Abstract
Saccharomyces cerevisiae cells lacking Dot1 exhibit a complete loss of H3K79 methylation and defects in heterochromatin-mediated silencing. To further understand the mechanism of Dot1-mediated methylation, the substrate requirement of Dot1 was determined. This analysis found that Dot1 requires histone H4 for in vitro methyltransferase activity and the histone H4 tail for Dot1-mediated methylation in yeast. Mutational analyses demonstrated that the basic patch residues (R17H18R19) of the histone H4 N-terminal tail are required for Dot1 methyltransferase activity in vitro as well as Dot1-mediated histone H3K79 methylation in vivo. In vitro binding assays show that Dot1 can interact with the H4 N-terminal tail via the basic patch residues. Furthermore, an acidic patch at the C terminus of Dot1 is required for histone H4 tail binding in vitro, histone H3K79 di- and trimethylation in vivo, and proper telomere silencing. Our data suggest a novel trans-histone regulatory pathway whereby charged residues of one histone are required for the modification of another histone. These findings not only provide key insights into the mechanism of Dot1 histone methylation but also illustrate how chromatin-modifying enzymes engage their nucleosomal substrates in vivo.
Keywords: Dot1, methylation, histone, telomere, silencing, chromatin
Histones have N- and C-terminal tails and histone fold domains that are subject to various post-translational modifications such as acetylation, phosphorylation, ubiquitination, and methylation (Vaquero et al. 2003; Mersfelder and Parthun 2006; Kouzarides 2007). These histone modifications can act as sites for recruiting other chromatin-associated factors (Maurer-Stroh et al. 2003; Kouzarides 2007; Ruthenburg et al. 2007). Together, histone modifications and the factors that bind to these modifications can influence the dynamic state of chromatin structure and regulate the chromatin environment and gene expression profiles of a cell.
In Saccharomyces cerevisiae, Set1, Set2, and Dot1 are histone lysine methyltransferases that methylate lysine residues in histones at H3K4, H3K36, and H3K79, respectively (Lee et al. 2005). Among the three sites of histone methylation, histone H3K79 is unique in that it is not located within the H3 N-terminal tail domain but in the histone core region. Specifically, this methylation occurs at the L1 loop of the histone fold domain that is exposed on the surface of the nucleosome (Luger et al. 1997; White et al. 2001). In addition, the yeast Dot1 and human Dot1L H3K79 methyltransferases are also unique in that they are the only histone lysine methyltransferases that do not have a SET domain. Instead, the catalytic core region of Dot1 and Dot1L shares sequence motifs that are similar to class I arginine methyltransferases (Dlakic 2001; Cheng et al. 2005).
Yeast Dot1 was initially identified as a high-copy disruptor of telomere silencing (Singer et al. 1998). However, loss of DOT1, mutations that disrupt the catalytic activity of Dot1, or mutations at histone H3K79 can all lead to loss of telomere silencing (Ng et al. 2002a; Park et al. 2002; van Leeuwen et al. 2002). In addition, H3K79 is abundantly methylated at euchromatin and is hypomethylated at heterochromatin (Ng et al. 2002a, 2003; van Leeuwen et al. 2002). Although the presence of H3K79 methylation in euchromatin seems counterintuitive to Dot1’s silencing phenotype, it has been suggested that the silencing defects caused by overexpression of Dot1, deletion of DOT1, or loss of H3K79 methylation are due to an indirect mechanism where protein factors necessary for direct heterochromatin silencing, such as Sir proteins (Sir2, Sir3, and Sir4), are titrated away from silent chromatin (van Leeuwen and Gottschling 2002). This is supported by the observation that deletion or overexpression of Dot1 leads to mislocalization of Sir proteins from heterochromatin (San Segundo and Roeder 2000; Ng et al. 2002a, 2003; van Leeuwen et al. 2002). In addition, loss of Sir proteins leads to increased H3K79 methylation at telomeres, and overexpression of Sir3 extends the heterochromatin region at telomeres and concomitantly reduces H3K79 methylation (van Leeuwen et al. 2002; Ng et al. 2003). Therefore, it is believed that a functional role for H3K79 methylation is to restrict Sir proteins at heterochromatin by preventing Sir proteins from nonspecifically binding euchromatin (van Leeuwen and Gottschling 2002).
Histone methylation in S. cerevisiae is also regulated by a trans-histone tail modification where ubiquitination of histone H2B by Rad6 is required for Set1-mediated H3K4 di- and trimethylation and Dot1-mediated histone H3K79 di- and trimethylation (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002b; Sun and Allis 2002; Shahbazian et al. 2005). However, Set1 and Dot1 can methylate core histones and recombinant nucleosomes in vitro that lack ubiquitylated H2B (Roguev et al. 2001; Santos-Rosa et al. 2002; Sawada et al. 2004). How H2B ubiquitination allows for Set1-mediated H3K4 and Dot1-mediated H3K79 di- and trimethylation in vivo is still unclear.
In this study, we have identified a trans-histone methylation event where the basic patch residues (R17H18R19) of histone H4 are required for Dot1-mediated H3K79 methylation but not H3K4 or H3K36 methylation. We also identify an acidic region on Dot1 that is required for binding to the histone H4 basic patch, histone H3K79 di- and trimethylation, and proper telomere silencing. This charge-based interaction between Dot1 and the H4 basic patch provides new mechanistic insights into Dot1-mediated H3K79 methylation and a novel trans-histone methylation event.
Results
Dot1 methyltransferase substrate specificity
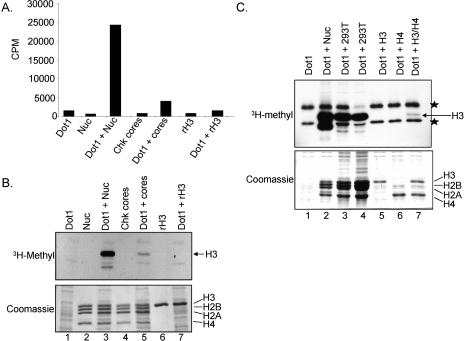
Previous reports have shown that Dot1 prefers nucleosomal substrates in in vitro histone methyltransferase (HMTase) assays (Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002; Sawada et al. 2004), whereas methylation of core histones by Dot1 has not been observed (Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002; Sawada et al. 2004). To investigate the substrate specificity of Dot1, in vitro HMTase assays were performed using purified recombinant Dot1 with nucleosomes, core histones, or recombinant histones substrates. All in vitro HMTase assays were analyzed by filter-binding assays and scintillation counting to measure total S-adenosyl-L-[methyl-3H]methionine (3H-SAM) incorporation as well as SDS-PAGE and fluorography to identify the specific histone labeled. As expected, Dot1 shows robust activity against nucleosomal substrates specific to histone H3 (Fig. 1A,B, lane 3). In addition to the robust activity of Dot1 on nucleosomal substrates, we did detect some incorporation of 3H-Methyl on histone H3 when Dot1 was incubated with chicken core histones (Fig. 1A,B, lane 5) or human 293T core histones substrates (Fig. 1C, lanes 3,4). Dot1 was unable to methylate recombinant histone H3 (Fig. 1A,B, lane 7) or HPLC-purified H3 isolated from 293T cells (Fig. 1C, lane 5). These data suggest that other histones within the nucleosome and core histone mix can contribute to Dot1 activity on histone H3. To further test this idea, Dot1 methyltransferase activity was assayed using H3 and H4 separately or in combination as substrates (Fig. 1C). Surprisingly, Dot1 is able to weakly methylate H3 in a mixture of H3 and H4 histones (Fig. 1C, lane 7). Since it appears that Dot1 needs histone H4 to methylate H3, we wanted to further investigate how H4 participated in Dot1-mediated H3K79 methylation and to assess if a new trans-histone methylation event was occurring.
Figure 1.
