Abstract
A new single molecule system – Transchip – was developed for analysis of transcription products at their genomic origins. The bacteriophage T7 RNA polymerase and its promoters were used in a model system, and resultant RNAs were imaged and detected at their positions along single template DNA molecules. This system, Transchip, has drawn from critical aspects of Optical Mapping, a single molecule system that enables the construction of high resolution, ordered restriction maps of whole genomes from single DNA molecules. Through statistical analysis of hundreds of single molecule template/transcript complexes, Transchip enables analysis of the locations and strength of promoters, the direction and processivity of transcription reactions and termination of transcription. These novel results suggest that the new system may serve as a high-throughput platform to investigate transcriptional events on a large, genome-wide scale.
Keywords: Single molecule, transcription, elongation complex, T7 RNA polymerase, promoter mapping, automated fluorescence microscopy
Introductory Statement
This study aims to develop an integrated system for transcriptional analysis of single template DNAs, with the ultimate goal being analysis of transcriptional regulation for genes on a whole genome basis. Large template DNA molecules can be queried to probe for regulatory elements and for transcriptional events. The Transchip system described in this manuscript aims to produce large and meaningful datasets for transcriptional analysis, all based around single molecule measurements. Experiments based on single molecules are direct and rapid, and can generate a read-out of thousands of data points in a short time period.
The development of single molecule approaches to biochemical analysis has enabled a broad range of techniques that visualize the details of and that elegantly probe individual steps involved in transcription [1–6]. These elegant biochemical studies performed on single molecules have brought numerous insights into the mechanisms of transcription, but they do not lend themselves to high throughout transcriptional analysis. Visualization and mapping of RNAs transcribed from single template molecules has long been possible by electron microscopy (for ex. [7–9]) but is also not amenable for high-throughput, single molecule analysis of transcription products. Analysis of gene expression on a genome-wide basis has revealed the “transcriptome” of organisms [10–17], with some approaches amenable to high-throughput analysis.
The ability to directly detect nascent RNAs at their site of synthesis along single template DNAs would allow genome-wide analysis of transcriptional events. Transchip is a new single molecule system designed with the goal of making single molecules practical substrates for experimental studies – enabling ensemble averages from a large number of parallel measurements and allowing a significantly more effective approach for single molecule studies. This system exploits integrated analysis of single DNA molecules stretched on modified glass surfaces with imaging of RNAs associated with the templates. Single molecule genome mapping approaches (Optical Mapping) have enabled the high-throughput generation and analysis of large molecular datasets and produced an array of tools amenable to a broad range of molecular studies [18–27]; these approaches were exploited for analysis of transcriptional events on single template molecules.
Transchip directly tracks in vitro transcriptional products (elongation complexes) on individual template DNA molecules by automated fluorescence microscopy. Detailed molecular data are extracted from images and provide large datasets for statistical analysis and biochemical modeling. The principal test system is based on the actions and sequences associated with T7 RNA polymerase (RNAP), which is the best characterized member of a family of RNAPs that includes most of the bacteriophage-encoded RNAPs as well as the mitochondrial RNAPs [28–31]. Previous experiments indicated that T7 RNA polymerase can actively incorporate fluorochrome labeled ribonucleoside triphosphates into nascent transcripts [32], most readily with rhodamine dyes (Rhodamine Green, Tetramethylrhodamine).
Templates were designed that carried a combination of T7 RNA polymerase promoters and terminator and that could be directly imaged following mounting and elongation on modified glass surfaces. We investigate whether RNAs could be imaged associated with their respective single template DNAs using fluorescence microscopy; whether their positions and that of their promoters could be mapped and whether the direction and extent of transcription could be determined. Additional experiments assessed the RNase and protease sensitivity of observed putative elongation complexes (ECs), as well as the ability of Transchip to detect ECs from single round transcription reactions. Furthermore, experiments varying time or adding chain terminators to transcription reactions were performed to control and modulate the positions of ECs. The results demonstrate that transcription data obtained from ensembles of single template DNA molecules, and collected with an automated image acquisition system, can locate single or multiple promoters, the extent and direction of transcription, and the relative strength of promoters in this model system.
Material and Methods
Template DNA Construction and Preparation
Multiple template DNAs were used in these studies, including a cosmid and a plasmid template DNA, bearing one promoter for T7 RNA polymerase; several plasmid DNAs with multiple promoters varying in strength and orientation, and T7 genomic DNA. The cosmid LANL-16c_380H5 (44,675 bp; accession number AC004233; with human chromosome 16 sequences and a synthetic T7 promoter [TAATACGACTCACTATAGGGCGA]) was obtained from Los Alamos National Laboratory. This template DNA was linearized with Sal I, extracted twice with a 1:1 mixture of phenol and chloroform/isoamyl alcohol (24:1, v/v), and ethanol precipitated. Template DNA concentrations were determined by UV spectrometry.
Multiple plasmids were engineered to contain different combinations of T7 RNA polymerase promoters and terminator to assay whether promoter locations, orientation and strengths could be distinguished (see Fig. 1).To construct 1SP_PE (25.231 kb; with one synthetic T7 promoter [SP]), a 22.010 kb Bgl II fragment was isolated from lambda DNA and inserted into BamH I cleaved parental vector, pGEM-11Zf (-) (Promega, Madison WI). 1SP_PE was modified to construct 1SP_Tφ (25.828 kb), with a promoter (SP) and a terminator. A T7 terminator region located between positions 24099 and 24691 of the T7 genome [33] was amplified with the primers Tφ_forward, GGA AGA TCT CAC CCG CGC TGC TAA CAA AG, and Tφ_reverse, CGC GGA TCC GGT GCC CCA AAG AAT C, digested with Bgl II and BamH I, and ligated into 1SP_PE that had been partially cleaved with Bam H I. Plasmids bearing the terminator fragment ~5.1 kb downstream of the T7 promoter in the correct orientation were selected.
Fig. 1.
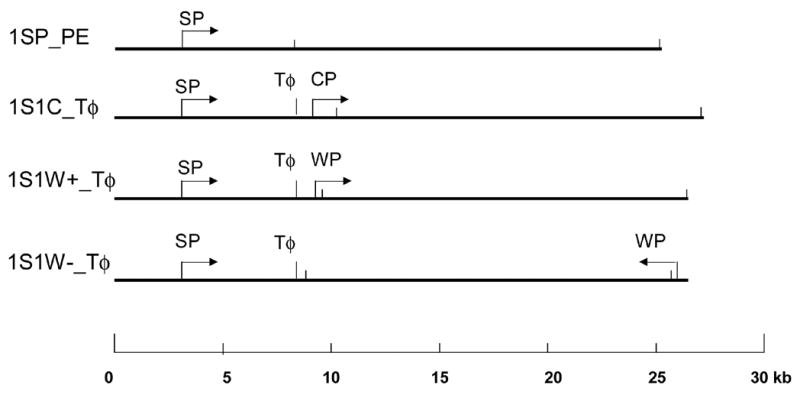
Diagram of plasmids - 1SP_PE, 1S1C_Tφ, 1S1W+_Tφ, and 1S1W-_Tφ - constructed with one or more of the following transcription elements: a synthetic T7 promoter, SP; a consensus T7 promoter, CP; a T7 class II promoter, WP; and a terminator (Tφ). The arrows show the direction of transcription. The short, vertical bars represent BamH I cleavage sites. The scale bar (bottom) is in kb.
Construction of 1S1C_ Tφ
This template was designed to contain two T7 promoters of similar strength. A region of T7 genomic DNA from 26976 to 28355 including the consensus T7 promoter φ13 (CP: TAA TAC GAC TCA CTA TAG GGA GA) was amplified with the primers CP_forward, GG A AG ATC TAG ACT CTG GTA CGT TTG ACA TTT and CP_reverse, CG CGG AT CCG CTT GAA GGG CAG AT, and cloned into pCR2.1-TOPO™ (Invitrogen). The ~1.4 kb Bgl II/BamH I fragment was isolated from the resultant plasmid, and ligated with the 8.985 kb BamH I fragment from 1SP_Tφ to form pSP- Tφ_CP. The 16.84 kb BamH I fragment from 1SP_PE was isolated and inserted into the BamH I site of pSP- Tφ -CP to form 1S1C_ Tφ (27.212 kb), with one synthetic T7 promoter (SP), one consensus T7 promoter (CP) and a T7 terminator, Tφ.
Construction of 1S1W+_Tφ and 1S1W-_Tφ
These templates were designed to contain two T7 promoters, one strong (SP) and the second a weak T7 promoter (WP). 1S1W-_Tφ contains WP in the opposite orientation and ~23 kb distant from SP. A region of the T7 genome from 12242 to 12973 that contains a class II T7 promoter φ4c (WP: CAATCCGACTCACTAAAGAGAGA [position 12654–12676]) was amplified with the primers WP_forward, GG AAG ATC TAG A TAA GGT ACG AGT GGC AGT T and WP_reverse, CGC GGA TCC TCT GGG TTC TTA AGG TGA CAA AT. The cloning procedure was analogous to that for 1S1C_Tφ. The two resultant plasmids, 1S1W+_Tφ and 1S1W-_Tφ, have different orientations and positions of the WP promoter (see Fig. 1; both are 26.489 kb). All plasmid DNAs were transformed into ElectroMax® DH 10B™ competent cells (GIBCO BRL®) by electroporation using a Bio-Rad Gene Pulser unit and purified with a QIAGEN-tip 100 (Qiagen, Inc., Valencia CA) following the manufacturer’s protocols. They were linearized with Hind III, extracted with phenol/chloroform and ethanol precipitated. The T7 RNA promoter SP is 3.136 kb from one end following linearization of templates with Hind III.
