Abstract
Mitochondrial fusion and fission play important roles for mitochondrial morphology and function. We identified Mdm30 as a novel component required for maintenance of fusion-competent mitochondria in yeast. The Mdm30 sequence contains an F-box motif that is commonly found in subunits of Skp1-Cdc53-F-box protein ubiquitin ligases. A fraction of Mdm30 is associated with mitochondria. Cells lacking Mdm30 contain highly aggregated or fragmented mitochondria instead of the branched tubular network seen in wild-type cells. Δmdm30 cells lose mitochondrial DNA at elevated temperature and fail to fuse mitochondria in zygotes at all temperatures. These defects are rescued by deletion of DNM1, a gene encoding a component of the mitochondrial division machinery. The protein level of Fzo1, a key component of the mitochondrial fusion machinery, is regulated by Mdm30. Elevated Fzo1 levels in cells lacking Mdm30 or in cells overexpressing Fzo1 from a heterologous promoter induce mitochondrial aggregation in a similar manner. Our results suggest that Mdm30 controls mitochondrial shape by regulating the steady-state level of Fzo1 and point to a connection of the ubiquitin/26S proteasome system and mitochondria.
INTRODUCTION
Mitochondria are highly dynamic organelles. In many eukaryotic cell types they continuously move along cytoskeletal tracks and frequently fuse and divide. Their shape varies depending on the cell type, physiological status, and nutritional conditions (Bereiter-Hahn and Vöth, 1994; Yaffe, 1999; Griparic and van der Bliek, 2001; Westermann, 2002). Recent studies have shown that mitochondrial dynamics is crucial for a variety of cellular functions. For example, fusion of mitochondria is important for sperm development in Drosophila (Hales and Fuller, 1997) and for maintenance of mitochondrial DNA (mtDNA) in yeast (Berger and Yaffe, 2000). Furthermore, fusion is required for the formation of intracellular mitochondrial networks that allow the dissipation of energy in the cell (Skulachev, 2001) and for complementation of mitochondrial gene products, a mechanism thought to constitute a defense against cellular aging (Ono et al., 2001). Mitochondrial fission is an important step in apoptosis (Frank et al., 2001) and is required for embryonic development in nematodes (Labrousse et al., 1999). Several of the proteins determining mitochondrial behavior have been identified in recent years (Hermann and Shaw, 1998; Yaffe, 1999; Jensen et al., 2000; Boldogh et al., 2001; Shaw and Nunnari, 2002; Westermann and Prokisch, 2002). Yet, many of the molecular processes regulating mitochondrial dynamics remain to be described.
The molecular components of mitochondrial fusion and fission have been most extensively studied in budding yeast. Yeast mitochondria form an extended tubular network located below the cell cortex (Hoffmann and Avers, 1973). Mitochondria undergo gross structural changes during adaptation to nutritional conditions and growth phase (Stevens, 1981; Pon and Schatz, 1991; Egner et al., 2002). During vegetative growth, the continuity of the mitochondrial network is maintained by a balanced frequency of opposing fusion and fission events (Nunnari et al., 1997).
Fzo1, a large GTPase located in the mitochondrial outer membrane, is a key component of the mitochondrial fusion machinery. Deletion of the FZO1 gene results in a fragmented mitochondrial morphology and loss of mtDNA (Hermann et al., 1998; Rapaport et al., 1998), and conditional fzo1 mutants are blocked in mitochondrial fusion during yeast mating (Hermann et al., 1998). Both the large N-terminal part of the Fzo1 protein and its smaller C-terminal tail are exposed to the cytosol. A short loop in the intermembrane space connects Fzo1 to as yet unknown factors in the inner membrane to coordinate fusion of the mitochondrial double membranes (Fritz et al., 2001). Other components involved in mitochondrial fusion are the outer membrane protein Ugo1 and the dynamin-related intermembrane space protein Mgm1. Similar to fzo1 mutants, cells lacking functional Ugo1 or Mgm1 harbor fragmented and/or aggregated mitochondria, they lose mtDNA, and their mitochondria fail to fuse in zygotes (Wong et al., 2000; Sesaki and Jensen, 2001). However, the precise role of these components during mitochondrial membrane fusion is unknown.
Mitochondrial fission depends on the dynamin-related GTPase Dnm1, which assembles on mitochondria at sites of constriction and fission (Otsuga et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999). Dnm1 interacts with the WD40 repeat protein Mdv1 (Fekkes et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001) and the outer membrane component Fis1 (Mozdy et al., 2000). Mutants of dnm1, mdv1, or fis1 harbor long tubular mitochondria or closed planar nets of interconnected mitochondria. Fragmentation of mitochondria in mutants defective in fusion can be suppressed by mutation of components of the outer membrane division machinery (Bleazard et al., 1999; Sesaki and Jensen, 1999; Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001; Sesaki and Jensen, 2001). This indicates that fusion and fission antagonistically regulate mitochondrial shape. However, the molecular mechanisms regulating fusion and fission activity are unknown.
Recently, we conducted a systematic genome-wide screen that identified a number of novel genes important for mitochondrial distribution and morphology, MDM (Dimmer et al., 2002). One of the genes isolated in this screen, MDM30, encodes a novel F-box protein of unknown function. Herein, we report on the functional characterization of Mdm30 and discuss a role of this protein in regulation of mitochondrial fusion.
MATERIALS AND METHODS
Constructions of Plasmids and Yeast Strains
Standard methods were used for cloning procedures (Sambrook et al., 1989). Polymerase chain reaction (PCR) was performed using Pfu polymerase (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The plasmid for expression of Mdm30 from the GAL1 promoter in yeast, pYES-MDM30, was generated by PCR amplification of the MDM30 coding region from genomic yeast DNA by using the primers 5′ AAA CCA TGG GTA CCA TGA CAA AGA GGA GAA ACC TC and 5′ AAA GGA TCC CTA TAA ATT ATG TAA AGG CTG TTC and cloning into the BamHI/KpnI sites of vector pYES2 (Invitrogen, Groningen, The Netherlands). The plasmid for assaying FZO1 promoter activity, YIpFZO1β2, was generated by PCR amplification of the FZO1 promoter by using the primers 5′ GGG AAG CTT ACT ACC ATC CTT CTA GCC and 5′ GGG GGA TCC AAA CAT CGT TAA ATG AGC CTA CCG and cloning into the HindIII and BamHI sites of vector YIpMELβ2 (Melcher et al., 2000).
Standard methods were used for growth and manipulation of yeast strains (Sherman et al., 1986; Gietz et al., 1992). Yeast cultures were grown at 30°C, if not indicated otherwise. All strains used in this study were isogenic to BY4741, BY4742, and BY4743 (Brachmann et al., 1998). Haploid deletion strains Δmdm30, Δdnm1, and Δfzo1 were obtained from EUROSCARF (Frankfurt, Germany). Double mutants Δmdm30/Δdnm1 and Δmdm30/Δfzo1 were generated by mating of haploid strains, sporulation, and tetrad dissection. Genotypes of haploid progeny were determined by PCR.
