Abstract
Secondary antibody responses are characterized by the rapid kinetics of the responding cells, including the production of larger amounts of serum Ig compared with the primary response. Memory B cells, which are responsible for this phenomenon, undergo greater proliferation and differentiation into Ig-secreting plasma cells than naïve B cells. We have found that memory cells rapidly enter cell division, irrespective of extrinsic stimuli. Microarray analysis of human splenic B cells revealed that naïve cells express higher levels than memory B cells of Krüppel-like factor (KLF) 4, KLF9, and promyelocytic leukemia zinc finger (PLZF), transcription factors important in maintaining cellular quiescence. These genes were down-regulated after activation through CD40 and the B cell receptor. Enforced expression of KLF4, KLF9, and PLZF in memory B cells delayed their entry into division and reduced the number of proliferating cells, such that the behavior of transfected memory cells resembled that of naïve B cells. Thus, the accelerated response of memory B cells correlates with reduced expression of KLF4, KLF9, and PLZF and the subsequent regulatory effects they exert on the cell cycle.
Keywords: human B cells, immunological memory, proliferation, cellular quiescence, lymphocyte activation
A primary Ab response is initiated when naïve B cells initially encounter foreign antigen (Ag). Two distinct populations of effector B cells arise from a germinal center during a primary response: plasma cells (PCs), which secrete neutralizing Ab, and memory cells, a reservoir of Ag-specific B cells, which mediate long-term serological immunity (1–3). This phenomenon is achieved by the ability of memory B cells to be rapidly activated after subsequent exposure to the immunizing Ag and their persistence in the absence of an ongoing immune response (2–6). These qualities facilitate the production of abundant quantities of Ag-specific PCs with enhanced kinetics compared with the primary immune response.
Several factors contribute to the rapid activation of memory B cells during secondary responses. First, B cells with increased Ag affinity are selected into the memory cell pool (3), and a greater number of these cells persist than corresponding naïve cells (5). Second, the localization of memory B cells near Ag-draining sites (2, 4, 7, 8) may facilitate earlier contact with Ag than naïve B cells. Third, memory B cells express higher levels of costimulatory molecules than naïve B cells, allowing them to elicit T cell help more rapidly (2, 4, 8). On the other hand, differences in primary and secondary Ab responses may result from qualitatively distinct signals elicited through the cytoplasmic domains of IgM and IgG B cell receptors (BcRs) (9, 10). Despite these physical, phenotypic, and biochemical differences between naïve and memory B cells, in vitro studies revealed that memory cell responses mirror those of in vivo secondary responses, with them proliferating and differentiating into PCs more efficiently than naïve cells to various stimuli (11–13). Consequently, these signaling differences do not fully explain the superior characteristics of memory B cell responses.
To investigate the mechanism(s) underlying enhanced secondary Ab responses in more detail, differences in responses of human splenic naïve and memory B cell subsets to defined stimuli in vitro, and differences in gene expression by these cells ex vivo, were investigated. Both IgM memory and isotype-switched memory cells entered their first division earlier than naïve cells, regardless of the type or dose of stimulus used, suggesting that intrinsic differences between resting naïve and memory B cells control their ability to enter division at different times. Gene expression profiles revealed that memory cells have down-regulated expression of the cell-cycle regulatory genes Krüppel-like factor (KLF) 4, KLF9, and promyelocytic leukemia zinc finger (PLZF). Reduced expression of these genes in quiescent memory B cells may allow them to enter division earlier and proliferate more rapidly than naïve cells, thus dominating the secondary immune response. Indeed, ectopic expression of KLF4 and KLF9 reduced the number of B cells being recruited into division and delayed their entry into division. Importantly, the effect of enforced expression of KLFs was substantially greater in memory than naïve B cells, consistent with reduced expression of these negative cell-cycle regulators in memory B cells. Thus, memory B cells appear to undergo a rewiring process such that their activation threshold is significantly reduced compared with naïve B cells, allowing them to enter division more quickly and produce a greater Ab response.
Results
Memory B Cells Enter Division Earlier Than Naïve B Cells Irrespective of Stimulus.
Various extrinsic factors can contribute to the elevated secondary, compared with primary, Ab response. To investigate whether such factors are wholly responsible for robust secondary responses, or whether the kinetics of naïve and memory B cell responses also are regulated by intrinsic properties in vitro, assays in which the time taken by B cells to enter their first division [time to first division (ttfd)] (13, 14) were utilized. The advantage of this system is that the behavior of ex vivo-isolated naïve and memory B cells can be tested under controlled conditions, independent of in vivo microenvironmental cues that may influence the final outcome of Ag stimulation.