Bacteriophage T7 genomic DNA was prepared by phage lysis with proteinase K digestion (50 μg/ml, Sigma-Aldrich, St. Louis MO) in 1× digestion buffer (10 mM Tris-HCl, 5 mM EDTA and 0.5% SDS, pH 7.8) at 55°C for 2 hrs. The lysate was extracted twice with a 1:1 mixture of phenol and chloroform/isoamyl alcohol (24:1, v/v) and ethanol precipitated.
Reagents
Fluorescein-12-UTP, Rhodamine Green-5-UTP, Tetramethylrhodamine (TMR)-6-UTP, Texas Red-5-UTP and BODIPY TMR-14-UTP were obtained from Molecular Probes (Eugene OR); NTPs from Roche Applied Science (Indianapolis, IN), and heparin (sodium salt) from Gibco-BRL (Gaithersburg, MD). Buffers and solutions were prepared RNase-free by standard methods and were filtered through Millex-GV filters (0.22 μm pore size, Millipore Corp., Billerica MA).
Surface Treatments
Two types of modified glass surfaces were used in this study for mounting, elongating and imaging DNA template/elongation complexes - APDEMS and trimethyl silane [19–21, 34]. The trimethyl silane surfaces proved more stable than the APDEMS surfaces and thus were used for most of the experiments; both had similar characteristics for DNA stretching and imaging. APDEMS surfaces (used for the experiment of Fig. 2 and 3) were prepared as follows. Glass cover slips (22 × 22 mm; Fisher’s Finest) were racked and cleaned by boiling in concentrated nitric acid (HNO3) for 12 h. They were rinsed extensively with high-purity, deionized water. The cleaning was repeated with concentrated hydrochloric acid (HCl). The cover slips were rinsed extensively with high-purity water. A stock solution (2% by weight) of 3-aminopropyldiethoxymethylsilane (APDEMS; Gelest Inc., Morrisville PA), distilled under argon, was prepared by dissolving APDEMS in deionized water and allowing it to hydrolyze on a shaker for 7.5 h. Then 45 to 60 μL of the hydrolyzed APDEMS was added to 250 ml ethanol with 30 racked, cleaned cover slips. The cover slips could be used for up to two weeks.
Fig. 2.
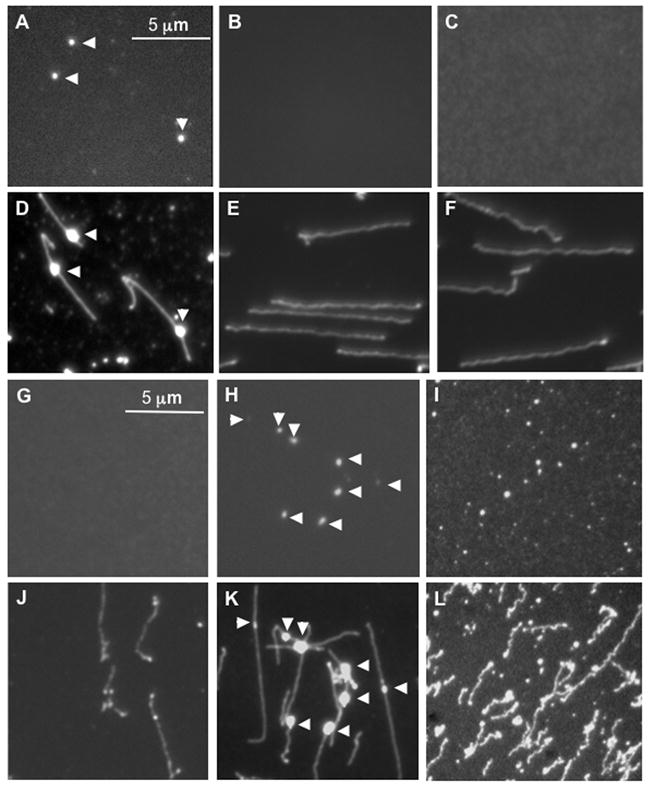
Identification and localization of fluorescently labeled transcripts on DNA template LANL-16c_380H5. In vitro transcription reactions were conducted and mounted on APDEMS OM surfaces, as described. A–C and G–I) Images were taken using XF-102 filter, which is suitable for TMR-6-UTP labeled RNA. A) In vitro transcription with T7 RNA polymerase, NTPs and TMR-6-UTP. B) Control transcription reaction, lacking NTPs. C) Control reaction, lacking T7 RNA polymerase). D–F) These images correspond to those of A–C, taken using a YOYO-1 filter following staining with YOYO-1. Elongation complexes were digested with RNase I (G) or with RNase H (H), or with proteinase K (I) on the surface. J–L) These images correspond to those of G–I, following staining with YOYO-1. The surfaces were APDEMS surfaces (see Materials and Methods).
Fig. 3.
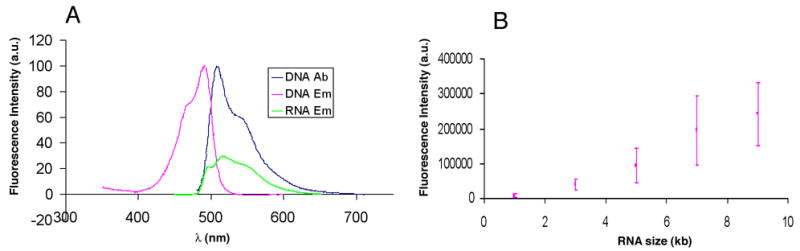
Analysis of YOYO-1 stained RNA. A) Comparison of spectrum of YOYO-1 stained DNA (DNA Em) with YOYO-1 stained RNA (RNA Em) excited by the maximum absorption wavelength for DNA (DNA Ab) 491 nm. B) Scatter plot of the mean integrated fluorescence intensity of single RNA molecules mounted on Optical Mapping surfaces (APDEMS), with error bars indicating the standard deviation. Acquisition time: 5000 ms; number of analyzed molecules: 100 to ~200 for each measurement; light power: ~ 1mW. The RNAs were diluted to an appropriate concentration (~ 50 pg/ml) before they were mounted on the surface.
To prepare trimethyl surfaces, glass cover slips were racked, cleaned by boiling in Nanostrip (Cyantek Corp., Fremont CA) for 50 min at 68°C, and rinsed with high purity, deionized water 5 times. The cover slips were then hydrolyzed in boiling concentrated hydrochloric acid at 98°C for 6 h, rinsed extensively with high purity water, and were individually rinsed three times in absolute ethanol and stored in absolute ethanol. Thirty acid hydrolyzed surfaces were placed in a flat Teflon block holder and treated in 250 ml silane solution (70 to 80 μl of trimethyl silane [N-trimethylsilyl-propyl-N,N,N-trimethylammonium chloride; Gelest Corp.] and 10 μl of vinyl silane [vinyltrimethoxysilane, Gelest Corp.] in 250 ml high purity water). These were incubated at 65°C with gentle shaking (50 rpm) for 17.5 h, and cooled to room temperature in a fume hood for 1 h. The solution was aspirated, and the surfaces were rinsed three times with high purity water, once with distilled absolute ethanol, and stored in distilled absolute ethanol.
In Vitro Transcription Assays
Transcription reactions (20 μl) were in transcription buffer (40 mM Hepes-HCl, 6 mM MgCl2, 10 mM DTT, pH 7.9 at 25 °C), 0.5 mM each in ATP, CTP, GTP and UTP, and included 10 units RNase inhibitor (20 units/μl, SUPERase-In™, Ambion, Austin TX). Varying amounts of T7 RNA polymerase (from 5 units to 50 units [N.E. Biolabs, Ipswich MA]) and linearized DNA template were present, as noted. For the fluorochrome labeling experiments (Fig. 2), the NTPs were 0.5 mM in each for ATP, GTP, and CTP, and 0.05 mM in UTP (total UTP/fluorochrome labeled UTP [e.g. tetramethylrhodamine (TMR)-6-UTP] = 10:1). (TMR-6-UTP had been chosen as the optimal fluorochrome-labeled NTP overall among the five tested -Fluorescein-12-UTP, Rhodamine Green-5-UTP, TMR-6-UTP, Texas Red-5-UTP and BODIPY TMR-14-UTP.) Reactions were preincubated for 10 min at 37°C, initiated by the addition of NTPs, incubated at 37°C for 15 min., and stopped. Prior to mounting a 1:100 dilution of transcription reaction products on the surface, unincorporated NTPs were removed using a CentriSpin10 column (Princeton Separations, Adelphia NJ) to reduce the background caused by the random binding of free TMR-6-UTP to the surface. For single-round transcription reactions, samples were preincubated 37 °C 10 min with all components except UTP. Heparin was added to 100 μg/mL, and reactions incubated 2 min. before the addition of UTP. Samples were processed as above.
Capture of Elongation Complexes and Fixation
In order to modulate, or control, the distribution of position and size of elongation complexes, 3′dNTP (3′-deoxynucleotide) was included in transcription reactions. We first assessed whether transcriptional elongation could be selectively stopped and captured by including different ratios of 3′dNTP to NTP in transcription reactions: 1:500, 1:1000, 1:2000, 1:2500, and 1:5000. Complexes were cross-linked with 1% HCHO at 4°C for 30 min to stabilize them and diluted 100 fold prior to on-surface mounting.
