Abstract
The giant protein titin, which is responsible for passive elasticity in muscle fibers, is built from ∼300 regular immunoglobulin-like (Ig) domains and FN-III repeats. While the soft elasticity derived from its entropic regions, as well as the stiff mechanical resistance derived from the unfolding of the secondary structure elements of Ig- and FN-III domains have been studied extensively, less is known about the mechanical elasticity stemming from the orientation of neighboring domains relative to each other. Here we address the dynamics and energetics of interdomain arrangement of two adjacent Ig-domains of titin, Z1, and Z2, using molecular dynamics (MD) simulations. The simulations reveal conformational flexibility, due to the domain-domain geometry, that lends an intermediate force elasticity to titin. We employ adaptive biasing force MD simulations to calculate the energy required to bend the Z1Z2 tandem open to identify energetically feasible interdomain arrangements of the Z1 and Z2 domains. The finding is cast into a stochastic model for Z1Z2 interdomain elasticity that is generalized to a multiple domain chain replicating many Z1Z2-like units and representing a long titin segment. The elastic properties of this chain suggest that titin derives so-called tertiary structure elasticity from bending and twisting of its domains. Finally, we employ steered molecular dynamics simulations to stretch individual Z1 and Z2 domains and characterize the so-called secondary structure elasticity of the two domains. Our study suggests that titin's overall elastic response at weak force stems from a soft entropic spring behavior (not described here), from tertiary structure elasticity with an elastic spring constant of ∼0.001–1 pN/Å and, at strong forces, from secondary structure elasticity.
INTRODUCTION
The ability of striated muscle to stretch and relax, in an elastic manner, is crucial for its role as a force-bearing component in higher organisms. The muscle sarcomere extends beyond its equilibrium length immediately after contraction, and a passive restoring force arises from a protein called titin, whose elastic response restores a muscle fiber to its resting length (1–6). Titin is the largest known protein, with a length >1 μm and a molecular weight of up to 4 MDa. It is built from a modular construct of ∼300 tandem repeats of predominantly immunoglobulin-like (Ig) and fibronectin type III (Fn-III) domains, and from flexible random coil-like PEVK (rich in proline, glutamate, valine, and lysine) regions (7); furthermore, single Ig-domain insertions exist that distinguish isoforms of titin in cardiac and skeletal muscle (8).
Titin is the third most abundant protein of vertebrate striated muscle, after actin and myosin. The N-terminal section of titin originates from the sarcomeric Z-disc (9–11) and its C-terminal region is an integral component of the sarcomeric M-line (12,13). Fig. 1 depicts a schematic view of titin's individual components, and its position within the muscle sarcomere.
FIGURE 1.
Schematic of the titin spring in muscle, and detailed view of titin's Z1Z2 domains. Shown is an electron micrograph image (courtesy R. Craig, University of Massachusetts) of the human muscle sarcomere and the schematic representation of titin along the sarcomere length. Titin is anchored with one of its ends (N-terminal) through the protein telethonin to the sarcomeric Z-disc and with its other end (C-terminal) to the M-line. Components of titin shown in this figure include the proximal Z1Z2 domains (circled), entropic PEVK region, the well-studied I27 (now numbered I91) domain in the I-band, and the Ig-Fn-III triad A168-170 of the A-band.
Extensive studies have characterized titin as a protein with many distinct functions. At the early stages of myofibril growth, titin is believed to play a crucial role as a molecular scaffold for organizing the various components of the sarcomere, as evidenced by exact matches between the pattern of repeat domains in titin and myosin (thick filament) periodicity (14,15). There is also evidence that the ends of titin not only anchor at, but interact with, signaling proteins at the Z-line and M-line, possibly mediated by passive stretching forces (6,7). Stretch-mediated signaling involving titin has recently been correlated with titin kinase activity at its C-terminus (16,17), and the titin kinase phosphorylation activity has also been observed at the N-terminus where the Z-line protein telethonin is located (18,19). The recently solved structure of the titin Z1Z2-telethonin complex (20) and subsequent investigation into the mechanical binding of Z1Z2 to telethonin (21) also suggest how the N-terminus of titin is anchored at the Z-disc.
Besides acting as a molecular scaffold and signaling platform, titin functions as a molecular spring that dampens sarcomeric extension forces. The I-band region of titin, which forms an elastic connection between the ends of thick filaments and the sarcomeric Z-disc, contains flexible PEVK domains interdigitated between Ig-domain repeats. These random coil PEVK domains are believed to reversibly unfold to permit extension, like an entropic spring, and to contribute to titin elasticity when weak forces are applied (22–25).
Investigations of the extensibility and elasticity of titin, both in situ and at the level of single molecules, have revealed that titin is not merely a soft entropic spring, but also exhibits a stiffer, structure-dependent elasticity (26). Fig. 2 shows a schematic presentation of titin elasticity across a wide range of extension and force as studied by optical tweezers (22,27) and atomic force microscopy (AFM) (28–34). This figure illustrates that a region of soft elasticity at weak forces (<50 pN) is followed by a region of nonlinear stiff elasticity at forces of >50 pN. While the stretching and unfolding of individual or linked PEVK, Ig-, and FN-III domains connected with the soft and stiff elasticity of titin have been extensively characterized through AFM (26,28–38), optical tweezers (22,27,39), and computational unfolding studies (21,40–53), an elasticity of titin associated with domain-domain extension has been less extensively investigated.
FIGURE 2.
Two regimes of titin elasticity—soft and stiff nonlinear. Shown schematically is how titin responds to stretching forces of various magnitudes (22,27–29,31,32,34). At weak forces (<50 pN), the primary contribution of titin's response arises from the PEVK domains and the straightening of a bent and twisted titin chain. At high forces (>50 pN) titin exhibits a highly nonlinear elastic response. In this study we will relate the non-PEVK soft elasticity of titin to the so-called tertiary structure elasticity and the nonlinear stiff elasticity to the secondary structure elasticity. These two types of elasticities are defined in the text.
The soft elasticity of titin is likely due, in part, to a generic behavior of random coils formed by PEVK and other similar domains, an entropic spring behavior in principle well understood (8,23–25,54). The studies of stiff elasticity were based on the then available structures of titin domains I1 (55) and I91 (formerly I27) (56) and revealed that the individual domains, when subject to sufficiently strong stretching forces, unravel their secondary structure, giving rise to titin's nonlinear stiff elasticity called “secondary structure elasticity”. The soft elasticity of titin is also due to a straightening out of titin's multidomain segments (22,57). Adjacent segments may exhibit a flexibility that leads a titin chain to assume in the relaxed state a locally compacted shape, and a straight shape when under tension. The question arises if the elasticity linked to such structural transition contributes to the soft elasticity of titin characterized through a spring constant that falls in a range of observed values.
Lacking until recently structures of two or more adjacent titin domains made it impossible to computationally investigate titin's soft elasticity associated with domain-domain rearrangement. The relative orientation of adjacent domains should be controlled through the respective linker as well as domain-domain interaction. The tertiary structure specifying the arrangement of adjacent domains should resist forces that stretch domain pairs apart, thereby giving rise to a local elastic property, to be referred to in this article as “tertiary structure elasticity”. Contributions of many adjacent domain pairs can add and confer to titin an overall elasticity. Fortunately, the structures of a tandem of two adjacent Ig-domains, Z1Z2 (58) and three consecutive domains A168-170 (59) have been recently elucidated by x-ray crystallography. In addition, data on the conformation and dynamics of Z1Z2 in solution have become available through small angle x-ray scattering (SAXS) and NMR spectroscopy (58). These data allow us now to explore the extent to which the dynamics of interdomain arrangement contributes to the elasticity of the titin chain.
Given the wealth of experimental data on Z1Z2, our study will use this tandem as a model system to explore the role of interdomain relations in titin elasticity. Crystallographic data revealed Z1Z2 in two conformational states, one in an overall V-shape shown in Fig. 3 A and one in an extended shape shown in Fig. 3 B. Z1Z2 models satisfying NMR residual dipolar coupling (RDC) data and SAXS measurements in solution are shown in Fig. 3 C (for further details, see (58)). The models correspond to a structure that is intermediate between those shown in Fig. 3, A and B. The three structures showing Z1Z2 in closed, intermediate, and wide-open V-shape suggest immediately that interdomain dynamics might contribute to the tertiary structure elasticity of titin. A highly schematic representation of this elasticity is shown in Fig. 3 D, in which the tandem domains become extended while preserving secondary structure integrity of titin's domains. However, it is yet to be established whether the multiple conformations accessible to Ig tandems in the filament are, or are not, energetically equivalent and how they contribute to elasticity in titin. This information is best obtained by means of molecular dynamics simulations.
FIGURE 3.
Experimentally derived conformers for titin Z1Z2 and a possible mode of tertiary structure elasticity. (A,B) Crystal structure (PDB code 2A38) for the N-terminal region of titin comprised of the tandem Z1 (purple) and Z2 (orange) domains in two crystallographically constrained conformations, termed closed and open, respectively. (C) NMR-RDC models of the experimentally observed semi-extended conformation for Z1Z2. The colors correspond to the NMR lowest (blue) and second lowest (red) energy conformers and the small angle x-ray scattering best-fit conformer (green) (58). (D) Highly schematic view of the tertiary structure elasticity of titin due to bending adjacent protein domains open and closed. In actual titin, not every linker may contribute high flexibility; some linkers might be short and stiff.
Accordingly, in this article, we employ MD simulations to study the conformational dynamics of the titin Z1Z2 tandem, focusing specifically on the forces governing domain-domain interactions and associated elastic properties. Extended timescale simulations of the open Z1Z2 conformer (see Materials and Methods) reveal, independently, a quasi-equilibrium structure that matches precisely the experimentally observed semi-extended NMR-RDC conformer. Using a combination of steered molecular dynamics (SMD) (60) and adaptive biasing force (ABF) (61–65) simulations, we calculate the work required to hinge apart the Z1 and Z2 domains without unfolding either Ig-domain, thereby exploring only the contribution from tertiary structure characteristics. We then extend our scope by using the simulation results to construct a schematic stochastic model for a system of titin Ig-domain repeats (an Ig-chain), in which changes in tertiary structure endows the chain with overall elasticity.
Finally, we also study the nonlinear stiff elasticity of titin. This elasticity is due to the interactions within the secondary structure elements of titin, namely Ig- and Fn-III -like domains, which strongly resist mechanical stretching forces and have been well characterized in the studies mentioned above. In this case, we probe the elasticity of individual Z1 and Z2 domains by means of simulations, as done earlier for I1 (67) and I91 (31,40,46). Taken together, our findings present Z1 and Z2 as generic building blocks establishing the overall secondary and tertiary structure elasticity of titin.