To estimate Ugo1 levels, wild-type and Δmdm30 strains were transformed with plasmid pHS55 encoding the Ugo1 protein fused to a triple hemagglutinin (HA) epitope (Sesaki and Jensen, 2001). To measure FZO1 promoter activity, wild-type and Δmdm30 strains were transformed with the integrating plasmid YIpFZO1β2. To construct a strain expressing Mdm30 with a C-terminal triple HA tag, Mdm30-3xHA, a DNA fragment containing the epitope-coding sequences and a kanMX6 transformation marker cassette was amplified from plasmid pFA6a-3HA-kanMX6 (Bähler et al., 1998) by using primers 5′ AGC TTT CCA ATC TAG CGA ATG ATG AAC AGC CTT TAC ATA ATT TAC GGA TCC CCG GGT TAA TTA A and 5′ CGG GCT GAT AAA AAA AGG TGT AAT AGA ATG TGT CAG GAT GCT ACT GAA TTC GAG CTC GTT TAA AC and transformed into wild-type yeast. Correct insertion into the MDM30 locus was confirmed by PCR.
To construct strains expressing Fzo1 under control of the GAL1 promoter, a DNA fragment containing the HIS3 marker gene and the GAL1 promoter was amplified from plasmid pTL26 (Lafontaine and Tollervey, 1996) by using primers 5′ GCG GTT TAT TGC TGT CTT TGA ATT GTT GTT TTC CTT CAG ACA TCG AAT TCC TTG AAT TTT CAA A and 5′ AAA AGT TGG TGC GCA GTC CGG GTA AAT ACA GCT TTT CA T GCT GAC TCT TGG CCT CCT CTA GT and transformed into yeast strains BY4742 and Δmdm30. Correct insertion into the genome was confirmed by PCR. For overexpression of Mdm30, yeast strains were transformed with pYES-MDM30 and grown on galactose-containing medium lacking uracil to select for the overexpressing plasmid. Induction of the GAL1 promoter for overexpression of Fzo1 or Mdm30 was by growth on galactose-containing medium overnight.
For visualization of mitochondria, yeast strains were transformed with plasmid pVT100U-mtGFP or pYX113-mtGFP, both encoding mitochondria-targeted GFP (mtGFP) (Westermann and Neupert, 2000) or pRS416-GAL1+PrF0ATP9-RFP, encoding mitochondria-targeted DsRed (mtRFP) (Mozdy et al., 2000). For visualization of the endoplasmic reticulum (ER), yeast strains were transformed with pWP1055, encoding ER-targeted GFP (Prinz et al., 2000).
Microscopy
Yeast cultures were grown at 30°C in liquid YPD (yeast extract-peptone-dextrose) medium to mid-logarithmic growth phase, if not indicated otherwise. Staining of the actin cytoskeleton with rhodamine-phalloidin (Molecular Probes, Eugene, OR) was performed as described previously (Amberg, 1998). Staining of the vacuole with 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (Molecular Probes) was performed according to the manufacturer's instructions. Staining of mtDNA with 4,6-diamidino-2-phenylindole (DAPI) in methanol-fixed cells was performed as described previously (Jones and Fangman, 1992). Epifluorescence microscopy was according to standard procedures (Westermann and Neupert, 2000). Quantification of mitochondrial morphology defects was performed without prior reference to strain identity.
Assay of Mitochondrial Fusion in Vivo
Mitochondrial fusion in vivo was examined essentially as described previously (Nunnari et al., 1997; Mozdy et al., 2000) with some minor modifications. Cells of opposite mating types harboring plasmids encoding mtGFP or mtRFP under control of the GAL1 promoter were precultured in synthetic raffinose-containing medium under selection for the plasmids. Cells were grown to log phase at 30°C in synthetic medium containing raffinose and galactose to induce expression of the fluorescent proteins. Then, yeast cells were incubated for 2 h at 30°C in YPD medium (pH adjusted to 3.5) to shut off expression of the fluorescent proteins and to synchronize the cell cycle. Cultures were mixed, cells were transferred to nitrocellulose, placed for 3 h at 30°C on YPD plates (pH 4.5) to allow mating, resuspended in water, and analyzed by fluorescence microscopy.
Miscellaneous
Loss of mtDNA was monitored as described previously (Duchniewicz et al., 1999). Mitochondria were isolated by differential centrifugation as described previously (Diekert et al., 2001). Fractionation of yeast cells and purification of mitochondria on a sucrose gradient was performed as described previously (Rowley et al., 1994). To prepare total cell extracts, 2 OD600 units of cells were vortexed with glass beads for 2 min in 20 μl of sample buffer (2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 60 mM Tris-HCl, pH 6.8). Then, 80 μl of sample buffer were added, and 10 μl of the sample was analyzed by SDS-PAGE and immunoblotting. Monoclonal antibody 12CA5 recognizing the HA epitope was obtained from Roche Diagnostics (Mannheim, Germany). SDS-PAGE and blotting of proteins to nitrocellulose were performed according to standard methods. The enhanced chemiluminescence detection system (Amersham Biosciences AB, Uppsala, Sweden) was used for Western blotting. β-Galactosidase activity in total cell extracts was measured as described previously (Melcher et al., 2000).
RESULTS
MDM30 Encodes a Mitochondria-associated F-Box Protein
The MDM30 gene (systematic name YLR368W) encodes a protein of 598 amino acid residues (ca. 70 kDa). At its N terminus, the Mdm30 protein contains a domain known as the F-box (Figure 1A). The F-box is a motif of ∼45 amino acid residues that functions as an interaction site with the Skp1 protein. Skp1 assembles together with Cdc53 (cullin) and various members of the F-box protein family to build Skp1-Cdc53-F-box protein (SCF) complexes, which comprise a particularly versatile class of ubiquitin protein ligases (Bai et al., 1996; Patton et al., 1998; Deshaies, 1999; Kipreos and Pagano, 2000). A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae by a systematic yeast two hybrid approach revealed interactions of Mdm30 with both Skp1 and Cdc53 (Uetz et al., 2000), indicating that Mdm30 is a bona fide SCF complex component. Outside of the F-box domain, Mdm30 does not show significant homology to any other protein sequence in the databases.
Figure 1.
