| Pic | 1 (6.7) | 3 (15.8) | 3 (23.1) |
| Sat | 3 (20) | 4 (21.05) | 3 (23.1) |
| Pic‐like | 1 (6.7) | 2 (15.4) | 1 (7.7) |
| EspI | 0 | 0 | 1 (7.7) |
| SepA | 2 (13.3) | 2 (10.5) | 1 (7.7) |
| SigA | 2 (13.3) | 2 (10.5) | 1 (7.7) |
| Total | 5 (33.3) | 9 (47.4) | 7 (53.8) |
| Adhesins | |||
| Ag43‡ | 4 (26.7) | 9 (47.4) | 7 (53.8) |
| AfaE3 | 0 | 2 (10.5) | 1 (7.7) |
| AIDA‐I | 3 (20) | 3 (15.8) | 4 (30.8) |
| F1C | 0 | 1 (5.3) | 0 |
| Sfa | 0 | 3 (15.8) | 2 (15.4) |
| PAI I | 1 (6.7) | 3 (15.8) | 3 (23.1) |
| Sfp | 1 (6.7) | 0 (5.3) | 0 |
| Total | 7 (46.6) | 12 (63.2) | 8 (61.5) |
CD, Crohn's disease; UC, ulcerative colitis.
*Table 2 gives the functions of each of the genotypic characteristics.
†Values in parentheses represent the frequency of items in patients as a percentage.
‡Antigen 43 (Ag43) is a self‐recognising surface adhesin found in most Escherichia coli strains.
E coli strains were also assayed for the presence of serine protease autotransporter proteins (SPATE). Alignments were made of a prominent group (Vat, Sat, Pic, EspI) of SPATE nucleic acid sequences, and primers to conserved regions were designed to amplify targets which, when digested with HaeIII, were diagnostic of the different groups of SPATE. The identity of each SPATE was confirmed by sequence analysis. One new SPATE, Pic‐like, was identified by sequence analysis and alignment with known SPATE. When all SPATE‐positive E coli isolates were totalled, patients with IBD had a higher number of SPATE‐positive isolates than the controls (table 4). Interestingly, this “new” SPATE has actually previously been identified in the enteropathogenic E coli E22 genome (Genbank: AAJV01000028), but no functions were assigned.
PCR analyses of E coli isolates were conducted for a range of adhesins commonly found in pathogenic E coli (tables 3 and 4). Isolates were positive only for agn43 and aidA, and gene clusters coding for AfaE‐3, F1C, Sfa, PAI I and Sfp. No E coli positive for cnf1, cnf2, eae, hlyA, ipgB, aggR, bmaE or genes coding for verotoxins, heat‐stable and heat‐labile toxins were found in biopsy specimens of patients with UC or CD.
χ2 test based on the Mantel–Haenszel method (table 5) was performed to verify relationships between disease, B2+D genotype, SPATE toxins and adhesins. We observed a significant difference (p = 0.04) in the number of E coli isolates from the B2+D genotype for both patients with UC and those with CD (table 5). The relationship between IBD and an adhesin (p = 0.07) or ⩾2 adhesins (p = 0.03) was significant, but not with a SPATE and an adhesin (p = 0.15). The relationship between the B2+D genotype and SPATE(s), adhesion(s) or APEC was significant (p<0.05).
Table 5 Statistical relationship between the presence of virulence factors in patients with inflammatory bowel disease and control subjects.
| Disease | Virulence factor | p Value |
|---|---|---|
| IBD | ⩾2 adhesins | 0.03* |
| IBD | B2+D | 0.04* |
| IBD | Adhesins | 0.07 |
| IBD | SPATE(s)+adhesin(s) | 0.15 |
| Genotype | Virulence feature | p Value |
|---|---|---|
| B2+D | SPATE(s)+adhesin(s) | 0.002** |
| B2+D | Adhesins | 0.04* |
| B2+D | SPATE | 0.02* |
| B2+D | APEC | 0.04* |
APEC, avian pathogenic E coli; IBD, inflammatory bowel disease; SPATE, serine protease autotransporters.
*p<0.05, **p<0.01.