MATERIALS AND METHODS
Simulated systems
Atomic coordinates of the titin Z1Z2 Ig-domain tandem were taken from the crystal structure (Protein Data Bank entry code 2A38). The structure consists of Z1Z2 arranged in two separate conformations: a compact form, termed the closed conformation in which Z1 and Z2 form the arms of a V (see Fig. 3 A), and an extended form, termed the open form, in which Z1 and Z2 are arranged in a stretched-out manner (see Fig. 3 B). The Z1Z2 tandem consists of a total of 194 residues; Z1 contains 98 residues; a three-residue linker connects it with the 93-residue Z2 domain. The topology of the complex and missing hydrogen atoms were generated using psfgen (68) with the topology file for CHARMM27 (69).
In the present MD study, four systems were investigated. The first two systems involved the two crystal conformers of Z1Z2, open and closed, that were solvated in a water box. The closed titin Z1Z2 conformer was placed in the center of a water box of dimension 122 × 79 × 66 Å3, consisting of 194 protein residues (2915 atoms) and 18,546 water molecules (55,638 atoms) with a total system size of 58,553 atoms. The open titin Z1Z2 conformer required a water box of dimension 329 × 62 × 64 Å3 for solvation, as shown in Fig. 5 A: the system consisted of 194 protein residues (2915 atoms) and 39,886 water molecules (119,658 atoms) with a total system size of 122,573 atoms. The second set of systems involved the domains Z1 and Z2 separately. Residues 1–99 of the 2A38 (the Z1Z2 structure) complex, which includes titin Z1 and the first residue of the linker, were placed into a water box of dimension 322 × 60 × 58 Å3 to allow room for subsequent stretching simulations. The Z1-only system consisted of 99 protein residues (1463 atoms) and 34,285 water molecules (102,855 atoms) with a total system size of 104,318 atoms. Residues 101–194 of 2A38, consisting of titin Z2 and one linker domain residue, were solvated in a 320 × 60 × 59 Å3 water box, the system consisting of 94 protein residues (1440 atoms) and 35,221 water molecules (105,663 atoms) with a total system size of 107,103 atoms.
FIGURE 5.
Equilibrium simulation of Z1Z2 and convergence to NMR conformers. (A) The simulation system for the open form of crystallographically resolved Z1Z2 (Z1 in green; Z2 in orange; water is blue). (B) RMSD plot for simA2 comparing the deviation of secondary structural elements from the three experimentally derived structures (ii–iv; see text) for Z1Z2 shown in Fig. 3. The colors, corresponding to those also used in Fig. 3 C, are selected as follows: the comparison of i, the simulated system, with ii is shown in blue, with iii in red, and with iv in green. The closest convergence of the MD model, i, to the experimental structures ii–iv, arises between 25 ns and 38 ns. (C) Snapshots of the simulated Z1Z2 segment (simulation simA2) overlaid with iii at 0 ns, at ∼11.5 ns, and at ∼27 ns. A movie showing the entire trajectory for simA2 can be found in Supplementary Material.
Molecular dynamics
Simulations were performed using NAMD 2.6 (68) with the CHARMM27 force field for proteins (69) and the TIP3P model (70) for water. Particle mesh Ewald (PME) summation was employed to calculate long-range electrostatic forces with a grid size of <1 Å. The van der Waals interactions employed a switching function starting at 10 Å and a cutoff of 12 Å. An integration time step of 1 fs was adopted, with a multiple time-stepping algorithm (71,72) employed to compute covalent bonds every time step, short-range nonbonded interactions every two time steps, and long-range electrostatic forces every four time steps. Multiple time-stepping increases the efficiency of simulation by a factor of two, making the nanosecond timescale more readily accessible for our simulations. Constant temperature control (T = 300 K) was employed using Langevin dynamics and NPT ensemble simulations. A Langevin coupling coefficient of 5 ps−1 was used for the temperature control; constant pressure was maintained at P = 1 atm (101.3 kPa) utilizing the Nosé-Hoover Langevin piston method with a decay period of 100 fs and a damping time constant of 50 fs.
SMD simulations (60,73) were carried out by either fixing the Cα atom of the N-terminus of either titin Z1 or Z2, and applying an external force to the C-terminal Cα atom (see figures in Results for stretching configurations); the stretching forces were directed along the t = 0 vector connecting the mentioned termini. Constant force SMD simulations applied a time-independent potential of F = kx to the specified atom(s), where x is the Cα-Cα distance for the two termini. For the spring constant, we chose a value of 3 kBT/Å2, which corresponds to a root mean-square deviation (RMSD) value of 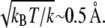
The adaptive biasing force (ABF) method, originally developed by Pohorille and co-workers (61–63) and recently adapted into NAMD (derived for the canonical ensemble NVT) as an executable library (64,65), was employed to calculate the reversible work, or potential of mean force (PMF), required to extend the Z1Z2 tandem from the closed conformation to the open conformation through rotation around a hinge axis. The ABF method uses a continuously updated estimate of a free energy profile along an a priori selected reaction coordinate to apply a bias that overcomes energetic barriers, eventually leading to a free diffusion type motion along the reaction coordinate. The free energy derivative can then be determined and the potential of mean force (PMF) recovered by thermodynamic integration (65). In the case of Z1Z2, the sampling space was confined to a separation of two centers of mass located at the opposing tips of the Z1Z2 tandem corresponding to the N-terminus of Z1 and the C-terminus of Z2, respectively. The region for the center of mass chosen for the tip of Z1 included the α-carbons of residues 1–6, 31–34, and 84–86. For Z2, the center of mass for the tip of Z2 included the α-carbons of residues 116–121, 164–171, and 192–194. A restraining harmonic potential, specified in the ABF simulation configuration script, was applied to the terminal α-carbons, residues 1 and 194, to constrain the motion along the reaction coordinate such that it adopted a rotation motion around the hinge axis. The distance range between the centers of mass defined above for each ABF run (simC1-C7) was 5 Å in each case, the exception being simC5 where a range of 10 Å was sampled. The size for each ABF sampling bin was 0.1 Å. All simulations were run long enough to observe convergence of the system for accurate sampling of the free energy landscape.
Analysis
System coordinates were saved every picosecond, and analysis was performed using the program VMD (74). The structural changes in the calculated models were monitored by calculating the RMSD of their backbone atoms. The end-to-end distance profile of a stretched system is based on the distance between the two stretched or between the stretched and fixed terminal Cα atoms. The change of the end-to-end distance is defined as the extension of the system. Salt bridges were calculated in VMD with an oxygen-nitrogen cutoff of 3.2 Å; hydrogen bonds were calculated with a distance cutoff of 3.0 Å and a donor-hydrogen-acceptor cutoff angle of 120° (equal to 60° out-of-line angle) for bonding pairs. The interdomain angle between the Z1Z2 Ig-tandem was calculated as the angle generated by connecting the center-of-mass regions at the N-terminus of Z1 (residues 1–6, 31–34, and 84–87), linker (residues 17–19, 98–104, and 131–132), and C-terminal of Z2 (residues 116–121, 164–171, and 192–194). By capturing the centers of mass for this calculation, we avoid fluctuations that arise from thermal motion of single atoms, a significant improvement over a previous approach (21).
Summary of simulations
The simulations carried out are listed in Table 1. The equilibrations of the four systems (simA1-4) employed an identical procedure. The systems were first minimized for 10,000 conjugate gradient steps, then equilibrated in a 300 K heat bath with constant volume for 25 ps. After this, the volume of the system was allowed to freely change under NPT conditions. The equilibration times varied from 5 ns to 40 ns (see Table 1). Steered molecular dynamics (SMD) simulations utilized the equilibrated closed Z1Z2, Z1, and Z2 structures from simA1, simA3, and simA4, respectively. In constant force simulations, forces of 50, 94, and 164.5 pN were applied to open the closed conformer (simB1-3), and forces of 350 and 500 pN were employed for unfolding studies of individual Z1 and Z2 domains (simD1-4, simE1-4). The SMD parameters are listed in Table 1. Seven sets of ABF simulations (simC1-7) were carried out to sample across the entire thermally feasible hinging motion of Z1Z2, beginning from the compact closed conformer, through the intermediate semi-extended state observed in simA2, and terminating with the open form.
TABLE 1.
Summary of simulations
| Name | Structure | Type | Ensemble | Atoms × 1000 | Size Å3 | Special parameters | Time ns |
|---|---|---|---|---|---|---|---|
| simA1 | Z1Z2-closed | EQ | NpT | 59 | 122 × 79 × 66 | — | 10.0 |
| simA2 | Z1Z2-open | EQ | NpT | 123 | 329 × 62 × 64 | — | 40.0 |
| simA3 | Z1 | EQ | NpT | 105 | 322 × 60 × 58 | — | 5.0 |
| simA4 | Z2 | EQ | NpT | 108 | 320 × 60 × 59 | — | 5.0 |
| simB1 | Z1Z2-closed | SCF | NV | 59 | 122 × 79 × 66 | 50 pN* | 2.0 |
| simB2 | Z1Z2-closed | SCF | NV | 59 | 122 × 79 × 66 | 94 pN† | 2.0 |
| simB3 | Z1Z2-closed | SCF | NV | 59 | 122 × 79 × 66 | 164.5 pN‡ | 2.0 |
| simC1 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 45–50 ŧ | 60.0 |
| simC2 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 50–55 ŧ | 10.0 |
| simC3 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 55–60 ŧ | 10.0 |
| simC4 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 60–65 ŧ | 10.0 |
| simC5 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 65–75 ŧ | 10.0 |
| simC6 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 75–80 ŧ | 20.0 |
| simC7 | Z1Z2-closed | ABF | NVT | 59 | 122 × 79 × 66 | 80–85 ŧ | 10.0 |
| simD1 | Z1 | SCF | NV | 105 | 322 × 60 × 58 | 500 pN¶ | 7.0 |
| simD2 | Z1 | SCF | NV | 105 | 322 × 60 × 58 | 350 pN¶ | 30.0 |
| simD3 | Z1 | SCF | NV | 105 | 322 × 60 × 58 | 350 pN¶ | 30.0 |
| simD4 | Z1 | SCF | NV | 105 | 322 × 60 × 58 | 350 pN¶ | 30.0 |
| simE1 | Z2 | SCF | NV | 108 | 320 × 60 × 59 | 500 pN‖ | 8.0 |
| simE2 | Z2 | SCF | NV | 108 | 320 × 60 × 59 | 350 pN‖ | 30.0 |
| simE3 | Z2 | SCF | NV | 108 | 320 × 60 × 59 | 350 pN‖ | 15.0 |
| simE4 | Z2 | SCF | NV | 108 | 320 × 60 × 59 | 350 pN‖ | 30.0 |
Type: EQ denotes equilibration; ABF denotes adaptive biasing force simulations, and SCF denotes constant force SMD simulations. Ensemble lists the variables held constant during the simulations; N, V, p, and T correspond to number of atoms, volume, pressure, and temperature, respectively. Footnotes under special parameters describe the ABF sampling range, and, in the case of SMD simulations, which atoms were fixed or had an external force applied to them.
Fixed all α-carbons of Z1 and applied force to C-terminal α-carbon of Z2.
Fixed all α-carbons of Z1 and applied force to of 1 pN to all α-carbons of Z2.
Fixed all α-carbons of Z1 and applied force to of 1.75 pN to all α-carbons of Z2.
Total end-to-end extension of sampling range.
