Abstract
Cohesin, which mediates sister chromatid cohesion, is composed of four subunits, named Scc1/Rad21, Scc3, Smc1, and Smc3 in yeast. Caenorhabditis elegans has a single homolog for each of Scc3, Smc1, and Smc3, but as many as four for Scc1/Rad21 (COH-1, SCC-1/COH-2, COH-3, and REC-8). Except for REC-8 required for meiosis, function of these C. elegans proteins remains largely unknown. Herein, we examined their possible involvement in mitosis and development. Embryos depleted of the homolog of either Scc3, or Smc1, or Smc3 by RNA interference revealed a defect in mitotic chromosome segregation but not in chromosome condensation and cytokinesis. Depletion of SCC-1/COH-2 caused similar phenotypes. SCC-1/COH-2 was present in cells destined to divide. It localized to chromosomes in a cell cycle-dependent manner. Worms depleted of COH-1 arrested at either the late embryonic or the larval stage, with no indication of mitotic dysfunction. COH-1 associated chromosomes throughout the cell cycle in all somatic cells undergoing late embryogenesis or larval development. Thus, SCC-1/COH-2 and the homologs of Scc3, Smc1, and Smc3 facilitate mitotic chromosome segregation during the development, presumably by forming a cohesin complex, whereas COH-1 seems to play a role important for development but unrelated to mitosis.
INTRODUCTION
In mitotic division, the replicated copies of each chromosome, namely, sister chromatids, are attached together until their segregation in anaphase. This cohesion between sister chromatids is crucial to establish the bipolar orientation of the paired chromatids relative to the mitotic spindle and to ensure accurate delivery of a complete set of chromosomes to each daughter cell. The sister chromatid cohesion is mediated by a multisubunit complex called cohesin (Cohen-Fix, 2001; Lee and Orr-Weaver, 2001). In budding yeast Saccharomyces cerevisiae, cohesin is composed of four proteins, namely, Scc1 (also called Mcd1), Scc3, Smc1, and Smc3 (Guacci et al., 1997; Michaelis et al., 1997; Toth et al., 1999). Equivalent complexes have been identified in fission yeast Schizosaccharomyces pombe, where Rad21 corresponds to budding yeast Scc1, and frog Xenopus laevis (Losada et al., 1998; Tomonaga et al., 2000). In addition, homologs of each cohesin component have been found in other organisms based on sequence similarities, although their function has not been clarified extensively (Hirano, 1999; Parisi et al., 1999; Toth et al., 1999; Dong et al., 2001).
Among the four kinds of cohesin subunits, Scc1/Rad21 plays a crucial role for the onset of anaphase. Scc1 is cleaved by an endopeptidase called separase, and this triggers the separation of sister chromatids (Ciosk et al., 1998; Uhlmann et al., 1999). Rad21 undergoes similar cleavage (Tomonaga et al., 2000). In budding and fission yeast meiosis, Scc1/Rad21 is replaced with its meiosis-specific counterpart called Rec8 (Klein et al., 1999; Watanabe and Nurse, 1999). Like Scc1/Rad21, Rec8 is cleaved by separase and gives a cue for the onset of meiotic anaphase I in budding yeast (Buonomo et al., 2000). Because the other three subunits of cohesin apparently function in both mitosis and meiosis, the replacement of Scc1/Rad21 with Rec8 seems to be crucial for establishing the reductional pattern of chromosome segregation specific to meiosis I.
In the nematode Caenorhabditis elegans, four Scc1/Rad21 family members have been reported (Parisi et al., 1999; Pasierbek et al., 2001). One of them, REC-8, was found to localize to synaptonemal complexes at the pachytene stage and to chromosomal axes at the diakinesis stage in meiotic prophase I (Pasierbek et al., 2001). In addition, depletion of REC-8 by RNA interference (RNAi) resulted in splitting of the chromosomes into sister chromatids and appearance of univalents at the diakinesis stage (Pasierbek et al., 2001). Thus, REC-8 seems to be the functional homolog of both budding and fission yeast Rec8. Two Scc1/Rad21 homologs, called COH-1 and COH-2, were found, respectively, to be essential for the viability of the worm, whereas no apparent phenotype was observed in an animal depleted of the fourth homolog, COH-3 (Pasierbek et al., 2001). The question has not been critically answered, however, whether they are involved in mitotic chromosomal cohesion. Furthermore, homologs of the other three kinds of cohesin subunits remain largely uncharacterized in C. elegans. Under these circumstances, we set out to explore possible function of cohesin component homologs of C. elegans during its development. Herein, we show that COH-2 and the homologs of Scc3, Smc1 and Smc3, are involved in proper chromosome segregation during mitosis, but COH-1 seems to have novel function necessary for development but unrelated to mitosis. Because the use of scc-1 instead of coh-2 as the main registered gene name has been recently agreed (Hodgkin, Meyer, and Loidl, unpublished data), we hereafter denote the gene product as SCC-1/COH-2 in this article.
MATERIALS AND METHODS
Strains
Maintenance and genetic manipulation of C. elegans were carried out as described previously (Brenner, 1974). The wild-type C. elegans var. Bristol strain N2 and AZ212 (unc-110(ed3) ruIs32[pAZ132:pie- 1/GFP/histone H2B] III) were used. N2 was maintained at 20°C and AZ212 was maintained at 25°C.
RNA Interference
As the templates to prepare double-stranded RNA (dsRNA), the following cDNA clones were used: yk226d1 (coh-1), yk256h5 (scc1/coh-2), yk97g6 (scc-3), yk52h10 (him-1/smc-1), yk72e8 (coh-3), and yk295d12 (smc-3). These templates were polymerase chain reaction amplified using either a pair of T7 and T3 primers (T7: 5′-GTA ATA CGA CTC ACT ATA GGG C-3′; T3: 5′-AAT TAA CCC TCA CTA TTG GG-3′), or a pair of M13-forward and M13-reverse primers (forward: 5′-GTA AAA CGA CGG CCA GT-3′; reverse: 5′-GGA AAC AGC TAT GAC CAT G-3′). RNA was synthesized in vitro with T7 and T3 RNA polymerases, and complementary strands were mixed to generate dsRNA to be used for RNAi (Fire et al., 1998). Delivery of dsRNA into worms was performed by either microinjection or soaking. In the case of microinjection, 1–3 μg/μl dsRNA was injected into either the gonad or the intestine of young adult hermaphrodites, and phenotypes of F1 progeny laid 12–30 h after the injection were characterized. In the case of RNAi by soaking (Maeda et al., 2001), L4 larvae were treated in a dsRNA solution (1 μg/μl in soaking buffer) for 24 h, and phenotypes of the F1 generation were observed. To examine postembryonic phenotypes caused by RNAi, L1 larvae were soaked in a dsRNA solution similarly, and abnormalities of the recovered worms were observed. In addition, worms that hatched from the embryos laid soon after dsRNA injection (6–12 h) were also examined to assess the postembryonic phenotypes.