Dot1 methylates nucleosomes and core histones in vitro. (A) HMTase reactions were performed using recombinant-purified Dot1 incubated with 3H-SAM and the following substrates: chicken oligonucleosomes (Nuc), chicken core histones (Chk cores), or recombinant histone H3 (rH3). Half of the reaction samples were analyzed by filter binding and scintillation counting. (B) The remaining reaction amounts (A) were resolved by SDS-PAGE and analyzed by Coomassie staining (bottom panel) and fluorography (3H-Methyl, top panel). Positions of the individual histones are indicated. (C) Dot1 HMTase assays were performed with the following substrates: chicken oligonucleosomes (lane 2), core histones from 293T cells (lanes 3,4), HPLC-purified H3 or H4 from 293T cells (lanes 5,6), and a mixture of HPLC-purified H3 and H4 (lane 7). (Top panel) Samples were run on SDS-PAGE and fluorographed (3H-Methyl). Two stars indicate the position of a nonspecific bacterial protein that was detected when overexposing the fluorographed gel. (Bottom panel) A Coomassie-stained gel of the HMTase assays is shown.
A basic patch of amino acids in histone H4 is required for H3K79 methylation
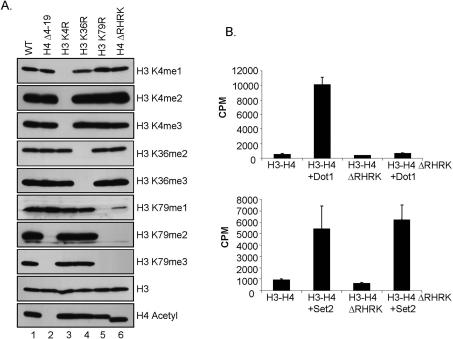
To further examine the apparent requirement of H4 for Dot1-mediated H3K79 methylation, we examined the histone methylation status of yeast strains expressing histone H3 and H4 N-terminal tail deletions. Yeast cells expressing wild-type or mutant histones were analyzed by Western blotting using antibodies that detect the various methylation states of H3 at K4, K36, and K79. Strikingly, global histone H3K79 di- and trimethylation is abolished in yeast strains expressing histone H4 lacking amino acids 4–19 (Fig. 2A, lane 2). Loss of H3K79 methylation is specific to deletion of the H4 N-terminal tail, as deletion of the N-terminal tail of H3 (amino acids 3–29) has no apparent effect on global H3K79 or H3K36 methylation (Supplementary Fig. S1, lane 3). Also, the H4 N-terminal tail deletion specifically results in loss of H3K79 di- and trimethylation but not H3K4 and H3K36 methylation (Fig. 2A, lane 2).
Figure 2.
A basic patch of amino acids in histone H4 is required for H3K79 methylation in vivo. (A) Western blots of whole-cell extracts using methyl-specific antibodies show the methylation status of H3K4, H3K36, and H3K79 in yeast cells expressing wild-type histones or cells expressing the indicated histone mutants. Antibodies directed against histone H3 and acetylated H4 serve as loading controls. Cells expressing mutant histones H3K4R, H3K36R, or H3K79R serve as negative controls for the indicated histone antibodies. (B) Dot1 is not active on yeast chromatin substrates isolated from cells lacking the basic patch of histone H4 in vitro. Recombinant-purified Dot1 (1.2 μg) was incubated with soluble chromatin substrates isolated from yeast cells expressing either wild-type histone H4, or from cells expressing histone H4 lacking the basic amino acids R17–K20 (H4ΔRHRK) in the presence of 3H-SAM in an in vitro HMTase assay. Incorporation of 3H-Methyl was measured by scintillation. Recombinant-purified Set2 (2.0 μg) was used as a control.
To further define the amino acids on histone H4 required for H3K79 methylation, we generated various H4 tail deletion mutants. This mutational analysis showed that amino acids 17–20, which correspond to the basic amino acids RHRK, is the region of H4 that is required for H3K79 methylation (Fig. 2A, lane 6). H3K79 monomethylation is greatly reduced in the H4ΔRHRK basic patch mutant, while H4 Δ4–19 N-terminal tail deletion has intact monomethylation (Fig. 2A, lanes 2,6). These amino acids in H4 have been defined as part of the histone H4 basic patch (Johnson et al. 1990, 1992; Luger et al. 1997). In the crystal structure of the Xenopus laevis nucleosome, these H4 basic patch residues interact with a patch of acidic residues contributed by histones H2A and H2B on an adjacent nucleosome (Luger et al. 1997). However, in the yeast nucleosome crystal structure, the H4 N-terminal tail forms a different conformation, poising the H4 basic patch to interact with the DNA of the neighboring nucleosome (White et al. 2001). Our data suggest the possibility of a new histone trans-tail pathway whereby the basic patch of the H4 N-terminal tail is required for H3K79 methylation.
Dot1 is not active on chromatin substrates lacking the H4 basic patch in vitro
Since there is a significant decrease in H3K79 monomethylation and no detectable H3K79 di- and trimethylation in yeast strains expressing the histone H4 basic patch mutant (H4ΔRHRK), we decided to examine if bacterially expressed Dot1 is active on yeast chromatin substrates lacking these residues. Soluble chromatin was isolated from the nuclei of yeast cells expressing the H4ΔRHRK mutant or wild-type chromatin. In vitro HMTase assays were performed using these chromatin substrates incubated with purified recombinant Dot1 and 3H-SAM. In support of our in vivo methylation data, when Dot1 is incubated with H4ΔRHRK chromatin as a substrate, little HMTase activity is observed as compared with a wild-type chromatin substrate (Fig. 2B). As a control, both of these chromatin substrates were assayed with the yeast HMTase Set2, a H3K36-specific methyltransferase, in order to determine if the H4ΔRHRK chromatin can serve as a viable substrate in an HMTase assay. As expected based on our in vivo H3K36 methylation results, purified recombinant Set2 can methylate wild-type or H4ΔRHRK chromatin equally well (Fig. 2B). Together, these data suggest that H4ΔRHRK chromatin can serve as a viable substrate for HMTase activity, and that the loss of H3K79 methylation in vivo and in vitro is specifically due to the absence of the basic patch of histone H4. Our in vitro results further suggest that Dot1 directly recognizes the H4 basic patch to mediate H3K79 methylation in vivo.
The basic amino acids of the basic patch of histone H4 are necessary for H3K79 methylation
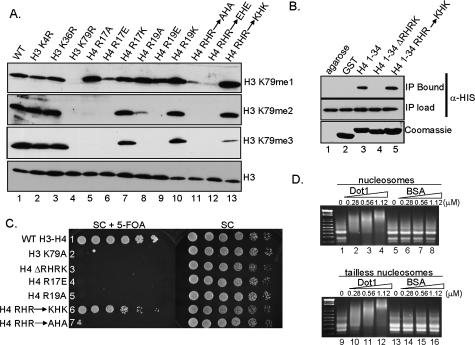
After determining that the basic patch of histone H4 is necessary for H3K79 methylation, we performed additional mutational analyses to further characterize the role of this region in Dot1-mediated H3K79 methylation. Yeast strains were engineered to express mutant histones in which the basic amino acids in the basic patch of H4 have been substituted by neutral, acidic, or basic amino acids. Histone H4 double mutants were generated with both R17 and R19 changed to either neutral or acidic amino acids. Cells expressing these mutants were analyzed by Western blotting, and the methyl state of H3K79 was examined. Interestingly, wild-type levels of H3K79 methylation are detected only when the substitutions made at R17 (H4R17K) and R19 (H4R19K) maintained the positive charge (Fig. 3A, lanes 7,10). Histone H4R17K,R19K double mutants also have wild-type levels of H3K79 mono- and dimethylation, but the levels of trimethylation are slightly reduced (Fig. 3A, lane 13). Yeast strains expressing neutral charge mutants, H4R17A and H4R19A, show near wild-type levels of H3K79 monomethylation but dramatically reduced or abolished amounts of H3K79 di- and trimethylation, respectively (Fig. 3A, lanes 5,8). Monomethylation is significantly reduced and di- and trimethylation of H3K79 are undetectable in the H4R17A,R19A and H4R17E,R19E double mutants (Fig. 3A, lanes 11,12). The reduction of H3K79 dimethylation in the single neutral amino acids substitution mutants is likely due to a partial reduction of the overall positive charge of this region of H4 (Fig. 3A, lanes 5,8). In contrast, H3K79 di- and trimethylation were completely abolished when H4R17, H4R19, or both residues were replaced by acidic amino acids (Fig. 3A, lanes 6,9,12).