DNA Mounting, Overlay, Digestion, and Staining
The mounting of template DNAs with associated elongation complexes on modified glass surfaces is a key step prior to their visualization. A cover layer of acrylamide helps maintain the molecules in place (used with trimethyl silane surfaces), and template DNAs can be digested with restriction enzyme prior to staining with fluorescent dye. In brief, DNA molecules bearing elongation complexes were mounted on derivatized glass surfaces (Optical Mapping [OM] surfaces; APDEMS, Fig. 2 and 3; trimethyl, Figs. 4–8) by capillary action as described by Lim et al. [22]. For trimethyl surfaces (Figs. 4–8), a thin layer of acrylamide (3.3%) was applied to the surface, and, upon curing, was washed with 200 μl HE (10 mM Hepes and 1 mM EDTA) for 2 min. For restriction digestion, 200 μl of digestion buffer (50 mM NaCl, 10 mM Hepes, 10 mM MgCl2, 1 mM DTT, pH 7.9 at 25°C), and 60 units BamH I or Xba I [New England Biolabs]) were added to the surface, which was incubated in a humidified chamber at 37°C for 30 to 60 min. The surface was washed twice with 200 μl HE and mounted onto a glass slide with 18 μl of 0.2 μM YOYO-1 solution (containing 5 parts of YOYO-1 [Molecular Probes] and 95 parts of β-mercaptoethanol in 20% [v/v] TE). The sample was sealed with nail polish and incubated at room temperature in the dark for 20 min or overnight before imaging.
Fig. 4.
Transcription reactions with template DNAs bearing one (A), two (B) or multiple (C) T7 promoters were surface-mounted (on trimethyl surfaces) and stained with YOYO-1. A) Representative image from a reaction supported by the cosmid LANL-16c_380H5, a 44.675 kb template with one promoter 16 kb from one end. B) Representative image from a reaction supported by 1S1C_Tφ, which is 27.212 kb and has two promoters at 3.675 kb and 9.665 kb from one end. C) Representative images from a reaction supported by T7 genomic DNA, which is 39.937 kb and has 17 promoters. All template DNAs were linearized prior to transcription.
Fig. 8.
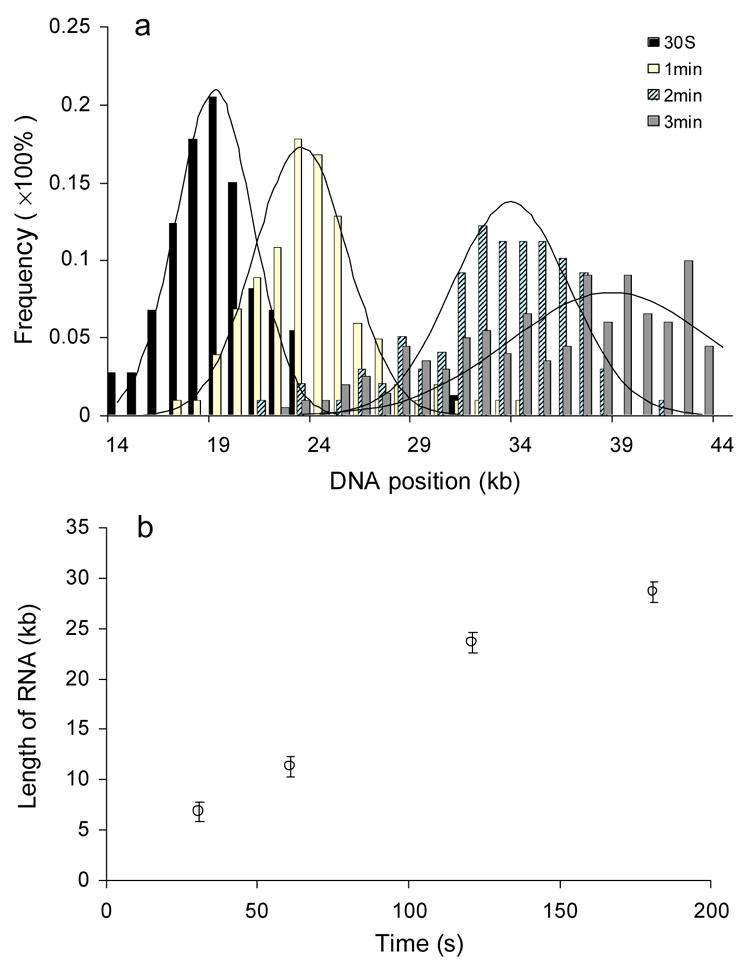
Analysis of the processivity of T7 RNA polymerase and its elongation rate. A) The distribution of positions of ECs on the template cosmid LANL-16c_380H5 is shown for single-round transcription reactions conducted for 30 sec, 1 min, 2 min and 3 min. The frequency of ECs is normalized with respect to the total number of measurements (73, 101, 98 and 199 measurements were included, respectively, for the 30 sec, 1 min, 2 min and 3 min time points). The X coordinate starts at the 14 kb position, since the promoter is at 15.9 kb. The solid plots are the Gaussian curve fit (analyzed using Origin software). The R2 values of regression fit are respectively 0.98 (30 sec), 0.94 (1 min), 0.92 (2 min) and 0.81 (3 min). Reactions were 20 ng/μl in template DNA and contained 50 units of T7 RNAP. They were preincubated 10 min before addition of NTPs and heparin (to 100 μg/mL). (B) The plot shows the estimated length of RNAs at different time points.
RNase and Proteinase Digestion of Surface-Mounted Reaction Products
Punctates that represent putative transcription elongation complexes should be sensitive to RNase as well as protease digestion. Two RNases were tested: RNase I, which cleaves single-stranded RNA to yield nucleoside 3′-monophosphates, and RNase H, which specifically degrades the RNA in an RNA:DNA hybrid. DNA templates bearing elongation complexes were mounted on the OM surface as described. The stretched DNAs were then incubated in RNase specific buffer for 2 min with, respectively, 10 units RNase I or 10 units RNase H (Epicentre Biotechnologies, Madison WI). For digestion with proteinase K, 200 μL 1× digestion buffer (10 mM Tris-HCl, 5 mM EDTA and 0.5% SDS, pH 7.8 at 25°C) containing 50 μg/ml proteinase K was loaded on the surface bearing elongated DNA templates with associated elongation complexes, and the surface was incubated at 37°C for 30 min.
Analysis of YOYO-1 Stained RNAs of Different Sizes on Optical Mapping Surfaces
We analyzed fluorescence measurements of YOYO-1 stained RNAs to determine whether they might be detected within ECs associated with single template DNAs. The emission spectrum of YOYO-1 stained RNAs was measured and compared to that of DNA, to analyze its fluorescence intensity and wavelength dependence. A standard RNA mixture (1 kb RNA ladder, N.E. Biolabs) was diluted to 10 μg/ml in 0.5 μM YOYO-1 solution, and its emission spectrum measured (the excitation wavelength was 491 nm; fluorescence spectrophotometer is a Hitachi F-4500). The spectrum was compared with the fluorescence spectrum of the DNA sample of the same mass and concentration in 0.5 μM YOYO-1 solution. In addition, the fluorescence intensity of individual YOYO-1 stained, surface-mounted RNAs was measured to determine dependence of intensity on the size of RNAs. RNAs of 1 kb, 3 kb, 5 kb, 7 kb and 9 kb were gel purified (from the 1 kb RNA ladder), extracted with phenol and chloroform, and ethanol precipitated. These RNAs were diluted to ~ 50 pg/ml, mounted on OM surfaces and stained with 5 μL of 0.2 μM fluorescence dye YOYO-1 solution. The stained molecules were illuminated by the same power of light source (~ 1 mW) and imaged for the same time period (5 s). About 200 individual molecules were analyzed for each size of RNA sample for integrated fluorescence intensity.
Image Acquisition and Processing
DNA samples were imaged by fluorescence microscopy as described previously [22] using a 100 × objective (N.A. 1.3, Plan-Neofluar, Zeiss) and a high-resolution digital camera with Kodak KAF-1400 chip (1317 × 1035 pixels, 6.8×6.8 μm2 pixel size, PentaMAX [Princeton Scientific Instruments, Monmouth Junction NJ]). For experiments containing fluorochrome labeled RNAs, images registered at the same position as DNA images were collected using different filter packs (XF 102 for Tetramethylrhodamine, XF 108 for Texas Red and BODIPY TMR, and XF 115 for YOYO-1, Fluorescein and Rhodamine Green). Each pair of images was then superimposed by a program written in MATLAB™ to find the correlation between the labeled RNA and DNA molecules.