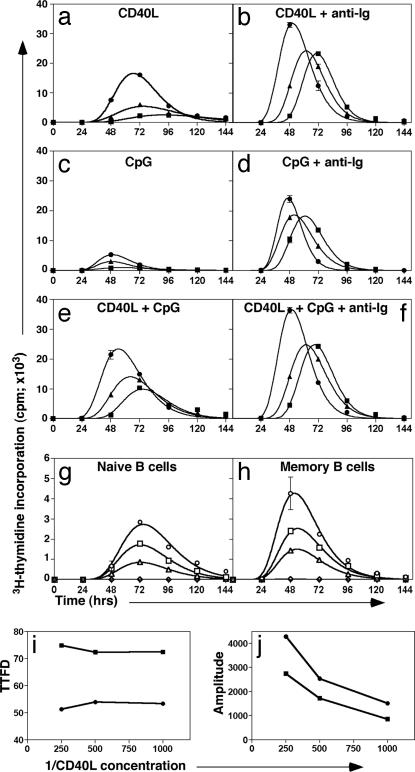
Different combinations of CD40L, CpG, and F(ab′)2 fragments of anti-Ig were used as stimuli to determine whether division kinetics of naïve, IgM-expressing, and isotype-switched memory cells can be distinguished on the basis of differential requirements for T cell help, BcR, and/or Toll-like receptor signaling. In the presence of CD40L alone, both memory subsets entered division ≈15–25 h earlier than naïve cells did, as shown by the ttfd (taken as the mean of the fitted curve; Fig. 1a and Table 1). Comparison of the amplitudes of the curves also revealed that ≈2- to 8-fold more memory cells were recruited into division than naïve cells (Fig. 1a and Table 1). F(ab′)2 anti-Ig was only weakly mitogenic when used alone. However, when combined with CD40L, it substantially reduced the ttfd of naïve B cells compared with cells stimulated with CD40L alone and increased the relative number of naïve cells entering division by ≈10-fold (Fig. 1b and Table 1). Thus, the ttfd of naïve B cells stimulated with CD40L and anti-Ig approximated that of memory cells stimulated with CD40L alone (compare Fig. 1 a and b), demonstrating the reduced requirements for activation of memory cells over naïve cells. Anti-Ig also shortened the ttfd of CD40L-stimulated IgM+ memory and isotype-switched memory B cells, thus their entry into division still was significantly earlier than naïve cells.
Fig. 1.
IgM and switched memory B cells enter division earlier than naïve B cells do regardless of the type or doses of stimulus used. (a–f) Sort-purified naïve (■), IgM memory (▴), and switched memory (●) B cells were cultured (1.5 × 105 per ml) with CD40L alone (a), CD40L plus F(ab′)2 fragments of anti-Ig (10 μg/ml) (b), CpG (1 μg/ml) (c), CpG plus anti-Ig (d), CD40L and CpG (e), or CD40L, CpG, and anti-Ig (f). Proliferation was assessed every 24 h by determining incorporation of [3H]thymidine during a 4-h pulse. The mitotic inhibitor demecolcine (10 ng/ml) was added at the onset of culture to measure entry of the cells into their first S phase and therefore ttfd. The center of the curve is the mean ttfd; the height is proportional to the number of cells entering division. Values represent the mean ± SEM of triplicate cultures. (g–j) Sort-purified naïve (g) and total memory (h) B cells were cultured (2.5 × 105 per ml) with 2-fold dilutions of CD40L (○, 1/250; □, 1/500; ▵, 1/1,000; ◇, no CD40L). The ttfd (i) and amplitude (j) of the responses of naïve (■) and memory (●) B cells were calculated. These results are representative of two or more independent experiments performed with B cells from different spleens.
