Abstract
Sulphate is a major macronutrient required for the synthesis of the sulphur (S)-containing amino acid cysteine and thus is critical for cellular metabolism, growth and development and response to various abiotic and biotic stresses. A recent genome-wide expression study suggested that several auxin-inducible genes were up-regulated by S deficiency in Arabidopsis. Here, we examined the relationship between auxin signaling and S deficiency. Investigation of DR5::GUS expression patterns indicates that auxin accumulation and/or response is suppressed by S deficiency. Consistently, S deficiency resulted in the suppression of lateral root development, but the axr1-3 mutant was insensitive to this response. Furthermore, the activation of the promoter for the putative thioglucosidase gene (At2g44460) by S deficiency was suppressed by auxin, cytokinin and abscisic acid (ABA). Interestingly, the activation of At2g44460 by S deficiency is regulated by the availability of carbon and nitrogen nutrients in a tissue-specific manner. These results demonstrate that auxin plays a negative role in signaling to S deficiency. Given that activation of the genes encoding the sulphate transporter SULTR1;2 and 5′-adenylylsulphate reductase APR2 are suppressed by cytokinin only, we hypothesize that while cytokinin may play an important role in general S deficiency response, auxin might be only involved in a subset of S deficiency responses such as the release of thiol groups from the S storage sources.
Keywords: Sulphate deficiency, Sulfur metabolism, Thioglucosidase, APR2, Auxin, axr1
Introduction
Sulphate, as a major macronutrient, is required for the biosynthesis of many sulphur (S)-containing compounds including amino acids (Cys and Met), proteins, glutathionine and secondary products such as glucosinolates (Leustek et al., 2000; Saito, 2004). Therefore, the availability of sulphate and its uptake and assimilation are essential for cellular metabolism, plant growth and development, and response to various biotic and abiotic stresses (Leustek et al., 2000; Saito, 2004; Rausch and Wachter, 2005).
Although numerous physiological, molecular and biochemical studies have shown that sulphate uptake and assimilation are controlled by the S availability, how plant cells sense the S status and transduce the signals to activate gene expression remains largely unknown. It is demonstrated that sulphate uptake and metabolism are feedback regulated by the intracellular S status (Leustek et al., 2000; Saito, 2004). Furthermore, they are tightly coordinated by carbon and nitrogen nutrients (Koprivova et al., 2000; Hesse et al., 2003, 2004; Kopriva and Rennenberg, 2004; Maruyama-Nakashita et al., 2004a), making it even more challenging to dissect the S nutrient status signaling pathway or network. Nevertheless, signaling mechanisms have begun to emerge through biochemical, genetic and genomic studies. A recent study using inhibitors has implicated an important role for protein phosphorylation and dephosphorylation in the transcriptional regulation of the high affinity sulphate transporter SULTR1;1 induction at the early stage of S deficiency (−S) response (Maruyama-Nakashita et al., 2004c). More convincing evidence for the involvement of hormone signaling components in −S response come from the genetic study of the cytokinin receptor mutant cre1 (Maruyama-Nakashita et al., 2004b). The potential role of cytokinin in −S response is first implicated by a study in which the −S-activated expression of a seed storage protein β-conglycinin β-subunit is shown to be promoted by applying cytokinin (Ohkama et al., 2002). Recently, cytokinin is shown to inhibit both the −S activated transcription of the gene encoding the high-affinity sulphate transporter SULTR1;2 and the sulphate uptake (Maruyama-Nakashita et al., 2004b). Importantly, the cre1 mutation reduced the cytokinin-mediated suppression of both the SULTR1;2 expression and the sulphate uptake (Maruyama-Nakashita et al., 2004b). This genetic evidence unequivocally demonstrates that cytokinin perception plays a negative role in −S response at least with regard to sulphate transport.
Recent genome-wide expression profiling studies have revealed many candidate genes that are likely involved in −S signaling (Hirai et al., 2003, 2004; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003). Among these, two groups of the −S activated genes are most interesting. One group includes several auxin-inducible genes such as IAA9, IAA17, IAA18 and IAA28 and genes potentially involved in tryptophan-IAA biosynthesis such as NIT3 (Nikiforova et al., 2003). The activation of NIT3 that encodes a nitrilase isoform involved in auxin biosynthesis was reported earlier, however, direct measurement did not reveal a statistically significant difference in IAA levels between seedlings treated by sufficient or deficient S (Kutz et al., 2002). Based on the integrative analyses of transcriptome and metabolome data, it has been further proposed that auxin influx and the IAA28-mediated auxin signaling circuit play an important role in modulating −S response (Nikiforova et al., 2005a, 2005b). Since most of the IAA proteins are negative regulators in auxin signaling (Weijers and Jurgens, 2004), these studies instead indicate that auxin might play a negative role in −S response. However, another study has shown that the 26S proteasome inhibitors (MG132 and clasto-lactacystin-β-lactone) had no effect in the induction of SULTR1;1 (Maruyama-Nakashita et al., 2004c). This seems to be in contrast to the demonstrated role of 26S proteasome in controlling −S response in filamentous fungi and yeast (Marzluf, 1997). It is possible that other plant genes activated by −S can be regulated by the 26S proteasome. However, given by the critical role of 26S proteasome-mediated protein degradation in auxin signaling, this implicates the role of auxin, if there is any, is likely restricted to certain genes or certain aspects of −S responses. Therefore, in order to determine the definite role of auxin in −S signaling, molecular and genetic approaches are necessary.
The other group of genes, such as At2g44460 and At3g60140, encode the putative thioglucosidase genes, and their transcription is induced most strongly by −S (Maruyama-Nakashita et al., 2003). Although their biochemical function remains to be determined, it has been proposed based on sequence homology that these genes might function in the release of thiol groups by breaking down gluconsinolates (Maruyama-Nakashita et al., 2003). The integrative analysis of transcriptome and metabolome studies has implicated that gluconsinolates may play an important role in −S response (Hirai et al., 2004, 2005). Thus, in response and subsequent adaptation to changes in S nutrient levels, plant cells must be able to sense both intracellular S nutrient status and extracellular S availability in order to execute an energy cost-effective program to increase either the transport of sulphate from soil or the release of thiols from the intracellular storage, or both.
In this study, we aimed to determine the relationship between −S and auxin response using molecular and genetic approaches. We first showed that the auxin response reporter DR5::GUS (Ulmasov et al., 1997) was down-regulated by −S. Consistent with this, lateral root formation was suppressed, but this growth and developmental response was abolished in the auxin signaling mutant axr1 (Leyser et al., 1993). Furthermore, external application of auxin down-regulated the activity of the At2g44460 gene promoter under −S. Taken together, our results demonstrate that auxin plays a negative regulatory role in modulating the −S response. Additionally, our results indicate that abscisic acid (ABA) might play a similar role in −S response. Interestingly, auxin did not suppress expression of the 5’-adenylylsulphate reductase gene (APR2) under −S, as observed for SULT1;2 (Maruyama-Nakashita et al., 2004b). This indicates that auxin might be involved in only a subset of −S response, such as the activation of At2g44460 expression and possibly as a consequence, the release of thiols from the intracellular S storage. The demonstration of the involvement of auxin signaling in −S response will advance our mechanistic understanding of how plants cope with the dynamic nutrient environments.
Materials and methods
Plant materials and nutrient and hormone treatments
Arabidopsis thaliana Columbia (Col), transgenic plants and axr1-3 in the Col background were used in this study. For nutrient and hormone treatments, seeds were sterilized and placed on either liquid or 1% PhytoBlend-solidified medium with or without sulphate, and cold-treated at 4°C for 2–4 days. After germination, seedlings were then grown in the growth chamber at 22°C with 16 h light and 8 h dark.