Image Analysis of Elongation Complexes and Template Molecules
To detect ECs on the YOYO-1 stained DNA backbone, a fluorescence intensity profile was automatically constructed using the software, Pathfinder, for each molecule [27, 35–38]. In brief, a group of images was first filtered to separate the image foreground (data) from the background, and connected pixels above the threshold point were saved as objects. Pixels in each object were evaluated one by one to find the pixel with maximum intensity (Gaussian blob peaks) along the linear object. Finally, these pixels were chained together to form line segments, which are the backbone of individual DNA molecules or fragments. The intensity variation along DNA molecules or fragments can be presented as a plot to show the intensity variation on a DNA backbone, and the potential ECs were detected as bright punctates based on the fluorescent profile. Here, bright punctates were identified when
| (1) |
where Imax is the maximum intensity of pixel in the punctates, Iaverage is the average pixel intensity of the DNA backbone excluding some dilated pixels with the intensity value two times as high as those of neighboring pixels, and SD is the standard deviation of pixel intensities defined by a mask that covers the DNA backbone and calculated on a per molecule basis. Marks were then manually assigned to these potential ECs using the image editing software, Omari [38]. The program produced contour length in pixels for each fragment (Li) and elongation complex (LECs) as well as integrated fluorescence intensity for each fragment (Ii) and ECs (IECs). Two important parameters were measured: the integrated fluorescence intensity of RNA in each EC (IRNA) and the fractional elongation of each molecule (FE %). IRNA was calculated by subtracting the integrated fluorescence intensity of the DNA backbone from the total intensity of ECs:
| (2) |
FE % was calculated by dividing the pixel length of each molecule by the full B-DNA polymer contour length (L0):
| (3) |
where LDNA is the imaged contour length of DNA molecules, and L0 was computed full B-DNA contour length based on the number of base pairs. Molecules with FE % > 70% were then chosen to find the positions of ECs since molecules with low FE % could be partially broken. Here, a simple data process program called Getmap (written in Java) was used to determine the polarity of the molecule based on the restriction digestion pattern and to compute the position of ECs. Getmap uses the following function to estimate the positions of ECs:
| (4) |
Here, PECs’ refers to the relative position of ECs on the specific restriction fragment i (in kb), DEC s-i is the distance from the center of ECs to the left end of the fragment i, and Li and Ni are the length of fragment in pixels and kb respectively. PECs is the absolute position of ECs on a specific molecule (in kb) and Si is the left end position of the fragment i in the molecule (in kb).
Poisson Curve Fitting
The distribution of ECs along the DNA templates was fitted to a Poisson:
| (5) |
where μ is the average position of all ECs along DNA templates and X is the distance of ECs from the promoter. The distribution of ECs is binned into 1 kb units; the bin containing a promoter was set as 0. μ and X are scaled on the bin. The fitting routine repeatedly searches different μ and finds the maximum regression R2, which is defined as
| (6) |
where Oi and Ei are the observed and expected frequency of ECs in the ith bin respectively. This analysis was done using a routine in Microsoft® Excel.
Results
Imaging of Elongation Complexes on Optical Mapping Surfaces
The initial test for determining whether transcription elongation products could be detected on single template DNAs mounted on derivatized glass surfaces utilized a large template DNA with a single T7 promoter. Transcription reactions supported by the 45 kb template DNA (cosmid LANL-16c_380H5), bearing a single synthetic T7 promoter, were catalyzed in vitro by T7 RNA polymerase. The fluorescently labeled NTP, TMR-6-UTP, was included to allow fluorescent imaging of labeled RNA. Transcription products were mounted on Optical Mapping (OM) surfaces and imaged, as described in Materials and Methods. Distinct punctates can be observed on surfaces bearing transcription reaction components when imaged with the filter that detects TMR (XF102; see Fig. 2A). Images from surfaces bearing control reactions lacking NTPs (Fig. 2B) or lacking T7 RNA polymerase (RNAP; Fig. 2C) did not contain any statistically abundant punctates. Furthermore, when YOYO-1 is subsequently added to the same surface to stain the DNA template backbone, the punctates are seen to coincide with the linear template DNA backbone (Fig. 2D, compare to Fig. 2A; Fig. 2K compare to Fig. 2H). In contrast, the template molecules from reactions lacking NTPs or RNAP apparent in Fig. 2E and 2F (the areas of Fig. 2B and 2C, respectively, following YOYO-1 staining) lack punctates. These results suggest that the punctates are potential elongation complexes containing TMR-6-UTP labeled RNA.
If the punctates are indeed ECs, they should be sensitive to RNase and protease digestion. The sensitivity of the punctates to RNase was assessed by digestion of surface-mounted transcription products with RNase I. This resulted in the virtual disappearance of the bright punctates (Fig. 2G and J), as expected if the punctates contain TMR-labeled RNA. Digestion with RNase H left the bright punctates intact (Fig. 2H and K). This could be due to RNAP inhibiting the RNase H digestion of the RNA in the RNA/DNA hybrid or release of the RNA following digestion, or due to TMR-labeled RNA binding the surface. Figs. 2J and 2K correspond to the same areas as in Fig. 2G and 2H, respectively, following YOYO-1 staining to visualize the template DNA. These results indicate that the punctates contain RNA which is accessible to RNase on the derivatized glass surface.
Proteinase K digestion was then performed directly on surface-mounted transcription reactions to investigate whether digestion of RNA polymerase affected observation of the punctates along the template DNAs (Fig. 2I, 2L). Comparison of Fig. 2I (imaged with the XF102 filter) with Fig. 2L (the same area imaged following YOYO-1 staining) reveals that punctates are present but that most are no longer associated with the DNA template after proteinase K digestion. The sensitivity of the punctates to digestion with RNase I and their disassociation from template DNA following proteinase K digestion suggest that they are complexes containing protein (T7 RNA polymerase) and nascent RNA associated with template DNA, i.e., are elongation complexes (ECs).
The observation that the punctates could be observed following YOYO-1 staining (Fig. 2D, 2K) led us to assess whether the elongation complexes might be visualized using a single fluorochrome stain (YOYO-1) and identified by their unique morphological signatures - bright, ball-like structures associated with the template backbone. This would simplify image collection and processing and involve analysis of just one set of images. Since YOYO-1 is reported to stain RNA in solution [39, 40], we compared the spectrum of YOYO-1 stained RNA to that of YOYO-1 stained DNA (Fig. 3A). YOYO-1 stained RNA was excited within the wavelength range of YOYO-1 bound to DNA (Fig. 3A), with the magnitude being about one-third of the excitation of YOYO-1 bound to DNA of the same mass.
Further measurements were conducted – in this case of individual, surface-mounted RNAs 1k, 3k, 5k, 7k and 9k bases in length – to investigate the fluorescence intensities of YOYO-1 stained RNAs and their contribution to the overall fluorescence intensity of imaged reaction products. The RNAs were surface-mounted, stained with YOYO-1, and imaged. The fluorescence intensities of approximately 100 to 200 single RNA molecules of each size were used to determine the average fluorescence intensity (Fig. 3B). The intensity was proportional to the size of the RNAs, although the variation is about 50% of the average intensity, much higher than for DNA molecules of the same size. This could be due to a variety of secondary structures that affect YOYO-1 staining of RNA and result in a range of intensities.
Imaging of Surface-Mounted Reactions with Templates Bearing One or Multiple T7 Promoters
The number of punctates corresponding to ECs along a template should increase with an increase in promoters. Thus, template DNAs were chosen that contained one, two or multiple T7 RNA promoters. Since we determined that the YOYO-1 stained RNAs were readily observable on OM surfaces (Fig. 3B), we next tested whether potential elongation complexes synthesized with unlabeled NTPs, mounted on OM surfaces and stained with YOYO-1 could be observed associated with template molecules. Reactions supported by the cosmid LANL-16c_380H5, with one promoter, revealed single DNA template molecules with one brightly fluorescent punctate (as seen in Fig. 4A). The template 1S1C_Tφ, which has two T7 RNA polymerase promoters, had two punctates associated with the template (as seen in Fig. 4B), while multiple punctates were apparent on T7 genomic DNA (Fig. 4C). Most of the images of template/transcription products supported by the cosmid LANL-16c_380H5 contained one punctate, and by 1S1C_Tφ one or two, and punctates were sensitive to RNase (as in Fig. 2G; data not shown). These results confirm that the punctates can be imaged following staining with YOYO-1 on the surface, that they contain RNA (and are ECs), and that the number observed increases with an increase in promoters in a template.
Evaluation of the Limitation of Detection of Surface-Mounted, YOYO-1 Stained ECs
It was critical to determine whether ECs could be called with confidence against noise that arose from variation in the density of DNA coil segments on the template DNA backbone, since ECs were characterized in terms of morphological and fluorescence signatures sharing a common fluorochrome stain (YOYO-1) with the template DNA. A signature for ECs was required that could be quantitatively defined, be incorporated into machine vision routines, and that would robustly operate over a large number of molecules. This would enable automatic generation of transcriptional data files. Differential labeling and two color imaging would obviate this issue, but might also perturb the transcription reactions in unpredictable ways.
Fluorescence intensity profiles of ~60 YOYO-1 stained cosmid template molecules were analyzed, as seen in Fig. 5 (and described in Materials and Methods). The template in Fig. 5A was transcribed with 5 units of T7 RNAP (“T5”) under multi-round conditions, while the template in Fig. 5B bearing a punctate was transcribed at a low concentration of RNAP (1 unit, “T1”) under single round transcription conditions (Fig. 5B). The control was not transcribed (Fig. 5C). The peaks in the profiles of Fig. 5A and 5B correspond to the bright punctates (ECs) seen in the corresponding images associated with template DNAs.
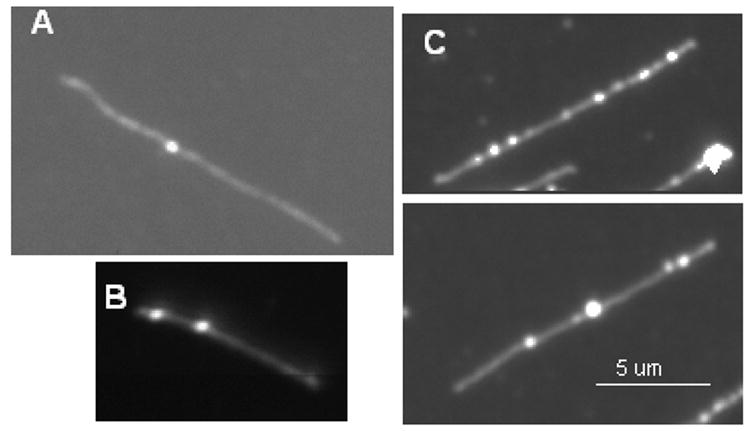
Fig. 5.