Discussion
Culture‐independent methods only provide evidence of the presence and abundance of a bacterium in a sample; they do not provide functional information. The most powerful method of understanding the function of a bacterium is to culture it and to study its physiology. The logic in our decision to culture the Enterobacteriaceae was to (a) use a culture‐independent method to identify DNA bands that are present in tissues of patients with IBD but not in controls, (b) cut the bands out of the gel and sequence them, (c) identify the bacterial species on the basis of the sequence composition, (d) specifically culture the bacterial group identified from the sequence information and (e) investigate potential virulence factors in the cultured bacteria. The RISA analysis (fig 1) we used provided the rationale for culturing E coli and other Enterobacteriaceae.
The numbers of Enterobacteriaceae in our biopsy tissues were 3–4 logs higher in the tissues of patients with IBD than in the controls (fig 2). Martin et al17 found a significant increase in the numbers of E coli only in specimens of patients with CD, but not in those with UC, after a mucin‐releasing step with dithiothreitol. Darfeuille‐Michaud et al20 isolated E coli in higher numbers from tissues of patients with IBD than from controls, but did not enumerate the bacteria on the epithelial tissue with techniques to determine numbers in a range >1 log. Mylonaki et al21 used fluorescence microscopy to show that the numbers of E coli were high in the rectal tissue of patients with UC but not of controls, but this methodology did not allow for decimal enumeration.
Conte et al22 showed that Gram‐negative bacteria, including E coli, have increased by 3–4 logs in tissues of patients with IBD, a result strikingly similar to ours. It is thus clear that the numbers of Enterobacteriaceae do increase on the epithelial tissue of patients with IBD, irrespective of the differences in techniques used. The major difference in our study was that we resuscitated tissue in buffered peptone water to ensure that bacteria, even at very low numbers, were given the maximum chance to grow. This was done because E coli in environmental samples enter into the viable but non‐culturable state, making them difficult to grow without a resuscitation step.23
Clermont et al15 developed a method to type pathogenic E coli using chuA, a gene required for haem transport in enterohaemorrhagic O157:H7 E coli, yjaA, a gene identified in E coli K‐12 but which has no known function, and TSPE4.C2, a cryptic fragment that was identified from subtractive libraries. These genes, when applied to 230 isolates, determined that type B2, and to a lesser extent type D, included virulent extraintestinal strains of E coli, but the prevalence of B2 and D in gastrointestinal isolates was not determined. E coli from stool samples from a range of geographically separate normal healthy human subjects determined that non‐pathogenic groups A and B1 were most prevalent, while group D made up only 15% and group B2 11% of isolates.24 A significant relationship (p<0.05) between the B2 genotype of “resident”, or adherence factor carrying E coli in infants25 and in human colonic cells of adults26, has been shown. We found a significant relationship (p = 0.04) between IBD and the B2+D genotype (table 5). Our data suggest a significant relationship between the presence of a SPATE (p = 0.02) or an adhesin (p = 0.04) in B2+D‐positive E coli strains.
The “resident” population that Nowrouzian et al25 referred to are strains that have adherence factors, in particular P fimbriae that promote adherence to enterocytes. This definition is of course somewhat arbitrary because there are a large number of cell factors that promote adhesin to intestinal tissue. E coli from our controls had relatively few adhesins (table 4), while IBD strains had a higher prevalence of Ag43, AIDA‐I, Sfa, AfaE‐3 and PAI I. Ag43 is a surface adhesin that promotes bacterial biofilm formation due to cell‐to‐cell aggregation,27 Sfa is one of a class of S‐fimbrial adhesins,16 and AIDA‐I is an adhesin‐like protein.16 Martin et al17 measured adherence and invasion of IBD‐derived E coli in tissue culture, and showed that E coli strains isolated from patients with CD possessed haemagglutinating ability to all red cells regardless of blood group. We did not measure adherence and invasion in a cell culture assay but all E coli isolates, both from tissues of patients with IBD and controls, were negative for the presence of bmaE gene encoding M‐agglutinin.