For nutrient and hormone treatments, seeds were sowed on the half-strength Murashige and Skoog (MS) medium supplemented with 1% sucrose and cold treated for 4 days. After 5 days of vertical growth, seedlings were transferred to 24-well plates that contain 1.5 ml liquid nutrient solutions and cultured for 2 days with gentle shaking (150 rpm). The C, N and S nutrient solutions were prepared by adjusting the original full-strength (1X) MS medium (about 60 mM N and 1.73 mM S) with various C, N or S. Briefly, sucrose was used as the C source unless specified, and 60 mM sucrose was designated 60 mM C. For the N sources, KNO3 and NH4NO3 were used based on the 1:1 molar ratio; for example, 60 mM N represents 20 mM KNO3 and 20 mM NH4NO3. When lower amounts of KNO3 were prepared, KCl was added to maintain the same final molar concentration of K+ as in 1XMS. For the medium involving various S levels, the minor salt MnSO4 · H2O was replaced by MnCl2 · 4H2O to maintain the same molar concentrations of Mn2+ as in 1XMS. For the full-strength (+S) that contained 1.6001 mM (instead of 1.7301 mM S in 1XMS), ZnSO4 · H2O was replaced by the same molar concentration of ZnCl2 · 4H2O while MgSO4 · 7H2O and FeSO4 · 7H2O were the same as in 1XMS. To prepare for the medium containing 0.0001 mM S that was from CuSO4 only, MgSO4 · 7H2O, FeSO4 · 7H2O, and ZnSO4 · H2O were respectively replaced by the same molar concentrations of MgCl2 · 6H2O, FeCl2 · 4H2O and ZnCl2 · 4H2O as in 1XMS. For the 0.0301 mM sulphate medium, only ZnSO4 · H2O (0.03 mM) and CuSO4 (0.0001 mM) were present, and MgSO4 · 7H2O and FeSO4 · 7H2O were respectively replaced by MgCl2 · 6H2O and FeCl2 · 4H2O at the same molar concentrations as in MS. For the medium containing 0.0001 mM sulphate (designated −S) that was from CuSO4 only, MgSO4 · 7H2O, FeSO4 · 7H2O, and ZnSO4 · H2O were respectively replaced by the same molar concentrations of MgCl2 · 6H2O, FeCl2 · 4H2O and ZnCl2 · 4H2O as in 1XMS.
For all promoter::GUS reporter lines for the purpose of GUS detection, seedlings were first grown vertically on the half-strength MS medium with 1% sucrose. Five days after cold treatment, seedlings were then transferred to 24-well plates that contained 1.5 ml liquid nutrient solutions as described above and cultured for 2 days with gentle shaking (150 rpm). In one experiment, DR5::GUS, Col and axr1-3 seedlings after 5 days of germination and vertical growth on the solidified medium as above were also transferred to the agar (Phytoblend)-solidified medium of either +S or −S for 10 days before determining GUS expression patterns or root development. For anthocyanin and chlorophyll measurement and RT-PCR analysis, seeds were directly placed in 50 ml of liquid medium of either +S or −S prepared as described above.
For the hormone treatments on the promoter::GUS transgenic lines, young seedlings were germinated and vertically grown on the agar-solidified half-strength MS medium (with 1% sucrose) for 5 days after cold treatment. They were then transferred to 24-well plates with liquid medium for 2 days, as described above. For all hormones, 1 μM was used, with the controls that did not supplement any hormones. These hormones include ABA, GA3, BA (cytokinin), ACC (ethylene precursor), BR (epibrassinolide), IAA (auxin), SA (salicylic acid), JA (methyl jasmonate). All chemicals were purchased from Sigma-Aldrich.
Promoter::β-glucuronidase (GUS) construction and GUS assay
For the putative thioglucosidase gene At2g44460 promoter::GUS construct, a 2.7 kb promoter fragments including 9 bp downstream of ATG was PCR amplified from genomic DNA using the high fidelity DNA polymerase Elongase® (Invitrogen) and the gene-specific primers, with the underlying bases indicating the introduced restriction enzyme sites for cloning: sense (DZP11: 5′-ATCCTGCAGCACA ACGAAACCCGATTGATG-3′) and antisense (DZ P12: 5′-ACAACCATGGTGAAAAAATGCATCT TCATATTCCT-3′). The amplified DNA fragment was digested by PstI and NcoI, and then cloned into PstI and NcoI sites of the binary vector pCAMBIA1301 (B4) that contains GUS and the CaMV35S terminator. For the APR2 (At1g62180) promoter::GUS construct, a 3.5 kb fragment including 11 bp downstream of ATG was similarly amplified using the two primers: DZP1, sense: 5′-CATCTGCAGAGATAGATGAAGCGATCACGA-3′ incorporating a PstI site; DZP2, antisense: 5′-CGAAGATCTACAGCTAAAGCCATTTCTAATC-3′ incorporating a BglII site. This fragment digested by PstI and BglII was then cloned into the PstI and BglII sites of pCAMBIA1301. The replacement of the CaMV35S promoter with At2g44460 and APR2 promoters resulted in the transcriptional and translational fusion of these promoters with GUS, giving rise to the DZ9 and DZ5 vectors, respectively. The identity of the PCR-amplified promoter fragments for both vectors was verified by DNA sequencing.
The DZ9 and DZ5 vectors were respectively introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis Col plants by the floral dip method (Clough & Bent, 1998). Homozygous transgenic lines that confer a single T-DNA insertion were obtained by hygromycin selection. Histochemical GUS activity assays were performed on homozygous transgenic seedlings as described (Jefferson et al., 1987), except that the substrate X-Gluc concentration was diluted by four folds.
Root growth and development assay
The lateral root primordia (LRP) that included Stage I to Stage VII and the lateral roots (LR) were counted under microscope, according to Malamy and Benfey (1997).
Anthocyanin and chlorophyll assays
Anthocyanins and chlorophylls were extracted from young seedlings germinated and grown in liquid medium of either +S or −S for 5 days after cold treatment. Briefly, for chlorophyll extraction and measurement, about 20 mg seedlings were placed in 700 μl N, N-dimethylformamide, wrapped with the foil and shaken gently at 4°C, and then the extracts were measured twice for the absorbance at 664 and 647 nm. Total chlorophyll levels were determined using the method of Moran (1982). For anthocyanins, seedlings of about 20 mg were placed in the extraction buffer (99% methanol and 1% concentrated HCl) and shaken as described for chlorophyll extraction, according to the procedure (Rabino and Mancinelli, 1986). The extracts were then measured twice for the absorbance at 530 and 657 nm, and anthocyanin levels were determined based on the formula, A530–0.25 × A657 (Rabino and Mancinelli, 1986). Three replicates for each genotype/treatment were performed for both chlorophyll and anthocyanin measurements.
Reverse transcription (RT)-PCR analysis
Total RNA was extracted with TRIzol (Invitrogen) from 5 to 7 days old seedlings treated by nutrients as above, and then reverse transcribed by Superscript III reverse transcriptase (Invitrogen). PCR analysis was performed using the Taq DNA polymerase (Gen-Script), with gene specific primers and ACT2 as the internal control as described (Xin et al., 2005). Gene specific primers were designed. For SULTR4;2, the sense primer DZP50 was 5′-TCCACCGCTTCATC CTCTTCATCT-3′, and antisense primer DZP51 was 5′-AGAGCCGATGTTGGAAGCAGTAA-3′. For the putative thioglucosidase gene/At2g44460, the sense primer YZP63 was 5′-AACGAGCTCTTGCCACT GAACT-3′ and the antisense primer YZP64 was: 5′-GAGATGGTCCTCATGGTAGCTT-3′.
Results
Sulphate deficiency reduces the activity of the auxin response marker DR5::GUS
To test whether auxin is involved in −S response, we first analyzed the auxin response using the well-characterized DR5::GUS line (Ulmasov et al., 1997). Treatments of young seedlings by −S (extremely low sulphate, 0.0001 mM) and intermediate low (0.0301 mM) concentrations of sulphate for 48 h resulted in different GUS staining patterns compared to the normal S concentration (1.6001 mM, designated +S). Decreasing S concentrations gradually reduced GUS staining in the cotyledons, primary root tips and LR. In the cotyledons, the strong expression of DR5::GUS at 1.6001 mM S in the tip and surrounding edges, where auxin is synthesized, was dramatically reduced under lower sulphate concentrations (Fig. 1). In primary root tips, emerged LR and LRP, the intensity and distribution were also reduced under extremely low and intermediate sulphate concentrations. These results suggest that auxin accumulation and/or sensitivity are suppressed by −S.