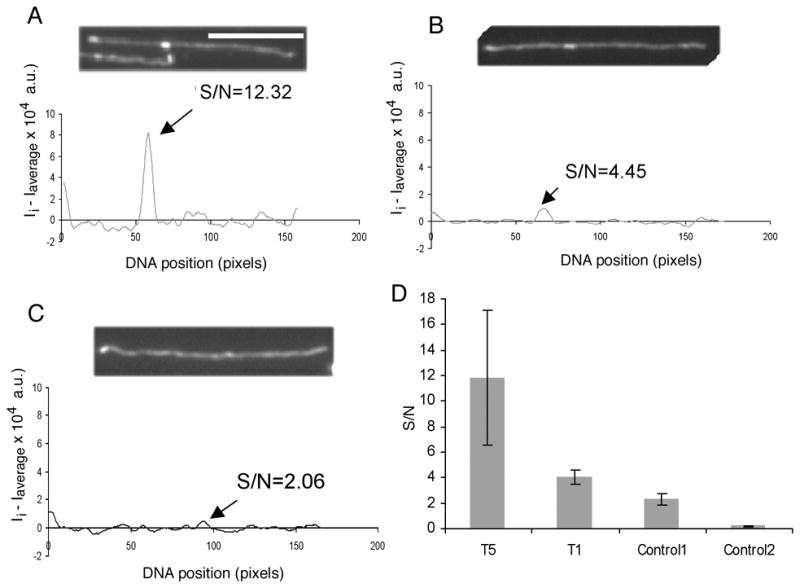
Fluorescence intensity profiles of single DNA molecules following transcription supported by cosmid LANL-16c_380H5 (at 20 ng/μl) with a punctate from transcription with 5 units of T7 RNAP, T5 (A), an EC from a single round with 1 unit of T7 RNAP, T1 (B) and no EC (C). The molecule analyzed in each of the panels (A), (B) and (C) is the top one in the image. The X coordinate represents the DNA length in pixels and the Y coordinate represents the normalized fluorescence intensity of each pixel along the DNA backbone. The arrows point to the pixel in the plot with the maximum intensity on the DNA backbone, labeled with the ratio of S/N (signal to noise). The white bar in the image represents 5 μm. D) The bar graph shows S/N levels for a punctate with multiple RNAs (T5, from a reaction with 5 units of T7 RNAP); for a punctate from a reaction T1, with 1 unit of T7 RNAP and heparin (both with a 3′dNTP:NTP ratio of 1:2000); pixels with maximum intensity on control DNA molecules (control 1), and every pixel on control DNA molecules (control 2). Each bar is calculated from 20 molecules, and error bars show the standard deviation.
A quantitative analysis of these profiles was performed by smoothing the individual pixel fluorescence intensity (Ii) of the DNA backbone using a Gaussian filter and scaling the individual pixel intensity of the DNA backbone through subtracting the average pixel intensity of the backbone (Ii − Iaverage). The noise associated with this profile was defined by computing the standard deviation of fluorescence intensity over the entire length of a molecule that stemmed from the variation of DNA segmental density. The fluorescence intensity of every pixel in the profile was then divided by noise to determine S/N, from which we identified loci of maximum S/N. For Fig. 5A and 5B, these corresponded positions of punctuates (ECs).
The bar graph (Fig. 5D) shows the S/N variations of the peak punctates along the template DNA from reactions with 5 units RNAP (T5), 1 unit of RNAP (T1; conducted under single round conditions) and with no enzyme (Control 1). For T5, the S/N ratio of the EC is approximately 12, while for T1, the average S/N of the most intense pixel in the DNA backbone was about 4, nearly twice as high as the most intense peak on a DNA backbone lacking ECs (S/N ~2; Control 1). Thus, a punctate corresponding to an EC from even a single round transcription reaction was distinguishable from a “peak” due to uneven stretching of template DNAs.
Assessment of the Direction of Transcription and Movement of Elongation Complexes
The experiment of Fig. 6 was designed to assess if the movement of ECs could be controlled and elongation complex movement blocked when reactions were conducted in the presence of the chain terminators, 3′dNTPs. T7 RNA polymerase does not differentiate 3′dNTPs from normal NTPs [41], and incorporated 3′dNTPs would result in chain termination. Four different ratios of 3′dNTP/NTP mixtures (1:500, 1:1000, 1:2000 and 1:5000) were used in transcription reactions supported by the cosmid LANL-16c_380H5. Following transcription and cross-linking of elongation complexes to template DNA, reactions were mounted on OM surfaces, digested with Xba I to allow orientation of the template, stained with YOYO-1 and imaged.
Fig. 6.
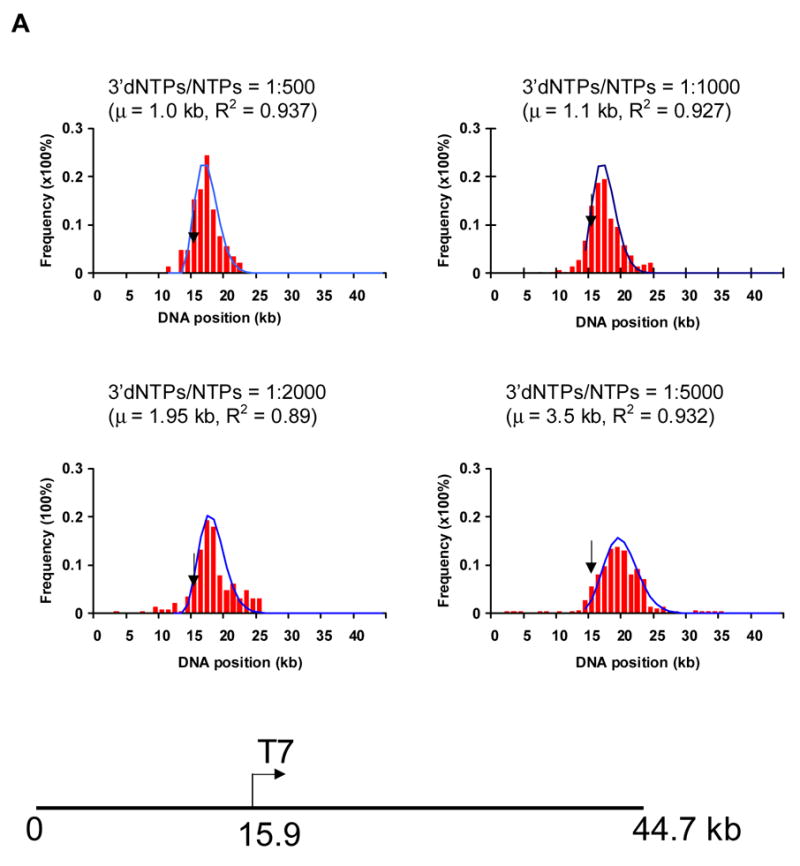
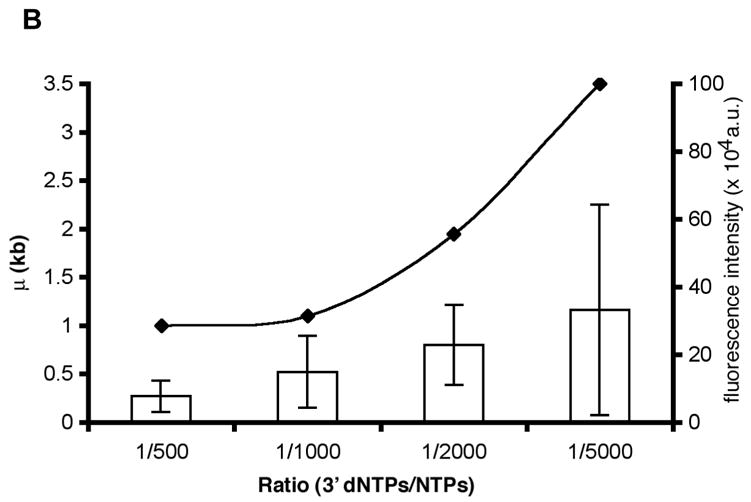
Assessment of positions of ECs along single template molecules in transcription reactions conducted at decreasing levels of 3′ dNTPs. A) The histograms show the distribution of ECs on the linear cosmid DNA (LANL-16c_380H5) from reactions conducted with ratios of 1:500, 1:1000, 1:2000 and 1:5000 of 3′dNTPs to NTPs. The arrow in each histogram indicates the location of the T7 promoter (at 15.9 kb; see diagram of LANL-16c_380H5 below). The frequency of the distribution of ECs is normalized with respect to the total number of ECs analyzed for each histogram. (Approximately 100 to 200 ECs were analyzed for each histogram.) The smooth curves represent fits to a Poisson, with regression fit (R2) and calculated mean value (μ) shown for each graph. μ corresponds to the average distance of ECs downstream of the promoter. B) The ratio of 3′dNTPs/NTPs is plotted vs μ (kb) and vs. the average integrated fluorescence intensity of ECs. The average distance of ECs downstream from the promoter (μ) increases with decreasing ratios of 3′dNTPs/NTPs. Error bars in the bar graph were calculated as the standard deviation on sets of ECs. (138, 100, 92 and 123 measurements were made for reactions conducted at 1:500, 1:1000, 1:2000 and 1:5000 ratios of 3′dNTPs/NTPs, respectively.) The integrated fluorescence intensity was calculated for each stalled EC. The arbitrary units represent the grey value from pixels of ECs.