Mdm30 contains an F-box and is partially associated with mitochondria. (A) The F-box motif of Mdm30 is aligned with an F-box consensus sequence according to Patton et al. (1998). Residues that match the consensus sequence are highlighted by black boxes. (B) Wild-type yeast cells (WT; lanes 1 and 2) and yeast cells expressing Mdm30 with a triple HA epitope (Mdm30-3xHA; lanes 3 and 4) were fractionated into cytosol and mitochondria. Mitochondria were further purified on a sucrose step gradient. Then 50 μg of each fraction was analyzed by immunoblotting. Markers were Tom70 for mitochondria and Bmh2 for cytosol. M, mitochondria (lanes 1 and 3); C, cytosol (lanes 2 and 4).
To investigate the subcellular location of Mdm30 we constructed a strain expressing a variant carrying a C-terminal triple HA epitope, Mdm30-3xHA, under control of its endogenous promoter. Cells were fractionated into cytosol and mitochondria and analyzed by immunoblotting. Mdm30-3xHA was detected both in the cytosolic fraction and in purified mitochondria (Figure 1B). We conclude that a subfraction of Mdm30 is associated with mitochondria.
Morphology and Distribution of Organelles in Cells Lacking Mdm30
On fermentable carbon sources, such as glucose, wild-type mitochondrial networks are relatively simple with rather few tubules and branches. On nonfermentable carbon sources, such as glycerol, mitochondria are much more elaborate and ramified (Figure 2A) (Egner et al., 2002). The Δmdm30 deletion mutant grown on glucose-containing medium harbors highly aggregated mitochondria with some fragmented organelles (Figure 2A and Table 1) (Dimmer et al., 2002). On the nonfermentable carbon source glycerol, Δmdm30 cells displayed many mitochondrial fragments that were evenly distributed below the cell cortex (Figure 2A). Mitochondrial morphology of Δmdm30 cells was indiscernible at 30 and 37°C (our unpublished observations). These results demonstrate that Mdm30 is essential for the establishment of wild-type-like mitochondrial morphology. However, mitochondria lacking Mdm30 are apparently still able to adapt to changes of the carbon source.
Figure 2.
Δmdm30 cells exhibit grossly altered mitochondrial morphology. (A) Mitochondrial morphology. Wild-type and Δmdm30 cells expressing mtGFP were grown to log phase in glucose-containing medium (YPD) or glycerol-containing medium (YPG) and examined by fluorescence (left) and phase contrast (right) microscopy. (B) Morphology of other cellular structures in wild-type and Δmdm30 cells. For staining of vacuoles, cells were grown to log phase in YPD medium and stained with 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate. For visualization of the ER, cells expressing ER-targeted GFP were grown to log phase in glucose-containing synthetic minimal medium lacking methionine. For staining of F-actin, cells were grown to log phase in YPD medium, fixed, and stained with rhodamine-phalloidin. Samples were analyzed by fluorescence and phase contrast microscopy.
Table 1.
Quantification of mitochondrial morphology in Δmdm30/Δfzo1 cells
|
Mitochondrial morphology (% of cells)
|
|||
|---|---|---|---|
| Strain | Wild type-like | Fragmented | Aggregated |
| Wild type | 95 | 1 | 4 |
| Δmdm30 | 0 | 8 | 92 |
| Δfzo1 | 0 | 90 | 10 |
| Δmdm30/Δfzo1 | 0 | 92 | 8 |
n = 100 cells
Next, we asked whether the morphology of intracellular structures other than mitochondria is affected in Δmdm30 cells. Staining of wild-type and Δmdm30 cells with fluorescent probes specific for vacuoles, the endoplasmic reticulum and actin filaments revealed that the morphology of these structures was not altered by deletion of the MDM30 gene (Figure 2B). These data suggest that organellar morphology defects in Δmdm30 cells are restricted to mitochondria.
Mdm30 Is Required for Maintenance of Mitochondrial DNA at Elevated Temperature
To test whether the MDM30 gene is required for normal growth, serial dilutions of wild-type and Δmdm30 yeast cultures were spotted onto plates containing either glucose or glycerol as carbon source and incubated at 30 or 37°C. Although growth of the Δmdm30 strain was almost like wild-type under most conditions, we observed a strong reduction of growth on the nonfermentable carbon source glycerol at elevated temperature (Figure 3A), suggesting that Mdm30 is required for maintenance of respiratory competence at elevated temperature.
Figure 3.
Deletion of the MDM30 gene leads to respiratory deficiency at elevated temperature and loss of mtDNA. (A) Growth phenotype of the Δmdm30 mutant. Wild-type and Δmdm30 cells were grown overnight in glucose-containing medium at 30°C. Then, 10-fold serial dilutions were spotted onto plates containing glucose (YPD) or glycerol (YPG) as carbon source. YPD plates were incubated for 2 d, and YPG plates were incubated for 3 d at the indicated temperatures. (B) Loss of respiratory competence in Δmdm30 cells grown at elevated temperature. Wild-type and Δmdm30 cells were precultured on YPG plates at 30°C to select for cells containing mtDNA. These colonies were used to inoculate liquid cultures in YPD medium to allow for loss of mtDNA. YPD cultures growing at 30 or 37°C were maintained at log phase. At the indicated time points, aliquots of these cultures were plated on glucose-containing medium. The next day, colonies were replica-plated to glycerol-containing medium, and the percentage of respiratory-competent colonies was determined. (C) Δmdm30 cells lose mtDNA. Wild-type and Δmdm30 cells harvested from YPD cultures growing for 20 h at 37°C were stained with DAPI and viewed by fluorescence (left) and phase contrast (right) microscopy. Arrowheads point to mtDNA chondrolites. Big fluorescent spots represent nuclei.
Given that many mutants with aberrant mitochondrial morphology tend to lose mtDNA (Berger and Yaffe, 2000; Contamine and Picard, 2000), we examined whether a defect in mtDNA inheritance was the reason for loss of respiratory competence in Δmdm30 cells. To test this, wild-type and Δmdm30 cells were grown at 30 and 37°C in liquid culture on a fermentable carbon source (glucose) to allow for loss of mtDNA. At several time points, aliquots were collected from logarithmically growing cultures, and cells were plated at an appropriate dilution onto glucose-containing medium. After an incubation at permissive temperature, colonies were replica-plated onto medium containing a nonfermentable carbon source (glycerol), and the percentage of colonies able to grow was determined. The numbers directly reflect the percentage of cells harboring functional mitochondria in liquid culture. When the Δmdm30 strain was grown at normal temperature, the number of cells maintaining their respiratory functions was close to 100% throughout the time course of the experiment. In contrast, when the cultures were shifted to elevated temperature, the number of cells containing functional mitochondria dropped immediately. After cultivation for 24 h at 37°C, <10% of the cells were able to grow on a nonfermentable carbon source, whereas wild-type cells remained respiratory-competent under all growth conditions (Figure 3B).