Fig. 1.
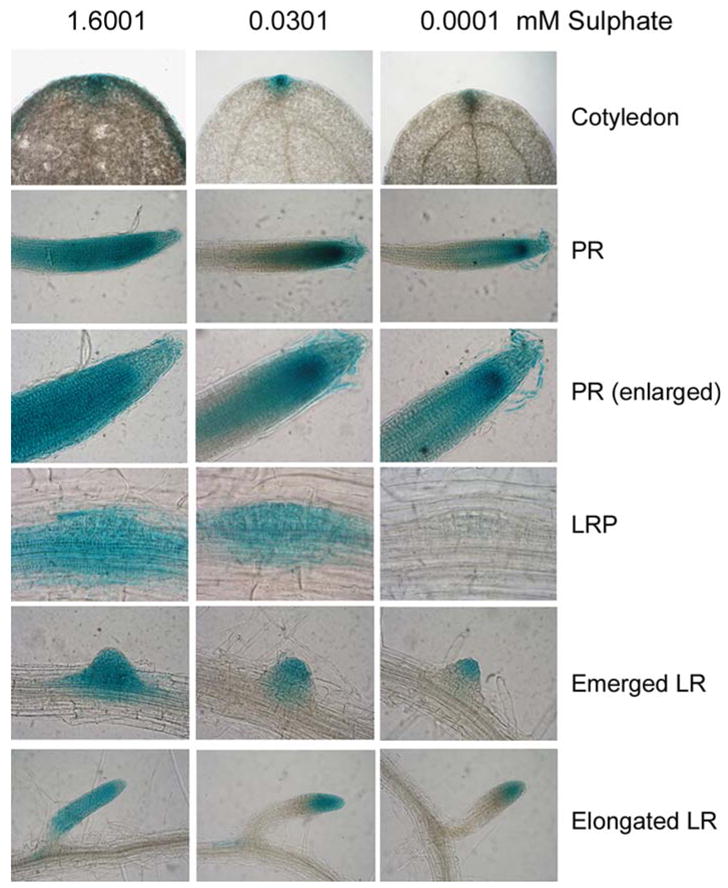
DR5::GUS expression is suppressed by sulphate deficiency. Shown are GUS staining patterns in cotyledons, primary roots (PR), lateral root primordia (LRP), and emerged or elongated long lateral roots (LR)
Sulphate deficiency suppresses the lateral root development
Auxin is critical for root growth and development, and therefore we investigated the lateral root development from the −S treated DR5::GUS plants. As shown in Fig. 2, −S only very weakly stimulated primary root elongation but strongly suppressed lateral root development. Because of the weak effect in the primary root growth (Fig. 2A), we focused our analysis on lateral root development. When the S concentration decreased, the total number of LRP on primary root for each seedling also decreased, although the number of emerged or elongated LR did not change (Fig. 2B). It is possible that these elongated LR already emerged before the −S treatments, or that −S was not sufficient to block the later stages of LRP from emerging. Nevertheless, when the densities of LR or LRP on the primary root were compared, both LR and LRP densities were reduced by decreasing sulphate to 0.0301 mM, although the LRP density was reduced slightly more than the LR density (Fig. 2C). The further reduction of LR or LRP density by extremely low S concentration (0.0001 mM) was not statistically significant (P=0.07). Therefore, our results clearly show that −S suppresses lateral root development, consistent with the decreased DR5::GUS activity under −S (Fig. 1).
Fig. 2.
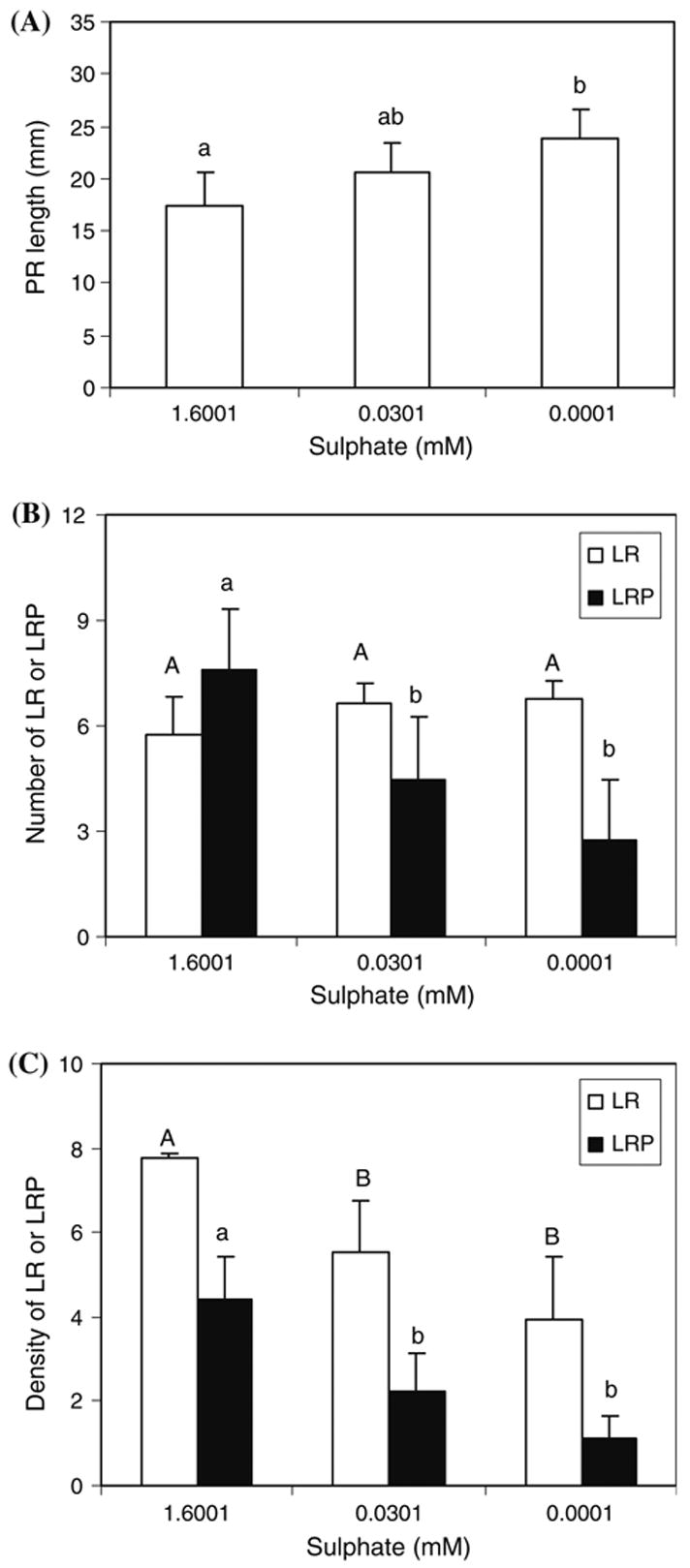
Sulphate deficiency suppresses lateral root development. DR5::GUS seedlings treated identically in Fig. 1 were used to measure the primary root (PR) length and count under microscope the number of the emerged or elongated lateral roots (LR) and the lateral root primordia (LRP). Various stages of LRP were determined according to Malamy and Benfey (1997). The average of 4–7 seedlings measured or counted were shown with the bar representing the standard deviations. (A) Primary root length. (B) Total number of lateral roots and LRP for each seedling. (C) Density of lateral roots or LRP on the primary roots. Density is expressed as the average number of LR or LRP on per cm primary root (PR). Different letters (in B and C, uppercase letters for the LR, and lowercase letters for the LRP) above the columns indicate statistically significant difference (P=0.05)
Sulphate deficiency-suppressed lateral root development is altered in the auxin signaling mutant axr1-3
To substantiate the involvement of auxin signaling in −S response, we subject the auxin signaling mutant, axr1-3, to two different growth conditions. The axr1-3 weak allele has a mutation in AXR1 encoding an enzymatic unit of the E1 RUB1-activating enzyme that is important for ubiquitin-mediated protein degradation (Leyser et al., 1993). When five-day-old seedlings grown vertically on agar plates of +S were transferred to the agar plates of either +S or −S, although there was no difference in the LR density after 10 days of vertical growth, the difference in the LRP density was observed (Fig. 3A). For Columbia (Col) wild-type seedlings grown under −S, the LRP density was reduced by 40% compared to that at +S. The axr1-3 seedlings had a lower density of LRP than Col under +S, but importantly, they did not show further reduction under −S. When the liquid culture condition was tested, a similar pattern was observed (Fig. 3B). Most of the young seedlings after 5 days of germination and growth in the liquid medium of either +S or −S did not develop LR (Fig. 3B). However, the density of LRP on the primary roots of Col seedlings under −S was reduced to 87% of that under +S (Fig. 3A). Although this difference was not very dramatic, it was statistically significant. The lack of difference in the LR density and the less dramatic difference in the LRP density compared to that in DR5::GUS seedlings might be due to different genotypes because DR5::GUS exhibited an almost identical GUS expression pattern under three growth and treatment conditions tested (data not shown). Importantly, we showed that under two different growth conditions, axr1-3 seedlings exhibited a similar insensitive pattern to changes in sulphate concentrations. Therefore, these results demonstrate that AXR1-mediated auxin signaling plays a negative role of −S response at least in the lateral root development.