Figure 3.
Basic amino acid residues within the basic patch of histone H4 are necessary for H3K79 methylation, Dot1 H4–tail interactions, and telomere silencing. (A) Western blots were performed using whole-cell extracts from cells expressing wild-type histones or the indicated histone mutations. The methylation status of H3K79 was examined using methyl-specific antibodies directed against H3K79me1, H3K79me2, and H3K79me3. Antibodies specific for histone H3 were used as a loading control. (B) In vitro binding assays were performed to test if Dot1 interacts with the H4 N-terminal tail. Bacterial cell extracts from cells expressing recombinant His6-Dot1 were incubated in the presence of a GST-H4 tail encoding residues 1–34 of histone H4 (GST-H41–34; lane 3), GST-H41–34 lacking the basic amino acids R17–K20 (GST-H41–34ΔRHRK; lane 4), and GST-H41–34 containing R17K and R19K mutations (GST-H41–34RHR–KHK; lane 5) bound to glutathione agarose beads. Bound His6-Dot1 was detected by α-HIS antibodies. (Top panel, lane 2) GST-bound beads or glutathione agarose beads alone were incubated with Dot1 extracts as negative controls. (Middle panel) Reaction inputs were probed with α-HIS antibodies to confirm equivalent amounts of Dot1 (IP load). GST-histone constructs were Coomassie-stained to indicate the amount of GST histone fusion protein loaded per lane. (C) Maintenance of charge of the H4 basic patch is required for telomere silencing. Strain UCC1369 (URA3-TEL-VIIL), expressing the indicated histone mutations, was grown to saturation, normalized to OD600, serially diluted (4×), and spotted on SC or SC + 5-FOA media. UCC1369 expressing wild-type histones and H3K79A served as controls for telomere silencing assays; cells were grown on SC media as a growth control. (D) Nucleosome-binding gel mobility assays were performed using chicken erythrocyte nucleosomes and recombinant-purified His6-Dot1. Wild-type nucleosomes (lanes 1–8) and tailless nucleosomes (lanes 9–16) were used as substrates. BSA was used as a control. The concentration of purified proteins in each reaction is indicated.
Surprisingly, H3K79 monomethylation seems to be relatively unaffected in yeast with neutral amino acid substitutions at H4R17 or H4R19 (Fig. 3A, lane 5,8), similar to what is observed with the H4 Δ4–19 N-terminal tail deletions (Fig. 2A, lane 2). However, a H4R17E mutation appears to have a more dramatic impact on H3K79 monomethylation than a H4R19E mutation (Fig. 3A, lanes 6,9). In addition, H4R17A mutants have a more substantial impact on H3K79 dimethylation than H4R19A (Fig. 3A, lanes 5,8). Double substitutions at R17 and R19 to either neutral or acidic amino acids greatly reduce H3K79 monomethylation to similar levels as H4R17E mutants and H4ΔRHRK mutants, suggesting the charge contributed by R17 is more important for H3K79 methylation. In addition, yeast chromatin isolated from a H4ΔRHRK mutant also has detectable, but reduced, H3K79 monomethylation, yet lacks H3K79 di- and trimethylation (Fig. 2A; Supplementary Fig. S2A). No defects in histone H3K79 methylation are observed with H4K16 or H4K20 mutations (Supplementary Fig. S2D; data not shown). However, histone H4H18 mutations also lose histone H3K79 methylation (data not shown). Together, these results suggest that maintenance of a small basic patch involving amino acid residues R17H18R19 in histone H4 is critical for Dot1-mediated H3K79 methylation.
Dot1 binds to the basic patch of histone H4 in vitro
The basic patch of histone H4 has been shown to be an important binding site for various proteins within yeast, including Iswi2 and the heterochromatin silencing proteins, Sir3 and Sir4 (Hecht et al. 1995; Fazzio et al. 2005). Therefore, we propose that Dot1 must directly interact with the basic patch of histone H4 to mediate histone H3K79 methylation.
To test this hypothesis, we constructed histone H4 N-terminal peptide GST fusion proteins, either coding for GST-H41–34 or GST-H41–34ΔRHRK. These constructs were expressed in Escherichia coli and purified with glutathione agarose. E. coli extracts expressing recombinant His6-Dot1 were incubated with either GST-H41–34 or GST-H41–34ΔRHRK fusion proteins bound to glutathione agarose. After removal of unbound lysates and washing, bound proteins were eluted with SDS sample buffer. Western blots using α-HIS antibodies were used to detect the presence of His6-Dot1. As predicted, we observe that Dot1 binds to GST-H41–34 in this assay (Fig. 3B, lane 3) but not to GST or agarose beads (Fig. 3B, lanes 1,2). Furthermore, Dot1 is no longer able to bind to GST-H41–34ΔRHRK, suggesting that Dot1–H4 tail interactions are mediated by this basic region of histone H4 (Fig. 3B, lane 4).
To determine if the H4–Dot1 interaction is charge-dependent, we constructed a GST-H41–34 fusion protein in which the amino acids H4R17 and H4R19 were substituted with lysine residues, thereby maintaining the positive charge. In our assay, Dot1 is still able to interact with a histone H4 mutant maintaining the charge status of the histone tail (Fig. 3B, lane 5), suggesting that Dot1 must interact with the basic amino acids of histone H4 to mediate histone H3K79 methylation.
The basic patch of histone H4 is important for telomere silencing
Previous reports have shown that Dot1 and H3K79 methylation are important for telomere silencing (Singer et al. 1998; Ng et al. 2002a; van Leeuwen et al. 2002). Interestingly, yeast cells expressing histone H4 basic patch mutations also have silencing defects in the mating-type loci (Johnson et al. 1990, 1992). Since Sir3 and Sir4 can bind to the basic patch of histone H4, it is believed that these mutations prevent Sir3 and Sir4 from binding and lead to loss of silencing (Johnson et al. 1990, 1992; Hecht et al. 1995).
To determine if the histone H4 basic patch is important for telomere silencing, we generated various histone mutants in a yeast strain containing an integrated URA3 gene at TEL-VII-L (UCC1369, kindly provided by D. Gottschling). Wild-type and histone mutants were plated on media with the toxic uracil analog, 5-fluoroorotic acid (5-FOA). In a wild-type cell, the URA3 gene, when placed at the telomere, is silenced, and cells will be able to grow on 5-FOA-containing media. In contrast, in cells that lose silencing, the URA3 gene will be expressed, and cells will not grow and will die in the presence of 5-FOA. Strains expressing the various histone mutants were grown to saturation, normalized for OD600, serially diluted, and spotted on either SC plates as a control or SC plates containing 5-FOA. We observe that cells lacking the H4 basic patch (H4ΔRHRK, Fig. 3C, row 3) exhibit a telomere silencing defect when grown in the presence of 5-FOA, similar to that of a H3K79A mutant positive control (Fig. 3C, row 2). Additionally, histone H4R17E and H4R19A mutants also exhibit defects in telomere silencing (Fig. 3C, rows 4,5).
To determine if maintaining proper charge at the basic patch is important in telomere silencing, we tested similar mutants as described previously. By maintaining the positive charge with conserved mutations (H4R17K,R19K), near wild-type levels of silencing are obtained (Fig. 3C, row 6). In contrast, cells expressing histone H4R17A,R19A mutants exhibit a telomere silencing defect (Fig. 3C, row 7). Although different H4 mutations were used, similar observations have been previously reported where maintaining charge at the histone H4 basic patch is important for mating-type silencing (Johnson et al. 1990, 1992). Our data suggest that the charge of the H4 basic patch must be maintained for proper histone H3K79 mono-, di-, and trimethylation as well as telomere silencing.