To confirm that failure of Δmdm30 cells to grow on glycerol was due to loss of mtDNA, aliquots harvested at the end of the experiment were stained with the DNA-specific dye DAPI. Wild-type cells exhibited several chondrolites, small fluorescent bodies consisting of mtDNA. In contrast, most Δmdm30 cells that had been grown at 37°C did not contain any detectable mtDNA (Figure 3C). However, at 30°C Δmdm30 cells maintained functional mitochondrial genomes because they were able to grow on nonfermentable carbon sources. We conclude that Δmdm30 cells fail to maintain mtDNA at elevated temperature. It is unknown whether this is due to a defect in replication, packaging, or partitioning of the genomes.
Genetic Interactions of MDM30 with FZO1 and DNM1
Establishment and maintenance of a wild-type–like mitochondrial morphology depend on the antagonistic action of the mitochondrial fusion and fission machineries. Block of mitochondrial fusion by deletion of either one of the FZO1 or UGO1 genes leads to a respiratory-deficient growth phenotype and the formation of highly fragmented mitochondria (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen, 2001). Block of mitochondrial fission by deletion of either one of the DNM1, MDV1, or FIS1 genes leads to the formation of long tubular mitochondria or closed mitochondrial nets without affecting respiratory competence (Otsuga et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999; Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001). To investigate whether Mdm30 is involved in these processes, we constructed Δmdm30/Δfzo1 and Δmdm30/Δdnm1 double mutants by genetic crosses and examined their phenotypes.
The Δmdm30/Δfzo1 double mutant was respiratory-deficient, like the Δfzo1 parent. In addition, the Δmdm30/Δfzo1 double mutant displayed a synthetic growth defect on glucose-containing medium. Growth of the double mutant was significantly reduced on glucose-containing medium at 30 and 37°C, when compared with the parental mutant strains (Figure 4A). This behavior was observed for each of four independent double-deleted progeny, indicating that it is specific for the deleted alleles. However, mitochondrial morphology of Δmdm30/Δfzo1 double mutants was indiscernible from the Δfzo1 parent. Mitochondria were highly fragmented, and aggregated mitochondria, which are predominant in the Δmdm30 parent, could only occasionally be observed (Figure 4B; Table 1). This indicates that the Δfzo1 mutation is epistatic to the Δmdm30 mutation in maintaining mitochondrial morphology; i.e., in the absence of Fzo1, deletion of the MDM30 gene does not have an additional effect on mitochondrial morphology. The synthetic growth phenotype suggests that Mdm30 might play an additional role in another pathway important for normal cell growth.
Figure 4.
Δfzo1 and Δdnm1 mutations are epistatic to Δmdm30. (A) Growth phenotypes of Δmdm30/Δfzo1 and Δmdm30/ Δdnm1 mutants. Growth of strains was assessed as described in Figure 3A. (B) Mitochondrial phenotypes of Δmdm30/Δfzo1 and Δmdm30/Δdnm1 mutants. Mitochondrial morphology of mtGFP-expressing cells grown in glucose-containing medium was analyzed as described in Figure 2A.
On the other hand, the temperature-sensitive respiratory deficiency of the Δmdm30 mutant was rescued by deletion of the DNM1 gene (Figure 4A). Moreover, mitochondrial morphology of Δmdm30/Δdnm1 double mutants was indiscernible from the Δdnm1 parent. Mitochondria formed elongated interconnected structures, which are characteristic of mutants defective in mitochondrial fission (Figure 4B). Fragmented or aggregated mitochondria could never be observed in Δmdm30/Δdnm1 double mutants. This indicates that the Δdnm1 mutation is epistatic to the Δmdm30 mutation, i.e., deletion of the DNM1 gene prevents mitochondrial aggregation in mdm30 mutants.
Mdm30 Is Required for Maintenance of Fusion-competent Mitochondria
Mitochondrial fragmentation or aggregation and mtDNA loss phenotypes are typically observed in mutants defective in mitochondrial fusion (Hermann et al., 1998; Rapaport et al., 1998; Wong et al., 2000; Sesaki and Jensen, 2001). As in the Δmdm30 mutant, these defects can be suppressed by deletion of the DNM1 gene (Bleazard et al., 1999; Sesaki and Jensen, 1999; Wong et al., 2000; Sesaki and Jensen, 2001). To determine whether Mdm30, like Fzo1, Ugo1, and Mgm1, has a role in mitochondrial fusion, we examined mitochondrial fusion in Δmdm30 cells in vivo.
Fusion was assayed by mating of haploid cells of opposite mating type harboring different fluorescent markers in the mitochondrial matrix (Nunnari et al., 1997). On mating of wild-type cells, the fluorescent labels immediately intermixed in zygotes, indicating mitochondrial fusion and content mixing (Figure 5A). In contrast, mitochondria of Δmdm30 cells failed to fuse, even though mitochondria of both parents entered the newly formed bud and were closely associated with one another (Figure 5B). This result indicates that Mdm30 function is important for mitochondrial fusion.
Figure 5.
Mitochondrial fusion is blocked in Δmdm30 cells and can be restored by deletion of the DNM1 gene. (A) Wild-type (WT) cells of opposite mating types containing mitochondria preloaded with mtGFP or mtRFP were mated and analyzed by fluorescence microscopy. The distribution of mtGFP and mtRFP in a representative zygote containing a medial bud (asterisk) is shown. (B) Δmdm30 zygotes and (C) Δmdm30/Δdnm1 zygotes were analyzed as described above.
Deletion of the DNM1 gene does not restore fusion activity in fzo1 and ugo1 mutants, suggesting that Fzo1 and Ugo1 are crucial components of the mitochondrial membrane fusion machinery (Bleazard et al., 1999; Sesaki and Jensen, 1999, 2001). The same is true for MGM1 when null alleles are analyzed (Shaw and Nunnari, 2002) instead of conditional mgm1 alleles (Wong et al., 2000). To determine whether Mdm30 is essential for mitochondrial membrane fusion, we examined mitochondrial fusion during mating of Δmdm30/Δdnm1 double mutant cells. We observed efficient mitochondrial content mixing in Δmdm30/Δdnm1 zygotes (Figure 5C). This indicates that the block of fusion in Δmdm30 cells can be suppressed by deletion of the DNM1 gene, i.e., Mdm30 is not required for mitochondrial fusion in the absence of Dnm1. Together, these observations demonstrate that Mdm30 does not play a direct role in mitochondrial fusion. However, it is required to maintain fusion-competent mitochondria in a wild-type background.
Mdm30 Controls the Steady-State Level of Fzo1
The phenotypic and genetic analyses described above suggested that Mdm30 is required to maintain the balance of mitochondrial fusion and fission and that Mdm30 itself is not an integral part of the fusion and fission machineries. We considered the possibility that Mdm30 might be a regulatory factor that controls the steady-state level of one or several protein(s) known to be involved in mitochondrial fusion and fission.