Fig. 3.
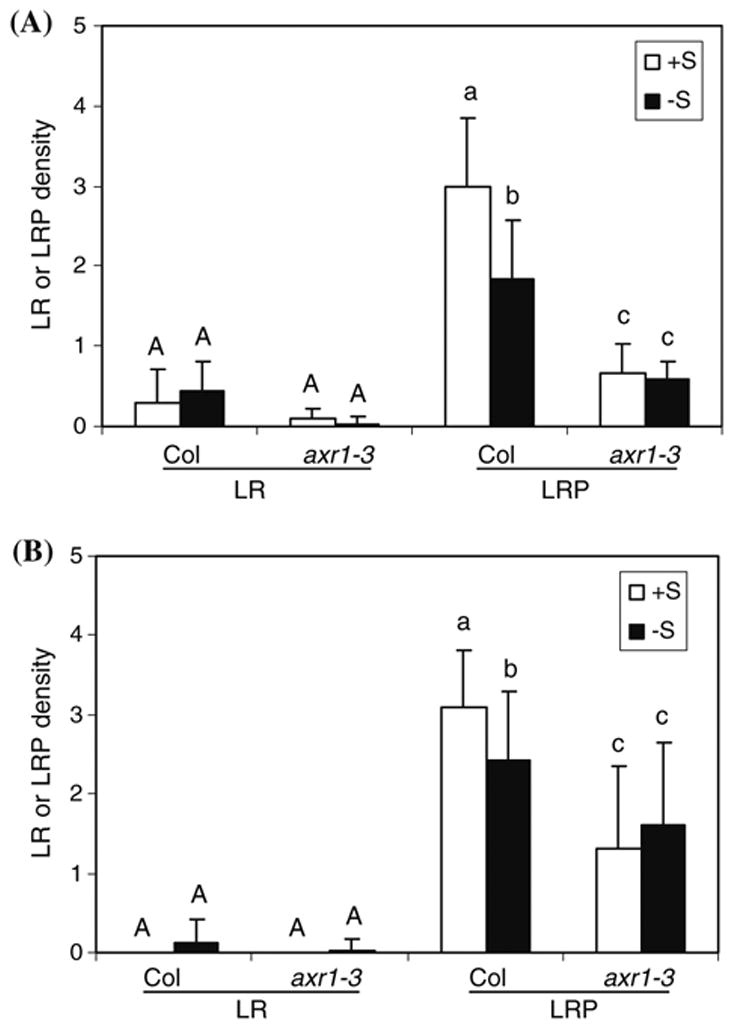
Altered sulphate deficiency stress response in lateral root development in axr1. LR, lateral roots; LRP, lateral root primordia. Density is expressed as the average number of LR or LRP on per cm primary root (PR), with the SD bars shown. (A) Seedlings were transferred to and grown vertically on the agar-solidified medium for 10 d after 5-d vertical growth on the normal, solidified medium (n=4–5). (B) Seedlings were continuously grown for 5 d in liquid medium (n=16). Different letters (uppercase letters for the LR density, and lowercase letters for the LRP density) above the columns indicate statistically significant difference (P=0.05)
Differential responses of axr1-3 to sulphate deficiency-affected accumulation of anthocyanins and chlorophylls
Another physiological aspect of −S response could potentially be the accumulation of anthocyanins, given that −S caused the strong activation of several MYB transcription factor genes including MYB75 and MYB90 (Nikiforova et al., 2003). MYB75 and MYB90 have been demonstrated to function in the regulation of anthocyanin biosynthesis (Borevitz et al., 2000). Therefore, we measured the contents of anthocyanins under −S and +S. Col seedlings clearly doubled the accumulation of anthocyanins when they were subjected to −S compared to +S (Fig. 4A). However, axr1-3 accumulated similar amounts of anthocyanins under +S as Col, and also had a similar increase by −S. This indicates that axr1-3 dose not alter the response to −S in the aspect of anthocyanin accumulation.
Fig. 4.

Altered sulphate deficiency stress response in chlorophyll contents in axr1. Anthocyanin (A) and chlorophyll (B) levels were measured for Col wild-type and axr1 after 5 days (for anthocyanins) or 3, 5, 7, and 9 days (for chlorophylls) of growth in +S or −S. The average of at least three replicates were shown, with the bar representing the SD. Different letters above the columns indicate a statistical significant difference (P=0.05) FW, fresh weight
Surprisingly, in response to −S, both Col and axr1-3 seedlings accumulated more chlorophylls around 5 days after cold treatment and then declined to the same level as +S at Day 7 before a significant decrease at Day 9 (Fig. 4B). Interestingly, at Day 3, axr1-3 accumulated more chlorophylls than Col under both +S and −S conditions, and its chlorophyll levels were similar to that in Col at Day 5. At Day 5, chlorophylls in axr1-3 under +S was similar to that in Col under −S and also increased by −S. Although at Day 7, there was no difference among all genotype/treatments, chlorophylls decreased dramatically in axr1-3 under −S at Day 9. Therefore, while axr1-3 still responded to −S by increasing chlorophyll accumulation transiently, it accumulated more chlorophylls during 3–5 days after cold treatment. These observations implicate that AXR1-mediated auxin signaling is at least part of the suppression mechanism involved in the transient chlorophyll accumulation in response to −S stress. This transient increase has not been reported before and its role needs to be further investigated. Given that excessive chlorophylls could lead to the generation of singlet oxygen as observed in high light or drought stresses (Krieger-Liszkay, 2005), it is possible that over-accumulation of chlorophylls at the early stage of −S might either be toxic to the −S-stressed plants or serve as a signal for adaptation to −S through singlet oxygen.