A novel finding of this study is the higher prevalence of E coli from the B2+D phylogenetic group in tissues of patients with IBD. Moreover, SPATE28 and adhesins, that have been primarily associated with E coli isolated from urinary tract infections, were also more prevalent in E coli isolates from patients with IBD. SPATE are a unique class of transporters found in the Enterobacteriaceae that direct their own transport across the outer cell membrane. These proteins have been implicated in virulence but their precise role is not known. We believe that the functional properties of SPATE make them potentially important in IBD because they exhibit functions like degradation of the barrier function of the gut, and cleavage of proteins in the enterocyte, all of which are phenotypes associated with IBD.28 For example, Vat, Pic and Pic‐like have haemaglutinin, mucinase and elastase activity,29,30 Sat has cytotoxic effects31,32,33 on cells as well as elastase activity,27,28,29,30 and EspP cleaves lipoproteins.27,28,29,30 More recently, functional studies on SPATE have demonstrated that Pic and Sat can cleave coagulation factor V, potentially linking it to haemorrhagic events in the gut. Additionally, Pic is thought be involved in colonisation of E coli to intestinal tissue.34
One means by which SPATE may be involved in promoting inflammation could be as an accessory protein in pathogenicity islands (PAI). EspC, a SPATE from enteroaggregative E coli,35 and the non‐SPATE Tsh autotransporter36 have been associated with PAI. Bidet et al37 suggested that PAI IJ96 were associated with hra, hlyA, cnf1 and pap. We determined the prevalence of hlyA and cnf1 and cnf2 in our isolates but they were not present. When we designed primers that were conserved for a range of PAI, only 23.1% of CD and 15.8% of UC were positive. If these were the only PAI, then <50% of our SPATE would be associated with PAI. Further work is needed to investigate the possibility of the association of other PAI with SPATE.
One might argue that IBD is caused by a dysfunctional immune system that results in an increase in E coli on the gut tissue, and that E coli has nothing whatever to do with initiating inflammation in UC or CD. If this was true then the genotypes of E coli isolated from tissue should be a consequence of random colonisation of gut mucosa. Consequently, the distribution of these genotypes should be equivalent among IBD and control tissues. This is not the case, with IBD having more (p = 0.04) putatively pathogenic bacteria than controls. What we do not know at this point is whether the increase in certain types of E coli in IBD is a consequence of inflammation, or the cause. One way forward is to describe the E coli population before inflammation sets in—for example, obtain biopsy tissue that ranges along the colon, distal and proximal to UC lesions.
Abbreviations
CD - Crohn's disease
IBD - inflammatory bowel disease
PAI - pathogenicity island
RFLP - restriction fragment length polymorphism
RISA - ribosomal intergenic spacer analysis
SPATE - serine protease autotransporters
UC - ulcerative colitis
Footnotes
Funding: This work was funded by the Crohn's and Colitis Foundation of Canada. CNB is supported in part by a Crohn's and Colitis Foundation of Canada Research Scientist Award.
Competing interests: None.
References
- 1.Koutroubakis I E. Therapy insight: vascular complications in patients with inflammatory bowel disease. Nat Clin Pract Gastroenterol Hepatol 20052266–272. [DOI] [PubMed] [Google Scholar]
- 2.Sartor R B. Role of commensal enteric bacteria in the pathogenesis of immune‐mediated intestinal inflammation: lessons from animal models and implications for translational research. J Pediatr Gastroenterol Nutr 200540(Suppl 1)S30–S31. [DOI] [PubMed] [Google Scholar]
- 3.Rath H C. The role of endogenous bacterial flora: bystander or the necessary prerequisite? Eur J Gastroenterol Hepatol 200315615–620. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein C N, Wajda A, Blanchard J F. The clustering of other chronic inflammatory diseases in inflammatory bowel disease: a population‐based study. Gastroenterol 2005129827–836. [DOI] [PubMed] [Google Scholar]
- 5.Garcia‐Rodriguez L A, Gonzalez‐Perez A, Johansson S.et al Risk factors for inflammatory bowel disease in the general population. Aliment Pharmacol Ther 200522309–315. [DOI] [PubMed] [Google Scholar]
- 6.Hume G, Radford‐Smith G L. The pathogenesis of Crohn's disease in the 21st century. Pathology 200234561–567. [PubMed] [Google Scholar]
- 7.Silverberg M S, Satsangi J, Ahmad T.et al Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 200519(Suppl A)5–36. [DOI] [PubMed] [Google Scholar]
- 8.Korzenik J R. Past and current theories of etiology of IBD: toothpaste, worms, and refrigerators. J Clin Gastroenterol Nutr 200540(Suppl 1)S30–S31. [DOI] [PubMed] [Google Scholar]