Dot1 binds nucleosomes independently of histone tails
In cells expressing H4 lacking the basic patch, we observe defects in H3K79 methylation. In addition, if the basic patch of H4 is deleted in the context of a H4 histone–peptide GST fusion, Dot1 no longer interacts with this fusion protein. To test whether Dot1–nucleosome interactions were affected by the loss of the H4 basic patch, nucleosomes lacking histone N-terminal tails were prepared by trypsin digestion (Supplementary Fig. S3B; Yang and Hayes 2004). This method has been reported to remove residues 1–23 of histone H4, residues 1–27 of histone H3, residues 1–42 of histone H2A, and residues 1–34 of histone H2B (Whitlock and Simpson 1977). Nucleosome-binding assays were performed using chicken erythrocyte nucleosomes or tailless nucleosomes incubated with purified recombinant Dot1 and analyzed by agarose gel electrophoresis followed by ethidium bromide staining (Min et al. 2003). We observed that Dot1 is able to bind and shift both nucleosomes and tailless nucleosomes (Fig. 3D). It did appear that there was a slight reduction in Dot1 binding to tailless nucleosomes as compared with wild-type (Fig. 3D, cf. lanes 2 and 10). From this data, we conclude that Dot1 activity, while dependent on the H4 tail, does not require the H4 tail for nucleosomal binding. As expected, in vitro methyltransferase assays show that there is little to no Dot1 activity on these tailless nucleosomes (Supplementary Fig. S3A). This is similar to what is observed in HMT assays using histone H4ΔRHRK chromatin substrates (Fig. 2B). These data also indicate that there must be other sites of interaction occurring between Dot1 and the nucleosome.
An acid patch on Dot1 is needed for interacting with the H4 basic patch and for histone H3K79 di- and trimethylation
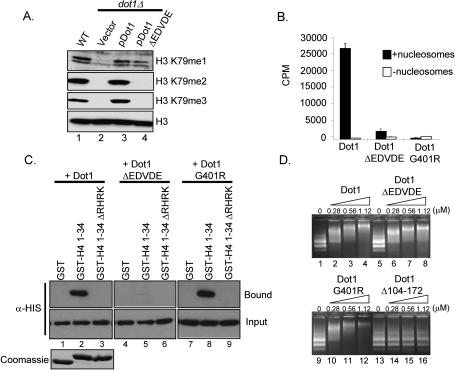
Since maintaining the positive charge of the histone H4 basic patch is necessary for histone H3K79 methylation and telomere silencing, we predict that Dot1 might have a corresponding region that interacts with the H4 basic patch. To identify this region, Flag epitope-tagged C-terminal Dot1 deletion constructs were generated and expressed in yeast in cells lacking Dot1 under the control of the ADH1 promoter (Supplementary Fig. S2B). Yeast whole-cell lysates were generated and analyzed for H3K79 methylation status by Western blot analysis. We observed that H3K79 methylation is rescued in yeast cells expressing wild-type Dot1 as well as a Dot1 C-terminal truncation mutant (pDot11–575) (Supplementary Fig. S2B, lanes 3,4). However, yeast cells expressing a Dot1 mutant that lacks an additional 20 amino acids at the C terminus (pDot11–555) have H3K79 monomethylation but are unable to rescue H3K79 di- and trimethylation (Supplementary Fig. S2B, lane 5). Based on structural analysis of yeast Dot1, this C-terminal region of Dot1 forms an exposed loop structure that would make it accessible for protein–protein interactions (Sawada et al. 2004). Upon further inspection of this region, we identified an acidic patch from amino acids 557–561 containing four acidic residues, EDVDE. To determine if this acidic patch on Dot1 is needed for H3K79 di- and trimethylation, we deleted the acidic patch (EDVDE) in both yeast and E. coli Dot1 expression vectors. Wild-type Flag epitope-tagged Dot1 or Dot1 lacking the acid patch (Dot1ΔEDVDE) were expressed in yeast cells under control of the ADH1 promoter, and the status of H3K79 methylation was examined by Western blot analysis. Similar to the Dot1 C-terminal truncation mutant (pDot11–555), we observe a loss of H3K79 trimethylation and a near complete loss of H3K79 dimethylation, as compared with a wild-type strain and a dot1 deletion strain expressing wild-type Dot1 (Fig. 4A). Interestingly, we do observe wild-type levels of H3K79 monomethylation in cells expressing the Dot1ΔEDVDE mutant.
Figure 4.
An acidic patch of amino acids in the C terminus of Dot1 is important for H3 methylation in vitro, K79 di- and trimethylation in vivo, and interaction with the H4 tail in vitro. (A) Western blots of whole-cell extracts using methyl-specific antibodies show the methyl status of H3K79 in wild-type or dot1Δ cells expressing full-length Flag epitope-tagged Dot1 or Dot1ΔEDVDE. Antibodies directed against histone H3 serve as a loading control. (B) In vitro methyltransferase assays using recombinant-purified His6-Dot1, His6-Dot1G401R, or His6-Dot1ΔEDVDE incubated with chicken nucleosomes and 3H-SAM. Incorporation of 3H-Methyl was measured by scintillation. (C) GST-H4 peptide fusion pull-downs were performed as described previously. Recombinant-purified His6-Dot1, His6-Dot1G401R, or His6-Dot1ΔEDVDE was incubated in the presence of a GST-H4 tail peptide fusion encoding residues 1–34 of histone H4 (GST-H41–34) or GST-H41–34 lacking the basic amino acids R17–K20 (GST-H41–34ΔRHRK). (Top panel) Bound His6-Dot1 and His6-Dot1 mutants were detected by α-HIS antibodies (Bound). (Middle panels) Reaction inputs were probed with α-HIS antibodies to confirm equivalent amounts of Dot1 protein (Input). (Bottom panel) GST-histone constructs were Coomassie-stained to indicate the amount of GST histone fusion protein loaded per lane. (D) Nucleosome-binding gel mobility assays were performed using chicken erythrocyte nucleosomes and recombinant-purified His6-Dot1, His6-Dot1G401R, His6-Dot1Δ104–172, or His6-Dot1ΔEDVDE at the indicated concentrations. Micromoles of Dot1 in each reaction are indicated.
To test if Dot1ΔEDVDE had in vitro methyltransferase activity, Dot1ΔEDVDE, wild-type Dot1, and Dot1G401R, a known catalytically inactive Dot1 mutant, were expressed in bacteria, purified, and assayed for HMTase activity using chicken erythrocyte nucleosomes as substrates. As indicated in Figure 4B, Dot1ΔEDVDE mutants show significant reduction in its activity against nucleosomal substrates as compared with wild-type Dot1. However, the Dot1ΔEDVDE mutant has some weak activity specific to H3, as compared with the catalytically inactive Dot1G401R mutant as indicated by filter and gel analysis (Fig. 4B; data not shown). Since Dot1ΔEDVDE is able to still monomethylate in vivo, we suspect this in vitro activity is likely specific for H3K79 monomethylation.
Finally, to determine whether the Dot1 acidic patch mediates its interaction with the N terminus of histone H4, we performed in vitro binding assays in which GST-H41–34 was incubated with recombinant wild-type Dot1, Dot1ΔEDVDE, or Dot1G401R. As compared with wild-type Dot1, binding of the Dot1 acidic patch mutant, Dot1ΔEDVDE, to GST-H41–34 is barely detectable (Fig. 4C, lanes 2,5). In addition, Dot1G401R, although catalytically inactive, interacts with GST-H41–34, suggesting that Dot1 activity is not a prerequisite for H4 binding (Fig. 4C, lane 8). Together, our data support a model whereby the acidic patch of Dot1 must interact with the H4 basic patch for H3K79 di- and trimethylation.
Previously, Dot1 has been shown to bind nucleosomes, and that the N terminus of Dot1 is required for binding and methyltransferase activity (Sawada et al. 2004). To determine if the Dot1ΔEDVDE mutant is able to bind nucleosomal substrates, nucleosomal binding assays were performed. As shown in Figure 4D, Dot1, Dot1ΔEDVDE, and Dot1G401R are all able to bind and shift nucleosomes as compared with lanes containing no Dot1 protein (Fig. 4D). As controls, a Dot1 mutant defective in nucleosome binding, Dot1Δ104–172, and BSA were used (Fig. 4D, lanes 13–16; data not shown). Our results indicate that the Dot1ΔEDVDE mutant is still capable of binding to nucleosomes, possibly to mediate monomethylation of H3K79. However, the nucleosome-binding ability of the Dot1ΔEDVDE mutant is not sufficient to mediate di- and trimethylation of H3K79.