Generation of COH-1 and SCC-1/COH-2 Antibodies
Rabbit and rat polyclonal antibodies against COH-1 and SCC-1/COH-2 were prepared as follows. The relatively unique region in each open reading frame (amino acids 173–559 for COH-1; amino acids 118–544 for SCC-1/COH-2) was polymerase chain reaction amplified from a cDNA clone and cloned into pGEX-KG vector (Guan and Dixon, 1991). Each glutathione S-transferase (GST) fusion protein was expressed from the resulting plasmid in Escherichia coli and used as an antigen after purification. COH-1- and SCC-1/COH-2–specific antibodies raised in rabbit and rat were affinity purified using histidine (His)-tagged COH-1 and SCC-1/COH-2 proteins, respectively. To create a His-tagged fusion construct, the same cDNA fragments as used above were cloned into pET19b vector (Novagen, Madison, WI). The affinity-purified antibodies, respectively, recognized COH-1 and SCC-1/COH-2 specifically. Both rabbit and rat antibodies were used in immunofluorescence analysis, and they gave the same staining patterns.
Immnofluorescence and DNA Staining
Embryos were processed for staining as described previously (Miller and Shakes, 1995). In brief, embryos permeabilized by the freezecrack method were fixed by placing in methanol for 2 min at -20°C and then in acetone for 4 min at -20°C, and rehydrated at room temperature. Rehydrated embryos were treated with a blocking solution (3% bovine serum albumin in phosphate-buffered saline containing 0.5% Tween 20) for 30 min at room temperature. They were then incubated with the primary antibody at 4°C overnight and then with the secondary antibody at room temperature for 1–2 h. 4,6-Diamidino-2-phenylindole was added to the final concentration of 2 μg/ml, and the sample was mounted for epifluorescence microscopy. To visualize DNA under confocal laser microscopy, fixed samples were treated with 1/10,000 diluted Sytox Green (Molecular Probes, Eugene, OR) for 10 min. The following antibodies were used: primary antibodies, anti-COH-1(this study), anti-SCC-1/COH-2 (this study), and anti-nuclear pore complexes mouse monoclonal antibody (MAb414) (Babco, Richmond, CA); and secondary antibodies, fluorescein isothiocyanate-conjugated sheep anti-mouse IgG antibody (Cappel Laboratories, Durham, NC), Cy3-labeled goat anti-rabbit IgG antibody (Chemicon International, Temecula, CA), and rhodamin-labeled anti-rat IgG antibody (Cappel Laboratories). To reduce nonspecific background signals, the secondary antibodies were treated with the C. elegans acetone powder before use.
For confocal imaging, the LSM510 system attached to an Axioplan 2 microscope (Carl Zeiss, Jena, Germany) was used. Other images were taken digitally by either of the following combinations: an AxioCam charge-coupled device camera attached to an Axioplan 2 microscope with the AxioVision software (Carl Zeiss); or a cooled charge-coupled device camera C4742–95-10NR (Hamamatsu Photonics) attached to a Zeiss Axioplan 2 microscope with the FISH Imaging Software (Hamamatsu Photonics, Bridgewater, NJ).
Live Observation of Embryos and Four-Dimensional Recording
Young adult hermaphrodites were dissected in M9 buffer and the collected embryos were mounted on a 2% agar pad under a coverslip. Four-dimensional recording of green fluorescent protein (GFP)-fluorescence and differential interference contrast (DIC) images was performed using the LSM510 system attached to an Axioplan 2 microscope (Carl Zeiss). Images were taken every 40 s, at five different focal planes at least.
RESULTS
Homologs of Cohesin Components in C. elegans
A search of the C. elegans genome database indicated that homologs of the four components of the cohesin complex were apparently conserved in this worm. A single homolog was found for each of Scc3, Smc1, and Smc3, which we hereafter call SCC-3 (open reading frame name F18E2.3), HIM-1/SMC-1 (F28B3.7), and SMC-3 (Y47D3A.26), according to their registered gene names. Allelism between him-1 and smc-1 (F28B3.7) has been established (Meyer, unpublished data). Four Scc1/Rad21 homologs (COH-1, SCC-1/COH-2, COH-3, and REC-8) were reported previously (Pasierbek et al., 2001). Comparison of their amino acid sequences has shown that COH-1 and SCC-1/COH-2 are highly similar to each other and both belong to the Scc1/Rad21 subfamily, as opposed to the Rec8 subfamily. REC-8 is the ortholog of yeast Rec8 and is specifically involved in meiotic chromosome cohesion, whereas the homology of COH-3 with the Scc1/Rad21 family members was less significant (Pasierbek et al., 2001). To examine whether these putative cohesin components in C. elegans were involved in chromosomal cohesion during mitosis, we depleted each protein by RNAi and monitored mitosis in embryos.
We first examined the RNAi phenotypes for the scc-3, him-1/smc-1, and smc-3 genes. Depletion of any of these gene products resulted in embryonic lethality with complete penetrance. To characterize the process of chromosome segregation in RNAi animals, chromosomes were visualized by GFP-tagged histone H2B, and time-lapse images of the fluorescence of H2B-GFP as well as DIC images of the embryos were collected using a confocal microscope. For each of these three genes, RNAi-affected embryos were all arrested during embryogenesis, and ∼30% of them displayed defective mitosis in early cell cycles, as described below.