The distribution of ECs along the DNA template can be seen in the histograms of Fig. 6. Since the average distance of ECs from the promoter is seen to increase in reactions with lower 3′dNTP concentrations, the direction of transcription can be determined. A comparison of the histograms for reactions conducted at a ratio of 1:500, 1:1000, 1:2000 and 1:5000 of 3′ dNTPs: NTPs (Fig. 6) reveals that the peak position for ECs transcribed with a 1:500 ratio was 1.0 kb downstream of the promoter, while it was 1.1 kb for 1:1000, 1.95 kb for 1:2000, and 3.5 kb for the lowest concentration of 3′dNTP (3′dNTPs: NTPs, 1:5000). The few complexes located upstream of the promoter may be due to non-specific transcription. The length (Fig. 6B), as well as the fluorescence intensity (see bar graph, Fig. 6B), of RNA contained in the punctates was also shown to increase at lower concentrations of 3′ dNTP. Thus, template molecules were oriented and the positions of ECs were mapped on the templates for parallel transcription reactions (~200 for each histogram).
Comparison of Elongation Complexes Produced by Templates Bearing Different T7 RNA Promoters and/or a Terminator
We next tested whether multiple promoters on a template could be distinguished in terms of position, orientation and strength. A template containing a single T7 promoter, 1SP_PE, and three templates containing two different T7 promoters, 1S1C_Tφ, 1S1W+_Tφ and 1S1W-_Tφ, were designed to assess whether Transchip analysis could determine the number of promoters each had, their orientation and their relative strengths (see Fig. 1).
The templates were transcribed in vitro by T7 RNA polymerase in the presence of two concentrations of 3′dNTP to NTPs: 1:1000 (Fig. 7) and 1:2500 (data not shown). Following mounting on OM surfaces, template DNAs were digested with BamH I to orient the templates. Representative images of digested template DNAs (where digestion sites are seen as gaps) and associated ECs are shown in Fig. 7A.
Fig. 7.
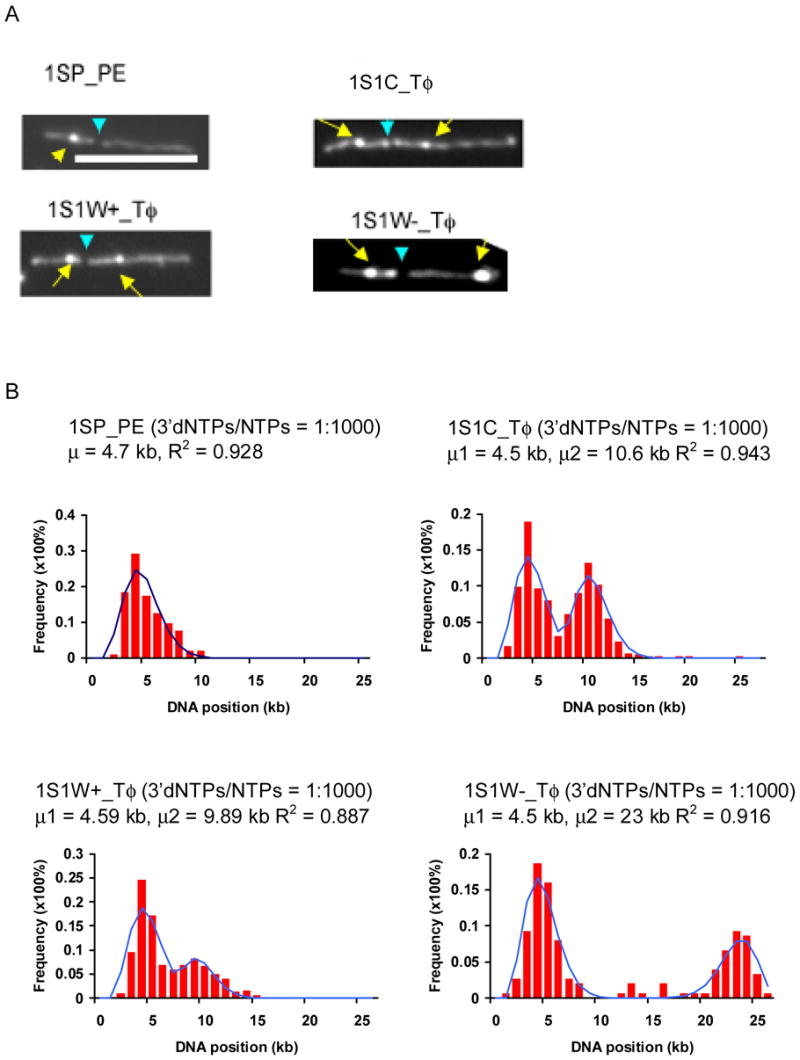
Localization of promoters on the template DNAs 1SP_PE, 1S1C_Tφ, 1S1W+_Tφ, and 1S1W-_Tφ. A) Representative images of surface-mounted transcription reactions supported by 1SP_PE, 1S1C_Tφ, 1S1W+_Tφ, and 1S1W-_Tφ. Yellow arrows point to prominent ECs, and blue arrows to BamH I restriction sites. The two discernible BamH I fragments are 8.3 and 16.8 kb for 1SP_PE; 10.3 and 16.8 (1S1C_Tφ); 9.6 and 16.8 (1S1W+_Tφ), and 8.9 and 16.8 (1S1W-Tφ). (White bar: 5 μm). B) Histograms show the distributions of ECs on template DNAs at ratios of 3′dNTP to NTP of 1:1000. The solid plots are the Poisson curve fits with shown regression fit (R2) and calculated mean value (μ) for each graph. (100–200 molecules were analyzed for each graph.)
By analyzing the distribution of ECs synthesized at the two different concentrations of 3′dNTPs on the templates 1SP_PE, 1S1C_Tφ, 1S1W+_Tφ, and 1S1W-_Tφ, the locations of the different promoters and the direction of transcription were determined. Most of the images of the template/reaction products from the template with one promoter, 1SP_PE, showed one bright punctate on the template DNA backbone (Fig. 7), and the average distance of ECs for 1SP_PE was 1.56 kb from the promoter, which is 3.14 kb from one end. The template 1S1C_Tφ is known to have two promoters, SP and CP, both of which are strong T7 RNA polymerase promoters (Fig. 1). Two peaks corresponding to positions with the most ECs are apparent in the plots of Fig. 7B (plots are shown for the 1:1000 ratio of 3′dNTP to NTPs). This is also observed in the plots of 1S1W+_Tφ, and 1S1W-_Tφ (Fig. 7B). The locations of promoters were estimated as described and compared with their known locations. As seen in Table 1, the results indicate that the one promoter of 1SP_PE and both promoters in all three templates 1S1C_Tφ, 1S1W+_Tφ, and 1S1W-_Tφ were mapped, and that the absolute error for their positions is about 500 bp, which is close to the resolution of light microscope. In addition, the orientation of the promoters was determined, and the WP promoter in 1S1W-_Tφ was shown to be the only one in the reverse orientation.
Table 1.
Comparison of Estimated Promoter Locations vs. Their Known Locations
| Template | 1SP_PE | 1S1C_Tφ | 1S1W+_Tφ | 1S1W-_Tφ | |||
|---|---|---|---|---|---|---|---|
| Promoter | SP | SP | CP | SP | WP | SP | WP |
| Estimated promoter location (kb) | 3.35 | 3.67 | 9.66 | 3.79 | 9.02 | 3.79 | 24.9 |
| Actual promoter location (kb) | 3.14 | 3.14 | 9.14 | 3.14 | 8.69 | 3.14 | 26.0 |
By computing the ratio of total fluorescence intensity of the ECs initiated from different promoters, the relative strength of each promoter can be evaluated.
| (7) |
where IECs_P1 and IECs_P2 are the integrated fluorescence intensities of each EC from promoter P1 and P2, respectively, and Sratio is the calculated ratio of the strength of promoter P1 to P2. Table 2 compares the promoter strength of three DNA templates we designed, each with two different promoters. The plasmid 1S1C_ Tφ has the two promoters, SP and CP, both strong promoters for T7 RNA polymerase [42]. They display a comparable promoter strength, as expected, with a ratio of 1.05 (see Table 2). Both templates 1S1W+_Tφ and 1S1W-_Tφ have a synthetic T7 promoter (SP) as well as a class II promoter (WP). A comparison of the number of ECs and average fluorescence intensity of ECs for these two indicate that more RNAs are contained in elongation complexes associated with the T7 synthetic promoter (SP) than with those for the class II promoter (WP). The SP promoter is reported to be about twice as strong as WP [42, 43], and SP was determined to be 2.69 times as strong as WP in 1S1W+_Tφ (Table 2). The relative strength of the SP and WP promoters is seen to be affected by the sequence context: when WP is downstream of SP in 1S1W+_Tφ, it is weaker (SP is 2.69 times as strong) than when located at the end of the template in 1S1W-_Tφ (where the ratio is 1.79; see Table 2).
Table 2.