Fixed N-terminal α-carbon and applied force to C-terminal α-carbon of Z1.
Fixed N-terminal α-carbon and applied force to C-terminal α-carbon of Z2.
Simulations were conducted on the Teragrid and SGI Altix supercomputer clusters, equipped with Intel Itanium 2 processors, at the National Center for Supercomputing Applications at the University of Illinois at Urbana-Champaign. A system of 100,000 atoms required one day of simulation for 3 ns on 64 1.6 GHz Itanium 2 processors. The total simulation time reported in this study included 146 ns for the closed Z1Z2 conformer (59,000 atoms), 40 ns for the open conformer (123,000 atoms), 105 ns for Z1 alone (105,000 atoms), and 90 ns for Z2 alone (108,000 atoms).
Modeling of multidomain tertiary structure elasticity
A key question raised in our study is how the tertiary structure of adjacent domain pairs endows a multidomain chain with mechanical elasticity. For this purpose, we devised a model for titin that is simple, yet incorporates the key properties of the mechanics of Z1Z2 as elucidated by our simulations. The aim of the model was to furnish a simple representation for titin's I-band that demonstrates the muscle protein's overall tertiary structure elasticity. In this model, we employ the free energy profiles determined for the angular motion of Z1Z2 to construct by N-fold replication a multidomain system as illustrated in Fig. 3 D. For the sake of simplicity we assume a planar system in which the tertiary structure characteristics of a domain pair is captured by a single variable angle α0 or the equivalent variable end-to-end distance x; we ignore in this description torsional degrees of freedom that likewise contribute to the elasticity of end-to-end distance motion. We will refer to this system as the multidomain model.
For the sake of illustration we will make first a further drastic simplification and assume that domain pair angles take on only two values corresponding to a closed and open state, and refer to this planar multidomain system as the two-state multidomain model. This model is illustrated in Fig. 4 A. The model (see Eqs. 1–7), despite the discrete character of the underlying simple domain pair motion, exhibits rather continuous overall extensions for large N as specified below. The model will be replaced by a fully continuous description further below (see Eqs. 16–32).
FIGURE 4.
Stochastic modeling of titin's tertiary structure elasticity. (A) Schematic representation of a two-state multidomain chain. (B) Plot of the time-dependent end-to-end distribution given by Eq. 14, for N = 100, p = 0.3, b = 1, τR = 1, α0 = 45°, and X0 = 1.2 l0, which corresponds to an equilibrium length l0 ≈ 81 and initial length X0 ≈ 98. The initially extended chain relaxes back to its equilibrium length after a time comparable to τR. (C) Plot of the quadratic potential (Eq. 7) for the same system parameters and temperature T = 300 K.
The two-state multidomain model links N pairs of identical hinged domains, as shown for the case N = 5 in Fig. 4 A. Each pair is in either the closed or the open state with associated probabilities p and q = 1−p, respectively. A pair in the open state makes an angle of 180° at the hinge, while in the closed state it makes a fixed angle α0. The length of each domain is b/2 (hence, a pair in the open state has length b). Fig. 4 A shows an example of such chain, with two domain pairs in the open state and three domain pairs in the closed state. The reader may note from Fig. 4 A that, in the present model, only every other angle between adjacent domain pairs is included. One can readily extend the model and include every domain-domain angle by doubling the number of two-state systems and, to keep the chain's overall length unchanged, halving the length b. Similarly, one can describe the scenario in which fewer domain-domain angles contribute elasticity, say only every fourth angle (the remaining angles remaining stiff), in which case one doubles b and halves N.
In equilibrium, the distribution of the end-to-end distance X can be derived readily. For a chain with N domain pairs, there are  possible conformations with n closed and N–n open domain pairs, and the probability for the occurrence of each conformation is pn qN–n. The probability of finding the chain with n closed pairs is then
possible conformations with n closed and N–n open domain pairs, and the probability for the occurrence of each conformation is pn qN–n. The probability of finding the chain with n closed pairs is then
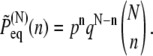 |
(1) |
The reader may verify that the total probability observes the normalization condition 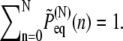
In view of our interest in the elastic behavior of the complete chain, the overall length X is a more relevant variable of choice than n. One can replace the dependence on n by one on X via the relation
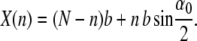 |
(2) |
Employing Stirling's approximation for the binomial coefficient in Eq. 1, one can derive the probability of finding the chain with overall length X
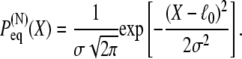 |
(3) |
Here 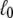 and σ are defined as
and σ are defined as
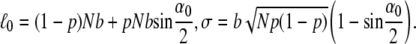 |
(4) |
The reader may verify again the normalization condition 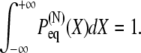
The Boltzmann relation
 |
(5) |
or, equivalently,
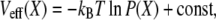 |
(6) |
permits one to associate the distribution defined in Eq. 3 with an effective potential
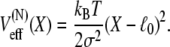 |
(7) |
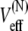 has the form of a harmonic potential with spring constant k = kBT/σ2, showing that the two-state multidomain chain behaves like a spring. One discerns also the physical significance of l0 and σ: l0 is the equilibrium length of the chain, while
has the form of a harmonic potential with spring constant k = kBT/σ2, showing that the two-state multidomain chain behaves like a spring. One discerns also the physical significance of l0 and σ: l0 is the equilibrium length of the chain, while 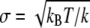 is the RMSD value of the spring in thermal equilibrium.
is the RMSD value of the spring in thermal equilibrium.
So far we have considered the multidomain chain in equilibrium when its end-to-end distribution obeys Eq. 3 and is related to the effective potential via Eq. 5. If the system is brought out of equilibrium, for example, by stretching or compressing the chain to an end-to-end distance X0, the system will start with a different distribution, but eventually return to the distribution given by Eq. 5. Indeed, for any time-dependent distribution of a multidomain chain with N domain pairs, P(N)(X, t), the following relation has to hold:
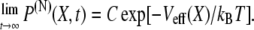 |
(8) |
Here C is a normalization constant. It is known that such distributions obey the Smoluchowski equation (75,76)
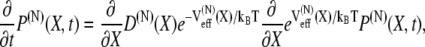 |
(9) |
where D(N)(X) is a (generally position-dependent) diffusion coefficient. Employing in this equation 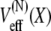 from Eq. 3 results in
from Eq. 3 results in
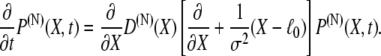 |
(10) |
The diffusion coefficient D(N)(X) was identified in the literature (77,78) to be
 |
(11) |
where 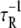 is the transition rate for a single two-state domain pair. Accordingly, the equation governing the evolution of P(N)(X, t) is
is the transition rate for a single two-state domain pair. Accordingly, the equation governing the evolution of P(N)(X, t) is
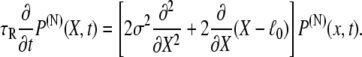 |
(12) |
We note that the diffusion coefficient D(N) scales with the number of domain pairs as
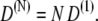 |
(13) |
The solution of Eq. 12 is expected to vanish for X → ±∞. The initial condition is P(N)(X, t = 0) = δ(X–X0) as stated before. The solution of Eq. 12 obeying these conditions is known to be (79)
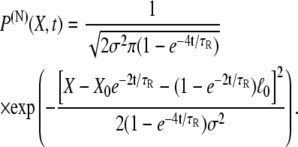 |
(14) |
The stochastic motion of the two-state multidomain chain in nonequilibrium is completely described by Eq. 14. The first notable property of the time-dependent distribution given by Eq. 14 is that it reduces, as required by Eq. 8, for t → ∞ to the equilibrium end-to-end distribution 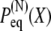 stated in Eq. 3.
stated in Eq. 3.
The elastic property of a two-state multidomain chain is embodied in Eq. 14, which states that an initially compressed or stretched chain relocates to its equilibrium length after a time comparable to the transition rate 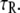 This property is depicted in Fig. 4 B for N = 100, p = 0.3, b = 1, τR = 1, α0 = 45° (which corresponds to
This property is depicted in Fig. 4 B for N = 100, p = 0.3, b = 1, τR = 1, α0 = 45° (which corresponds to  ), and an initial end-to-end length
), and an initial end-to-end length  This initially stretched chain starts then at X0 ≈ 98 and, after a time comparable to τR, the distribution decays back to the equilibrium distribution. The motion is that of a Brownian harmonic oscillator governed by the potential shown in Fig. 4 C.
This initially stretched chain starts then at X0 ≈ 98 and, after a time comparable to τR, the distribution decays back to the equilibrium distribution. The motion is that of a Brownian harmonic oscillator governed by the potential shown in Fig. 4 C.
So far we have considered a stochastic description for the idealized two-state multidomain model. However, our description can be extended to multidomain systems that replicate the elastic behavior of domain pairs like Z1Z2. It should be noted, though, that the description assumes that the angular degrees of freedom are independent of each other along the chain, i.e., the opening and closing motion of one domain pair does not affect the opening and closing of any other domain pair. Similar models are widely employed in polymer physics, at least at the qualitative level of description that we adopt here (80).
In biological multidomain systems like titin (7,46) and fibronectin (46) the individual domains along multidomain chains are similar, yet distinctively different. Accordingly, the individual domain pair lengths xj in a multidomain system with overall length
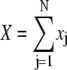 |
(15) |
can have distinct individual distributions related to their potentials of mean force 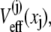 namely,
namely,
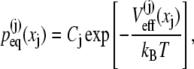 |
(16) |
where Cj is the normalization constant
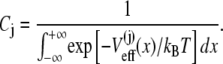 |
(17) |
If the xj degrees of freedom are assumed again to behave independently, then the probability for the multidomain system with a length configuration {x1, x2, x3, …, xN}, in which the jth domain pair has length xj, is
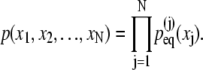 |
(18) |
However, we are interested in the probability of the chain assuming an end-to-end distance of length X. This probability is given by
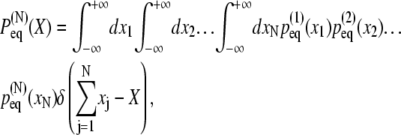 |
(19) |
where δ(…) is the Dirac δ-function.