- 9.Bull T J, McMinn E J, Sidi‐Boumedine K.et al Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J Clin Microbiol 2003412915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckburg P B, Bik E M, Bernstein C N.et al Diversity of the human intestinal microbial flora. Science 20053081635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riesenfeld C S, Schloss P D, Handelsman J.et al Metagenomics: genomic analysis of microbial communities. Annu Rev Genet 200438525–552. [DOI] [PubMed] [Google Scholar]
- 12.Rappe M S, Giovannoni S J. The uncultured microbial majority. Annu Rev Microbiol 200457369–394. [DOI] [PubMed] [Google Scholar]
- 13.Cardinale M, Brusetti L, Quatrini P.et al Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol 2004706147–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein C N, Nayar G, Hamel A.et al Study of animal‐borne infections in the mucosas of patients with inflammatory bowel disease and population‐based controls. J Clin Microbiol 2003414986–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clermont O, Bonacorsi S, Bingen E.et al Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000664555–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 20041714–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin H M, Campbell B J, Hart C A.et al Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 200412780–93. [DOI] [PubMed] [Google Scholar]
- 18.Machado J, Grimont F, Grimont P A.et al Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res Microbiol 2000151535–546. [DOI] [PubMed] [Google Scholar]
- 19.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, eds. Nucleic acid techniques in bacterial systematics. New York, NY: Wiley, 1991115–175.
- 20.Darfeuille‐Michaud A, Boudeau J, Bulois P.et al High prevalence of adherent‐invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004127412–421. [DOI] [PubMed] [Google Scholar]
- 21.Mylonaki M, Rayment N B, Rampton D S.et al Molecular characterization of rectal mucosa‐associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis 200511481–487. [DOI] [PubMed] [Google Scholar]
- 22.Conte M P, Schippa S, Zamboni I.et al Gut‐associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006551760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nystrom T. Not quite dead enough: on bacterial life, culturability, senescence, and death. Arch Microbiol 2001176159–164. [DOI] [PubMed] [Google Scholar]
- 24.Duriez P, Clermont O, Bonacorsi S.et al Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 20011471671–1676. [DOI] [PubMed] [Google Scholar]
- 25.Nowrouzian F L, Wold A E, Adlerberth I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis 20051911078–1083. [DOI] [PubMed] [Google Scholar]
- 26.Nowrouzian F L, Adlerberth I, Wold A E. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect 20068834–840. [DOI] [PubMed] [Google Scholar]
- 27.Henderson I R, Navarro‐Garcia F, Desvaux M.et al Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 200468692–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson I R, Nataro J P. Virulence functions of autotransporter proteins. Infect Immun 2001691231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parham N J, Pollard S J, Desvaux M.et al Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol 2005434076–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta P R, Cappello R, Navarro‐Garcia F.et al Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect Immun 2002707105–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betis F, Brest P, Hofman V.et al The Afa/Dr adhesins of diffusely adhering Escherichia coli stimulate interleukin‐8 secretion, activate mitogen‐activated protein kinases, and promote polymorphonuclear transepithelial migration in T84 polarized epithelial cells. Infect Immun 2003711068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taddei C R, Fasano A, Ferreira A J.et al Secreted autotransporter toxin produced by a diffusely adhering Escherichia coli strain causes intestinal damage in animal model assays. FEMS Microbiol Lett 2005250263–269. [DOI] [PubMed] [Google Scholar]
- 33.Guignot J, Chaplais C, Coconnier‐Polter M H.et al The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell Microbiol 20079204–221. [DOI] [PubMed] [Google Scholar]
- 34.Henderson I R, Czeczulin J, Eslava C.et al Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun 1999675587–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellies J L, Navarro‐Garcia F, Okeke I.et al EspC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect Immun 200169315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dozois C M, Dho‐Moulin M, Bree A.et al Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect Immun 2000684145–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bidet P, Bonacorsi S, Clermont O.et al Multiple insertional events, restricted by the genetic background, have led to acquisition of pathogenicity island IIJ96‐like domains among Escherichia coli strains of different clinical origins. Infect Immun 2005734081–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]