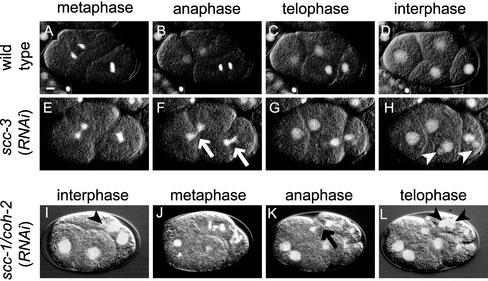
In wild-type embryos, condensed chromosomes aligned on the metaphase plate and then they separated at once in anaphase, giving a view of splitting two parallel discs (Figure 1, A–D, movie WT.mov). After cytokinesis, chromosomes were decondensed and nuclear membrane was reassembled around them. In contrast, embryos depleted of either SCC-3, or HIM-1/SMC-1, or SMC-3 behaved differently from the wild-type, with these three kinds of RNAi embryos showing phenotypes indistinguishable from each other (Figure 1, E–H, movie scc-3.mov; our unpublished data). Chromosomes were condensed at prometaphase in these embryos, but the metaphase plate often looked diffuse, or was completely missing in some cases. Then, a mass of chromosomes staying in the middle of the bipolar spindle frequently seemed to be divided by a cleavage furrow (Figure 1F, arrows). This might mean the lack of anaphase, or alternatively, the progression of aberrant anaphase with extensive chromosome bridging, which were not distinguishable at the current resolution. Chromosomes were decondensed after cytokinesis, as in wild-type embryos, but multiple (mainly two to four) nuclei of variable size were often formed in daughter cells (Figure 1H, arrowheads). In the case of wild-type embryos, nuclear membrane has been shown to reassemble around subsets of decondensed chromosomes at telophase, but they fuse until they form a single nucleus that encloses the whole chromosome set (Newport, 1987). We speculate that nuclear membrane could reassemble around each chromosome or a subset of a few chromosomes but failed to fuse to form a single nucleus in cells devoid of either SCC-3, or HIM-1/SMC-1, or SMC-3. This is probably because chromosomes were not separated synchronously in these cells and hence were not close enough to each other to compose a unified nucleus. Subsequent cell cycles were also aberrant similarly in the RNAi animals. Nuclear membrane of the multiple small nuclei generated in the previous cell cycle broke down simultaneously and chromosomes were condensed, but only a loose metaphase plate was formed again, and a mass of chromosomes was divided irregularly by a cleavage furrow. These observations suggest that the three homologs of cohesin components, namely, SCC-3, HIM-1/SMC-1, and SMC-3, are essential for proper chromosome segregation, but unlikely to be involved in other aspects of the cell cycle.
Figure 1.
Depletion of each cohesin component homolog causes defect in chromosome segregation. Time-lapse images of early embryos of wild-type (A–D), scc-3 (RNAi) (E–H), and scc-1/coh-2 (RNAi) (I–L) embryos. These images can be seen also in video movies (WT.mov, scc-3.mov, and coh-2.mov, respectively). Fluorescence of GFP-tagged histone H2B and the DIC images are overlaid. The arrows in F and K indicate anaphase chromosomes with abnormal morphology. The arrowheads in H, I, and L indicate small extranuclei generated by abnormal chromosome segregation. Bar, 5 μm.
COH-1 and SCC-1/COH-2 Play Distinct Roles in Development
Among the four C. elegans Scc1/Rad21 homologs, REC-8 is involved specifically in meiosis, and no detectable phenotype results from RNAi for coh-3 (Pasierbek et al., 2001). Therefore, we focused on the two remaining Scc1/Rad21 homologs, namely, COH-1 and SCC-1/COH-2, and examined their possible function in mitosis. When we disrupted function of the scc-1/coh-2 gene by RNAi, F1 embryos exhibited embryonic lethality with complete penetrance (Figure 2A), as described previously (Pasierbek et al., 2001). We carefully observed the cell division process in scc-1/coh-2(RNAi) embryos, and found similar chromosome segregation defects in early cell cycles to the scc-3(RNAi), him-1/smc1(RNAi), or smc-3(RNAi) embryos, although the penetrance was relatively low (Figure 1, I–L, movie coh-2.mov). This low penetrance (∼10% of the RNAi-affected embryos) might be due to either gene-specific inefficiency of RNAi reaction or relatively high stability of the SCC-1/COH-2 protein compared with other cohesin components. The scc-1/coh-2(RNAi) embryos showed loose metaphase plates, asynchronous separation of chromosomes at anaphase, and formation of multiple nuclei after decondensation of chromosomes. Thus, we speculated that SCC-1/COH-2 was likely to function in the same process as the three other cohesin component homologs described above, at least in early embryogenesis.
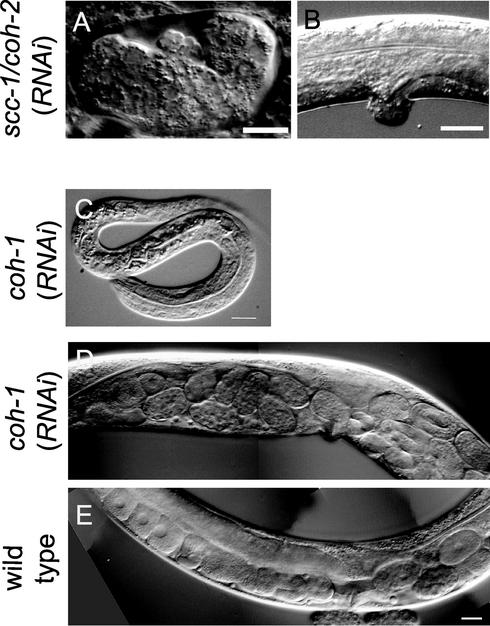
Figure 2.
SCC-1/COH-2 and COH-1 play distinct roles in development. (A and B) SCC-1/COH-2 is required for mitotic division throughout development. Depletion of SCC-1/COH-2 in embryos resulted in embryonic lethality. A terminally arrested embryo is shown in A. When the function of SCC-1/COH-2 was depleted postembryonically, some worms exhibited abnormal morphology of the vulva (B). (C–E) COH-1 is essential for larval development and the egg-laying ability. Some coh-1(RNAi) F1 progeny arrested as L1 larvae (C). When the function of COH-1 was disrupted in L1 worms by RNAi by soaking, coh-1(RNAi) adults showed an egg-laying defect, accumulating more eggs in the gonad (D) than wild-type hermaphrodites (E). Bar, 10 μm (A and C), 20 μm (B), and 20 μm (D and E).