The expression Eq. 19, as is well known (81), can be evaluated numerically through Fourier transform. For this purpose one writes Eq. 19 more succinctly as
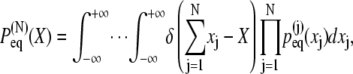 |
(20) |
and expresses the δ-function
 |
(21) |
which allows one to rewrite Eq. 20
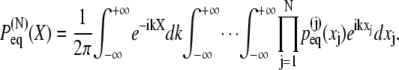 |
(22) |
Denoting the Fourier transform of 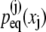 by
by 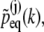
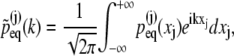 |
(23) |
one obtains the general expression for the end-to-end distribution
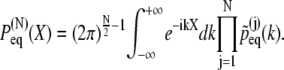 |
(24) |
In case that all distributions 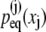 are Gaussian, namely,
are Gaussian, namely,
 |
(25) |
the 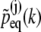 can be readily identified
can be readily identified
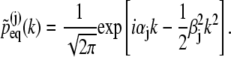 |
(26) |
Employing this result in Eq. 24 yields
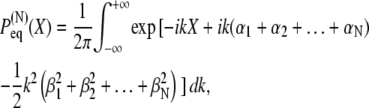 |
(27) |
or
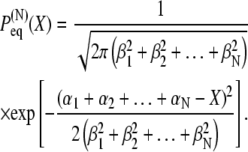 |
(28) |
Therefore, linking together N domain pairs that obey Gaussian distributions results in a chain with an end-to-end distribution 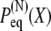 that is also Gaussian. Moreover, the mean of the total distribution is the sum of the individual Gaussian means and the width of the total distribution is the sum of the individual Gaussian widths. This is, of course, a well-known result.
that is also Gaussian. Moreover, the mean of the total distribution is the sum of the individual Gaussian means and the width of the total distribution is the sum of the individual Gaussian widths. This is, of course, a well-known result.
The distribution in Eq. 28 is associated with an effective harmonic potential
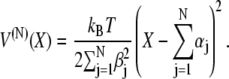 |
(29) |
In the simplest case that all distributions are identical, i.e., αj = α and βj = β for every domain pair j corresponding to a distribution
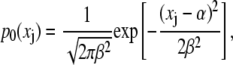 |
(30) |
one obtains the end-to-end distribution
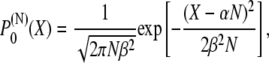 |
(31) |
which is associated with the effective harmonic potential
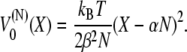 |
(32) |
Accordingly, the time evolution of this system in nonequilibrium situations is characterized again by Eq. 10 and the resulting dynamics is governed by the expression in Eq. 14.
To calculate the motion of the stretched or compressed spring one needs the diffusion coefficient D(N). For the individual degrees of freedom the diffusion coefficients D(j) can be determined along with the free energy profile by means of the ABF method (61,63–65) introduced below. The overall diffusion coefficient governing the motion of X should be 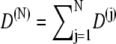 in analogy to Eq. 13, but this is a conjecture.
in analogy to Eq. 13, but this is a conjecture.
RESULTS
This section is based on multidomain models for titin and on the simulations summarized in Table 1. We first characterize the elastic properties of Z1Z2 and then extend the results to a multidomain model of titin elasticity involving replicated Z1Z2 units. We finally investigate the mechanical stability of Z1 and Z2 separately and relate the results also to the overall elastic behavior of titin.
Titin Z1Z2 equilibration and conformational state change
Equilibration (simulation simA2) of solvated tandem titin Z1Z2 (see Fig. 5 A) in the open configuration for 40 ns was accompanied by a spontaneous conformational change in which the termini of Z1Z2 rotated around the linker region to assume a semi-extended conformation. At the start of the simulation, the end-to-end distance between the N-terminus of Z1 and the C-terminus of Z2 measured 82.7 Å and the interdomain angle (defined in Materials and Methods) measured 149.4°. Over the course of the simulation, the Z1 and Z2 domains rotated toward each other, bending at the linker region, settling into a quasi-equilibrium state with end-to-end distances of 73–75 Å and interdomain angles of 128–130°.
Previously reported NMR-RDC conformers for Z1Z2 suggested that the preferential conformation of Z1Z2 in solution is, in fact, an intermediate state between the open and closed forms observed in the crystal structure (58), termed the semi-extended state. Indeed, simA2 approached this semi-extended state as clearly revealed in a superposition of the NMR-RDC structure and the MD trajectory, shown in Fig. 5 C.
The root mean-square deviation (RMSD) between i), the backbone α-carbons of the semi-extended secondary structural elements of the MD model; ii), the lowest energy NMR-RDC conformer; iii), the second lowest energy NMR-RDC conformer; and iv), the RDC conformer best-fitting SAXS data was calculated to assess the structural fit between the computational model and the molecule in solution. Models ii–iv represent the experimentally observed conformation of Z1Z2 in solution. Although all fits were comparable, the closest agreement was found with model iii. The result, plotted in Fig. 5 B, reveals that the fit is achieved after 22 ns of simulation, at which point the two ends of Z1Z2 begin to hinge toward each other, achieving remarkable structural alignment with the mentioned NMR conformer as shown in Fig. 5 C, illustrating the gradual bending of Z1Z2 from its crystallographic open state to the semi-extended state. At the beginning of the simulation, the open crystallographic form of Z1Z2 aligns poorly with the NMR conformer (Fig. 5 C-i); however, extended simulation lead to an intermediate fit within ∼2.5 Å at 11.5 ns (Fig. 5 C-ii) followed by a nearly total alignment of all β-strands within ∼1.5 Å at 27 ns (Fig. 5 C-iii). It should be noted that the MD simulations were performed by us before the NMR structures became available, and the convergence of both the MD model, in which the semi-extended state for Z1Z2 remained stable for ∼10 ns, and the observed NMR conformers suggests strongly that the energetically viable semi-extended conformation reported here represents a significant quasi-equilibrium state for solvated tandem Z1Z2 Ig-domains.
Tertiary structure elasticity of Z1Z2
The finding that titin Z1Z2 bends at its linker region and, thereby, adopts several stable tertiary structures has important ramifications for the overall elasticity of titin chains. Indeed, the elasticity depends on (entropic or enthalpic) energy stored in titin when its overall length is changed, and our findings above suggest that the Z1Z2 domain pair alters its enthalpic energy content when the end-to-end distance is altered. Titin is composed of many domains and tertiary structure transitions among any adjacent domain pair like Z1Z2 can contribute to the overall titin length and its associated energetics.
Both experimental and computational studies have examined previously the energetic requirements to stretch and unfold titin Ig-domains, both for single domains and for tandem chains (7,26,29,30,35,36,40,43,82–87), but most investigations focused on stretching involving strong forces that actually unraveled the secondary structure of individual domains. Here we investigate whether stretching with weak forces can lead to changes in tertiary structure and, thereby, contribute to the intermediate elasticity of stretched titin.
To characterize this type of elasticity, we ask what force is required to alter the arrangement of tandem Ig-domains, i.e., to stretch the elements of the chain out by reorienting neighboring domains, but without unfolding the domains. For this purpose, a series of low force SMD simulations were performed on the closed Z1Z2 conformer, in which Z1 was held fixed and Z2 permitted to rotate at the linker region, to open the tandem complex. A force of 50 pN (sim B1) applied to the C-terminus of Z2 over 2 ns was found not to be sufficient to open Z1Z2; however, forces of 94 pN and 164.5 pN (see Materials and Methods for simulation parameters) opened the pair within 2 ns without unfolding either domain. This suggests that bending at the Z1Z2 hinge is possible with rather small forces (comparatively, at MD timescales). It is desirable, then, to determine the free energy profile that governs the mechanical domain-domain behavior.
Before continuing, we note that forces mentioned, i.e., 90–160 pN, are stronger than forces known to extend titin chains using the most sensitive single molecule force spectroscopy techniques such as optical tweezers (4,22,88). We will show below that for a chain of many domains, forces on the order of 50 pN, which are comparable to those employed with optical tweezers, can stretch chains by ∼600 Å; this is possible since every individual domain is stretched by only a few Å, while the 90–160 pN forces lead to larger stretching and bending, e.g., stretching of ∼40 Å in one domain pair and of 1.2 μm for a 300 domain chain. Naturally the effect of strong forces is relatively easy to discern in simulations. To describe the effect of weak forces is relatively difficult and requires the calculation of the potential of mean force (free energy profile) linked to domain-domain bending.
The calculation, by means of MD simulations, of a free energy profile for molecular motions, such as opening Z1Z2 around its centered hinge, requires significant sampling. Although sampling through multiple SMD simulations to reconstruct the potential of mean force (PMF) along a reaction coordinate has been previously achieved (89–93) for systems involving conduction of small molecules through membrane channels, this method has not been employed for either the present system or similar systems. While the potential of mean force linked to stretch an α-helix in vacuum has been successfully determined by means of SMD (90), an analogous application to solvated helices presently is beyond the reach of the SMD approach. However, an application of the ABF method to stretch a solvated helix and determine the corresponding PMF has been successful (65). We employ, therefore, the ABF method (61–65).
ABF simulations (simC1-simC7) were performed on Z1Z2 to calculate the energy required to move through the observed conformers of Z1Z2. Beginning with the closed form as a reference state with ΔE = 0 (see Fig. 3 A), the ABF simulations exerted a force along a predefined reaction coordinate to move Z1 and Z2 apart through rotation around the linker, extending the structure to the semi-extended form (see Fig. 3 C) and terminating in the open form (see Fig. 3 B). Each simulation (simC1-simC7) sampled conformations within a narrow range of hinge angles, corresponding to a 5 Å separation between the N-terminus of Z1 and the C-terminus of Z2, and was carried out long enough for the adaptive biasing force to sample the entire 5 Å range of movement. An exception is simC4, in which case the sampling window covered 10 Å. Together, simC1-simC7 cover the end-to-end distance between termini of Z1Z2 from 45 Å to 85 Å, a range including the minimum and the maximum extension of two linked domains. Fig. 6 A illustrates the simulations carried out, i.e., simC1-simC7, showing positions i–viii of the tip of Z2 during its opening motion, corresponding to an extension range of 40 Å. The free energy profile (PMF) for the arc motion encompassed by simC1-C7 is plotted in Fig. 6 B and reveals two distinct plateaus, denoted (I and II) in Fig. 6 B, which correspond to intermediate transition states observed in simA2, also shown in Fig. 5 C at t = 11.5 and 27 ns. The plateau (II) for the PMF at Fig. 6 B is particularly significant in that it provides strong corroborating evidence (in addition to simA2) that the semi-extended conformer for Z1Z2 (see Figs. 3, C, and 5, B and C) is energetically stable. The metastable nature of this conformation, however, is evident both from the shallow, but well-defined, local minimum seen in Fig. 6 B and its appearance in the unconstrained simulation (see Fig. 5 B). Furthermore, the calculated PMF is found to increase by 20 kcal/mol upon forcing Z1Z2 from the closed to the open conformation, indicating that domain-domain rearrangement requires a measurable energy input and can buffer external forces. Indeed, this ΔE of 20 kcal/mol corresponds to the binding energy of approximately four hydrogen bonds, i.e., fewer hydrogen bonds than needed to initiate unraveling of the Z1 and Z2 domains individually, as shown above.
FIGURE 6.