Auxin suppresses the sulphate deficiency-activated, putative thioglucosidase gene (At2g44460)
The insensitivity to −S in lateral root development and the altered chlorophyll accumulation observed in axr1-3 led us to test whether auxin can directly suppress the activation of genes by −S. However, RT-PCR analysis using young seedlings grown in the liquid medium failed to show the suppression by IAA of expression of APR2, SULTR4;2,and At2g44460, the putative thioglucosidase gene that is most strongly induced by −S in a DNA microarray study (Maruyama-Nakashita et al., 2003), under −S (data not shown). As RT-PCR might not reveal any tissue-specific patterns of gene expression, we decided to investigate the GUS expression patterns using the At2g44460 promoter::GUS and APR2 promoter::GUS reporter systems. GUS staining in several independent lines showed that the promoters of both genes were strongly activated by −S with a preference in the roots (Fig. 5 and data not shown). The consistency between the promoter activity and the gene expression studies (Maruyama-Nakashita et al., 2003; Hira et al., 2003, 2004) thus allows us to use these promoter::GUS lines to investigate the effects of auxin and other hormones in the transcriptional regulation of At2g44460 and APR2 genes. Consistently, treatments of 1 μM IAA for 2 days did suppress the activity of the At2g44460 gene promoter in the primary root but not in the shoots under −S (Fig. 5A). This tissue-specific suppression may explain why we did not detect the obvious difference of transcript levels using RNA extracted from the whole seedlings (data not shown). The suppression of At2g44460 is probably triggered by −S because IAA did not suppress the GUS activities under the +S condition. Actually, IAA seemed to slightly activate the promoter activity in the emerging LR and the cotyledons under +S. In contrast, the APR2 promoter activity was not affected by auxin treatment under −S (Fig. 5B). Therefore, the auxin-mediated suppression of gene expression is probably specific to certain genes and/or certain tissue types. Surprisingly, the At2g44460 promoter was also suppressed by ABA and cytokinin (BA), but not by other hormones except brassinosteroids (BR) that seemed to slightly activate the promoter activity (Fig. 5A). The slight activation of At2g44460 by BR is not surprising given by the known antagonistic interactions between BR and ABA/auxin (Ephritikhine et al., 1999). In contrast to the At2g44460 promoter, APR2 was only suppressed by cytokinin (Fig. 5B), similar to SULTR1;2 (Maruyama-Nakashita et al., 2004b). The differential regulation by IAA, ABA and BA for At2g44460 and APR2 was similarly observed by independently transformed promoter::GUS reporter lines (Supplemental Fig. 1). Therefore, on the one hand, cytokinin has a similar inhibitory effect on these three genes likely involved in different aspects of sulphate transport and S metabolism. On the other hand, auxin and ABA seem to have more specific effects on certain genes such as At2g44460.
Fig. 5.
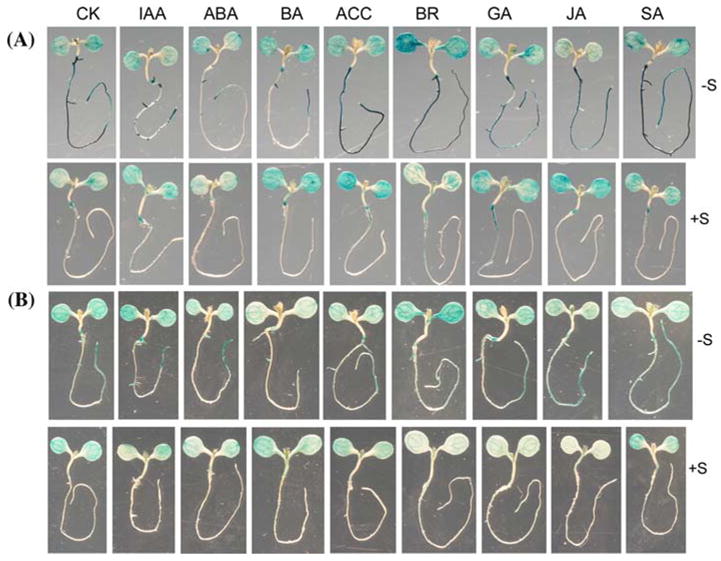
Promoter activities of the putative thioglucosidase gene At2g44460 and APR2 are differentially suppressed by hormones. Five-day old seedlings grown identically as in Fig. 1 were treated for 48 h under either −S or +S supplemented with 1 μM hormones. CK, no hormone control; IAA, auxin; BA, Cytokinin; ACC, ethylene precursor; BR, brassinosteroids; JA, methyl jasmonic acid; SA, salicylic acid. (A) GUS staining pattern of the putative thioglucosidase gene At2g44460 promoter. (B) GUS staining pattern of the APR2 gene promoter
If ABA suppresses the activity of the At2g44460 promoter like auxin, we would expect that −S might also suppress ABA accumulation or response. To gain some supporting evidence for the possible involvement of ABA in −S response, we treated the pRD29B::GUS reporter line under various sulphate levels as in the case of DR5::GUS (Fig. 1). pRD29B::GUS has been demonstrated to reflect the physiologically active pools of ABA (Christmann et al., 2005). Not surprisingly, decreasing sulphate amount from the normal concentration (1.6001 mM) to the intermediate sulphate concentration (0.0301 mM) caused an obvious reduction of GUS staining in the cotyledons (Supplemental Fig. 2). When sulphate was further reduced to 0.0001 mM, GUS staining in the hypocotyl-root junction almost disappeared, resulting in a very weak GUS staining in the seedlings. We did not observe any GUS staining difference in the primary root tips of the seedlings (data not shown). Together with the ABA-mediated suppression of the putative thioglucosidase gene At2g44460 promoter under −S, this result indicates that ABA might also participate in modulating −S responses.
The putative thioglucosidase gene (At2g44460) exhibits a carbon and nitrogen nutrient-dependent regulatory pattern in activation by sulphate deficiency
To test whether the auxin- and ABA-suppressed putative thioglucosidase gene (At2g44460) is specifically involved in −S response, we decided to determine whether this gene is also activated by the deficiency of carbon (C) and nitrogen (N) nutrients. Furthermore, it has been shown C and N nutrients regulate the expression of certain genes under −S (Koprivova et al., 2000; Hesse et al., 2003, 2004; Kopriva and Rennenberg, 2004; Maruyama-Nakashita et al., 2004a), but it remains unknown whether C and N nutrients have differential or overlapping effects on −S activation of gene expression. Therefore, we used a combinatorial design to assess the promoter activities of At2g44460 under various combinations of C, N and S nutrients (Fig. 6A, B).
Fig. 6.