Table 1.
ttfd analysis of naïve and memory B cell subsets
| ttfd, h |
Amplitude, cpm × 103 |
|||||
|---|---|---|---|---|---|---|
| Naïve | IgM memory | Switched memory | Naïve | IgM memory | Switched memory | |
| CD40L | 92 ± 1.9 | 75 ± 2.0 | 67 ± 0.6 | 2.55 ± 0.13 | 5.4 ± 0.36 | 16.6 ± 0.5 |
| CD40L/anti-Ig | 71 ± 0.4 | 62 ± 0.2 | 50 ± 1.6 | 23.4 ± 0.3 | 24.2 ± 0.66 | 33.5 ± 1.1 |
| CpG | 57 ± 3.8 | 49 ± 1.9 | 49 ± 1.1 | 0.9 ± 0.1 | 3.0 ± 0.01 | 5.3 ± 0.1 |
| CpG/anti-Ig | 61 ± 0.3 | 52 ± 0.8 | 47 ± 2.6 | 18.3 ± 0.8 | 18.6 ± 0.5 | 24.2 ± 0.9 |
| CD40L/CpG | 75 ± 1.6 | 64 ± 0.6 | 54 ± 0.9 | 9.9 ± 0.6 | 14.0 ± 0.45 | 23.4 ± 0.8 |
| CD40L/CpG/anti-Ig | 69 ± 0.3 | 62 ± 0.2 | 50 ± 1.0 | 24.8 ± 0.4 | 24.9 ± 0.4 | 36.6 ± 0.6 |
B cell subsets were cultured with different combinations of CD40L, F(ab′)2 fragments of anti-Ig, and/or CpG. The ttfd and amplitude of the response were calculated as described in Fig. 1. Values represent the mean ± SEM of triplicate cultures. Similar results were obtained in several independent experiments with different donors.
The ttfd of B cells after activation with a T cell-independent stimulus also was investigated. As occurred in T cell-dependent responses, memory B cells entered division earlier than naïve B cells did in response to CpG (Fig. 1c and Table 1). The amplitude of the response of naïve and both memory subsets to CpG stimulation was increased by costimulation through the BcR or CD40, whereas the response of cells to CD40L, CpG, and anti-Ig resembled that induced by CD40L/anti-Ig (Fig. 1 d–f and Table 1). In all cases, the memory B cell response exceeded that of corresponding naïve B cells.
Naïve and Memory B Cell Responses Are Regulated by Intrinsic Differences.
To determine whether memory B cells exhibit greater sensitivity to stimuli than naïve B cells do, these cells were cultured with serial dilutions of CD40L. Surprisingly, lowering the dose of CD40L did not reduce the ttfd of naïve or memory (Fig. 1 g–j) B cells. Rather, it decreased the relative number of cells recruited into division, as revealed by a lower amplitude of the response (Fig. 1 g–j). Furthermore, the fold difference in amplitude between naïve and memory cells remained constant across the concentration range examined (Fig. 1j), suggesting that entry into division is not regulated by extrinsic signals but by preexisting cellular physiology. Therefore, memory B cells differ fundamentally in their intrinsic capacity to enter division earlier than naïve B cells, independent of the strength or type of extrinsic stimulatory signal.
Gene Expression Analysis of Human Splenic B Cell Subsets.
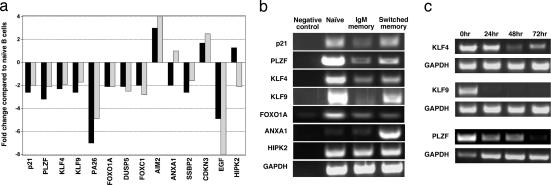
Microarray analysis was performed to identify genes capable of heightening memory B cell responses. The results indicated that many genes involved in the negative regulation of the cell cycle were differentially expressed between naïve and memory B cells (Fig. 2a). Specifically, genes that inhibit the G1/S phase transition were higher in naïve than memory cells, suggesting the cell cycle of naïve B cells is under tighter regulatory control than that of memory B cells. These genes included PLZF, p21, KLF4, KLF9, and the forkhead transcription factor FOXO1A (15–19). The expression differences for these genes, and ANXA1, were confirmed by semiquantitative (sq) PCR (Fig. 2b). This analysis revealed that KLF4, KLF9, and PLZF were expressed at ≈10-, 8-, and 2- to 4-fold higher levels in naïve B cells than in memory B cells [Fig. 2b and supporting information (SI) Fig. 5]. The greater differences in the levels of expression of these genes as revealed by sqPCR compared with the microarray data are consistent with increased sensitivity of sqPCR (20). Assessment of expression of these proteins in B cell subsets also was attempted; however, the sensitivity and specificity of available reagents was insufficient to demonstrate confidently increased levels in naïve B cells. Despite this limitation, we propose that the ability of memory B cells to respond with greater and accelerated kinetics than naïve B cells results from small changes in expression of a suite of genes involved in maintaining B cells in a quiescent state.