Adaptive biasing force (ABF) simulations of Z1Z2. The ABF simulations of titin Z1Z2 are depicted schematically (A); in the simulations, the crystallographic structure for the closed form of Z1Z2 (green) is hinged toward the open conformation and the potential of mean force (PMF) profile for this motion is calculated. The ABF simulations step through the states i–viii. SimC1 determines the PMF between states i and ii, corresponding to an extension of 45–50 Å. Likewise, simC2 determines the PMF between states ii and iii with extension 50–55 Å, simC3 between states iii and iv with extension 55–60 Å, simC4 between states iv and v with extension 60–65 Å, simC5 between states v and vi with extension 65–75 Å, simC6 between states vi and vii with extension 75–80 Å and, finally, simC7 between vii and viii with extension 80–85 Å. The PMF profile over the extension range 45–85 Å, i.e., for transitioning between the closed and open state, is plotted (B), with I and II denoting intermediate states along the trajectory describing the stretched Z1Z2. The trough at II corresponds to the semi-extended conformer for Z1Z2 observed both in experiment and simulation (simA2). The distribution 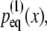 defined through Eq. 33, is shown (C, red stars) along with the Gaussian distribution
defined through Eq. 33, is shown (C, red stars) along with the Gaussian distribution  defined in Eq. 34 (blue line). The dashed line at 45 Å denotes the beginning of the simulation range.
defined in Eq. 34 (blue line). The dashed line at 45 Å denotes the beginning of the simulation range.
Which interactions are responsible for the response of Z1Z2 to stretching or compression? Equilibrium and steered molecular dynamics simulations presented above (simA1 and simB1-3) on the closed crystal conformer for Z1Z2 reveal several domain-domain contacts that could be responsible for stabilizing Z1Z2 in its closed conformation. Key stabilizing factors are surface contacts between Z1 and Z2 involving the pairs Ser14:Asp157, Gln13:Arg126, and Lys98:Ile130. A salt bridge involving Lys98 and Glu100 at the linker region was observed to form in both the closed conformer of simA1 and simB1-3, and also in the hinging motion observed for the open conformer in simA2 (shown in Fig. 7 B). While Z1 and Z2 in the semi-extended transition state observed in simA2 are too far apart for the above-mentioned VDW contacts, the charged interaction involving the Lys98:Glu100 salt bridge does appear to play a role in stabilizing this intermediate state. The closed crystal structure features a cadmium ion within the cleft between Z1 and Z2, forming a coordination site, shown in Fig. 7 A, between the two domains, with Glu26 and His28 of Z1 and Glu155 of Z2. This bound cadmium is likely the result of crystallization conditions, and given that the telethonin-bound structure for Z1Z2 (20) did not exhibit any bound metal ions, we excluded the cadmium ion from our simulation.
FIGURE 7.
Charge-charge interactions within the titin Z1Z2 Ig-tandem. (A) The metal binding site for cadmium between Z1 and Z2 in the closed crystallographic conformer. The coordination between cadmium and Glu26, His128 of Z1, and Glu155 of Z2, stabilizes the constrained compact arrangement of tandem Ig-domains. Numerous weak interactions such as a Lys98:Glu100 salt bridge at the linker, captured during simA2 and shown (B), stabilize further the equilibrium conformation of Z1Z2.
Replicating Z1Z2 into a multidomain chain
We now seek to replicate the tertiary structure elasticity of Z1Z2, i.e., how this domain pair stores mechanical energy through stretching and compression as shown in Fig. 4, into a multidomain chain that provides a rudimentary model for the I-band of titin. For this purpose we connect linearly N domain pairs, labeled j = 1, 2 … N, each domain j contributing a length xj to the overall chain length X (see Eq. 15), the elastic properties being described through the equilibrium distributions 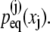 The model is described in Materials and Methods through Eqs. 15–32. Here we assume a model that replicates Z1Z2 N times, i.e., we assume that all distributions
The model is described in Materials and Methods through Eqs. 15–32. Here we assume a model that replicates Z1Z2 N times, i.e., we assume that all distributions 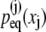 are equal to the one determined for Z1Z2. We also assume, in contrast to the chain shown in Fig. 4 A, that every angle is contributing to X. For such model we will compute the overall end-to-end distribution
are equal to the one determined for Z1Z2. We also assume, in contrast to the chain shown in Fig. 4 A, that every angle is contributing to X. For such model we will compute the overall end-to-end distribution 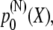 where N is the actual number of domains in the multidomain chain.
where N is the actual number of domains in the multidomain chain.
The multidomain model was constructed by connecting N domains, with each angle characterized by the Z1Z2 PMF 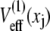 (Fig. 4 B) generated from simC1-simC7. This profile corresponds to an equilibrium distribution (see Eq. 16)
(Fig. 4 B) generated from simC1-simC7. This profile corresponds to an equilibrium distribution (see Eq. 16)
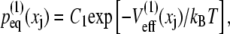 |
(33) |
as shown in Fig. 6 C. The calculated distribution was matched to a Gaussian
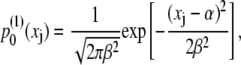 |
(34) |
with mean value α and width β.
The distributions 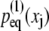 and
and 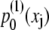 are matched in Fig. 6 C, yielding α = 45.7 Å, β = 1.26 Å, and an effective spring constant k = kBT/β2 = 0.63 kBT/Å2. Following the description provided in Materials and Methods, the distribution of the overall chain length X is given by Eq. 31 and the associated potential by Eq. 32.
are matched in Fig. 6 C, yielding α = 45.7 Å, β = 1.26 Å, and an effective spring constant k = kBT/β2 = 0.63 kBT/Å2. Following the description provided in Materials and Methods, the distribution of the overall chain length X is given by Eq. 31 and the associated potential by Eq. 32.
The effective potential, 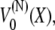 in Eq. 32 is harmonic, with spring constant
in Eq. 32 is harmonic, with spring constant
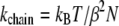 |
(35) |
and equilibrium length Nα/2, the factor 1/2 stemming from the inclusion of every angle in the multidomain chain. Comparing the potential for the multidomain model 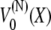 with that of the two-state system model given by Eq. 7 shows that the two models are actually mathematically equivalent and the behavior of a stretched chain is again described by the time-dependent distribution in Eq. 14.
with that of the two-state system model given by Eq. 7 shows that the two models are actually mathematically equivalent and the behavior of a stretched chain is again described by the time-dependent distribution in Eq. 14.
For the sake of illustration we assume a multidomain chain with N = 300 domains. This chain has an equilibrium length 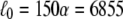 Å and overall distribution width
Å and overall distribution width 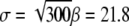 Å. If one applies a force f0 to the chain, the resulting extension is ΔX = f0/kchain, i.e.,
Å. If one applies a force f0 to the chain, the resulting extension is ΔX = f0/kchain, i.e.,
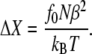 |
(36) |
Expressing f0 in units of pN, i.e., f0 = [f0] pN, and using 1 pN = 0.0241 kBT/Å for T = 300 K, one can write
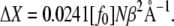 |
(37) |
In the case that a force of f0 = 50 pN, typical for experimentally stretching titin as tested in the literature (22,27), is applied to the multidomain chain, one predicts, from Eq. 37, an extension of ∼600 Å, which is ∼9% of the equilibrium length l0.
This extension corresponds to a spring constant kchain = f0/ΔX ≈ 0.1 pN/Å. This value, given the simplicity of our model, falls in the range of the observed soft elasticity (estimated from (22,27) and indicated in Fig. 2) of ∼0.001–1 pN/Å. The prediction of kchain should be improved further by including the twisting degrees of freedom that would add further mean-square deviations 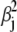 to the distribution in Eq. 28 and, thereby, soften the chain's overall spring constant.
to the distribution in Eq. 28 and, thereby, soften the chain's overall spring constant.
The chain's time evolution after stretching it to a length 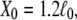 described through Eq. 14, is shown in Fig. 4 B for time in units τR. For a comparison with dynamics one needs to establish τR. This can be achieved through determination of the diffusion constant D1, for example, in the course of ABF simulations; this is feasible (61,63–65), but unfortunately, not straightforward and could not be done in the present case. The parameter τR appearing in Eq. 14 is related to D1 through the relationship τR = 2β2/D1.
described through Eq. 14, is shown in Fig. 4 B for time in units τR. For a comparison with dynamics one needs to establish τR. This can be achieved through determination of the diffusion constant D1, for example, in the course of ABF simulations; this is feasible (61,63–65), but unfortunately, not straightforward and could not be done in the present case. The parameter τR appearing in Eq. 14 is related to D1 through the relationship τR = 2β2/D1.
Secondary structure elasticity of Z1Z2
When forces exceeding 50 pN are applied to titin, the random coils of the PEVK segment are completely stretched out and the titin chain has become so straight that titin's soft and intermediate elasticity are both exhausted, rendering the chain completely stiff. At this point, titin's secondary structure elasticity comes into play, involving the sequential unfolding of its Ig- and Fn-III -like domains. The role of titin Ig-domain stability in resisting stretching forces by unfolding their secondary structure have been well characterized by force spectroscopy techniques such as atomic force microscopy (26,28–34,37,38), optical tweezers (27,39), as well as computational methods such as steered molecular dynamics (40,67,94,95). The majority of these studies have focused on the mechanical properties of I-band titin domains. It is well known that the individual Ig-domains in the I-band are far from homogenous, and that even small differences in Ig-domain folds influence their unfolding properties and, therefore, their response to mechanical stress.
Here we probe the mechanical resilience of the unique Z1 and Z2 Ig-folds using SMD simulations by applying stretching forces individually to the termini of the two domains in SMD simulations (simD1-4, simE1-4). The response is then compared to those of titin's Ig-domains I1 and I91 reported in the studies mentioned above. Based on fold topology, Z1 and Z2 belong to the same Ig subclass as I1, but not to that of I91 (96). However, previous MD stretching studies performed on Z1Z2 reveal that the force-bearing elements for Z1 and Z2 are the same terminal β-strands as in I1 and I91 (21). These previous studies on Z1Z2 did not investigate the mechanical properties of Z1 and Z2 individually. Left open were questions whether, for example, unfolding intermediates such as those seen in titin I91 exist in Z1 or Z2.
Our previous work (21) had shown that Z1Z2 unfolds within nanoseconds when stretched at a constant force of 750 pN. The simulations gave no evidence of intermediate states during stretched unfolding. To capture any intermediates, we performed simulations at lower stretching forces, namely, at 500 pN and 350 pN. In simD1 and simE1, a constant force of 500 pN applied to the C termini of Z1 and Z2, while the N-termini were held fixed, unfolded both Z1 and Z2 within 8 ns in a fashion identical to that observed in the previously reported stretching simulations (21).
In simD2-4 and simE2-4, the stretching force was lowered to 350 pN. In this case Z1 and Z2 did not unfold exactly in the same manner. In simD2-3 and simE2-3, Z1 unfolded as before, while Z2 exhibited an intermediate state during unfolding. Neither Z1 nor Z2 unfolded within the 30 ns of simD4 or simE4 (results not shown). Fig. 8, A–C, shows a snapshot of Z1 at t = 23.1 ns of stretching revealing simultaneous detachment of the domain's A and A′ strands; a 23.5 ns snapshot shows the subsequent extension of the N-terminal region. Fig. 8, D–F, illustrate the unfolding of Z2 in simulation simE2 with snapshots at t = 0 ns, at ∼19.9 ns (showing the detachment of Z2's A strand), and at ∼22.3 ns (showing the subsequent detachment of the A' strand). Once both the A and the A′ strands for titin Z1 and Z2 have separated from the main fold, the entire domain unravels quickly with little resistance as shown in Fig. 8 G.