When we chased possible postembryonic phenotypes of scc-1/coh-2(RNAi) animals by examining F1 worms laid soon after the injection of dsRNA (see MATERIALS AND METHODS), they either arrested as larvae or became sterile adults. Some arrested larvae were thin at their posterior part and showed a severe Unc (uncoordinated) phenotype unable to move backward. Variable defects were seen in germ cells of the sterile adults. Some animals had very few germ cells with no mature gametes, whereas others had normal-looking sperm and >100 undifferentiated germ cells in the gonad. In addition, some sterile adults showed a Pvl (protruding vulva) phenotype (Figure 2B). Thus, the cell types affected in these worms, namely, ventral nerve cord, vulva, and germ cells, apparently corresponded to the ones that are known to divide postembryonically. These observations suggest that the SCC-1/COH-2 function is essential for normal cell division during larval development, in addition to embryogenesis.
We also disrupted function of the coh-1 gene by RNAi. Whereas ∼8% of the F1 progeny arrested as unhatched embryos, the majority of progeny arrested as larvae in this case (37% L1-L2 arrest, and 22% L3-L4 arrest, n = 328). The variable phenotypes observed were probably due to genedependent inefficacy inherent of RNAi experiments. These larvae showed no apparent morphological abnormality (Figure 2C), but were affected severely in locomotion (our unpublished data). Even F1 progeny that arrested as unhatched embryos seemed to have completed normal morphogenesis. Because expression of COH-1 became detectable at the mid-embryogenesis stage (see below), and the RNAi-arrested embryos showed a significantly reduced level of COH-1, it seemed that the most rigorous loss-of-function phenotype for coh-1 would be late embryonic lethality after completion of morphogenesis. We next disrupted the function of COH-1 postembryonically by soaking L1 worms to a dsRNA solution and found that some coh-1(RNAi) adults were defective in egg laying (Figure 2, D and E). The observed defects in locomotion and egg laying implicated that the function of COH-1 might be important for proper function of muscle and/or the nervous system. In summary, the function of COH-1 seems to be dispensable for embryonic development but essential for viability of the organism. So far, no indication of defects in cell division has been observed in coh-1(RNAi) worms.
We then performed double and triple RNAi experiments for coh-1, scc-1/coh-2, and coh-3. When the function of coh-1 and scc-1/coh-2 was simultaneously disrupted by RNAi, the F1 embryos displayed phenotypes similar to those of scc-1/coh-2(RNAi) worms, with no apparent enhancement in the phenotypes (our unpublished data). Two double RNAi experiments involving coh-3, namely, coh-1(RNAi); coh-3(RNAi), and scc-1/coh-2(RNAi); coh-3(RNAi), resulted in the same phenotypes as single RNAi for each of coh-1 and scc-1/coh-2, respectively (our unpublished data). In addition, the triple RNAi [coh-1(RNAi); scc-1/coh-2(RNAi); coh-3(RNAi)] caused essentially the same embryonic phenotypes as scc-1/coh-2(RNAi) (our unpublished data). Therefore, we speculate that the function of coh-1 and that of scc-1/coh-2 is neither synergistic nor redundant and that coh-3 may contribute little to the physiology of the worm.
Cell Cycle-dependent Localization of SCC-1/COH-2 in Early Embryos
To visualize expression and localization of COH-1 and SCC-1/COH-2 during worm development, we generated polyclonal antibodies against the central region of each protein, because this region showed no significant similarity between the two. We confirmed that the affinity-purified anti-COH-1 and anti-SCC-1/COH-2 antibodies specifically recognized each protein, COH-1 as an apparent 95-kDa band and SCC-1/COH-2 as an apparent 110-kDa band, in Western blotting against the worm extract (our unpublished data). Both proteins migrated slower than the expected size of 75 kDa. This abnormal migration of the worm Scc1/Rad21 family members has been described previously, although the observed apparent molecular masses are somewhat different from our estimation (Pasierbek et al., 2001). The specificity of the antibodies was further confirmed by immunostaining of RNAi embryos depleted of either COH-1 or SCC-1/COH-2 (see below).
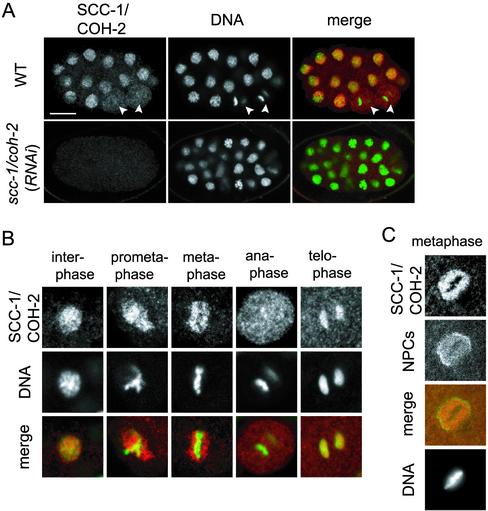
We analyzed localization of SCC-1/COH-2 in early embryos by using the specific antibodies (Figure 3A). SCC-1/COH-2 seemed to localize to the chromosomes in a cell cycle-dependent manner (Figure 3B). In interphase, SCC-1/COH-2 was seen throughout the nucleus, overlapping largely with DNA. At mitotic prophase, SCC-1/COH-2 started to separate from condensing chromosomes, and it was not detected on the chromosomes at prometaphase and metaphase. At metaphase, the SCC-1/COH-2 signal seemed as if surrounding the metaphase plate, although it was possible that a small amount of SCC-1/COH-2 was remaining on the metaphase chromosomes but escaped detection, because cohesin is reported to become detectable on metaphase chromosomes only after detergent extraction of soluble background in other metazoans (Warren et al., 2000). The SCC-1/COH-2 signal was then dispersed in the cytoplasm at anaphase. At telophase, the SCC-1/COH-2 protein began to reaccumulate on the chromosomes.
Figure 3.