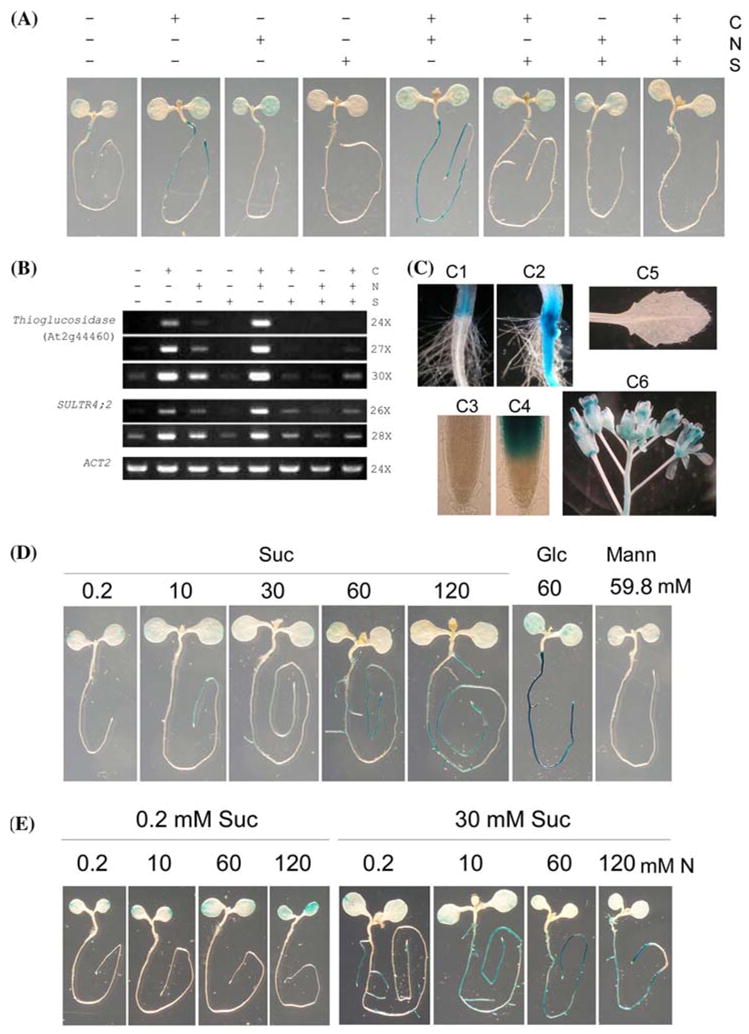
The putative thioglucosidase gene (At2g44460) promoter-driven GUS expression is differentially regulated by C, N and S nutrients in a tissue-specific manner. (A) GUS staining patterns under various C, N and S nutrient conditions. Five-day old seedlings were grown under half-strength MS basal salts supplemented with 1% sucrose and 1% agar were treated with various combinations of C, N and S liquid medium for 48 h and then stained for GUS activity for 3–5 h. +C, +N and +S indicate the presence of 60 mM sucrose, 60 mM total N (20 mM KNO3 and 20 mM NH4NO3), and 1.6001 mM respectively. −C, −N and −S represent the deficiencies of C (0.2 mM sucrose), N (0.2 mM total N), and 0.0001 mM (from CuSO4), respectively. (B) RT-PCR analysis of SULTR4;2 and the putative thioglucosidase gene expression under various C, N and S nutrient conditions. Five-day old seedlings were similarly treated by various combinations of C, N and S for 48 h (see legend in Fig. 6A) before RNA extraction. ACT2 was used as the internal control. PCR using 24, 27 and 30 cycles for the putative thioglucosidase gene and 26 and 28 cycles for SULTR4;2 were performed to differentiate the transcript abundance for every treatment. (C) Tissue-specific GUS expression patterns. Both the root-hypocotyl junctions (C1 and C2) and the root tips (C3 and C4) were shown to compare the effects of +S (C1 and C3) and −S (C2 and C4) treatments. A true leaf (C5) and flowers (C6) from a 30–day-old plant grown in the normal soil in the growth chamber were also shown. (D) GUS staining patterns under −S with various C levels. (E) GUS staining patterns under −S with various N levels at both low C and high C conditions. For (D) and (E), five-day old seedlings grown identically as in Fig. 6A were treated for 48 h before GUS staining. Suc, sucrose; Glc, glucose; Mann, mannitol
Similar to the observation shown in Fig. 5A, GUS activity in the seedlings grown at +C +N −S increased dramatically in the roots, while it did not change in the cotyledons, compared to the normal condition +C+N+S (Fig. 6A). This indicates that the −S activation of the promoter mainly occurs in the root system. In strong contrast to +C +N −S, deficiencies of C (−C +N +S) or N (+C −N +S) alone did not lead to obvious changes in the GUS staining patterns compared to +C+N+S (Fig. 6A). The lack of activation by −C or −N alone indicates that the activation of the At2g44460 promoter is relatively specific to −S.
Interestingly, the strong −S activation of At2g44460 promoter activity observed at +C +N −S disappeared if both N and C were also deficient (comparing −C −N −S with +C +N −S or −C −N +S; Fig. 6A). This indicates an absolute requirement for C and N in the activation of At2g44460 by −S. However, while the presence of N (−C +N −S) only slightly activated the induction by −S, the presence of C (+C −N −S) dramatically activated the −S activation, compared to −C−N−S. This indicates that under the conditions tested, N alone only has a minor effect for the −S activation, while C plays a predominant role in activating gene expression under −S. Nevertheless, +C +N −S exhibited a stronger induction than +C −N −S, suggesting a synergistic interaction between C and N on the −S activation of these two genes.
To assess where the observed changes of At2g44460 promoter activity reflected endogenous gene expression patterns in response to various C, N and S combinations, RT-PCR analysis using young seedlings was performed. The −S-activated sulphate transporter gene SULTR4;2 (Kataoka et al., 2004) was used as a control. As shown in Fig. 6B, both SULTR4;2 and At2g44460 transcripts exhibited a very similar pattern in response to C, N and S nutrients. Interestingly, there was even a very slight decrease of SULTR4;2 and At2g44460 expression by the deficiency of either C or N alone compared to that of S deficiency alone. In most cases, both RT-PCR and the At2g44460 promoter-driven GUS expression showed very similar patterns, demonstrating that the 2.7 kb promoter fragment with five SURE motifs responsible for −S response (Maruyama-Nakashita et al., 2005) contains all of necessary cis-regulatory elements in directing gene expression in response to various nutrients. Therefore, the At2g44460 promoter::GUS line was used to further reveal their spatial expression patterns in particular in response to C, N and S nutrient interactions.
The putative thioglucosidase gene At2g44460 clearly exhibited a tissue-specific expression patterns (Fig. 6C). First, −S activated its expression in the vascular tissues and epidermal cells but not in the root hairs (Fig. 6, C1 and C2). Second, GUS was very strongly activated by −S in the root regions close to the tip but not in the tip (Fig. 6, C3 and C4). Third, in adult plants grown in the soil where sulphate supply was normal, GUS activity was absent in young leaves (Fig. 6, C5) but strong in both male and female reproductive organs (Fig. 6, C6).
To gain further insights into how C and N nutrients regulate the expression of the putative thioglucosidase gene expression in response to −S, we performed dose–response studies for the At2g44460 promoter::GUS line. We first studied the effects of various sucrose levels in the induction of the promoter. At the −N −S background, the promoter activity increased with sucrose levels (Fig. 6D). Interestingly, when 60 mM glucose was used, the −S activation was slightly stronger than 60 mM sucrose, indicating that glucose is more potent than sucrose in activating the −S response. The effect of sucrose or glucose on the −S activation is unlikely due to osmosis because 59.8 mM mannitol plus 0.2 mM sucrose did not affect the putative thioglucosidase gene promoter activity, compared to 60 mM sucrose. Careful observations of the GUS staining patterns revealed that the −S activation appeared to occur only in the roots but not in the cotyledons (Fig. 6D). Interestingly, decreasing C levels caused the −S activation to be limited to the regions closer and closer to the root tip.
The indication of the synergism between C and N observed above (Fig. 6A, B) led us to further investigate the effects of N under both low C and high C conditions. At the −C−S background (0.2 mM Suc plus 0.0001 mM ), the presence of N from 10 to 120 mM only very slightly increased the promoter activity (Fig. 6E), consistent with our previous studies (Fig. 6A, B). At the very high N level, 120 mM, which is twice as that of the full strength of the MS medium, the At2g44460 promoter activity increased only slightly in both the cotyledons and the root tip region. However, when C increased to 30 mM, the strong activation was clearly observed even at the minimal N level (0.2 mM). This activation seemed to be restricted to the tip of primary or LR. Furthermore, 10 mM N already strongly activated the promoter throughout the whole root system, and 60 mM N seemed to have already saturated the response. These results clearly support that while C plays a predominant role in −S activation of the putative thioglucosidase gene expression, N has a synergistic effect with C. It is not known why at the low C level, the −S activation modulated by N also occurred in the cotyledons, while it happened exclusively to the roots at the higher C level. Importantly, as observed for the C effect (Fig. 6D), decreasing N from 60 to 0.2 mM under 30 mM C resulted in the suppression of the activation by −S in the root regions to be expanded away from the root tip (Fig. 6E).
Discussion
S nutrition is essential for various cellular activities and, therefore, sulphate uptake and S metabolism must be tightly coordinated in order to optimize cellular metabolism, growth and development. However, the mechanism by which plants constantly and robustly monitor the dynamic changes of S nutrients remains largely unknown. In this report, using a combination of genetic, molecular and physiological approaches, we have shown that auxin plays a negative regulatory role in −S response. Furthermore, the auxin-suppressed putative thioglucosidase gene (At2g44460) exhibits a C and N nutrient-dependent activation by −S in a tissue-specific manner.
A negative regulatory role of auxin in sulphate deficiency responses
Several lines of molecular and genetic evidence presented here convincingly show that auxin plays a negative regulatory role in −S response. First, using DR5::GUS as an auxin response marker, we have revealed that −S likely suppresses auxin level or sensitivity. Second and most importantly, the suppression of lateral root development is not observed in the auxin signaling mutant, axr1. The transient activation of chlorophyll accumulation by −S is also altered in axr1-3. Third, the activation by −S of the At2g44460 gene promoter is strongly suppressed by auxin in the roots.