FIGURE 8.
Stretching individual Ig-domains of Z1 and Z2. (A–C) Snapshots of the unfolding trajectory for titin Z1 under 350 pN constant force: (A) at t = 0 ns; (B) at t = 23.1 ns, i.e., the moment of simultaneous detachment of A and A′ strands (denoted by red and blue circles corresponding also to the strand color); and (C) at t = 23.5 ns, i.e., when the N-terminal region of Z1 extends. (D,E) Snapshots of the unfolding trajectory for titin Z2 under 350 pN constant force: (D) at t = 0 ns; (E) at t ∼ 19.9 ns when strand A detaches (red circle); and (F) at t ∼ 22.3 ns when strand A′ detaches (blue circle). An unfolding intermediate, in which the A′G strand holds briefly for ∼3 ns (see also Fig. 9 B) after the AB strand detaches, is seen during this unraveling of Z2, but not during the unraveling of Z1. After steps C and F, both Z1 and Z2 unfold quickly with little resistance, toward the elongated form shown in panel G.
The time evolution of the end-to-end distance for the 500 pN simulations (simD1 and simE1) is shown in Fig. 9 A. The slight initial increase in end-to-end distance, due to the extension of the flexible terminal linkers, is followed by a flat plateau in which the stretching force must overcome key force bearing interactions holding together β-strands A and B and A′ and G, both near the protein termini. The force-bearing interactions include a total of 10 interstrand AB (6) and A′G (4) hydrogen bonds in case of both Z1 and Z2. Once the strands are separated, though, the end-to-end distance rapidly increases as the domains unfold completely with little hindrance. Fig. 9 A depicts also the end-to-end distance for stretching with a 350 pN force (simD2-3 and simE2-3). Fig. 9 B shows an enlarged view of the unfolding plateau of Z2 that reveals the characteristic stable intermediate as the A and A′ strands detach one after the other, but with a delay (as opposed to simultaneous A and A′ strand rupture in Z1). An analysis into why Z1 unfolds differently from Z2 reveals mainly differences regarding the residues comprising the A′ β-strand. The A′ strand of Z1 contains a Val-Val-Val sequence, whereas the same strand in Z2 contains the sequence Met-Thr-Val. This suggests that the irregular peptide sequence at the Z2 A′ strand likely reinforces its interactions with the opposing G β-strand, leading to the characteristic delay in unfolding.
FIGURE 9.
Time evolution of the end-to-end distances of stretched Z1 and Z2. (A) Shown are the end-to-end distances of Z1 and Z2 for simulations simD1, simD2, simD3, simE1, simE2, and simE3. The traces demonstrate that unfolding occurs in a rupturelike event, i.e., rather suddenly, with a time delay after the onset of stretching that is longer for weaker forces. In particular, unfolding of Z1 and Z2 with a stretching force of 500 pN requires ∼7 ns simulation time, and with a stretching force of 350 pN requires ∼15 ns in simE2 and ∼30 ns in simD1-2 as well as simE1. Not shown here are the end-to-end distance traces from simD3 and simE3, in which Z1 and Z2 did not start to unfold within the 30-ns simulation period. (B) Shown is a detailed view of the unfolding event in simD1 (black) for Z1 and in simE1 (green) for Z2. The stepwise unraveling representative of an Ig unfolding intermediate (region circled) for Z2 (see also Fig. 8, D and E) reveals characteristic force plateaus corresponding to terminal β-strand rupture also seen for titin I91. Movies for the unfolding of Z1 and Z2 in simD2 and simE2, respectively, can be found in Supplementary Material.
The unfolding of Z1 and Z2 are remarkably similar to the unfolding of titin domains I1 and I91. Specifically, Z1 unfolds without intermediate through simultaneous rupture of its A and A′ strands as observed in I1 (67,95). Conversely, Z2 unfolds like I91 with the A strand rupturing first and the A′ strand second (43,44,97), i.e., exhibiting an unfolding intermediate.
DISCUSSION
The elasticity of titin is a key attribute of muscle function. This elasticity stretches over a wide range of titin extension, beginning at extension due to weak forces, a regime characterized through the entropic elasticity due to disordered PEVK segments and domain-domain rearrangement (22–24,26,29,57), and ending at extension due to high forces, a regime of secondary structure elasticity due to the unraveling of individual titin domains studied both by single molecule force spectroscopy techniques (22,26–39,86,97–99) and unfolding simulations (21,31,40,47–53,60,67,94,95,100,101). The key issue of this article is titin's mechanical response to weak forces due to tertiary structure elasticity (see Fig. 2). “Tertiary structure elasticity” is unfortunately a bit of a misnomer since it is not always linked to stretching tertiary structure elements of mechanical proteins apart, but in some cases (like titin) to stretching covalently-linked domains apart.
Pure tertiary structure elasticity is a key property of repeat proteins, i.e., proteins made of many homologous subunits of α-helices and β-sheets. A prime example is ankyrin (102,103), a protein linked to the cytoskeleton and exhibiting tertiary structure elasticity that alters the arrangement of its subunits made of a pair of α-helices. In case of ankyrin, a 100 Å long 24-repeat doubles its length under a force of 50 pN as predicted by simulation and measured by AFM (104,105). Titin's type of tertiary structure elasticity involving domain-domain motion is also found in the adhesion protein cadherin (106,107), where the elasticity seems to be strongly controlled by Ca2+ ions (104).
Tertiary structure elasticity in titin is due to the ability of adjacent domain pairs of titin to change their conformation through bending and twisting around the domain-domain link. Our study considered, for the sake of simplicity, solely elasticity due to bending; twisting could likewise contribute elasticity. Titin, due to its multidomain chain architecture, assumes a cordlike conformation that can extend and contract due to bending and twisting of adjacent domains. The titin cord can be stiff in some segments and flexible in others, deriving from the motion an overall elasticity measured by the ratio of extension/force. Our study made two contributions to advance the understanding of tertiary structure elasticity: it characterized the tertiary structure elasticity of a domain-domain building block, namely of Z1Z2, and it provided a theoretical framework to link building blocks into an overall elastic cord.
The most natural approach to study Z1Z2 tertiary structure elasticity is to apply stretching forces to the termini and to measure the resulting extension. As mentioned above, this could be done by AFM and by molecular dynamics simulations. Unfortunately, simulations can presently still cover only ∼100 ns timescales and to view sizable extensions in such a brief time requires forces stronger than expected in vivo. It is difficult to circumvent this problem and no accepted solution exists. One of the most promising solutions is the ABF method (61–65), applied in our study, that yields the potential energy surface and associated Boltzmann distribution linked to the bending/stretching of Z1Z2. Approximating the Boltzmann distribution by a Gaussian suggests that Z1Z2 can be stretched out by ∼2 Å due to bending by a 50 pN force.
If one assumes that titin, segmentwise, is a cord of 300 building blocks with elastic properties similar to those of Z1Z2, then one can easily imagine what happens when one applies a force of 50 pN to the cord's end. The locally compacted cord geometry will straighten a bit, starting in the first domain-domain angle that stretches likewise by ∼2 Å, then in the second domain-domain angle that stretches ∼2 Å, etc., until the 300th domain-domain angle is reached and the entire cord is stretched out by ∼600 Å. While this is an oversimplified picture, the more systematic theoretical model provided in this study leads essentially to the same result, namely that 50 pN extends the titin model by ∼600 Å, which is ∼9% of its length; doubling the force would double the extension in the description given. An estimate of the spring constant connected with titin's calculated tertiary structure elasticity yielded a value of ∼0.1 pN/Å, which falls within the range of values observed for titin's soft elasticity (22,27). From the agreement, which should improve further if twisting degrees of freedom are accounted for, we conclude that the chain energetics associated with titin's tertiary structure elasticity indeed contributes to the soft elasticity of titin.
The scenario presented above explains the range of titin elasticity shown in Fig. 2 in which forces below 50 pN act. If stretching forces exceed this value, secondary structure elasticity sets in. This secondary structure elasticity has been studied extensively for titin's domains I1 and I91 (22,26–29,31,32,34–40,47–53,60,67,86,94,95,97–101). Since this concept is quite familiar due to the celebrated prior work mentioned above, we probed this type of elasticity for Z1 and Z2 as well, to contrast the behavior with tertiary structure elasticity. In doing so we took advantage of technological development that permitted us to stretch Z1 and Z2 by forces weaker than ever attempted before computationally for titin Ig-domains, namely by 350 pN. The difficulty in applying weak forces stems from the fact that the weaker the force, the longer it takes for titin domains to rupture and unravel, thus increasing the computational cost of the simulation. In the present case we had to wait for 40 ns for unraveling to materialize. The investment in computing time has been well worth the effort, since we discerned that at the new low force Z2 exhibits a stretching intermediate not resolved computationally before. We note, however, that other than this intermediate, the stretching scenario seen is close to the ones found in earlier simulations of titin domains I1 and I91.
Our study coincides with an important experimental development of titin structure analysis, the first elucidations of entire titin segments. After the structure of Z1Z2 was reported in isolation and in complex with telethonin, the structure of the three domain segment A168-A170 of titin has been reported very recently (59) and the structure of a six-Ig tandem segment of titin will be reported soon (unpublished). These structures will permit further investigation into the tertiary structure elasticity of titin. For example, the structures will permit one to investigate the role of twist degrees of freedom as well as the role of heterogeneous domain-domain bending and twisting elasticity. One can expect that eventually the entire titin cord will be structurally resolved and described in terms of the underlying physical characteristics. One can safely expect that the resulting picture of titin elasticity will include tertiary structure elasticity suggested here as a key functional component.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Mu Gao, Chris Chipot, and Marcos Sotomayor for helpful discussions. We thank Claudia Muhle-Goll for kindly making available the NMR conformers used for comparison in this study.
This work was supported by the National Institutes of Health (NIH grant No. P41-RR05969 and grant No. R01-GM073655). Computer time was provided through the National Resource Allocation Committee grant (No. NRAC MCA93S028) from the National Science Foundation.
Editor: John E. Straub.