Fig. 2.
Differential expression of genes involved in the negative regulation of the cell cycle. (a) Data generated from naïve and memory B cell GeneChips was mined for genes differentially expressed by >2-fold. Genes with distinct roles in cell-cycle regulation and that differ in expression between memory and naïve B cells are shown. The results are presented as fold change for IgM memory (black bars) and switched memory (gray bars) B cells relative to naïve B cells. (b) Differential expression of genes involved in inhibiting the cell cycle, as revealed by microarray analysis, was confirmed by sqPCR using GAPDH as a standardizing control. (c) Naïve B cells were cultured with CD40L and anti-Ig for 72 h. Cells were harvested every 24 h, and expression of KLF4, KLF9, or PLZF was determined by sqPCR.
We chose to focus our investigation on the KLFs, because members of this family have important roles in maintaining quiescence of hemopoietic (21–24) and nonhemopoietic cells (25). Furthermore, the role of KLFs in human B cells has not been studied. We investigated whether signaling through CD40 and BcR modulates expression of these genes. Naïve B cells were stimulated with CD40L/anti-Ig for 3 days, and expression of KLF4, KLF9, and PLZF was determined every 24 h (Fig. 2c). KLF4 and PLZF were reduced after 24 h and remained at low but detectable levels for 72 h. This finding is consistent with reduced expression of KLF4 in activated murine B cells (22, 26). In contrast, KLF9 expression was extinguished within 24 h of activation (Fig. 2c). Therefore, expression of these genes can be down-modulated by stimulation through CD40 and the BcR, which then would allow cell division to occur.
The Number of Proliferating Cells Is Reduced When B Cells Overexpress KLF4/KLF9/PLZF.
The relationship between increased expression of cell-cycle regulators in naïve B cells and the reduced proliferation of these cells compared with memory B cells was examined by transfecting B cells with candidate genes. Efficient infection of human B cells with lentiviruses is achieved only when the cells are induced to proliferate (27), which precludes assessment of ttfd. Therefore, chimeric proteins containing the gene of interest fused to GFP were transiently transfected into resting B cells, and GFP+ cells subsequently were sorted for further analysis.
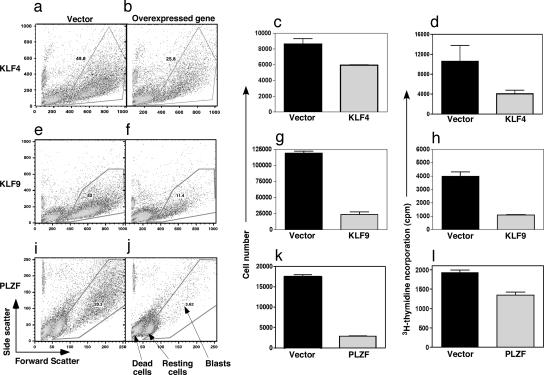
Initial studies demonstrated that overexpression of KLF4, KLF9, and PLZF modulated proliferation of activated B cells. Ectopic expression of KLF4, KLF9, and PLZF dramatically decreased the percentage of cells undergoing blastogenesis in response to CD40L (≈50–80%; Fig. 3 a, b, e, f, i, and j). Enumeration of viable cells also confirmed this decrease (Fig. 3 c, g, and k). It is important to note that the number of dead cells does not increase in these cultures. Instead, overexpression of these genes maintains B cells in a resting state, as shown by the presence of a population of nonblasting but viable cells (Fig. 3).
Fig. 3.
Cell-cycle regulators inhibit B cell blastogenesis. B cells expressing GFP after nucleofection with plasmids encoding KLF4 (a–d), KLF9 (e–h), PLZF (i–l), or the vector alone were cultured in duplicate with CD40L. Responses were assessed after 4–5 days by determining scatter profiles (a, b, e, f, i, and j), cell number (c, g, and k), and [3H]thymidine incorporation during the final 18 h (d, h, and l). Data are the mean ± SEM. and represent two or more experiments.