The role of hormones in nutrient signaling has received increasing attentions. Cytokinin is the major hormone that has been implicated or demonstrated in signaling to a number of nutrients, including nitrogen, phosphorus and sulphur (Takei et al., 2002; Ohkama et al., 2002; Franco-Zorrilla et al., 2005; Maruyama-Nakashita et al., 2004b). In this study, we show that cytokinin seems to have a similar effect in suppressing the activation by −S of the two genes that encode 5’-adenylylsulphate reductase (APR2) and the putative thioglucosidase (At2g44460), respectively. Together with the demonstration of the cytokinin suppression of the sulphate transporter (SULTR1;2) gene and sulphate transporter activity (Maruyama-Nakashita et al., 2004b), these results suggest that cytokinin likely plays an important role in mediating or modulating the general −S responses ranging from sulphate uptake to S metabolism and possibly thiol release from the intracellular S storage. How cytokinin perception and signaling are regulated by −S and subsequently trigger the physiological responses remain to be revealed.
The involvement of auxin in −S response seems to be more restricted to certain genes and/or specific tissue types. Availability of various nutrients differentially modifies root development and architecture (Lopez-Bucio et al., 2003). Part of our physiological observations seems to differ from the general perception that −S stimulates root branching (Lopez-Bucio et al., 2003) that is derived from an earlier study and a recent observation (Kutz et al., 2002; Nikiforova et al., 2003). While there were no quantitative analyses on the LR and LRP densities in these studies to support the promotion of lateral root development by a relatively long term (from 10 days to 4 or 6 weeks) −S stress, we did not observe a dramatic difference in the LR density in DR5::GUS and Col. Instead, we found a significant difference in the LRP density after 2–10 days of −S stress (Figs. 2–3). Similar discrepancies have also been observed in several studies that addressed the relationship between auxin and phosphate-starvation (Hardtke, 2006). These include the opposite effects of phosphate-starvation on lateral root density, and the contrasting results of DR5::GUS expression patterns in response to phosphate starvation (such as Linkohr et al., 2002; Lopez-Bucio et al., 2002; Al-Ghazi et al., 2003; Lopez-Bucio et al., 2005; Nacry et al., 2005). It is likely that these are caused by experimental variations given that plants are very sensitive to their surrounding environments and different developmental stages may show different responses (Nacry et al., 2005). In our study, we have found that the LRP density is suppressed by −S under all of the three different growth conditions (Figs. 2, 3). The consistency in the suppression of the LRP formation with the DR5::GUS staining pattern and the axr1 phenotypes allows us to conclude that the suppression of lateral root development is a physiological response to −S stresses at least in our experimental conditions. Together with the observed transient activation of chlorophyll accumulation, the suppression of the LRP formation is an early response to the −S stress so that plants would optimize cellular metabolism in other parts of the plant by reducing cell division in the roots. If the −S stress persists, plants might sense this as a sustained stress and therefore increase root branching and growth in order to compete for the uptake of S nutrients.
What is the role of auxin in regulating the −S response? The involvement of auxin in −S response has been implicated by earlier studies, although there is no convincing evidence to support it. First, the NIT3 gene has been shown to be activated by −S in a study using the promoter::GUS reporter system (Kutz et al., 2002), and several genes likely involved in tryptophan-IAA pathway (such as putative myrosinase and NIT3 genes) have been revealed to be up-regulated by −S in a DNA microarray study (Nikiforova et al., 2003). The NIT3-encoded nitrilase isoform 3 is proposed to convert the IAA precursor indole-3-actetonitrile, which results from the degradation of glucobrassicin (one of major types of glucosinolates), into IAA (Vorwerk et al., 2001). There seemed to be a correlation between the NIT3 promoter activation and the decline in total inorganic sulphate and thiols in root tissues of −S stressed plants (Kutz et al., 2002). However, the measurement of root IAA levels did not show a clear difference between plants treated with −S and +S (Kutz et al., 2002). Furthermore, the DNA microarray study also revealed the activation of several auxin-inducible genes such as IAA9, IAA17, IAA18 and IAA28 (Nikiforova et al., 2003). Although the precise roles for most of the auxin-induced genes in auxin signaling remain to be determined, they are generally transcriptional repressors (Weijers and Jurgens, 2004), indicating a negative role for auxin signaling in −S response. More recently, an auxin signaling circuit was proposed to explain the −S effect in root development (Nikiforova et al., 2005a) based on the integrative network analysis of both transcriptome (Nikiforova et al., 2003) and metabolome data (Nikiforova et al., 2005b). In this proposed circuit, IAA28 is induced to suppress auxin-induced genes, ultimately leading to the tight regulation of a balanced auxin response. Our results show that the DR5::GUS activity is suppressed by decreasing sulphate levels. Furthermore, the axr1 mutant did not show further suppression of lateral root development. Together with the observation that −S does not change the auxin levels (Kutz et al., 2002), we conclude that it is more likely that auxin sensitivity, instead of auxin level, is suppressed by −S.
The role of ABA in response to −S is intriguing. ABA is critical for plant response to a wide array of abiotic and biotic stresses, yet it inhibits the activation of the putative thioglucosidase gene At2g44460 in response to the −S stress. Future work using various ABA signaling mutants will help to determine which branch of ABA signaling is actually involved in−S response. Promoter scan of At2g44460 revealed many cis-regulatory elements (such as ABRE and MYB/ MYC recognition sites) that are involved in ABA response (data not shown). This gene also contains five SURE motifs that are important for −S activation (Maruyama-Nakashita et al., 2005). Therefore, it will be interesting to determine how the −S signal leads to the suppression of ABA accumulation or response so that At2g44460 is preferentially activated by transcriptional regulators recognizing the SURE motifs. The possibility that ABA and auxin signaling pathways interact with each other in response to −S cannot be ruled out. It has been shown that ABA stimulates lateral root primordium formation through auxin and inhibits lateral root growth independent of auxin (De Smet et al., 2003). In our study, we show that ABA and auxin affect the expression of the putative thioglucosidase gene in the roots with a similar pattern of retaining the expression in the primary or lateral root tips (Fig. 5A). However, decreasing sulphate levels lead to distinct tissue-specific patterns of DR5::GUS and pRD29B::GUS although both are suppressed (Figs. 1, 6). Therefore, if the cross-talk between auxin and ABA signaling exists in −S response, it is likely a complex interaction.
Role of the auxin-suppressed putative thioglucosidase gene (At2g44460) in sulphate deficiency response and its regulation by C and N nutrients
In this study using a combinatorial design of C, N and S nutrient treatments, we have revealed novel differential transcription patterns in response to S deficiency for genes encoding the auxin-suppressed, putative thioglucosidase (At2g44460) and a sulphate transporter SULTR4;2. We not only confirm that the −S activation requires both C and N availability, but show that the promotive effect of C and N is dependent upon the S availability because the deficiency of either C or N alone did not activate expression of these genes. Furthermore, N alone does not greatly affect gene expression in response to −S if the C level is kept low. However, when the C level increases, N shows a dramatic effect in activating gene expression under −S. Therefore, these results convincingly show that these genes are first responding to the S availability but the realization of the −S activation depends on the C and N nutrients. In this regulatory process, C plays a predominant role and N synergistically interacts with C.
Although the biochemical function of the protein encoded by the putative thioglucosidase gene remains to be determined, it has been hypothesized to act in the release of thiol groups from glucosinolates in response to −S (Maruyama-Nakashita et al., 2003). Glucosinolates are a large group of secondary compounds that contain C, N and S and are important for biotic and abiotic stress responses either as defense compounds or S storage sources (Rausch and Wachter, 2005). In an integrative transcriptome and metabolome study (Hirai et al., 2004, 2005), a regulatory link between glucosinolate metabolism, primary metabolism and S and N nutrition has been revealed. This implicates the importance of glucosinolates as an alternative S source when plants are −S stressed. The lack of promoter activity in the root hairs (Fig. 6C) implicates that this gene is not involved in sulphate uptake. It is possible that the enzyme encoded by this gene acts to recycle S by hydrolyzing either generally all or specifically some of glucosinolates.