Estimate of Promoter Strengths
| # ECs | Intensity of ECs | # ECs | Intensity of ECs | Ratio | |
|---|---|---|---|---|---|
| Promoter | SP | CP | |||
| 1S1C_Tφ | 209 | 788776 | 202 | 775923 | 1.05 |
| Promoter | SP | WP | |||
| 1S1W+_Tφ | 218 | 252394 | 98 | 208529 | 2.69 |
| 1S1W-_T φ | 90 | 153613 | 51 | 150914 | 1.79 |
Analysis of the Processivity of T7 RNA polymerase and Its Elongation Rate
In order to further analyze the position of elongation complexes and estimate the size of the transcribed RNAs, single-round transcription reactions supported by the cosmid (LANL-16c_380H5) template were conducted in the presence of heparin. The reaction times were 0 s, 30 s, 1 min, 2 min, and 3 min, and reactions were mounted and imaged as described. The distributions of ECs captured at different time points were fitted into a Gaussian curve (Fig. 8A), and the lengths of the RNAs were estimated by subtracting the position of the promoter location from the peak position of ECs furthest from the promoter, using the following equation
| (8) |
where μi and Si are the mean and standard deviation of the distribution of ECs for the time point i estimated from Gaussian curve fitting, respectively; P0 is the position of the promoter and Li is the estimated length of RNA (in kb) produced in time point i. As seen in the scatter plot of Fig. 8B (where the estimated position of ECs is plotted vs. transcription time), the peak positions of ECs move downstream with increasing time of transcription, until a plateau is reached near the 3 min time point. By 3 min, many ECs have moved to the end of their respective templates. The data were used to estimate the elongation rate for T7 RNA polymerase in the single-round reactions: 201 ± 18 nucleotides per second.
Discussion
While the Optical Mapping system has been developed into a robust, high throughput, single molecule system able to construct physical maps of whole genomes, including microbial, fungal, plant and mammalian genomes [18, 19, 34, 44–49], it has now been extended to analyze the positions and fluorescence intensities of in vitro transcription products associated with surface-bound single template DNA molecules. This new method allows rapid analysis of important transcription caveats on OM surfaces. Elongation complexes labeled by fluorochrome conjugated UTP or stained with YOYO-1 can be clearly detected through conventional fluorescence microscopy and mapped to their position on single template DNA molecules.
Our observation that ECs are readily detectable under fluorescence microscopy without alternatively labeling the RNA ([50] and Fig. 2) eliminates the challenging step of synthesizing RNAs with fluorescently labeled NTPs. The predominant morphology of ECs visualized following YOYO-1 staining is a circular punctate located on the DNA backbone, and these punctate-like ECs can be readily detected on stretched linear DNA (Figs. 2, 4, 5, and 7 [punctates were also seen on “combed” DNA] [50]), as well as on the surface when released from the DNA backbone following proteinase K digestion (Fig. 2L), suggesting that the morphology of ECs is determined by the secondary structure of single-stranded RNA.
Single ECs can be readily detected when transcription is conducted at a low concentration of RNA polymerase or in the presence of heparin to block subsequent initiation. Furthermore, the fluorescence intensity of punctates increases at higher concentrations of RNA polymerase, suggesting that most punctates contain multiple ECs.
Our single molecule detection of oriented transcription templates and their associated elongation complexes enables the mapping of promoter locations and determination of the direction of transcription and of promoter strength through careful statistical analysis of ensembles of single DNA molecules. The addition of 3′dNTPs to in vitro transcription reactions was a significant improvement that controls the movement of ECs along DNA backbone and facilitates the detection of promoters. In reactions with dNTPs (along with NTPs), only one peak (for ECs) is generally detected along templates with one T7 promoter (1SP_PE) and two apparent peaks for templates with two T7 promoters (1S1C_Tφ, 1S1W+_ Tφ and 1S1W-_ Tφ). This modification significantly reduced the possibility of false positive cases and was shown to improve the accuracy for the determination of promoter position.
A statistical analysis of the positions of ECs on surface mounted template DNA/transcription products following digestion with restriction enzyme allows promoters to be mapped within approximately 500 bp. The restriction enzyme digestion of surface-mounted template DNAs results in orientation of the DNA molecule; it also improves the accuracy of estimates of promoter loci. Furthermore, comparison of the positions of ECs on template DNAs at different transcription times (Fig. 8A) or from reactions conducted at different ratios of 3′dNTP and NTPs (Fig. 6, Fig. 7) allows the direction of transcription to be determined.
We also show that promoter strength can be estimated by measuring the fluorescence intensity of ECs and determining the frequency of ECs at specific template loci. The results obtained are consistent with those obtained by conventional methods.
Automated and high throughput application of Transchip to whole genome expression analysis promises to provide a new, direct method that detects elongation complexes in place on single template molecules and that reveals promoter sites, strength of promoters, direction of transcription, and termination of transcription with ensembles of single molecules. A schematic for genome-wide Transchip analysis is diagrammed in Fig. 9. Key improvements required for high throughput analysis include advances in adaptation to image processing, as well as improvements and optimization of the presentation of template/elongation complexes on surfaces or by other means. Perfect stretching of template DNAs with associated ECs on the surface (with little background of uneven stretching) would result in improvements in the calling of ECs. Another approach would be to use FRET (fluorescence resonance energy transfer) imaging or two-color labeling to clearly define the ECs. The linking of transcriptional data from single genomic DNA templates to sequence and/or map data promises to provide a powerful tool for probing expression patterns across genomes under defined conditions.
Fig. 9.
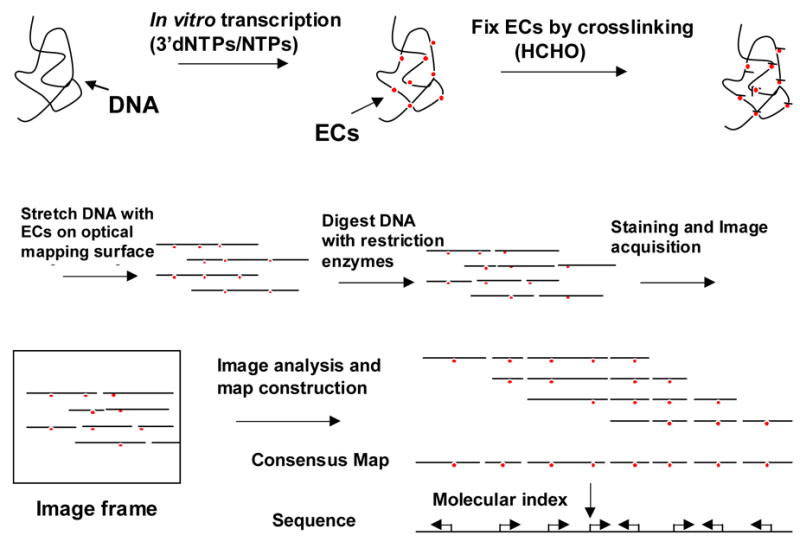
Analysis of genome-wide transcription products at the single molecule basis. For Transchip analysis across a genome, single genomic template DNAs are transcribed with the appropriate RNA polymerase and factors at different ratios of 3′dNTPs to NTPs. Elongation complexes are fixed to genomic DNA by crosslinking with formaldehyde (HCHO) prior to the mounting and elongation of the reaction products on Optical Mapping surfaces. Following restriction enzyme digestion of surface-mounted template molecules, the template molecules with associated elongation complexes (ECs) are stained with fluorescent dye, imaged, and positions of ECs determined. Molecular barcodes (single molecule restriction maps) are used to index individual DNA molecules to the consensus map or in silico map of the genome constructed from sequence. Aligned molecules and positions and directionality of promoters determined from Transchip analysis are used to construct an in vitro transcription profile for the whole genome.