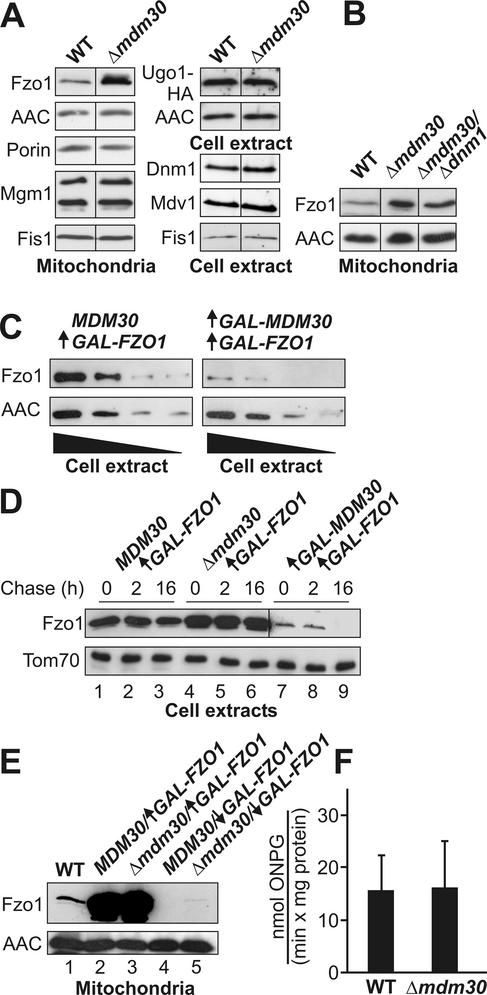
To test this idea, mitochondria isolated from the Δmdm30 strain and its isogenic wild-type were analyzed by Western blotting. As expected, the outer membrane protein porin and the inner membrane protein ADP/ATP carrier (AAC) were present in mitochondria of both strains at the same level. Also, the levels of Mgm1 and Fis1 were not altered in Δmdm30 mitochondria. In contrast, Fzo1 was present at a significantly higher level in Δmdm30 mitochondria (Figure 6A). Dnm1 and Mdv1 are cytosolic proteins that peripherally associate with mitochondria to mediate outer membrane fission (Otsuga et al., 1998; Tieu and Nunnari, 2000). To exclude effects due to an altered efficiency of binding to mitochondria, these proteins were analyzed by immunoblotting of total cell extracts. As with Fis1, Dnm1 and Mdv1 also were present at the same level in Δmdm30 and wild-type cells (Figure 6A). To test the steady-state levels of Ugo1, extracts of wild-type and Δmdm30 cells expressing an epitope-tagged version, Ugo1-HA (Sesaki and Jensen, 2001), were analyzed by immunoblotting. Deletion of the MDM30 gene had no effect on the level of Ugo1 (Figure 6A). Thus, the steady-state level of Fzo1 is influenced by Mdm30, whereas other components involved in fusion or fission are not affected.
Figure 6.
The protein level of Fzo1 depends on Mdm30. (A) Steady-state levels of mitochondrial proteins in wild-type (WT) and Δmdm30 cells. Equal amounts of mitochondria or total cell extracts isolated from wild-type and Δmdm30 cells were analyzed by immunoblotting by using specific antisera directed against the indicated proteins. For Ugo1, cells expressing an epitope-tagged protein, Ugo1-HA (Sesaki and Jensen, 2001), were analyzed. Note that Mgm1 is present in mitochondria in two forms of different size (Shepard and Yaffe, 1999). (B) Steady-state levels of Fzo1 in Δmdm30/Δdnm1 cells. Mitochondria of wild-type, Δmdm30, and Δmdm30/Δdnm1 cells were analyzed as described above. (C) Level of overexpressed Fzo1 in the absence or presence of Mdm30 overexpression. Cells carrying a chromosomal insertion of the GAL1 promoter upstream of the FZO1 coding region (left; MDM30/↑GAL-FZO1) and cells that in addition harbored a plasmid with an MDM30 allele under control of the GAL1 promoter (right; ↑GAL-MDM30/↑GAL-FZO1) were grown overnight in galactose-containing minimal medium under selection for the plasmid. Total cell extracts were prepared from log phase cultures, and twofold serial dilutions were analyzed by immunoblotting by using antisera against Fzo1 and, as a loading control, AAC. (D) Mdm30-dependent turnover of Fzo1. Cells carrying a chromosomal insertion of the GAL1 promoter in front of the FZO1 coding region in MDM30 wild-type (lanes 1–3), Δmdm30 (lanes 4–6), or GAL1-controlled MDM30 overexpression (lanes 7–9) backgrounds were grown overnight in galactose-containing medium. The next day, cultures were diluted with fresh medium and allowed to grow logarithmically for two generation times. Then, 7.5 mg/ml cycloheximide was added to stop cytosolic protein synthesis. After 0, 2, and 16 h, aliquots were harvested and cell extracts were analyzed as described above. Tom70 served as a loading control. Note that for lanes 7–9, a longer exposure of the Western blot was used to visualize the Fzo1 signal. (E) Fzo1 levels in mitochondria of GAL1-regulated strains. Wild-type cells (lane 1) and cells carrying a chromosomal insertion of the GAL1 promoter upstream of the FZO1 coding region in MDM30 (lanes 2 and 4) and Δmdm30 (lanes 2 and 5) backgrounds were grown in galactose-containing medium (lanes 2 and 3; MDM30/↑GAL-FZO1 and Δmdm30/↑GAL-FZO1) or in glucose-containing medium (lanes 4 and 5; MDM30/↓GAL-FZO1 and Δmdm30/↓GAL-FZO1). Mitochondria were isolated and equal amounts of mitochondrial protein were analyzed by immunoblotting by using antisera against Fzo1 and, as a loading control, AAC. (F) FZO1 promoter activity in wild-type and Δmdm30 cells. Cell extracts were prepared from logarithmically growing cultures of wild-type and Δmdm30 cells carrying a β-galactosidase reporter gene under control of the FZO1 promoter. β-Galactosidase activity was determined as nanomoles of o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolyzed/(min × mg protein). Error bars indicate standard deviations of 10 measurements obtained with five independently isolated clones. Strains lacking the reporter construct did not exhibit any significant activity.
Because mitochondrial fusion was restored in Δmdm30/Δdnm1 zygotes, we considered the possibility that deletion of the DNM1 gene might restore wild-type steady-state levels of Fzo1 in the absence of Mdm30. However, immunoblotting of isolated mitochondria revealed that Fzo1 levels are similarly elevated in Δmdm30 and Δmdm30/Δdnm1 cells (Figure 6B). This suggests that the Fzo1 protein level does not depend on the presence or absence of Dnm1.