Interestingly, the regions of the roots in which the putative thioglucosidase gene is activated by the −S stress seem to be controlled by C and N nutrients. Decreasing the C or N levels causes the −S activation of the promoter to be limited to the root regions closer and closer to the root tip. This indicates that some sort of signal derived from the root tip is required for −S activation of gene expression and that this signal is likely controlled by the availability of either C or N nutrients. This novel tissue-specific expression pattern by C and N nutrients implies that both C and N may specify the root regions in which gene expression can be activated by −S.
Although the regulatory mechanisms by which C and N nutrients are sensed and the signals are subsequently transduced and integrated to control −S activation remain largely unknown, the regulatory pattern of C, N and S interactions we observed seems to be a general phenomenon of gene expression in response to S deficiency stress. This is because similar GUS expression patterns were observed for the genes involved in the S storage release (At2g44460), sulphate uptake (SULTR4;2) and reduction (APR2 and APR3; see promoter::GUS activities in Supplemental Fig. 3). To further test this phenomenon with the Arabidopsis full genome chip using our combinatorial design will help reveal the regulatory network in C, N and S nutrient signaling pathways and their cross-talk.
In summary, our results provide molecular and genetic evidence that auxin plays a negative regulatory role in −S response. When the S nutrient status is perceived as being at very low levels, plants then down-regulate the auxin sensitivity, which would in turn release the suppression of gene expression and inhibit the lateral root development. Given that auxin suppresses the promoter activity of the putative thioglucosidase gene At2g44460 but not that of SULTR1;2 and APR2, we hypothesize that auxin might participate in only a subset of −S responses such as remobilizing S from the intracellular storage. Future work should test the biochemical and physiological functions of the putative thioglucosidase gene product and investigate which auxin signaling branch is involved in the response to sulphur nutrient status.
Acknowledgments
This work was supported by the NIH-SCORE grant (S06GM008225, Project Number 12 to Z-L Z) and in part by the City University of New York and the PSC-CYNY program (68436-00-37 to Z-L Z). We are grateful to Haiyang Wang (Boyce Thompson Institute) for providing the axr1-3 seeds, Erwin Grill (Tubingen University) for the pRD29B::GUS line, and Zhenbiao Yang (University of California at Riverside) for the DR5::GUS line with the permission of the use from Thomas Guilfoyle (University of Missouri-Columbia). We thank members of the Zheng Laboratory at the CUNY Lehman College for helpful discussion and technical assistance. We appreciate David Cain (CUNY Lehman College) for assistance with plant growth. We also thank anonymous reviewers for their constructive comments.
Footnotes
Electronic Supplementary Material Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s11103-006-9084-0 and is accessible for authorized users.
Contributor Information
Hanbin Dan, Department of Biological Sciences, Lehman College, City, University of New York, Bronx, NY 10468, USA, Plant Sciences PhD Subprogram, Graduate School and University Center, City University of New York, New York, NY 10016, USA.
Guohua Yang, Department of Biological Sciences, Lehman College, City, University of New York, Bronx, NY 10468, USA.
Zhi-Liang Zheng, Department of Biological Sciences, Lehman College, City, University of New York, Bronx, NY 10468, USA, Plant Sciences PhD Subprogram, Graduate School and University Center, City University of New York, New York, NY 10016, USA e-mail: zhiliang.zheng@lehman.cuny.edu.
References
- Al-Ghazi Y, Muller B, Pinloche S, Tranbarger TJ, Nacry P, Rossignol M, Tardieu F, Doumas P. Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signaling. Plant Cell Environ. 2003;26:1053–1066. [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Hoffmann T, Teplova I, Grill E, Muller A. Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 2005;137:209–219. doi: 10.1104/pp.104.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inze D, Foyer CH, Zhang H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003;33:543–555. doi: 10.1046/j.1365-313x.2003.01652.x. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 1999;18:303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martin AC, Leyva A, Paz-Ares J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005;138:847–857. doi: 10.1104/pp.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS. Root development—branching into novel spheres. Curr Opin Plant Biol. 2006;9:66–71. doi: 10.1016/j.pbi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Hesse H, Nikiforova V, Gakiere B, Hoefgen R. Molecular analysis and control of cysteine biosynthesis: integration of nitrogen and sulphur metabolism. J Exp Bot. 2004;55:1283–1292. doi: 10.1093/jxb/erh136. [DOI] [PubMed] [Google Scholar]
- Hesse H, Trachsel N, Suter M, Kopriva S, von Ballmoos P, Rennenberg H, Brunold C. Effect of glucose on assimilatory sulphate reduction in Arabidopsis thaliana roots. J Exp Bot. 2003;54:1701–1709. doi: 10.1093/jxb/erg177. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 2003;33:651–663. doi: 10.1046/j.1365-313x.2003.01658.x. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell. 2004;16:2693–2704. doi: 10.1105/tpc.104.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H. Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. J Exp Bot. 2004;55:1831–1842. doi: 10.1093/jxb/erh203. [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 2000;122:737–746. doi: 10.1104/pp.122.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A. Singlet oxygen production in photosynthesis. J Exp Bot. 2005;56:337–346. doi: 10.1093/jxb/erh237. [DOI] [PubMed] [Google Scholar]
- Kutz A, Muller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW. A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 2002;30:95–106. doi: 10.1046/j.1365-313x.2002.01271.x. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Ann Rev Plant Physiol Plant Mol Biol. 2000;51:141–165. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002;29:751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Perez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 2005;137:681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 2003;132:597–605. doi: 10.1104/pp.102.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. Regulation of high-affinity sulphate transporters in plants: towards systematic analysis of sulphur signalling and regulation. J Exp Bot. 2004a;55:1843–1849. doi: 10.1093/jxb/erh175. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 2004b;38:779–789. doi: 10.1111/j.1365-313X.2004.02079.x. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Yamaya T, Takahashi H. Induction of SULTR1;1 sulfate transporter in Arabidopsis roots involves protein phosphorylation/dephosphorylation circuit for transcriptional regulation. Plant Cell Physiol. 2004c;45:340–345. doi: 10.1093/pcp/pch029. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005;42:305–314. doi: 10.1111/j.1365-313X.2005.02363.x. [DOI] [PubMed] [Google Scholar]
- Marzluf GA. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- Moran R. Formulae for determination of chlorophyllous pigments extracted with N,N-dimethylformamide. Plant Physiol. 1982;69:1376–1381. doi: 10.1104/pp.69.6.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R. Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J Exp Bot. 2005a;56:1887–1896. doi: 10.1093/jxb/eri179. [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 2003;33:633–650. doi: 10.1046/j.1365-313x.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. Systems re-balancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 2005b;138:304–318. doi: 10.1104/pp.104.053793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama N, Takei K, Sakakibara H, Hayashi H, Yoneyama T, Fujiwara T. Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:1493–1501. doi: 10.1093/pcp/pcf183. [DOI] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986;81:922–924. doi: 10.1104/pp.81.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Wachter A. Sulfur metabolism: a versatile platform for launching defence operations. Trends Plant Sci. 2005;10:503–509. doi: 10.1016/j.tplants.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Saito K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 2004;136:2443–2450. doi: 10.1104/pp.104.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Takahashi T, Sugiyama T, Yamaya T, Sakakibara H. Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot. 2002;53:971–977. doi: 10.1093/jexbot/53.370.971. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Muller A, Weiler EW, Piotrowski M. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta. 2001;212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- Weijers D, Jurgens G. Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol. 2004;7:687–693. doi: 10.1016/j.pbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL. Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 2005;139:1350–1365. doi: 10.1104/pp.105.068064. [DOI] [PMC free article] [PubMed] [Google Scholar]