Cell cycle-dependent localization of SCC-1/COH-2 in early embryos. (A) Wild-type and scc-1/coh-2(RNAi) embryos were stained with anti-SCC-1/COH-2 antibodies (SCC-1/COH-2, red) and Sytox Green (DNA, green). The white arrowheads indicate anaphase cells, in which SCC-1/COH-2 is dissociated from the chromosomes and dispersed in the whole cytoplasm. The SCC-1/COH-2 signal was not detected in scc-1/coh-2(RNAi) embryos. Bar, 10 μm. (B) Magnified images of nuclei in wild-type early embryos. SCC-1/COH-2 accumulated in the interphase nuclei but was not detectable on condensed metaphase chromosomes. SCC-1/COH-2 associated again with chromosomes at telophase. (C) Wild-type nuclei in an early embryo were stained with anti-SCC-1/COH-2 antibodies (SCC-1/COH-2, red), an anti-nuclear pore complexes antibody (NPCs, green), and 4,6-diamidino-2-phenylindole (DNA). It is noticeable that few SCC-1/COH-2 molecules associate chromosomes at metaphase but they stay in the nucleoplasm surrounded by the nuclear membrane.
Whereas nuclear envelope breakdown normally occurs at prometaphase of mitosis in higher eukaryotes, nuclear membrane persists until the beginning of anaphase in early embryos in C. elegans (Lee et al., 2000; Liu et al., 2000). Therefore, the SCC-1/COH-2 signal observed around the metaphase plate might represent the protein molecules that were dissociated from the chromosomes and trapped by the nuclear envelope, as has been demonstrated for small nuclear ribonucleoprotein particles (Lee et al., 2000). Alternatively, it could be the molecules adhering to the side of condensed chromosomes. To distinguish these possibilities, we stained the embryos with both anti-SCC-1/COH-2 antibodies and an antibody against a component of the nuclear pore complexes. The SCC-1/COH-2 signal was evenly distributed within the nuclear envelope except for the chromosomal region (Figure 3C), suggesting that SCC-1/COH-2 molecules dissociated from the chromosomes at metaphase were trapped by the nuclear envelope. Consistently with this interpretation, the SCC-1/COH-2 staining around the metaphase plate was no longer seen at later stages of embryogenesis involving >30 cells, where nuclear envelope is known to break down before metaphase (Lee et al., 2000). SCC-1/COH-2 was dispersed into the whole cytoplasm of metaphase cells at these stages (our unpublished data).
SCC-1/COH-2 Is Expressed in Dividing Cells throughout Development
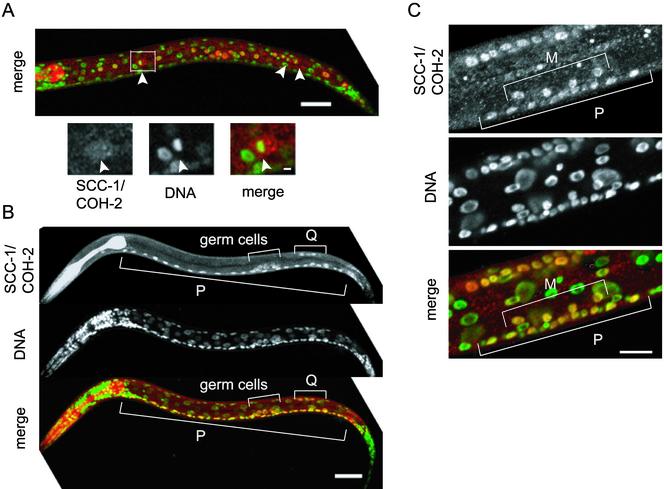
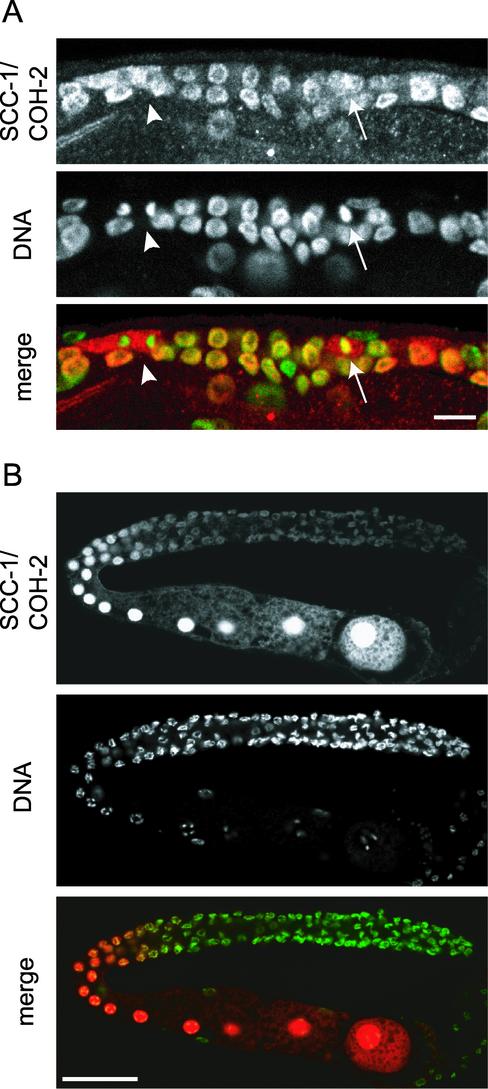
The developmental profile of SCC-1/COH-2 expression was examined by staining embryos and worms at various stages with anti-SCC-1/COH-2 antibodies. SCC-1/COH-2 was strongly expressed in virtually all cells in early embryos, but its expression was gradually weakened, and the signal could hardly be detected in late embryos, in which cell division was ceased almost completely (our unpublished data). Strong nuclear signals of SCC-1/COH-2 reappeared in larvae, though they were limited to a subset of cells (Figure 4). The cell lineage throughout the development has been elucidated in C. elegans, and the timing as well as the number of cell divisions is known to be invariant (Sulston and Horvitz, 1977). This enabled us to identify which types of cells expressed SCC-1/COH-2 postembryonically. We found that SCC-1/COH-2 was detectable only in cells that were going to divide. For example, in an L1 larva, intense SCC-1/COH-2 signals were detected in the 14 hypodermal V lineage cells, which divide synchronously (Figure 4A). Figure 4A illustrates the SCC-1/COH-2 signal dispersed and not detectable on condensed chromosomes, as observed in embryos of an intermediate stage. In a slightly older L1 larva, expression of SCC-1/COH-2 was seen in 22 P lineage cells to constitute the ventral nerve cord and in four Q lineage cells to produce posterior neuronal cells, all of which divide at the same time (Figure 4B). In this L1 larva, no signal was detected in the V lineage cells (Figure 4B), suggesting that the SCC-1/COH-2 protein is present only for a short time in the cell cycle, and likely to be degraded quickly after cell division. Larvae of later stages also expressed SCC-1/COH-2 in dividing cells: in an L3 larva, SCC-1/COH-2 was detected in four M lineage cells to produce the uterine and vulval muscle cells and in 10 P lineage vulval precursor cells, which divide concurrently (Figure 4C).