The acidic patch of Dot1 is important for telomere silencing
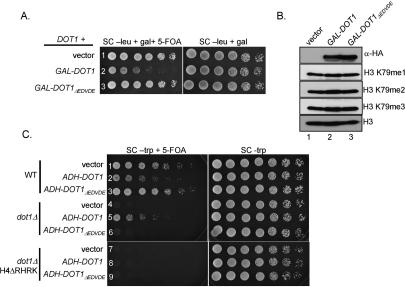
Previous studies have shown that yeast cells overexpressing Dot1 can disrupt telomere silencing (Singer et al. 1998; van Leeuwen et al. 2002). It has been shown that overexpression of Dot1 will cause H3K79 methylation to spread into regions of silent chromatin and displace proteins important for silencing (i.e., Sir3) away from telomere regions, resulting in silencing defects (Singer et al. 1998; van Leeuwen et al. 2002). This silencing defect is not observed if a catalytically inactive Dot1 is overexpressed (van Leeuwen et al. 2002). We wanted to determine if overexpressing Dot1ΔEDVDE, which lacks the ability to di- and trimethylate H3K79 and bind to the H4 N-terminal tail, would phenocopy the catalytically inactive Dot1 in telomere silencing assays. A yeast strain (UCC3503, kindly provided by D. Gottschling) expressing wild-type Dot1 and containing an integrated URA3 gene at TELVII-L was transformed with a plasmid encoding 3× hemagglutinin (HA3)-tagged Dot1 (GAL-DOT1) or Dot1ΔEDVDE (GAL-DOT1ΔEDVDE) under the control of a galactose-inducible promoter. Cells were grown in selective liquid media supplemented with glucose, normalized by OD600, serially diluted, and spotted on selective media containing galactose or galactose and 5-FOA to monitor telomere silencing. Consistent with previous reports, we observe a telomere silencing defect when we overexpress Dot1 in a yeast strain expressing endogenous Dot1 (Fig. 5A, row 2; Singer et al. 1998; van Leeuwen et al. 2002). However, Dot1ΔEDVDE overexpression did not result in a silencing defect in this strain, as growth on plates containing galactose and 5-FOA was similar to cells expressing vector only (Fig. 5A, cf. rows 3 and 1). Based on these data, Dot1ΔEDVDE behaves as a catalytically inactive mutant with respect to telomere silencing, suggesting that H3K79 di- and trimethylation are important for silencing, whereas H3K79 monomethylation is not. Western blots show that Dot1 and Dot1ΔEDVDE are equally expressed, and no global histone H3K79 methylation defects are observed in cells overexpressing Dot1 or Dot1ΔEDVDE (Fig. 5B). Since the acidic region of Dot1 is important for binding to the basic patch of H4, loss or weakening of this interaction, as we have shown in vitro, may prevent overexpressed Dot1ΔEDVDE from impacting silencing.
Figure 5.
Dot1ΔEDVDE overexpression does not result in a telomere silencing defect or detectable changes in H3K79 methylation in a strain expressing endogenous Dot1. (A) Yeast strain UCC3503 containing a plasmid encoding HA3-tagged galactose-inducible Dot1 or Dot1ΔEDVDE was tested for telomere silencing after galactose induction by plating on galactose containing media with or without 5-FOA. (B) Western blots were performed on whole-cell extracts after galactose induction to confirm that the HA-tagged Dot1 proteins were expressed upon induction. The methylation status of H3K79 upon Dot1 overexpression was also examined using H3K79 methyl-specific antibodies. (C) Cells expressing Dot1ΔEDVDE exhibit a telomere silencing defect similar to a dot1Δ strain. Yeast strain UCC1369 expressing the indicated histone mutations and Dot1 constructs was examined for defects in telomere silencing. Cells were grown to saturation, normalized to OD600, serially diluted, and spotted on selective media with or without 5-FOA.
Dot1ΔEDVDE was also tested for telomere silencing defects in strains lacking Dot1. Expression of HA3-tagged Dot1 (ADH-DOT1) from a constitutive ADH1 promoter imparts a silencing defect similar to what is observed using galactose-driven Dot1 expression in a wild-type strain, but partially complements a dot1Δ mutant (Fig. 5C, rows 2,5). However, expression of HA3-Dot1ΔEDVDE (ADH-DOT1ΔEDVDE) does not complement a dot1Δ mutant (Fig. 5C, row 6). These results are in agreement with the GAL-DOT1 and GAL-DOT1ΔEDVDE overexpression data (Fig. 5A). Furthermore, expression of Dot1 or Dot1ΔEDVDE in a strain lacking endogenous Dot1 does not rescue the telomere silencing defect observed in a histone H4ΔRHRK mutant strain (Fig. 5C, rows 8,9). Similar results are observed in cells expressing Dot1 or Dot1ΔEDVDE using the endogenous Dot1 promoter (data not shown). These data support a model whereby the acidic patch of Dot1 interacts with the basic patch of histone H4. Therefore, the loss of this Dot1–H4 interaction prevents proper H3K79 methylation, resulting in the observed silencing defect.
Sir3 inhibits Dot1-mediated H3K79 methylation in vitro
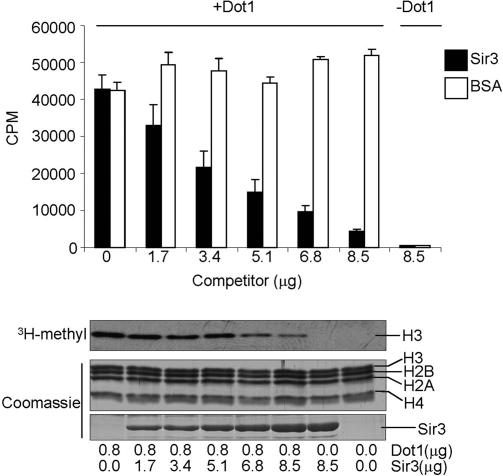
Sir3 has been show to bind to the basic patch of histone H4, and in vivo results suggest a competition between Sir3 and Dot1 (Johnson et al. 1990; Hecht et al. 1995; van Leeuwen et al. 2002; Ng et al. 2003). Therefore, based on our observations that Dot1 can also bind to the histone H4 basic patch, it is possible that Sir3 and Dot1 compete for the same binding site. We tested this idea in an in vitro HMTase assay in which we examined Dot1 HMTase activity in the presence of increasing amounts of recombinant-purified Sir3 C terminus (amino acids 502–978) (Fig. 6). A similar C-terminal fragment of Sir3 has been shown to be sufficient for binding to the H4 N-terminal tail (Hecht et al. 1995). We observe that the presence of Sir3 can dramatically inhibit Dot1 activity, while the addition of a nonspecific competitor (BSA) showed no inhibition (Fig. 6). Based on our current data, we suggest that Sir3 displaces Dot1 from interacting with the H4 basic patch and thus prevents Dot1 from methylating nucleosomal substrates. Alternatively, since Dot1 and Sir3 can interact with tailless nucleosomes, Sir3 may globally displace Dot1 from a similar binding site within the nucleosome core, which would ultimately prevent Dot1 from interacting with the H4 basic patch. Nonetheless, we provide the first evidence of a direct mechanism of competition between Dot1 and Sir3, thus supporting known in vivo observations.
Figure 6.
Sir3 inhibits Dot1-mediated methylation of nucleosomes. In vitro HMTase reactions were performed using recombinant-purified Dot1 incubated in the presence of increasing amounts of the recombinant-purified C terminus of Sir3 with nucleosomal substrates. Half of the reaction samples were analyzed by filter binding and scintillation counting. The remaining reaction amounts were resolved by SDS-PAGE and analyzed by fluorography (3H-Methyl). (Bottom panels) Coomassie-stained gels show the protein inputs in each reaction. The amounts of purified protein in each reaction and positions of the individual histones are indicated.