References
- 1.Wang, K. 1996. Titin/connectin and nebulin: giant protein ruler of muscle structure and function. Adv. Biophys. 33:123–134. [PubMed] [Google Scholar]
- 2.Erickson, H. 1997. Stretching single protein modules: titin is a weird spring. Science. 276:1090–1093. [DOI] [PubMed] [Google Scholar]
- 3.Maruyama, K. 1997. Connectin/titin, a giant elastic protein of muscle. FASEB J. 11:341–345. [DOI] [PubMed] [Google Scholar]
- 4.Linke, W. 2000. Stretching molecular springs: elasticity of titin filaments in vertebrate striated muscle. Histol. Histopath. 15:799–811. [DOI] [PubMed] [Google Scholar]
- 5.Tskhovrebova, L., and J. Trinick. 2003. Titin: properties and family relationships. Nat. Rev. Mol. Cell Biol. 4:679–689. [DOI] [PubMed] [Google Scholar]
- 6.Granzier, H. L., and S. Labeit. 2004. The giant protein titin a major player in myocardial mechanics, signaling and disease. Circ. Res. 94:284–295. [DOI] [PubMed] [Google Scholar]
- 7.Granzier, H. L., and S. Labeit. 2006. The giant muscle protein titin is an adjustable molecular spring. Exerc. Sport Sci. Rev. 34:50–53. [DOI] [PubMed] [Google Scholar]
- 8.Bang, M. L., T. Centner, F. Fornoff, A. J. Geach, M. Gotthardt, M. McNabb, C. C. Witt, D. Labeit, C. C. Gregorio, H. Granzier, and S. Labeit. 2001. The complete gene sequence of titin, expression of an unusual approximate to 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 89:1065–1072. [DOI] [PubMed] [Google Scholar]
- 9.Gautel, M., D. Goulding, B. Bullard, K. Weber, and D. O. Furst. 1996. The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J. Cell Sci. 109:2747–2754. [DOI] [PubMed] [Google Scholar]
- 10.Mues, A., P. F. M. van der Ven, P. Young, D. O. Furst, and M. Gautel. 1998. Two immunoglobulin-like domains of the z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett. 428:111–114. [DOI] [PubMed] [Google Scholar]
- 11.Pyle, W. G., and R. J. Solaro. 2004. At the crossroads of myocardial signaling—the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94:296–305. [DOI] [PubMed] [Google Scholar]
- 12.Furst, D., M. Osborn, R. Nave, and K. Weber. 1988. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immuno-electron microscopy—a map of 10 non-repetitive epitopes starting at the Z-line extends to the M-line. J. Cell Biol. 106:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obermann, W., M. Gautel, K. Weber, and D. Furst. 1997. Molecular structure of the sarcomeric M-band: mapping of titin and myosin binding domains in myomesin and the identification of potential regulatory phosphorylation site in myomesin. EMBO J. 16:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whiting, A., J. Wardale, and J. Trinick. 1992. The protein-ruler model of titin control of thick-filament assembly. J. Mol. Biol. 205:262–268. [Google Scholar]
- 15.van der Ven, P. F., E. Ehler, J. C. Perriard, and D. O. Furst. 1999. Thick filament assembly occurs after formation of a cytoskeletal scaffold. J. Muscle Res. Cell Mot. 20:569–579. [DOI] [PubMed] [Google Scholar]
- 16.Gräter, F., J. Shen, H. Jiang, M. Gautel, and H. Grubmüller. 2005. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 88:790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange, S., F. Xiang, A. Yakovenko, A. Vihola, P. Hackman, E. Rostkova, J. Kristensen, B. Brandmeier, G. Franzen, B. Hedberg, L. Gunnarsson, S. Hughes, S. Marchand, T. Sejersen, I. Richard, L. Edström, B. Udd, and M. Gautel. 2005. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 308:1599–1603. [DOI] [PubMed] [Google Scholar]
- 18.Gregorio, C. C., K. Trombitas, T. Centner, B. Kolmerer, G. Stier, K. Kunke, K. Suzuki, F. Obermayr, B. Herrmann, H. Granzier, H. Sorimachi, and S. Labeit. 1998. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kDa ligand (T-cap) is required for sarcomeric integrity. J. Cell Biol. 143:1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayans, O., P. F. M. van der Ven, M. Wilm, A. Mues, P. Young, D. O. Furst, M. Wilmanns, and M. Gautel. 1998. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 395:863–869. [DOI] [PubMed] [Google Scholar]
- 20.Zou, P. J., N. Pinotsis, M. Marino, S. Lange, A. Popov, I. Mavridis, M. Gautel, O. M. Mayans, and M. Wilmanns. 2006. Palindromic assembly of the giant muscle protein in the sarcomeric Z-disk. Nature. 439:229–233. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E. H., M. Gao, N. Pinotsis, M. Wilmanns, and K. Schulten. 2006. Mechanical strength of the titin Z1Z2/telethonin complex. Structure. 14:497–509. [DOI] [PubMed] [Google Scholar]
- 22.Leake, M., D. Wilson, M. Gautel, and R. Simmons. 2004. The elasticity of single titin molecules using a two-bead optical tweezers assay. Biophys. J. 87:1112–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy, A., L. Grama, T. Huber, P. Bianco, K. Trombitas, H. L. Granzier, and M. S. Z. Kellermayer. 2005. Hierarchical extensibility in the PEVK domain of skeletal-muscle titin. Biophys. J. 89:329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkar, A., S. Caamano, and J. Fernandez. 2005. The elasticity of individual titin PEVK exons measured by single molecular atomic force microscopy. J. Biol. Chem. 280:6261–6264. [DOI] [PubMed] [Google Scholar]
- 25.Duan, Y., J. DeKeyser, S. Damodaran, and M. Greaser. 2006. Studies on titin PEVK peptides and their interaction. Arch. Biochem. Biophys. 454:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, H., W. Linke, A. F. Oberhauser, M. Carrion-Vazquez, J. G. Kerkvliet, H. Lu, P. E. Marszalek, and J. M. Fernandez. 2002. Reverse engineering of the giant muscle protein titin. Nature. 418:998–1002. [DOI] [PubMed] [Google Scholar]
- 27.Tskhovrebova, L., J. Trinick, J. Sleep, and R. Simmons. 1997. Elasticity and unfolding of single molecules of the giant protein titin. Nature. 387:308–312. [DOI] [PubMed] [Google Scholar]
- 28.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 29.Rief, M., M. Gautel, A. Schemmel, and H. Gaub. 1998. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by AFM. Biophys. J. 75:3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linke, W. A., M. R. Stockmeier, M. Ivermeyer, H. Hosser, and P. Mundel. 1998. Characterizing titin's I-band Ig domain region as an entropic spring. J. Cell Sci. 111:1567–1574. [DOI] [PubMed] [Google Scholar]
- 31.Marszalek, P. E., H. Lu, H. Li, M. Carrion-Vazquez, A. F. Oberhauser, K. Schulten, and J. M. Fernandez. 1999. Mechanical unfolding intermediates in titin modules. Nature. 402:100–103. [DOI] [PubMed] [Google Scholar]
- 32.Carrion-Vazquez, M., A. Oberhauser, S. Fowler, P. Marszalek, S. Broedel, J. Clarke, and J. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 96:3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minajeva, A., M. Kulke, J. M. Fernandez, and W. A. Linke. 2001. Unfolding of titin domains explains the viscoelastic behavior of skeletal myofibrils. Biophys. J. 80:1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fowler, S. B., R. B. Best, J. L. T. Herrera, T. J. Rutherford, A. Steward, E. Paci, M. Karplus, and J. Clarke. 2002. Mechanical unfolding of a titin Ig domain: structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J. Mol. Biol. 322:841–849. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, B., and J. S. Evans. 2001. Modeling AFM-induced PEVK extension and the reversible unfolding of Ig/FNIII domains in single and multiple titin molecules. Biophys. J. 80:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linke, W. A., M. Kulke, H. Li, S. Fujita-Becker, C. Neagoe, D. J. Manstein, M. Gautel, and J. M. Fernandez. 2002. PEVK domain of titin: an entropic spring with actin-binding properties. J. Struct. Biol. 137:194–205. [DOI] [PubMed] [Google Scholar]
- 37.Li, H. B., and J. M. Fernandez. 2003. Mechanical design of the first proximal Ig domain of human cardiac titin revealed by single molecule force spectroscopy. J. Mol. Biol. 334:75–86. [DOI] [PubMed] [Google Scholar]
- 38.Williams, P. M., S. B. Fowler, R. B. Best, J. L. Toca-Herrera, K. A. Scott, A. Steward, and J. Clarke. 2003. Hidden complexity in the mechanical properties of titin. Nature. 422:446–449. [DOI] [PubMed] [Google Scholar]
- 39.Kellermayer, M., S. Smith, H. Granzier, and C. Bustamante. 1997. Folding-unfolding transition in single titin modules characterized with laser tweezers. Science. 276:1112–1116. [DOI] [PubMed] [Google Scholar]
- 40.Lu, H., B. Isralewitz, A. Krammer, V. Vogel, and K. Schulten. 1998. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 75:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu, H., and K. Schulten. 1999. Steered molecular dynamics simulations of force-induced protein domain unfolding. Proteins: Struct., Func. Gen. 35:453–463. [PubMed] [Google Scholar]
- 42.Paci, E., and M. Karplus. 2000. Unfolding proteins by external forces and temperature: the importance of topology and energetics. Proc. Natl. Acad. Sci. USA. 97:6521–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao, M., D. Craig, V. Vogel, and K. Schulten. 2002. Identifying unfolding intermediates of FN-III10 by steered molecular dynamics. J. Mol. Biol. 323:939–950. [DOI] [PubMed] [Google Scholar]
- 44.Gao, M., D. Craig, O. Lequin, I. D. Campbell, V. Vogel, and K. Schulten. 2003. Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proc. Natl. Acad. Sci. USA. 100:14784–14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craig, D., M. Gao, K. Schulten, and V. Vogel. 2004. Tuning the mechanical stability of fibronectin type III modules through sequence variation. Structure. 12:21–30. [DOI] [PubMed] [Google Scholar]
- 46.Gao, M., M. Sotomayor, E. Villa, E. Lee, and K. Schulten. 2006. Molecular mechanisms of cellular mechanics. Phys. Chem. Chem. Phys. 8:3692–3706. [DOI] [PubMed] [Google Scholar]
- 47.Best, R. B., S. B. Fowler, J. L. T. Herrera, A. Steward, E. Paci, and J. Clarke. 2003. Mechanical unfolding of a titin Ig domain: structure of transition state revealed by combining atomic force microscopy, protein engineering and molecular dynamics simulations. J. Mol. Biol. 330:867–877. [DOI] [PubMed] [Google Scholar]
- 48.Li, P., and D. Makarov. 2003. Theoretical studies of the mechanical unfolding of the muscle protein titin: bridging the timescale gap between simulation and experiment. J. Chem. Phys. 119:9260–9268. [Google Scholar]
- 49.Cieplak, M., T. Hoang, and M. Robbins. 2004. Thermal effects in stretching of Gō-like models of titin and secondary structures. Proteins. 56:285–297. [DOI] [PubMed] [Google Scholar]
- 50.Duff, N., N. Duong, and D. Lacks. 2006. Stretching the immunoglobulin 27 domain of the titin protein: the dynamic energy landscape. Biophys. J. 91:3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geierhaas, C., R. Best, E. Paci, M. Vendruscolo, and J. Clarke. 2006. Structural comparison of two alternative transition states for folding of titin I27. Biophys. J. 91:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, M., C. Hu, D. Klimov, and D. Thirumalai. 2006. Multiple stepwise refolding of immunoglobulin domain I27 upon force quench depends on initial conditions. Proc. Natl. Acad. Sci. USA. 103:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pabon, G., and L. Amzel. 2006. Mechanism of titin unfolding by force: insight from quasi-equilibrium molecular dynamics simulations. Biophys. J. 91:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma, K., L. S. Kan, and K. Wang. 2001. Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry. 40:3427–3438. [DOI] [PubMed] [Google Scholar]
- 55.Mayans, O., J. Wuerges, S. Canela, M. Gautel, and M. Wilmanns. 2001. Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin. Structure. 9:331–340. [DOI] [PubMed] [Google Scholar]
- 56.Improta, S., A. Politou, and A. Pastore. 1996. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure. 4:323–337. [DOI] [PubMed] [Google Scholar]
- 57.Trombitas, K., M. Greaser, S. Labeit, J. Jin, M. Kellermayer, M. Helmes, and H. Granzier. 1998. Titin extensibility in situ: entropic elasticity of permanently folded and permanently unfolded molecular segments. J. Cell Biol. 140:853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marino, M., P. Zou, D. I. Svergun, P. Garcia, C. Edlich, B. Simon, M. Wilmanns, C. Muhle-Goll, and O. Mayans. 2006. The Ig doublet Z1Z2: A model system for hybrid analysis of conformational dynamics in Ig tandems from titin. Structure. 14:1437–1447. [DOI] [PubMed] [Google Scholar]
- 59.Mrosek, M., D. Labeit, S. Witt, H. Heerklotz, E. von Castelmur, S. Labeit, and O. Mayans. 2007. Molecular determinants for the recruitment of the ubiquitin-ligase MuRF-1 onto M-line titin. FASEB J. Epub ahead of publication. [DOI] [PubMed]
- 60.Isralewitz, B., M. Gao, and K. Schulten. 2001. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 11:224–230. [DOI] [PubMed] [Google Scholar]
- 61.Darve, E., and A. Pohorille. 2001. Calculating free energies using average force. J. Chem. Phys. 115:9169–9183. [Google Scholar]
- 62.Darve, E., D. Wilson, and A. Pohorille. 2002. Calculating free energies using a scaled-force molecular dynamics algorithm. Mol. Sim. 28:113–144. [Google Scholar]
- 63.Rodriguez-Gomez, D., E. Darve, and A. Pohorille. 2004. Assessing the efficiency of free energy calculation methods. J. Chem. Phys. 120:3563–3578. [DOI] [PubMed] [Google Scholar]
- 64.Hénin, J., and C. Chipot. 2004. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J. Chem. Phys. 121:2904–2914. [DOI] [PubMed] [Google Scholar]
- 65.Henin, J., K. Schulten, and C. Chipot. 2006. Conformational equilibrium in alanine-rich peptides probed by reversible stretching simulations. J. Phys. Chem. B. 110:16718–16723. [DOI] [PubMed] [Google Scholar]
- 66.Reference deleted in proof.