Figure 4.
SCC-1/COH-2 is expressed in dividing cells throughout the development. (A) An L1 larva stained with anti-SCC-1/COH-2 antibodies (SCC-1/COH-2, red) and Sytox Green (DNA, green). The SCC-1/COH-2 signal was intense in the 14 hypodermal V lineage cells, which divide synchronously. The magnified view shows a cell at metaphase in which the SCC-1/COH-2 signal is dispersed in the cytoplasm. Arrowheads indicate condensed chromosomes. Bar, 20 μm. (B) A slightly older L1 larva, stained as described above. In addition to the dividing P lineage and Q lineage cells, germ cells were stained specifically with anti-SCC-1/COH-2 antibodies. Bar, 20 μm. (C) The ventral region of an L3 larva. The SCC-1/COH-2 signal could be detected in 4 M lineage and 10 P lineage cells, which divide concurrently. Bar, 10 μm.
SCC-1/COH-2 was expressed in germ cells throughout the development, including the adult stage. Germ cells of C. elegans continue to divide at the distal part of the gonad, from the L1 stage through adulthood, and we detected SCC-1/COH-2 in virtually all mitotic germ nuclei. Similarly to somatic cells in embryos, SCC-1/COH-2 was dispersed in the cytoplasm at mitotic prometaphase and was absent from the condensed anaphase chromosomes in germ cells (Figure 5A). In female germ cells that entered meiotic prophase in adult hermaphrodites, SCC-1/COH-2 was observed uniformly in the nuclei. It was unclear whether SCC-1/COH-2 localized to the condensed meiotic chromosomes, because of the strong SCC-1/COH-2 signal emitted from the nucleoplasm (Figure 5B). The localization of SCC-1/COH-2 we have assigned in female germ cells is somewhat different from that of a previous report (Pasierbek et al., 2001), in which SCC-1/COH-2 has been observed as distinct spots in pachytene nuclei but not detected in diplotene or later nuclei. The reason for this apparent difference is unclear. SCC-1/COH-2 was detected also in male germ cells at mitosis and meiosis, but it was not detectable in mature sperm (our unpublished data). Together, we conclude that expression of SCC-1/COH-2 is coupled strictly with progression of the cell division cycle throughout the development.
Figure 5.
SCC-1/COH-2 localizes to nuclei in germ cells. Hermaphrodite germ cells were stained with anti-SCC-1/COH-2 antibodies (SCC-1/COH-2, red) and Sytox Green (DNA, green). (A) Mitotic germ cells in an L3 larva. As in somatic cells, SCC-1/COH-2 was dispersed in the cytoplasm of germ cells at prometaphase (arrows), and was not detected on condensed anaphase chromosomes (arrowheads). Bar, 10 μm. (B) In a gonad of an adult hermaphrodite, SCC-1/COH-2 was weakly detected on the chromosomes of mitotic germ cells. The SCC-1/COH-2 signal became very intense in maturing oocytes and was spread evenly in the nuclei. Bar, 50 μm.
Localization of COH-1 in Embryos and Larvae
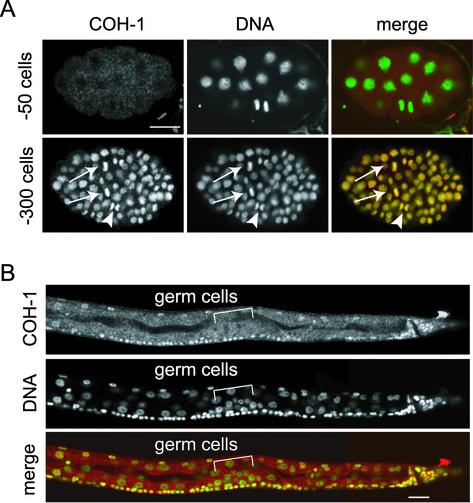
The temporal and spatial expression pattern of COH-1 was totally different from that of SCC-1/COH-2. Unlike SCC-1/COH-2, COH-1 was not detected in early embryos (Figure 6A). The COH-1 protein emerged in virtually all cells in embryos after the ∼100-cell stage and seemed to be expressed most strongly in embryos undergoing morphogenesis. During larval development, COH-1 could be seen apparently in all somatic cells, although the signal in the intestinal cells was somewhat weaker compared with other cells. Interestingly, however, COH-1 was not detected in germ cells throughout the larval development (Figure 6B). Moreover, in striking contrast to SCC-1/COH-2, which showed a cell cycle-dependent dynamic change in subcellular localization, COH-1 was found in association with chromosomes regardless of the stage of the cell cycle in all the cells expressing it. We note that the COH-1 localization we describe herein is again somewhat different from the previous observation (Pasierbek et al., 2001), in which COH-1 has been reported to be absent from the condensed mitotic chromosomes. This disagreement may be due to the difference of the antibody preparation and/or the staining procedures used. Together, we conclude that COH-1 and SCC-1/COH-2 are likely to be regulated in distinct manners with respect to their subcellular localization and developmental expression.
Figure 6.
Localization of COH-1 in embryos and larvae. (A) Wild-type embryos stained with anti-COH-1 antibody (COH-1, red) and Sytox green (DNA, green). COH-1 was not detectable in an early embryo (∼50 cells). In an embryo of a later stage (∼300 cells), COH-1 associated chromosomes in every cell, regardless of the phase of the cell cycle, including metaphase (arrows) and anaphase (arrowheads). Bar, 10 μm. (B) The posterior half of an L2 larva, stained as described above. COH-1 localized to chromosomes of virtually all somatic cells, but was missing from the germ cell nuclei. Bar, 10 μm.