Next, we asked whether Fzo1 protein overexpressed from a heterologous promoter is turned over in an Mdm30-dependent manner. A yeast strain was constructed that contained a chromosomal insertion of the strong and regulatable GAL1 promoter upstream of the FZO1 coding region. This strain received a plasmid harboring the MDM30 coding region under control of the GAL1 promoter. Expression of the GAL1-regulated genes was induced by growth in galactose-containing medium overnight. Then, cells were harvested from logarithmically growing cultures, and Fzo1 protein levels were analyzed by immunoblotting of total cell extracts. We found that the strain that simultaneously overexpressed Fzo1 and Mdm30 contained approximately five-fold less Fzo1 protein than a control strain that overexpressed Fzo1 in the presence of normal Mdm30 levels (Figure 6, C and D). This result points to a role of Mdm30 in the regulation of the turnover of Fzo1.
To test the Mdm30-dependent turnover of Fzo1 more directly we measured its degradation in vivo after expression from the GAL1 promoter. Expression of Fzo1 was induced in cells containing wild-type Mdm30 levels (MDM30), lacking Mdm30 (Δmdm30), or overexpressing Mdm30 (GAL-MDM30). Then, cycloheximide was added to stop protein synthesis. Aliquots of the cultures were harvested at different time points and analyzed by immunoblotting. Fzo1 levels remained relatively constant during the chase period in MDM30 (Figure 6D, lanes 1–3) and Δmdm30 cells (Figure 6D, lanes 4–6), indicating that Fzo1 turnover is relatively slow under these conditions. However, upon simultaneous overexpression of Mdm30, Fzo1 was completely degraded during the chase (Figure 6D, lanes 7–9). We suggest that Mdm30 controls degradation of Fzo1.
To estimate the relative amounts of Fzo1 protein in strains harboring GAL1-regulated FZO1 alleles, mitochondria were isolated from induced and repressed cultures and analyzed by immunoblotting. Induction of the GAL1 promoter led to a very high overexpression of Fzo1, when compared with wild-type mitochondria (Figure 6E, lanes 1 and 2). GAL1-controlled Fzo1 levels were similar in MDM30 wild-type and Δmdm30 mutant mitochondria (Figure 6E, lanes 2 and 3). Similarly, they were only slightly reduced in extracts of MDM30 cells compared with Δmdm30 cells (Figure 6D, compare lanes 1 and 4). Thus, wild-type Mdm30 levels might be so low that they soon become limiting for the regulation of Fzo1 turnover. On the other hand, upon repression of the GAL1 promoter by growth on glucose, Fzo1 expression was down-regulated to a level that the protein was not detectable any more by Western blotting (Figure 6E, lane 4). In contrast, trace amounts of Fzo1 protein could be detected on mitochondria isolated from glucose-grown cells harboring the Δmdm30 allele (Figure 6E, lane 5). This might indicate that a low amount of Fzo1 protein, which is expressed from the GAL1 promoter even under repressing conditions, is stabilized in Δmdm30 cells.
Our results suggest that Mdm30 regulates the Fzo1 level posttranscriptionally, because Fzo1 levels depend on Mdm30 also when Fzo1 is expressed from the heterologous GAL1 promoter (Figure 6, C–E). To confirm this, we expressed the reporter protein β-galactosidase (Melcher et al., 2000) under control of the FZO1 promoter in MDM30 wild-type and Δmdm30 mutant backgrounds. Deletion of the MDM30 gene had no effect on FZO1 promoter activity (Figure 6F). We conclude that the activity of the FZO1 promoter is controlled independently of Mdm30.
Aggregation of Mitochondria Depends on FZO1 and MDM30
Deletion of the MDM30 gene leads to elevated Fzo1 levels and mitochondrial aggregation. We tested whether GAL1-regulated overexpression of Fzo1 has a similar effect on mitochondrial morphology. Induction of the GAL1 promoter resulted in aggregation of mitochondria in MDM30 wild-type and Δmdm30 mutant backgrounds (Figure 7, A and B, and Table 2). Mitochondrial aggregation was also observed when Fzo1 and Mdm30 were simultaneously up-regulated by GAL1 induction, conditions that still lead to a high overexpression of Fzo1 (our unpublished observations). These data demonstrate that mitochondrial aggregation occurs independently of Mdm30 when Fzo1 is highly up-regulated from a heterologous promoter.
Figure 7.
Mitochondrial morphology depends on FZO1 and MDM30. Cells carrying a chromosomal insertion of the GAL1 promoter upstream of the FZO1 coding region in MDM30 (A and C) and Δmdm30 (B and D) backgrounds were grown in galactose-containing medium (YPGal; A and B) or in glucose-containing medium (YPD; C and D). Mitochondrial morphology of mtGFP-expressing cells was analyzed in log phase cultures as described in Figure 2A.
Table 2.
Quantification of mitochondrial morphology in MDM30 and Δmdm30 cells expressing the FZO1 gene under control of the GAL1 promoter
|
Mitochondrial morphology (% of cells)
|
||||
|---|---|---|---|---|
| Strain | Medium | Wild type-like/some tubules | Fragmented | Aggregated |
| MDM30/GAL-FZO1 | YPGal | 0 | 2 | 98 |
| Δmdm30/GAL-FZO1 | YPGal | 6 | 1 | 93 |
| MDM30/GAL-FZO1 | YPD | 0 | 96 | 4 |
| Δmdm30/GAL-FZO1 | YPD | 22 | 10 | 68 |
n = 100 cells
Repression of GAL1-regulated Fzo1 expression in MDM30 cells resulted in highly fragmented mitochondria (Figure 7C and Table 2) resembling Δfzo1 cells (compare Figure 4B). This is consistent with the absence of an Fzo1 signal in Western blots of mitochondria under these conditions (Figure 6E, lane 4). Interestingly, repression of GAL1-regulated Fzo1 expression in Δmdm30 cells partially restored a tubular mitochondrial morphology (Figure 7D and Table 2). Under these conditions, some Fzo1 protein was detectable on mitochondria (Figure 6E, lane 5), which might be sufficient to counteract mitochondrial fission and partially restore a tubular mitochondrial morphology. Together, our data suggest that Mdm30 plays an important role to regulate the steady state level of Fzo1 and thereby maintain normal mitochondrial morphology.