- 67.Gao, M., M. Wilmanns, and K. Schulten. 2002. Steered molecular dynamics studies of titin I1 domain unfolding. Biophys. J. 83:3435–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Phillips, J. C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale, and K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.MacKerell, A. D., Jr., D. Bashford, M. Bellott, R. L. Dunbrack, Jr., J. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, I. W. E. Reiher, B. Roux, M. Schlenkrich, J. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 70.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 71.Grubmüller, H., H. Heller, A. Windemuth, and K. Schulten. 1991. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Sim. 6:121–142. [Google Scholar]
- 72.Schlick, T., R. Skeel, A. Brünger, L. Kalé, J. A. Board, Jr., J. Hermans, and K. Schulten. 1999. Algorithmic challenges in computational molecular biophysics. J. Comput. Phys. 151:9–48. [Google Scholar]
- 73.Izrailev, S., S. Stepaniants, B. Isralewitz, D. Kosztin, H. Lu, F. Molnar, W. Wriggers, and K. Schulten. 1998. Steered molecular dynamics. In Computational Molecular Dynamics: Challenges, Methods, Ideas, Vol. 4 of Lecture Notes in Computational Science and Engineering. P. Deuflhard, J. Hermans, B. Leimkuhler, A. E. Mark, S. Reich, and R. D. Skeel, editors. Springer-Verlag, Berlin.
- 74.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 75.Schulten, K., Z. Schulten, and A. Szabo. 1980. Reactions governed by a binomial redistribution process. The Ehrenfest urn problem. Phys. 100A:599–614. [Google Scholar]
- 76.Risken, H. 1989. The Fokker-Planck Equation: Methods of Solution and Applications, 2nd Ed. Springer, New York.
- 77.Schulten, K. 1982. Magnetic field effects in chemistry and biology. In Festkörperprobleme, Vol. 22. J. Treusch, editor. Vieweg, Braunschweig, Germany.
- 78.Kac, M. 1947. Random walk and the theory of Brownian motion. Am. Math. Mon. 54:369–391. [Google Scholar]
- 79.Chandrasekhar, S. 1943. Stochastic problems in physics and astronomy. Rev. Mod. Phys. 15:1–89. [Google Scholar]
- 80.Flory, P. J. 2003. Statistical Mechanics of Polymer Chains. Hanser Publishers, Cincinnati.
- 81.Bird, R. B., C. F. Curtiss, R. C. Armstrong, and O. Hassager. 1987. Dynamics of Polymer Liquids. Wiley InterScience, New York.
- 82.Kellermayer, M., and H. Granzier. 1996. Elastic properties of single titin molecules made visible through fluorescent F-actin binding. Biochem. Biophys. Res. Commun. 221:491–497. [DOI] [PubMed] [Google Scholar]
- 83.Politou, A. S., M. Gautel, S. Improta, L. Vangelista, and A. Pastore. 1996. The elastic I-band region of titin is assembled in a “modular” fashion by weakly interacting Ig-like domains. J. Mol. Biol. 255:604–616. [DOI] [PubMed] [Google Scholar]
- 84.Improta, S., J. K. Krueger, M. Gautel, R. A. Atkinson, J.-F. Lefevre, S. Moulton, J. Trewhella, and A. Pastore. 1998. The assembly of immunoglobulin-like modules in titin: implications for muscle elasticity. J. Mol. Biol. 284:761–777. [DOI] [PubMed] [Google Scholar]
- 85.Scott, K. A., A. Steward, S. B. Fowler, and J. Clarke. 2002. Titin; a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 315:819–829. [DOI] [PubMed] [Google Scholar]
- 86.Kawakami, M., K. Byrne, D. J. Brockwell, S. E. Radford, and D. A. Smith. 2006. Viscoelastic study of the mechanical unfolding of a protein by AFM. Biophys. J. 11:1710–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vazina, A. A., N. F. Lanina, D. G. Alexeev, W. Bras, and I. P. Dolbnya. 2006. The structural principles of multidomain organization of the giant polypeptide chain of the muscle titin protein: SAXS/WAXS studies during the stretching of oriented titin fibers. J. Struct. Biol. 155:251–262. [DOI] [PubMed] [Google Scholar]
- 88.Kellemayer, M., S. Smith, C. Bustamante, and H. Granzier. 2001. Mechanical fatigue in repetitively stretched single molecules of titin. Biophys. J. 80:852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gullingsrud, J., R. Braun, and K. Schulten. 1999. Reconstructing potentials of mean force through time series analysis of steered molecular dynamics simulations. J. Comp. Phys. 151:190–211. [Google Scholar]
- 90.Park, S., F. Khalili-Araghi, E. Tajkhorshid, and K. Schulten. 2003. Free energy calculation from steered molecular dynamics simulations using Jarzynski's equality. J. Chem. Phys. 119:3559–3566. [Google Scholar]
- 91.Park, S., and K. Schulten. 2004. Calculating potentials of mean force from steered molecular dynamics simulations. J. Chem. Phys. 120:5946–5961. [DOI] [PubMed] [Google Scholar]
- 92.Wang, Y., K. Schulten, and E. Tajkhorshid. 2005. What makes an aquaporin a glycerol channel: a comparative study of AqpZ and GlpF. Structure. 13:1107–1118. [DOI] [PubMed] [Google Scholar]
- 93.Yin, Y., M. Ø. Jensen, E. Tajkhorshid, and K. Schulten. 2006. Sugar binding and protein conformational changes in lactose permease. Biophys. J. In press. [DOI] [PMC free article] [PubMed]
- 94.Lu, H., and K. Schulten. 2000. The key event in force-induced unfolding of titin's immunoglobulin domains. Biophys. J. 79:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao, M., H. Lu, and K. Schulten. 2002. Unfolding of titin domains studied by molecular dynamics simulations. J. Muscle Res. Cell Mot. 23:513–521. [DOI] [PubMed] [Google Scholar]
- 96.Marino, M., D. I. Svergun, L. Kreplak, P. V. Konarev, B. Maco, D. Labeit, and O. Mayans. 2005. Poly-Ig tandems from I-band titin share extended domain arrangements irrespective of the distinct features of their modular constituents. J. Muscle Res. Cell Mot. 26:355–365. [DOI] [PubMed] [Google Scholar]
- 97.Oberhauser, A., C. Badilla-Fernandez, M. Carrion-Vazquez, and J. Fernandez. 2002. The mechanical hierarchies of fibronectin observed with single molecule AFM. J. Mol. Biol. 319:433–447. [DOI] [PubMed] [Google Scholar]
- 98.Higgins, M., J. Sadler, and S. Jarvis. 2006. Frequency modulation AFM reveals individual intermediates associated with each unfolding I27 titin domain. Biophys. J. 90:640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sarkar, A., S. Caamano, and J. Fernandez. 2007. The mechanical fingerprint of a parallel polyprotein dimer. Biophys. J. 92:36–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu, H., A. Krammer, B. Isralewitz, V. Vogel, and K. Schulten. 2000. Computer modeling of force-induced titin domain unfolding. In Elastic Filaments of the Cell, Chapt. 1. J. Pollack and H. Granzier, editors. Kluwer Academic/Plenum Publishers, New York. [DOI] [PubMed]
- 101.Gao, M., H. Lu, and K. Schulten. 2001. Simulated refolding of stretched titin immunoglobulin domains. Biophys. J. 81:2268–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Michaely, P., D. R. Tomchick, M. Machius, and R. G. W. Anderson. 2002. Crystal structure of a 12 ANK repeat stack from human ankyrin-R. EMBO J. 21:6387–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sotomayor, M., D. P. Corey, and K. Schulten. 2005. In search of the hair-cell gating spring: elastic properties of ankyrin and cadherin repeats. Structure. 13:669–682. [DOI] [PubMed] [Google Scholar]
- 105.Lee, G., K. Abdi, Y. Jiang, P. Michaely, V. Bennett, and P. E. Marszalek. 2006. Nanospring behavior of ankyrin repeats. Nature. 440:246–249. [DOI] [PubMed] [Google Scholar]
- 106.Boggon, T. J., J. Murray, S. Chappuis-Flament, E. Wong, B. M. Gumbiner, and L. Shapiro. 2002. C- cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 296:1308–1313. [DOI] [PubMed] [Google Scholar]
- 107.Haussinger, D., T. Ahrens, H. J. Sass, O. Pertz, J. Engel, and S. Grzeslek. 2002. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation, and mapping of contact regions. J. Mol. Biol. 324:823–839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.