DISCUSSION
Mitotic Cohesin Complex in C. elegans
We have analyzed function of the C. elegans homologs of cohesin components in this study. Effects of depletion of each cohesin subunit have been previously examined in budding and fission yeast, in a Xenopus extract, and in cultured vertebrate cells (Guacci et al., 1997; Losada et al., 1998, 2000; Michaelis et al., 1997; Tomonaga et al., 2000; Sonoda et al., 2001). Our study seems unique in that it characterizes mitotic cohesin in a multicellular animal at the organismal level. The results we obtained suggest that SCC-1/COH-2, SCC-3, HIM-1/SMC-1, and SMC-3 are essential for proper segregation of chromosomes during mitosis. Depletion of any of these four proteins caused similar deficiency in chromosome segregation. The homologs of these proteins in yeast and frog have been shown to form a cohesin complex (Losada et al., 1998, 2000; Toth et al., 1999; Tomonaga et al., 2000). Thus, it is presumable that these four proteins, which are homologous with Scc1/Rad21, Scc3, Smc1, and Smc3, respectively, constitute a mitotic cohesin complex and function in sister chromatid cohesion in C. elegans, although it needs to be critically confirmed by biochemical analysis. Our findings also suggest that the function of cohesin components has been conserved among eukaryotic organisms.
In budding and fission yeast, mutants of cohesin components exhibit premature sister chromatid separation, and they consequently arrest at metaphase due to operation of the spindle checkpoint mechanism (Guacci et al., 1997; Michaelis et al., 1997; Tomonaga et al., 2000). Similarly, Scc1-deficient chicken DT40 cells result in premature sister chromatid separation and show a mitotic delay at prometaphase or metaphase, with only a small proportion of cells undergoing anaphase (Sonoda et al., 2001). In C. elegans embryos lacking any of the aforementioned cohesin subunit homologs, condensed chromosomes failed to form a tight metaphase plate, as in yeast and DT40-defective cells, but the following cell cycle events could proceed in them, probably because the spindle checkpoint is not operative in early embryos. This allows us to evaluate the extent of the direct involvement of C. elegans cohesin components in the cell cycle progression: SCC-1/COH-2, SCC-3, HIM-1/SMC-1, and SMC-3 are all dispensable for cytokinesis, nuclear disassembly/reassembly, and chromosome condensation/decondensation. It has been shown recently that homologs of the SMC-type subunits of the condensin complex are involved in chromosome condensation in C. elegans (Hagstrom et al., 2002). Thus, unlike budding yeast, in which the cohesin complex affects both chromosome condensation and cohesion, the two chromosomal events are likely to be regulated by distinct complexes in C. elegans, as in vertebrates (Michaelis et al., 1997; Sonoda et al., 2001).
The cell cycle-dependent localization of Scc1/Rad21 homologs has been reported in several organisms, though there seem to be considerable differences from organism to organism. In budding yeast, Scc1 is localized to the chromatin in late G1 and S phase. Scc1 dissociates from chromosomes as the cell enters mitotic anaphase, triggered by its cleavage by separase, a proteolytic enzyme activated by the anaphase-promoting complex (Guacci et al., 1997; Michaelis et al., 1997). In contrast, the fission yeast Rad21 protein is detectable on the chromosome throughout the cell cycle, and the cleavage of only a small fraction of Rad21 seems to be sufficient for the onset of anaphase in this organism (Tomonaga et al., 2000). The situation is further different in vertebrates and Drosophila. In these organisms, Scc1 is presumed to be removed from chromosomes in a two-step mechanism: First, Scc1 is removed from the chromosomal arms in a cleavage-independent manner during prophase, and then the protein staying in the centromeric region is removed at metaphase–anaphase transition, triggered by the cleavage by separase (Losada et al., 1998; Waizenegger et al., 2000; Warren et al., 2000). The chromosomes of C. elegans are holocentric, that is, nonlocalized kinetochores spread along the length of each chromosome (Albertson and Thomson, 1982). Therefore, if SCC-1/COH-2 persists on the centromeric regions until the onset of anaphase, as in higher animals, we can expect that it may constitute a longitudinal band on each chromosome. However, we could not detect SCC-1/COH-2 on the condensed chromosomes from prophase through anaphase. Two types of interpretation of the observation seem to be possible. Difficulties have been reported in detecting vertebrate Scc1/Rad21 homologs on metaphase chromosomes by immunofluorescence under certain conditions, probably because of low antigen accessibility to the condensed chromosomes (Losada et al., 2000). Therefore, it is possible that we technically failed to detect a small amount of SCC-1/COH-2 persisting at the centromeric regions. Alternatively, the possibility may remain as well that SCC-1/COH-2 indeed dissociates from chromosomes during prophase, and that another unidentified protein may withhold the chromosomal cohesion until the onset of anaphase in C. elegans.
The cleavage of Scc1/Rad21 by separase triggers metaphase–anaphase transition in yeast and vertebrates (Uhlmann et al., 1999; Hauf et al., 2001). In C. elegans, a homolog of separase, called SEP-1, has been shown to be essential for sister chromatid separation (Siomos et al., 2001). Thus, the simplest hypothesis is that SCC-1/COH-2 is cleaved by SEP-1 during mitosis. Indeed, there are several sequences that match the separase cleavage consensus (E/DxxR) in the SCC-1/COH-2 protein. Because we have not detected SCC-1/COH-2 on metaphase chromosomes, it seems intriguing to address whether SCC-1/COH-2 is really a target of SEP-1, and whether the cleavage of SCC-1/COH-2 is a trigger for the metaphase–anaphase transition.
Distinct Roles for COH-1 and SCC-1/COH-2
Our study has indicated that COH-1 and SCC-1/COH-2 play distinct function in the development of C. elegans. As described above, SCC-1/COH-2 is most likely to function as a component of the mitotic cohesin complex, whereas COH-1 does not seem to play any significant role in chromosomal cohesion, judging from its loss-of-function phenotypes and subcellular localization. COH-1 was present on chromosomes in all somatic cells, including nonmitotic cells, from the stage of ∼100-cell embryos through adulthood, and its depletion did not cause any cell cycle-related abnormalities. The coh-1(RNAi) embryos and animals looked morphologically normal, although they had a severe defect in locomotion, implying that COH-1 may be unrelated to cell division and cell fate determination. It may be that COH-1 is essential for the function or maintenance of differentiated cells. For example, because COH-1 is present on chromosomes of all somatic cells, it may regulate general gene expression by affecting the chromatin organization. Another possibility may be that COH-1 is involved in DNA repair, as has been suggested for fission yeast Rad21 (Tatebayashi et al., 1998).