DISCUSSION
Our studies establish that Mdm30 is a novel factor important for maintenance of fusion-competent mitochondria. Similar to other mutants defective in mitochondrial fusion, Δmdm30 cells harbor aggregated mitochondria, show a defect in mtDNA maintenance, and are blocked in mitochondrial fusion in vivo. Several lines of genetic and biochemical evidence suggest that Mdm30 is a regulatory factor for the mitochondrial fusion machinery. First, mitochondrial fusion is restored in Δmdm30/Δdnm1 zygotes, suggesting that Mdm30 is not an essential part of the fusion machinery. Second, the protein level of Fzo1, a key component of the mitochondrial fusion machinery, depends on Mdm30 expression. And third, MDM30-dependent changes of Fzo1 levels are concomitant with changes of mitochondrial morphology.
The fact that both the growth defect and the mitochondrial aggregation phenotype of the Δmdm30 mutant can be completely rescued by deletion of the DNM1 gene implies that regulation of mitochondrial fusion is the main function of Mdm30. However, two lines of evidence point to additional functions of Mdm30. Deletion of the MDM30 gene in a Δfzo1 background leads to a synthetic growth defect, and maintenance of mtDNA in Δmdm30 cells is temperature sensitive and not strictly correlated to mitochondrial morphology defects. Thus, also the maintenance of mtDNA and possibly other yet unknown processes might be modulated by Mdm30.
What might be the molecular role of Mdm30 in mitochondrial morphogenesis? The presence of an F-box motif and the observed interactions of Mdm30 with Skp1 and Cdc53 in the yeast two-hybrid system (Uetz et al., 2000) suggest that Mdm30 functions as part of an SCF-like ubiquitin protein ligase. One possibility is that the turnover of Fzo1 is regulated by Skp1-Cdc53-Mdm30–dependent ubiquitination and subsequent degradation by the 26S proteasome. In the simplest model, Fzo1 itself would be the target for ubiquitination. Yet, an involvement of additional factors cannot be excluded. It was not possible to show directly that Fzo1 is a substrate for the 26S proteasome. For example, we could not demonstrate an accumulation of ubiquitinated Fzo1 in a conditional proteasomal mutant, mpr1-1 (our unpublished observations). However, this was possibly due to the pleiotropic nature of this mutant in which many other processes in addition to mitochondrial functions are affected (Rinaldi et al., 1998).
In the absence of Mdm30, surplus Fzo1 accumulates. Because mitochondrial aggregation can be induced both by deletion of the MDM30 gene and by overexpression of Fzo1 from a heterologous promoter, we consider it likely that mitochondrial aggregation is a direct consequence of Fzo1 accumulation. Interestingly, overexpression of human mitofusins, homologs of yeast Fzo1, induces perinuclear clustering of mitochondria in mammalian cells in a similar manner (Santel and Fuller, 2001; Rojo et al., 2002). Clustered mitochondria in mammalian cells display a disorganized inner membrane and close contacts of apposing mitochondria without merging, suggesting that fusion might be blocked at the stage of adherence or docking of the organelles (Rojo et al., 2002). Thus, the accumulation of Fzo1 or mitofusins induces mitochondrial aggregation in different cellular systems, apparently by accumulation of unproductive fusion intermediates.
Two possible roles of Mdm30 in mitochondrial fusion may be proposed, which are not mutually exclusive. First, the numbers of fusion and fission events must be tightly balanced to maintain a tubular mitochondrial morphology (Nunnari et al., 1997). It is conceivable that Mdm30 is required to regulate the steady-state level of Fzo1 to maintain this balance. Second, mitochondrial aggregation upon overexpression of Fzo1 implies an accumulation of unproductive fusion complexes. A certain amount of such intermediates might be generated as dead-end products also under normal conditions. In this case, Mdm30 might be required to clear these complexes and thereby counteract mitochondrial aggregation.
The involvement of an F-box protein in mitochondrial fusion adds to previously reported evidence pointing to a connection of the ubiquitin/26S proteasome system and mitochondria. It has been reported that overexpression of a mutant form of ubiquitin, which is unable to build polyubiquitin chains, induces pronounced mitochondrial aggregation (Fisk and Yaffe, 1999). This phenotype is strikingly similar to that of the Δmdm30 strain. Thus, it is possible that deletion of the MDM30 gene and overexpression of mutant ubiquitin variants affect the same pathway. However, there are additional ubiquitin-dependent processes that influence mitochondrial morphology. Mutants defective in an essential ubiquitin ligase, Rsp5, harbor aberrant mitochondria (Fisk and Yaffe, 1999) due to a block of transcription of the OLE1 gene (Hoppe et al., 2000), which encodes a fatty acid desaturase crucial for mitochondrial inheritance (Stewart and Yaffe, 1991). Furthermore, conditional mutants of the MPR1 gene, which encodes an essential regulatory subunit of the 26S proteasome, display pleiotropic defects including impaired growth on glycerol, aberrant mitochondria and overreplication of mtDNA (Rinaldi et al., 1998). Finally, mutation of the YNT1 gene, which encodes another regulatory proteasome subunit, suppresses mitochondrial morphology defects in mutants lacking Yme1, an ATP-dependent protease of the mitochondrial intermembrane space (Campbell et al., 1994).
There are many open questions related to proteolytic processes in mitochondria. These include the identification of authentic proteolytic substrates with regulatory functions in mitochondrial biogenesis, and the elucidation of the role of the 26S proteasome for mitochondrial function (Käser and Langer, 2000). The regulation of Fzo1 by an F-box protein described herein may constitute the first link between ubiquitination processes and an authentic mitochondrial target protein. It is a challenge for the future to further analyze the role of ubiquitination in mitochondrial inheritance, to identify additional components involved in ubiquitination and to reveal their targets. This will contribute to our understanding of the dynamic behavior of mitochondria, and finally might also shed light on processes such as ubiquitination of mammalian sperm mitochondria, a mechanism proposed to ensure strictly maternal inheritance of mitochondria and mtDNA (Sutovsky et al., 1999, 2000).
Acknowledgments
We thank Walter Neupert for continuous support, the members of our laboratory for many stimulating discussions, Hannes Herrmann for critical comments on the manuscript, and Jürg Bähler, Ben Bleazard, Robert E. Jensen, Amy Mozdy, Jodi Nunnari, Will Prinz, Tom Rapoport, Hiromi Sesaki, Janet Shaw, and Quinton Tieu for making antibodies and plasmids available. This work was supported by the Deutsche Forschungsgemeinschaft through grants WE 2174/2-3 and SFB 413/B3.