Acknowledgments
This work was supported in part by Grant No. 5R01HG000225. Rod Runnheim, Dan Forrest, Shiguo Zhou and Hua Yu are thanked for their contributions and suggestions, and Louise Pape for editing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 2.Kasas S, Thomson NH, Smith BL, Hansma HG, Zhu X, Guthold M, Bustamante C, Kool ET, Kashlev M, Hansma PK. Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry. 1997;36:461–468. doi: 10.1021/bi9624402. [DOI] [PubMed] [Google Scholar]
- 3.Rich A. The rise of single-molecule DNA biochemistry. Proc Natl Acad Sci USA. 1998;95:13999–14000. doi: 10.1073/pnas.95.24.13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO Journal. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 6.Bai L, Santangelo TJ, Wang MD. Single-molecule analysis of RNA polymerase transcription. Annu Rev Biophys Biomol Struct. 2006;35:343–360. doi: 10.1146/annurev.biophys.35.010406.150153. [DOI] [PubMed] [Google Scholar]
- 7.French SL, Miller OL., Jr Transcription mapping of the Escherichia coli chromosome by electron microscopy. J Bacteriol. 1989;171:4207–16. doi: 10.1128/jb.171.8.4207-4216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKnight SL, Miller OL., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Miller OL., Jr The nucleolus, chromosomes, and visualization of genetic activity. J Cell Biol. 1981;91(3 Pt 2):15s–27s. doi: 10.1083/jcb.91.3.15s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnology. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 12.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 13.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 14.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, Heisterkamp S, van Kampen A, Versteeg R. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 15.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 16.Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, Bussemaker HJ, van Kampen AH. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashizaki Y, Kanamori M. Dynamic transcriptome of mice. Trends Biotechnol. 2004;22:161–167. doi: 10.1016/j.tibtech.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz DC, Li X, Hernandez LI, Ramnarain SP, Huff EJ, Wang YK. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 19.Meng X, Benson K, Chada K, Huff EJ, Schwartz DC. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat Genet. 1995;9:432–438. doi: 10.1038/ng0495-432. [DOI] [PubMed] [Google Scholar]
- 20.Aston C, Hiort C, Schwartz DC. Optical mapping: an approach for fine mapping. Methods Enzymol. 1999;303:55–73. doi: 10.1016/s0076-6879(99)03006-2. [DOI] [PubMed] [Google Scholar]
- 21.Aston C, Mishra B, Schwartz DC. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 1999;17:297–302. doi: 10.1016/s0167-7799(99)01326-8. [DOI] [PubMed] [Google Scholar]
- 22.Lim A, Dimalanta ET, Potamousis KD, Yen G, Apodoca J, Tao C, Lin J, Qi R, Skiadas J, Ramanathan A, Perna NT, Plunkett G, III, Burland V, Mau B, Hackett J, Blattner FR, Anantharaman TS, Mishra B, Schwartz DC. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 2001;11:1584–1593. doi: 10.1101/gr.172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou S, Kile A, Kvikstad E, Bechner M, Severin J, Forrest D, Runnheim R, Churas C, Anantharaman TS, Myler P, Vogt C, Ivens A, Stuart K, Schwartz DC. Shotgun optical mapping of the entire Leishmania major Friedlin genome. Mol Biochem Parasitol. 2004;138:97–106. doi: 10.1016/j.molbiopara.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Kile A, Bechner M, Place M, Kvikstad E, Deng W, Wei J, Severin J, Runnheim R, Churas C, Forrest D, Dimalanta ET, Lamers C, Burland V, Blattner FR, Schwartz DC. Single-molecule approach to bacterial genomic comparisons via optical mapping. J Bacteriol. 2004;186:7773–7782. doi: 10.1128/JB.186.22.7773-7782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reslewic S, Zhou S, Place M, Zhang Y, Briska A, Goldstein S, Churas C, Runnheim R, Forrest D, Lim A, Lapidus A, Han CS, Roberts GP, Schwartz DC. Whole-genome shotgun optical mapping of Rhodospirillum rubrum. Appl Environ Microbiol. 2005;71:5511–5522. doi: 10.1128/AEM.71.9.5511-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou S, Deng W, Anantharaman TS, Lim A, Dimalanta ET, Wang J, Wu T, Chunhong T, Creighton R, Kile A, Kvikstad E, Bechner M, Yen G, Garic-Stankovic A, Severin J, Forrest D, Runnheim R, Churas C, Lamers C, Perna NT, Burland V, Blattner FR, Mishra B, Schwartz DC. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl Environ Microbiol. 2002;68:6321–6331. doi: 10.1128/AEM.68.12.6321-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimalanta ET, Lim A, Runnheim R, Lamers C, Churas C, Forrest DK, de Pablo JJ, Graham MD, Coppersmith SN, Goldstein S, Schwartz DC. A microfluidic system for large DNA molecule arrays. Anal Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 28.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kochetkov SN, Rusakova EE, Tunitskaya VL. Recent studies of T7 RNA polymerase mechanism. FEBS Letters. 1998;440:264–267. doi: 10.1016/s0014-5793(98)01484-7. [DOI] [PubMed] [Google Scholar]
- 30.Sousa R. T7 RNA polymerase. Uirusu. 2001;51:81–94. doi: 10.2222/jsv.51.81. [DOI] [PubMed] [Google Scholar]
- 31.Sousa R, Mukherjee S. T7 RNA polymerase. Progress in Nucleic Acid Res and Mol Biol. 2003;73:1–41. doi: 10.1016/s0079-6603(03)01001-8. [DOI] [PubMed] [Google Scholar]
- 32.Waters JJ, Barlow AL, Gould CP. Demystified ... FISH. Mol Pathol. 1998;51:62–70. doi: 10.1136/mp.51.2.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 34.Cai W, Jing J, Irvin B, Ohler L, Rose E, Shizuya H, Kim UJ, Simon M, Anantharaman T, Mishra B, Schwartz DC. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc Natl Acad Sci USA. 1998;95:3390–3395. doi: 10.1073/pnas.95.7.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S, Deng W, Anantharaman TS, Lim A, Dimalanta ET, Wang J, Wu T, Tao C, Creighton R, Kile A, Kvikstad E, Bechner M, Yen G, Garic-Stankovic A, Severin J, Forrest D, Runnheim R, Churas C, Lamers C, Perna NT, Burland V, Blattner FR, Mishra B, Schwartz DC. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Applied and Environmental Microbiol. 2002;68:6321–6331. doi: 10.1128/AEM.68.12.6321-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou S, Kvikstad E, Kile A, Severin J, Forrest D, Runnheim R, Churas C, Hickman JW, Mackenzie C, Choudhary M, Donohue T, Kaplan S, Schwartz DC. Whole-genome shotgun optical mapping of Rhodobacter sphaeroides strain 2.4.1 and its use for whole-genome shotgun sequence assembly. Genome Res. 2003;13:2142–2151. doi: 10.1101/gr.1128803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou S, Schwartz DC. The Optical Mapping of microbial genomes. ASM News. 2004;70:323–330. [Google Scholar]
- 38.Zhou S, Kile A, Bechner M, Place M, Kvikstad E, Deng W, Wei J, Severin J, Runnheim R, Churas C, Forrest D, Dimalanta ET, Lamers C, Burland V, Blattner FR, Schwartz DC. A single molecule approach to bacterial genomic comparisons via Optical Mapping. J Bacteriol. 2004;186:7773–7782. doi: 10.1128/JB.186.22.7773-7782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura Y, Ichikawa Y, Ishikawa T, Ogura M, de Fries R, Shimada H, Mitsuhashi M. Fluorometric determination of total mRNA with oligo(dT) immobilized on microtiter plates. Clin Chem. 1996;42:1758–64. [PubMed] [Google Scholar]
- 40.Hamaguchi Y, Aso Y, Shimada H, Mitsuhashi M. Direct reverse transcription-PCR on oligo(dT)-immobilized polypropylene microplates after capturing total mRNA from crude cell lysates. Clin Chem. 1998;44:2256–2263. [PubMed] [Google Scholar]
- 41.MacDonald LE, Durbin RK, Dunn JJ, McAllister WT. Characterization of two types of termination signal for bacteriophage T7 RNA polymerase. J Mol Biol. 1994;238:145–58. doi: 10.1006/jmbi.1994.1277. [DOI] [PubMed] [Google Scholar]
- 42.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikeda RA, Warshamana GS, Chang LL. In vivo and in vitro activities of point mutants of the bacteriophage T7 RNA polymerase promoter. Biochemistry. 1992;31:9073–9080. doi: 10.1021/bi00152a051. [DOI] [PubMed] [Google Scholar]
- 44.Cai W, Aburatani H, Stanton VP, Jr, Housman DE, Wang Y, Schwartz DC. Ordered restriction endonuclease maps of yeast artificial chromosomes created by Optical Mapping on surfaces. Proc Natl Acad Sci USA. 1995;92:5164–5168. doi: 10.1073/pnas.92.11.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jing J, Reed J, Huang J, Hu X, Clarke V, Edington J, Housman D, Anantharaman TS, Huff EJ, Mishra B, Porter B, Shenker A, Wolfson E, Hiort C, Kantor R, Aston C, Schwartz DC. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc Natl Acad Sci USA. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai Z, Jing J, Aston C, Clarke V, Apodaca J, Dimalanta ET, Carucci DJ, Gardner MJ, Mishra B, Anantharaman TS, Paxia S, Hoffman SL, Venter JC, Huff EJ, Schwartz DC. A shotgun optical map of the entire Plasmodium falciparum genome. Nature Genetics. 1999;23:309–313. doi: 10.1038/15484. [DOI] [PubMed] [Google Scholar]
- 47.Lin J, Qi R, Aston C, Jing J, Anantharaman TS, Mishra B, White O, Daly MJ, Minton KW, Venter JC, Schwartz DC. Whole-genome shotgun optical mapping of. Deinococcus radiodurans, Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 48.Giacalone J, Delobette S, Gibaja V, Ni L, Skiadas Y, Qi R, Edington J, Lai Z, Gebauer D, Zhao H, Anantharaman T, Mishra B, Brown LG, Saxena R, Page DC, Schwartz DC. Optical mapping of BAC clones from the human Y chromosome DAZ locus. Genome Res. 2000;10:1421–1429. doi: 10.1101/gr.112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, Abouelleil A, Allen NR, Bi W, Bloom T, Borowsky ML, Bugalter BE, Butler J, Chang JL, Chen CK, Cook A, Corum B, Cuomo CA, de Jong PJ, DeCaprio D, Dewar K, FitzGerald M, Gilbert J, Gibson R, Gnerre S, Goldstein S, Grafham DV, Grocock R, Hafez N, Hagopian DS, Hart E, Norman CH, Humphray S, Jaffe DB, Jones M, Kamal M, Khodiyar VK, LaButti K, Laird G, Lehoczky J, Liu X, Lokyitsang T, Loveland J, Lui A, Macdonald P, Major JE, Matthews L, Mauceli E, McCarroll SA, Mihalev AH, Mudge J, Nguyen C, Nicol R, O’Leary SB, Osoegawa K, Schwartz DC, Shaw-Smith C, Stankiewicz P, Steward C, Swarbreck D, Venkataraman V, Whittaker CA, Yang X, Zimmer AR, Bradley A, Hubbard T, Birren BW, Rogers J, Lander ES, Nusbaum C. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature. 2006;440:1045–1049. doi: 10.1038/nature04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gueroui Z, Place C, Freyssingeas E, Berge B. Observation by fluorescence microscopy of transcription on single combed DNA. Proc Natl Acad Sci USA. 2002;99:6005–6010. doi: 10.1073/pnas.092561399. [DOI] [PMC free article] [PubMed] [Google Scholar]