Discussion
In this report, we determined that Dot1 requires three basic residues (R17H18R19) in the basic patch of histone H4 for H3K79 methylation. We also have determined that a charge-based interaction between an acidic domain of Dot1 and the basic patch of histone H4 is required for proper H3K79 methylation. Finally, we determined that the C-terminal acidic domain of Dot1 and the maintenance of the appropriate charge of the histone H4 basic patch are both required for telomere silencing. Altogether, we have identified a new trans-histone pathway that uses a charge-based mechanism to specifically facilitate histone H3K79 methylation.
Trans-histone pathways
Until now, only one other trans-histone pathway affecting histone lysine methylation has been identified in budding yeast. In S. cerevisiae, loss of histone H2B monoubiquitination or Rad6, the ubiquitin-conjugating enzyme that catalyzes this modification, results in a loss of H3K4 and H3K79 di- and trimethylation (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002b; Sun and Allis 2002; Shahbazian et al. 2005). Monomethylation of H3K4 and H3K79 appears to be unaffected in these mutants (Shahbazian et al. 2005). It is still unclear how H2B ubiquitination allows methylation of H3K4 and H3K79 to proceed from a monomethylated to a di- and trimethylated state. It is interesting to note that in cells expressing particular H4 basic patch mutants, or expressing a Dot1 acidic patch mutant that no longer interacts with the basic patch of histone H4, H3K79 monomethylation is still present. Therefore, it is quite possible that histone H2B ubiquitination affects the nucleosome structure to allow Dot1 access to the H4 basic patch region. Subsequently, this may allow Dot1 to mediate di- and trimethylation of H3K79. It is also likely that additional HMTases in yeast and other eukaryotes will have mechanisms that require another histone for trans-methylation.
Dot1-mediated histone H3K79 methylation
Yeast Dot1 and human Dot1L were the first non-SET domain histone lysine methyltransferases to be identified that specifically modify nucleosomal substrates (Feng et al. 2002; Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002). However, our Dot1 acid patch mutants are still capable of binding to nucleosomes, yet lack the ability to di- and trimethylate H3K79. This demonstrates that nucleosome binding alone is not sufficient to mediate di- and trimethylation of H3K79. Since we do not see dramatic defects in H3K79 monomethylation in cells expressing Dot1ΔEDVDE, it suggests that there are other regions of Dot1 that may be important for nucleosome interactions that are necessary for this modification. Furthermore, H4ΔRHRK mutants show a reduction in H3K79 monomethylation, yet abolished di- and trimethylation, which also implies that there are other potential points of interaction between Dot1 and histone H4.
The H4 basic patch interactions with Dot1 may contribute to the ability of Dot1 to methylate H3K79, possibly through facilitating Dot1 and H3K79 interactions prior to successive methylation events. It is also worth noting that the H4 tail is in close proximity to the H3K79 methylation site in the nucleosome crystal structure (Luger et al. 1997). From the Dot1 structure and Dot1–nucleosome docking model proposed in Sawada et al. (2004), the C-terminal acidic patch of Dot1 forms an exposed surface that is favorable for protein–protein interactions. Additionally, it appears that the acidic patch of Dot1 is in close proximity to the H4 tail and may accommodate the H4 tail basic patch. Although more structural analysis is needed to determine how Dot1 engages the nucleosome, our results indicate there are multiple modes by which Dot1 can uniquely interact with the nucleosome that are either permissive for nucleosome binding or methyltransferase activity.
Yeast Dot1 and human Dot1L both methylate H3K79, but Dot1L contains a C-terminal extension not found in Dot1 (Feng et al. 2002; Min et al. 2003). By sequence analysis, it appears that Dot1L does not possess the critical acidic patch of Dot1 that interacts with the H4 basic patch. However, there are stretches of acidic amino acids in the C-terminal extension of Dot1L, suggesting that a similar mechanism could exist for Dot1L. In humans, histone H4K20 is a major site of methylation, while little to no H4K20 methylation is present in S. cerevisiae (Fang et al. 2002; Nishioka et al. 2002; Garcia et al. 2007). Therefore, human Dot1L may have evolved slightly differently to deal with this additional chromatin modification.
Histone H4 basic patch
Our results indicate that there are three main amino acids (R17H18R19) in histone H4 that are critical for H3K79 methylation, as mutations in H4K16 or H4K20 do not disrupt H3K79 methylation (Supplementary Fig. S2D; data not shown). Intriguingly, H3K79 monomethylation is greatly reduced in the H4ΔRHRK basic patch mutant, while the H4 Δ4–19 N-terminal tail deletion has intact monomethylation. We believe this difference is because Arg 3 in the H4 N-terminal tail essentially replaces R19 in the H4 Δ4–19 deletion mutant. These results are consistent with the observation that cells expressing only single-arginine mutants in the basic patch region exhibit wild-type levels of monomethylation (Fig. 3A). In addition, it is possible that other unknown amino acids in the H4 tail are needed for monomethylation.
The histone H4 basic patch is a site of interaction for chromatin-associated proteins such as Sir3, Sir4, Drosophila ISWI, and yeast Isw2 (Hecht et al. 1995; Clapier et al. 2001, 2002; Fazzio et al. 2005). Therefore, other factors that bind to or modify residues around the H4 basic patch may modulate Dot1’s function. However, yeast lacking Sas2, a H4K16 histone acetyltransferase, Sir1, Sir2, Sir3, or Sir4 do not show any changes in global H3K79 methylation (Supplementary Fig. S2C,E). It has also been demonstrated that the chromatin remodeling activity of ISWI and Isw2 also needs the same three amino acids (R17H18R19) in the histone H4 basic patch that Dot1 requires for H3K79 methylation (Clapier et al. 2001, 2002; Fazzio et al. 2005). However, yeast cells lacking Isw1, Isw2, or Itc1, an Isw2 complex member, have wild-type levels of H3K79 methylation (Supplementary Fig. S2C). Further investigation is needed to determine if other protein factors, or unknown histone modifications, regulate Dot1 binding to the H4 basic patch.
Balance of Sir proteins in maintaining proper telomere silencing
Since H3K79 methylation was first observed to be highly enriched in euchromatin, it was unclear why deleting or overexpressing Dot1 disrupts heterochromatin silencing (Ng et al. 2002a, 2003; van Leeuwen et al. 2002). It has been proposed that loss or overexpression of Dot1 disrupts telomere silencing by an indirect mechanism, where protein factors necessary for direct heterochromatin silencing, such as Sir proteins (Sir2, Sir3, and Sir4), are titrated away from heterochromatin to euchromatin (van Leeuwen and Gottschling 2002). Our observation that Dot1 can interact with the basic patch of histone H4 suggests that Dot1 and Sir3 and/or Sir4 are likely competing for binding to the histone H4 basic patch. Furthermore, we demonstrate that Sir3 can effectively compete Dot1 activity from nucleosomal substrates in an in vitro HMTase assay (Fig. 6). This competition could explain why decreases in H3K79 methylation are observed when Sir3 is overexpressed and why Dot1 overexpression displaces Sir3, leading to silencing defects.
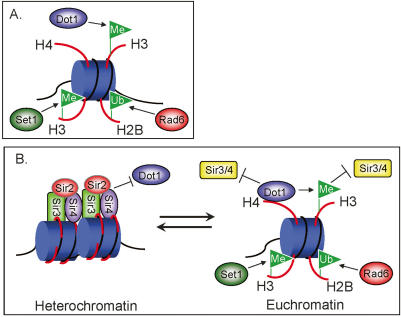
Interestingly, in the context of the nucleosome, histones H3 and H4 contribute residues to form a patch of amino acids surrounding H3K79 that are important for heterochromatin silencing (Park et al. 2002). It has been proposed that the region surrounding H3K79 is a direct site for Sir3 and/or Sir4 binding since mutations of these residues disrupt silencing specifically at telomeres and mating type loci (Park et al. 2002). This is also supported by data showing that Sir3 can interact with nucleosomes lacking histone tails (Georgel et al. 2001). Therefore, H3K79 methylation may prevent Sir3 and/or Sir4 from binding to this region of the nucleosome surrounding H3K79. As illustrated in Figure 7, the presence of Sir3 and/or Sir4 at heterochromatin prevents Dot1 binding to the H4 basic patch and in turn prevents histone H3K79 methylation at the Sir3/Sir4 silenced heterochromatin. Conversely, at euchromatin, the presence of Dot1 prevents Sir3 and/or Sir4 from binding to the basic patch of histone H4. Dot1-mediated H3K79 methylation further excludes binding of Sir3 and/or Sir4, and thus antagonizes the spreading of heterochromatin (Fig. 7). Altogether, it appears that a delicate balance between proper levels of heterochromatin and euchromatin protein factors, mediated by charge-based interactions and histone modifications, is critical for the establishment and maintenance of these specialized chromatin regions in yeast.