The majority of organisms examined so far have two Scc1/Rad21-related proteins, one of which is a member of the Scc1/Rad21 subfamily for mitosis and the other is a member of the Rec8 subfamily for meiosis (Parisi et al., 1999). C. elegans and Arabidopsis are the only exceptions thus far reported to have multiple Scc1/Rad21 subfamily members (Bai et al., 1999; Dong et al., 2001; Pasierbek et al., 2001). Arabidopsis has at least three Scc1/Rad21-related proteins. One of them, SYN1, has been suggested to be the ortholog of Rec8 (Bai et al., 1999). SYN2 and SYN3, whose function remains unspecified, are known to be transcribed in tissues undergoing mitosis, although not limited to them (Dong et al., 2001). Therefore, C. elegans COH-1 may be currently the only Scc1/Rad21 family member that is implicated to have a function unrelated to chromosomal cohesion. This may sound more interesting if we consider the fact that COH-1 and SCC-1/COH-2 are the most similar two among the worm Scc1/Rad21 family members.
Two types of Scc3 subunit(s) have been identified in Xenopus and human. These subunits, named SA1 and SA2, constitute separate cohesin complexes in the cell, and it is suggested that association of cohesinSA1 and cohesinSA2 to the chromatin may be differently regulated, although their functional difference in vivo is as yet unclear (Losada et al., 2000). This apparently raises an intriguing possibility that some organisms may have acquired multiple variants for a single cohesin subunit and may carry distinct cohesin complexes, each of which plays a unique function. It is therefore of interest to see whether COH-1 fulfills its function by forming a complex with other cohesin subunits in C. elegans. The unusual characteristics of COH-1 revealed in this study seem to make further analysis of this protein promising.
Supplementary Material
Acknowledgments
We thank Yoshinori Watanabe and other members of our laboratory for helpful discussion and comments. We are grateful to Barbara Meyer for communication of results before publication and to Yuji Kohara for providing cDNA clones. Some of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the National Institutes of Health. This work was supported by a Grant-in-Aid for Specially Promoted Research (to M.Y.) and a Grant-in-Aid for Scientific Research (to A.S.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
References
- Albertson, D.G., and Thomson, J.N. (1982). The kinetochores of Caenorhabditis elegans. Chromosoma 86, 409-428. [DOI] [PubMed] [Google Scholar]
- Bai, X., Peirson, B.N., Dong, F., Xue, C., and Makaroff, C.A. (1999). Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11, 417-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S.B., Clyne, R.K., Fuchs, J., Loidl, J., Uhlmann, F., and Nasmyth, K. (2000). Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387-398. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Zachariae, W., Michaelis, C., Shevchenko, A., Mann, M., and Nasmyth, K. (1998). An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O. (2001). The making and breaking of sister chromatid cohesion. Cell 106, 137-140. [DOI] [PubMed] [Google Scholar]
- Dong, F., Cai, X., and Makaroff, C.A. (2001). Cloning and characterization of two Arabidopsis genes that belong to the RAD21/REC8 family of chromosome cohesin proteins. Gene 271, 99-108. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- Guacci, V., Koshland, D., and Strunnikov, A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K.L., and Dixon, J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262-267. [DOI] [PubMed] [Google Scholar]
- Hagstrom, K.A., Holmes, V.F., Cozzarelli, N.R., and Meyer, B.J. (2002). C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16, 729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., Waizenegger, I.C., and Peters, J.M. (2001). Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320-1323. [DOI] [PubMed] [Google Scholar]
- Hirano, T. (1999). SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 13, 11-19. [DOI] [PubMed] [Google Scholar]
- Klein, F., Mahr, P., Galova, M., Buonomo, S.B., Michaelis, C., Nairz, K., and Nasmyth, K. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 98, 91-103. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., and Orr-Weaver, T.L. (2001). The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17, 753-777. [DOI] [PubMed] [Google Scholar]
- Lee, K.K., Gruenbaum, Y., Spann, P., Liu, J., and Wilson, K.L. (2000). C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell 11, 3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Ben-Shahar, T.R., Riemer, D., Treinin, M., Spann, P., Weber, K., Fire, A., and Gruenbaum, Y. (2000). Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11, 3937-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada, A., Hirano, M., and Hirano, T. (1998). Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12, 1986-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada, A., Yokochi, T., Kobayashi, R., and Hirano, T. (2000). Identification and characterization of SA/Scc3 subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150, 405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. (2001). Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11, 171-176. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35-45. [DOI] [PubMed] [Google Scholar]
- Miller, D.M., and Shakes, D.C. (1995). Immunofluorescence microscopy. Methods Cell Biol. 48, 365-394. [PubMed] [Google Scholar]
- Newport, J. (1987). Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell 48, 205-217. [DOI] [PubMed] [Google Scholar]
- Parisi, S., McKay, M.J., Molnar, M., Thompson, M.A., van der Spek, P.J., van Drunen-Schoenmaker, E., Kanaar, R., Lehmann, E., Hoeijmakers, J.H., and Kohli, J. (1999). Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19, 3515-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek, P., Jantsch, M., Melcher, M., Schleiffer, A., Schweizer, D., and Loidl, J. (2001). A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15, 1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomos, M.F., Badrinath, A., Pasierbek, P., Livingstone, D., White, J., Glotzer, M., and Nasmyth, K. (2001). Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11, 1825-1835. [DOI] [PubMed] [Google Scholar]
- Sonoda, E., et al. (2001). Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell. 1, 759-770. [DOI] [PubMed] [Google Scholar]
- Sulston, J.E., and Horvitz, H.R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. [DOI] [PubMed] [Google Scholar]
- Tatebayashi, K., Kato, J., and Ikeda, H. (1998). Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148, 49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga, et al. (2000). Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14, 2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., and Nasmyth, K. (1999). Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., Lottspeich, F., and Nasmyth, K. (1999). Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37-42. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I.C., Hauf, S., Meinke, A., and Peters, J.M. (2000). Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399-410. [DOI] [PubMed] [Google Scholar]
- Warren, W.D., et al. (2000). The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 10, 1463-1466. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and Nurse, P. (1999). Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400, 461-464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.