References
- Amberg, D.C. (1998). Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol. Biol. Cell 9, 3259-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., 3rd, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263-274. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn, J., and Vöth, M. (1994). Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27, 198-219. [DOI] [PubMed] [Google Scholar]
- Berger, K.H., and Yaffe, M.P. (2000). Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8, 508-513. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I.R., Yang, H.-C., and Pon, L.A. (2001). Mitochondrial inheritance in budding yeast. Traffic 2, 368-374. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Campbell, C.L., Tanaka, N., White, K.H., and Thorsness, P.E. (1994). Mitochondrial morphology and functional defects caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol. Biol. Cell 5, 899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DNM1-interacting protein, Net2p. Mol. Biol. Cell 12, 309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine, V., and Picard, M. (2000). Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64, 281-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435-467. [DOI] [PubMed] [Google Scholar]
- Diekert, K., de Kroon, A.I., Kispal, G., and Lill, R. (2001). Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37-51. [DOI] [PubMed] [Google Scholar]
- Dimmer, K.S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W., and Westermann, B. (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchniewicz, M., Germaniuk, A., Westermann, B., Neupert, W., Schwarz, E., and Marszalek, J. (1999). Dual role of the mitochondrial chaperone Mdj1p in inheritance of mitochondrial DNA in yeast. Mol. Cell. Biol. 19, 8201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner, A., Jakobs, S., and Hell, S.W. (2002). Fast 100 nm resolution 3D-microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. USA 99, 3370-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes, P., Shepard, K.A., and Yaffe, M.P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151, 333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, H.A., and Yaffe, M.P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S., Gaume, B., Bergmann-Leitner, E.S., Leitner, W.W., Robert, E.G., Catez, F., Smith, C.L., and Youle, R.J. (2001). The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1, 515-525. [DOI] [PubMed] [Google Scholar]
- Fritz, S., Rapaport, D., Klanner, E., Neupert, W., and Westermann, B. (2001). Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J. Cell Biol. 152, 683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., Jean, A.S., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic, L., and van der Bliek, A.M. (2001). The many shapes of mitochondrial membranes. Traffic 2, 235-244. [DOI] [PubMed] [Google Scholar]
- Hales, K.G., and Fuller, M.T. (1997). Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121-129. [DOI] [PubMed] [Google Scholar]
- Hermann, G.J., and Shaw, J.M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265-303. [DOI] [PubMed] [Google Scholar]
- Hermann, G.J., Thatcher, J.W., Mills, J.P., Hales, K.G., Fuller, M.T., Nunnari, J., and Shaw, J.M. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, H.-P., and Avers, C.J. (1973). Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science 181, 749-750. [DOI] [PubMed] [Google Scholar]
- Hoppe, T., Matuschewski, K., Rape, M., Schlenker, S., Ulrich, H.D., and Jentsch, S. (2000). Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102, 577-586. [DOI] [PubMed] [Google Scholar]
- Jensen, R.E., Aiken Hobbs, A.E., Cerveny, K.L., and Sesaki, H. (2000). Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc. Res. Tech. 51, 573-583. [DOI] [PubMed] [Google Scholar]
- Jones, B.A., and Fangman, W.L. (1992). Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 6, 380-389. [DOI] [PubMed] [Google Scholar]
- Käser, M., and Langer, T. (2000). Protein degradation in mitochondria. Semin. Cell Dev. Biol. 11, 181-190. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. 1, REVIEWS3002. [DOI] [PMC free article] [PubMed]
- Labrousse, A.M., Zappaterra, M.D., Rube, D.A., and van der Bliek, A.M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815-826. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D., and Tollervey, D. (1996). One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24, 3469-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, K., Sharma, B., Ding, W.V., and Nolden, M. (2000). Zero background yeast reporter plasmids. Gene 247, 53-61. [DOI] [PubMed] [Google Scholar]
- Mozdy, A., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fusion is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari, J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, T., Isobe, K., Nakada, K., and Hayashi, J.I. (2001). Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 28, 272-275. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B.R., Brisch, E., Thatcher, J.W., Hermann, G.J., Bleazard, W., and Shaw, J.M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: don't skip the F-box hypothesis. Trends Genet. 14, 236-243. [DOI] [PubMed] [Google Scholar]
- Pon, L., and Schatz, G. (1991). Biogenesis of yeast mitochondria. In: The Molecular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, ed. J.R. Broach, J.R. Pringle, and E.W. Jones, Cold Spring Harbor, NY: Cold Spring Harbor Press, 333-406.
- Prinz, W.A., Grzyb, L., Veenhuis, M., Kahana, J.A., Silver, P.A., and Rapoport, T.A. (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., Brunner, M., Neupert, W., and Westermann, B. (1998). Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150-20155. [DOI] [PubMed] [Google Scholar]
- Rinaldi, T., Ricci, C., Porro, D., Bolotin-Fukuhara, M., and Frontali, L. (1998). A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol. Biol. Cell 9, 2917-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, M., Legros, F., Chateau, D., and Lombes, A. (2002). Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115, 1663-1674. [DOI] [PubMed] [Google Scholar]
- Rowley, N., Prip-Buus, C., Westermann, B., Brown, C., Schwarz, E., Barrell, B., and Neupert, W. (1994). Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77, 249-259. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Santel, A., and Fuller, M.T. (2001). Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867-874. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (2001). UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J.M., and Nunnari, J. (2002). Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12, 178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard, K.A., and Yaffe, M.P. (1999). The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 144, 711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J. (1986). Methods in Yeast Genetics: A Laboratory Course, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Skulachev, V.P. (2001). Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 26, 23-29. [DOI] [PubMed] [Google Scholar]
- Stevens, B. (1981). Mitochondrial structure. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, ed. E.W. Strathern, E.W. Jones, and J.R. Broach, Cold Spring Harbor, NY: Cold Spring Harbor Press, 471-504.
- Stewart, L.C., and Yaffe, M.P. (1991). A role for unsaturated fatty acids in mitochondrial movement and inheritance. J. Cell Biol. 115, 1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky, P., Moreno, R.D., Ramalho-Santos, J., Dominko, T., Simerly, C., and Schatten, G. (1999). Ubiquitin tag for sperm mitochondria. Nature 402, 371-372. [DOI] [PubMed] [Google Scholar]
- Sutovsky, P., Moreno, R.D., Ramalho-Santos, J., Dominko, T., Simerly, C., and Schatten, G. (2000). Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 63, 582-590. [DOI] [PubMed] [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 601-603. [DOI] [PubMed] [Google Scholar]
- Westermann, B. (2002). Merging mitochondria matters. Cellular role and molecular machinery of mitochondrial fusion. EMBO Rep. 3, 527-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, B., and Neupert, W. (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421-1427. [DOI] [PubMed] [Google Scholar]
- Westermann, B., and Prokisch, H. (2002). Mitochondrial dynamics in filamentous fungi. Fungal Genet. Biol. 36, 91-97. [DOI] [PubMed] [Google Scholar]
- Wong, E.D., Wagner, J.A., Gorsich, S.W., McCaffery, J.M., Shaw, J.M., and Nunnari, J. (2000). The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P. (1999). The machinery of mitochondrial inheritance and behavior. Science 283, 1493-1497. [DOI] [PubMed] [Google Scholar]