Figure 7.
A newly identified trans-histone pathway. (A) A known trans-histone pathway important for histone methylation. Rad6-mediated ubiquitination of H2B required for H3K4 and H3K79 di- and trimethylation. (B) In heterochromatin, silencing is established and maintained by the presence of the Sir proteins (Sir2, Sir3, and Sir4). The presence of Sir3 and/or Sir4 can prevent Dot1 binding to the same target site on the histone H4 N-terminal tail, impeding Dot1 binding and H3K79 methylation at silent chromatin regions. In euchromatin, Dot1 interactions with the basic patch of the histone H4 N-terminal tail allows for H3K79 methylation, excluding the Sir3 and Sir4 proteins. It is likely that the presence of H3K79 methylation and the presence of Dot1 at active regions of chromatin both play a role in the exclusion of silencing factors at euchromatin.
Materials and methods
Yeast strains and plasmids
Plasmids and strains used in this study are listed in Supplementary Tables 1 and 2. See the Supplemental Material for details of plasmid and strain construction.
Yeast extract preparation, Western blotting, and antibodies
For histone analysis, yeast whole-cell extracts were prepared as described previously (Fingerman et al. 2005). Western blotting to detect modified histones was performed as described previously (Briggs et al. 2001). Extracts were run on 15% SDS-PAGE gels, transferred to PVDF, and immunoblotted with the indicated histone antibodies (see the Supplemental Material for the histone antibodies used).
In vitro HTMase assays
In vitro HMTase assays were performed as described previously (Briggs et al. 2002; Milne et al. 2002) using recombinant-purified Dot1 (1.2 μg) or Set2 (2.0 μg) incubated with 6 μg of purified chicken nucleosomes and 1.0 μCi of 3H-SAM. Yeast chromatin substrates were prepared as follows: Yeast strains expressing the desired histone mutants were grown to OD600 = 1.5–1.8, in a volume of 1 L in YPD medium. Cells were harvested and nuclei were prepared as described (Edmondson et al. 1996). To prepare soluble chromatin, 400 μL of nuclei/NP buffer suspension were centrifuged for 15 min at 5000g in a microfuge at 4°C to pellet nuclei. Supernatant was removed and nuclei were resuspended in sonication buffer (van Leeuwen et al. 2002) (20 mM HEPES at pH 7.0, 200 mM NaCl, 10 mM MgCl2, 10% glycerol, 0.1% Triton X-100, 5 mM β-mercaptoethanol) and sonicated to shear and solubilize the chromatin. The sample was then centrifuged at 14,000 rpm for 40 min at 4°C. The supernatant contained sheared, soluble chromatin. Sonicated chromatin samples were normalized, and 5 μg of each substrate were used in in vitro HMTase assays.
GST-H4-binding assays
BL21 cells expressing either GST-H4 tails (pGEX-2T H41–34, pGEX-2T H41–34ΔRHRK, or pGEX-2T H41–34R17K, R19K) or His6-Dot1 (pET28b-His6-Dot1, His6-Dot1G401R, or His6-Dot1ΔEDVDE) were grown to mid-log, induced with 0.4 mM IPTG for 4 h at 23°C, and harvested. Cell pellets were lysed by sonication at 4°C in 200 μL of lysis buffer (50 mM Tris-Cl at pH 8.0; 300 mM NaCl; 1 mM PMSF; 1 μg/mL each leupeptin, aprotinin, pepstatin). Cell lysates were clarified by centrifugation at 14,000 rpm for 5 min at 4°C. The supernatants contained soluble protein. For GST-H4 tail isolation, 10 μL of a 50% slurry of glutathione agarose (Sigma G4510) in PBS were added to the soluble fraction and incubated for 1 h at 4°C. After incubation, GST-H4 tail-bound agarose was pelleted and washed three times for 5 min each in lysis buffer. After washing, GST-H4 tail-bound beads were resuspended in 15 μL of lysis buffer. Of this final slurry, 1.5 μL was analyzed by SDS-PAGE for normalizing GST-H4 protein levels for binding reactions. Binding reactions were carried out as follows: Four microliters of GST-H4 tail-bound glutathione agarose beads were incubated with 20 μL of His6-Dot1, His6-Dot1G401R, or His6-Dot1ΔEDVDE lysates in a final reaction volume of 200 μL, with final buffer conditions being 50 mM Tris-Cl (pH 8.0); 150 mM NaCl; 1 mM PMSF; and 1 μg/mL each leupeptin, aprotinin, and pepstatin. Ten microliters of each reaction were removed to be used as a control for Dot1 input. Binding reactions were incubated with rotation for 2 h at 4°C. Beads were washed with a modified RIPA buffer (50 mM Tris at pH 7.4; 75 mM NaCl; 1% NP-40; 0.5% deoxycholate; 0.1% SDS; supplemented with 1 mM PMSF; 1 μg/mL each leupeptin, aprotinin, pepstatin) three times for 5 min each. After washing, beads were resuspended in 12 μL of 2× SDS-PAGE sample buffer. Samples were loaded onto a 12% SDS-PAGE gel and probed for His6-Dot1 using a 1:5000 dilution of α-HIS antibody (Santa Cruz Biotechnology).
Telomere silencing assays
Telomere silencing assays were performed as described previously (Fingerman et al. 2005). Strain UCC3503 (see Supplementary Table 2 for genotype [Singer et al. 1998]) expressing the indicated histone was grown for 3 d to saturation in SC-Leu, normalized for OD600, serially diluted (fourfold), and spotted (6 μL per spot) on SC-leu + galactose and SC-leu + galactose + 5-FOA (100 μg/mL) plates. Cell growth was monitored at 30°C over time. SC–galactose plates were photographed at 36 h, and SC–galactose–5-FOA were photographed at 48 h. Silencing assays using the ADH overexpression constructs were performed similarly, with the exception that cells were plated on SC-trp or SC-trp + 5-FOA (100 μg/mL).
Nucleosome-binding assays
Nucleome-binding assays were performed as described in Min et al. (2003). Briefly, equivalent amounts of recombinantpurified His6-Dot1, His6-Dot1G401R, Dot1Δ104–172, or His6-Dot1ΔEDVDE were incubated with nucleosomes isolated from chicken erythrocytes (1.45 μM). Reactions were set up as for in vitro HMTase reactions, incubated for 1 h at 30°C, and resolved by agarose gel electrophoresis (2.0% agarose in 0.5× TBE). Gels were stained with ethidium bromide and visualized with UV light.
Preparation of tailless nucleosomes
Tailless nucleosomes were prepared as described in Yang and Hayes (2004). Briefly, isolated chicken erythrocyte nucleosomes were incubated with trypsin–agarose beads (Sigma). After the indicated times, the trypsin–agarose was removed by centrifugation. Tailless nucleosomes were confirmed by both Western blotting and SDS-PAGE, followed by Coomassie staining.
Acknowledgments
We thank Ann Kirchmaier, Joe Ogas, and Harry Charbonneau for helpful discussions; Peter Cheung and Brian Strahl for critically reading our manuscript; and Bradley Wilson and Dah-Eun Jeong for technical assistance. We also thank Dan Gottschling for the UCC1369 and UCC3503 yeast strains; Sharon Roth Dent for yeast strains WZY42, WZY114, and WZY115; Brian Strahl for the pCAL-n-Set2-Flag plasmid construct; and Jacques Cote for helpful discussions and sharing unpublished results. The Purdue University Agricultural Experiment Station number is 2007-18131. This research was supported by grants from the National Institutes of Health to S.D.B. (GM74183) and I.M.F. (Purdue Cancer Center NCI Training Grant CA09634).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1560607
References
- Briggs S.D., Bryk M., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Bryk M., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D., Davie J.K., Dent S.Y., Winston F., Allis C.D., Dent S.Y., Winston F., Allis C.D., Winston F., Allis C.D., Allis C.D. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Hunt D.F., Allis C.D., Strahl B.D., Allis C.D., Strahl B.D., Strahl B.D. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Cheng X., Collins R.E., Zhang X., Collins R.E., Zhang X., Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Langst G., Corona D.F., Becker P.B., Nightingale K.P., Langst G., Corona D.F., Becker P.B., Nightingale K.P., Corona D.F., Becker P.B., Nightingale K.P., Becker P.B., Nightingale K.P., Nightingale K.P. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Nightingale K.P., Becker P.B., Nightingale K.P., Becker P.B., Becker P.B. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. Chromatin silencing protein and pachytene checkpoint regulator Dot1p has a methyltransferase fold. Trends Biochem. Sci. 2001;26:405–407. doi: 10.1016/s0968-0004(01)01856-4. [DOI] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A., Wood A., Dean K., Johnston M., Shilatifard A., Dean K., Johnston M., Shilatifard A., Johnston M., Shilatifard A., Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Smith M.M., Roth S.Y., Smith M.M., Roth S.Y., Roth S.Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Fang J., Feng Q., Ketel C.S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Feng Q., Ketel C.S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Ketel C.S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y., Tempst P., Simon J.A., Zhang Y., Simon J.A., Zhang Y., Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Gelbart M.E., Tsukiyama T., Gelbart M.E., Tsukiyama T., Tsukiyama T. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 2005;25:9165–9174. doi: 10.1128/MCB.25.21.9165-9174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Wang H., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y., Wang H., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y., Tempst P., Struhl K., Zhang Y., Struhl K., Zhang Y., Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Fingerman I.M., Wu C.L., Wilson B.D., Briggs S.D., Wu C.L., Wilson B.D., Briggs S.D., Wilson B.D., Briggs S.D., Briggs S.D. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia B.A., Hake S.B., Diaz R.L., Kauer M., Morris S.A., Recht J., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Hake S.B., Diaz R.L., Kauer M., Morris S.A., Recht J., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Diaz R.L., Kauer M., Morris S.A., Recht J., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Kauer M., Morris S.A., Recht J., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Morris S.A., Recht J., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Recht J., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Shabanowitz J., Mishra N., Strahl B.D., Allis C.D., Mishra N., Strahl B.D., Allis C.D., Strahl B.D., Allis C.D., Allis C.D., et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Georgel P.T., Pietz G., Fox C.A., Hansen J.C., Pietz G., Fox C.A., Hansen J.C., Fox C.A., Hansen J.C., Hansen J.C. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. 2001;98:8584–8589. doi: 10.1073/pnas.151258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S.M., Grunstein M., Laroche T., Strahl-Bolsinger S., Gasser S.M., Grunstein M., Strahl-Bolsinger S., Gasser S.M., Grunstein M., Gasser S.M., Grunstein M., Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Kayne P.S., Kahn E.S., Grunstein M., Kayne P.S., Kahn E.S., Grunstein M., Kahn E.S., Grunstein M., Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Fisher-Adams G., Grunstein M., Fisher-Adams G., Grunstein M., Grunstein M. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992;11:2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lacoste N., Utley R.T., Hunter J.M., Poirier G.G., Cote J., Utley R.T., Hunter J.M., Poirier G.G., Cote J., Hunter J.M., Poirier G.G., Cote J., Poirier G.G., Cote J., Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Lee D.Y., Teyssier C., Strahl B.D., Stallcup M.R., Teyssier C., Strahl B.D., Stallcup M.R., Strahl B.D., Stallcup M.R., Stallcup M.R. Role of protein methylation in regulation of transcription. Endocr. Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J., Richmond R.K., Sargent D.F., Richmond T.J., Sargent D.F., Richmond T.J., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S., Dickens N.J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C.P., Dickens N.J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C.P., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C.P., Kouzarides T., Eisenhaber F., Ponting C.P., Eisenhaber F., Ponting C.P., Ponting C.P. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Mersfelder E.L., Parthun M.R., Parthun M.R. The tale beyond the tail: Histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T.A., Briggs S.D., Brock H.W., Martin M.E., Gibbs D., Allis C.D., Hess J.L., Briggs S.D., Brock H.W., Martin M.E., Gibbs D., Allis C.D., Hess J.L., Brock H.W., Martin M.E., Gibbs D., Allis C.D., Hess J.L., Martin M.E., Gibbs D., Allis C.D., Hess J.L., Gibbs D., Allis C.D., Hess J.L., Allis C.D., Hess J.L., Hess J.L. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Min J., Feng Q., Li Z., Zhang Y., Xu R.M., Feng Q., Li Z., Zhang Y., Xu R.M., Li Z., Zhang Y., Xu R.M., Zhang Y., Xu R.M., Xu R.M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K., Tempst P., Zhang Y., Struhl K., Zhang Y., Struhl K., Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes & Dev. 2002a;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Xu R.M., Zhang Y., Struhl K., Xu R.M., Zhang Y., Struhl K., Zhang Y., Struhl K., Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002b;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Ciccone D.N., Morshead K.B., Oettinger M.A., Struhl K., Ciccone D.N., Morshead K.B., Oettinger M.A., Struhl K., Morshead K.B., Oettinger M.A., Struhl K., Oettinger M.A., Struhl K., Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K., Rice J.C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Rice J.C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Chuikov S., Valenzuela P., Tempst P., Steward R., Valenzuela P., Tempst P., Steward R., Tempst P., Steward R., Steward R., et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Park J.H., Cosgrove M.S., Youngman E., Wolberger C., Boeke J.D., Cosgrove M.S., Youngman E., Wolberger C., Boeke J.D., Youngman E., Wolberger C., Boeke J.D., Wolberger C., Boeke J.D., Boeke J.D. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F., Schaft D., Shevchenko A., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F., Shevchenko A., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F., Wilm M., Aasland R., Stewart A.F., Aasland R., Stewart A.F., Stewart A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A.J., Allis C.D., Wysocka J., Allis C.D., Wysocka J., Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- San Segundo P.A., Roeder G.S., Roeder G.S. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schreiber S.L., Mellor J., Kouzarides T., Mellor J., Kouzarides T., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sawada K., Yang Z., Horton J.R., Collins R.E., Zhang X., Cheng X., Yang Z., Horton J.R., Collins R.E., Zhang X., Cheng X., Horton J.R., Collins R.E., Zhang X., Cheng X., Collins R.E., Zhang X., Cheng X., Zhang X., Cheng X., Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J. Biol. Chem. 2004;279:43296–43306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M.D., Zhang K., Grunstein M., Zhang K., Grunstein M., Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Singer M.S., Kahana A., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E., Kahana A., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E., Goggin C., Mahowald M., Gottschling D.E., Mahowald M., Gottschling D.E., Gottschling D.E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.W., Allis C.D., Allis C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F., Gottschling D.E., Gottschling D.E. Genome-wide histone modifications: Gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 2002;14:756–762. doi: 10.1016/s0955-0674(02)00393-9. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken P.R., Gottschling D.E., Gafken P.R., Gottschling D.E., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Loyola A., Reinberg D., Loyola A., Reinberg D., Reinberg D. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003;2003:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- White C.L., Suto R.K., Luger K., Suto R.K., Luger K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J.P., Simpson R.T., Simpson R.T. Localization of the sites along nucleosome DNA which interact with NH2-terminal histone regions. J. Biol. Chem. 1977;252:6516–6520. [PubMed] [Google Scholar]
- Yang Z., Hayes J.J., Hayes J.J. Large scale preparation of nucleosomes containing site-specifically chemically modified histones lacking the core histone tail domains. Methods. 2004;33:25–32. doi: 10.1016/j.ymeth.2003.10.017. [DOI] [PubMed] [Google Scholar]