The reduction in the number of blasting cells and proliferation were confirmed by [3H]thymidine uptake by CD40L-stimulated transfected B cells. Proliferation of B cells overexpressing KLF4, KLF9, and PLZF was dramatically reduced compared with that of control-transfected B cells (Fig. 3 d, h, and l). The reduction in proliferation was examined further by determining the frequency of cells incorporating BrdU during in vitro activation. This approach allowed dead and nonblasting cells to be excluded from the analysis. Only small differences in the uptake of BrdU were observed by naïve or memory B cell blasts transfected with the candidate genes versus the control vector (SI Table 2). Thus, although the frequency of lymphoblasts was reduced by overexpression, comparable frequencies of these cells continued to enter S phase. Therefore, the reduction in cell number and [3H]thymidine incorporation resulted from a decrease in the number of cells becoming activated to enter division and/or their time of entry into the first division.
ttfd Analysis of B Cells Overexpressing Cell-Cycle Regulators.
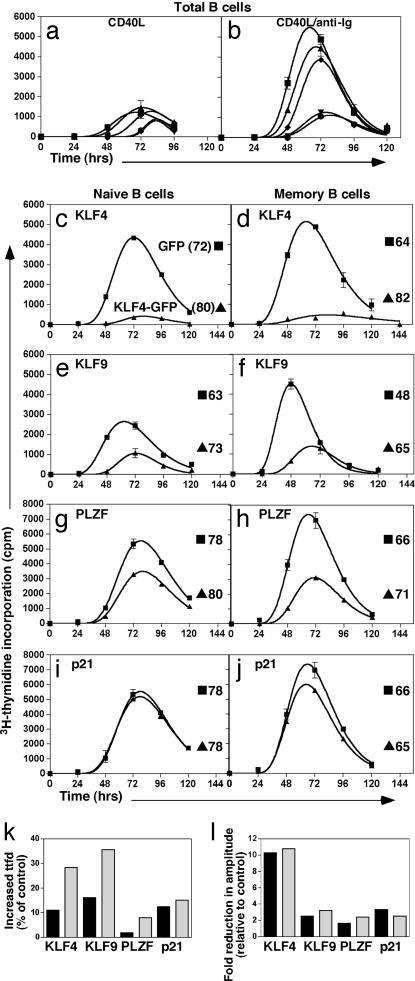
To investigate whether KLF4, KLF9, or PLZF indeed were able to affect entry of B cells into their first division, ttfd analysis was performed (Fig. 4). As the candidate genes can regulate p21 transcription, the effect of p21 on ttfd also was examined. In response to CD40L, overexpression of KLF4 or KLF9 caused a small reduction in the proportion of cells entering division (i.e., amplitude) compared with the control vector (Fig. 4a). There was no difference in the proportion of cells entering division between the control and cells transfected with either p21 or PLZF. In contrast, ttfd of cells transfected with any of the candidate genes was increased compared with the control vector. p21 had the smallest effect when compared with the vector control, increasing ttfd by ≈5 h. PLZF delayed the entry of CD40L-stimulated B cells by ≈11 h, whereas KLF4 and KLF9 had the greatest effect, increasing ttfd by 15 h (Fig. 4a).
Fig. 4.
ttfd analysis of subsets of B cells transfected with genes differentially expressed by naïve and memory B cells. (a and b) B cells were transfected with plasmids encoding KLF4-GFP (▾), KLF9-GFP (●), PLZF-GFP (◆), p21-GFP (▴), or GFP alone (■). GFP+ cells were isolated and then cultured with CD40L alone (a) or CD40L plus anti-Ig (b) in the presence of demecolcine. ttfd was determined as described for Fig. 1. (c–j) Transfected naïve (■; Left) and memory (■; Right) B cells expressing KLF4-GFP (c and d), KLF9-GFP (e and f), PLZF-GFP (g and h), p21-GFP (i and j), or GFP alone (▴) were isolated and cultured with CD40L and anti-Ig in the presence of demecolcine. ttfd was determined as described for Fig. 1. The values in c–j represent the ttfd for transfected B cells. (k) The relative effect (% change compared with control cultures) of overexpressing KLF4, KLF9, and PLZF on ttfd of naïve (black bars) and memory B cells (gray bars). (l) The fold change in amplitude of the response of naïve (black bars) and memory (gray bars) B cells overexpressing KLF4, KLF9, or PLZF relative to naïve and memory cells transfected with the vector control. Similar results were obtained in at least two independent experiments.
A more dramatic decrease in recruitment of cells into division occurred when transfected B cells were stimulated with CD40L/anti-Ig (Fig. 4b). KLF4 and KLF9 diminished the amplitude such that the height of the curve was ≈20% of control transfectants. Unlike CD40L-stimulated cells, the amplitude of p21-transfected B cells was slightly lower than that of control cultures. The proportion of PLZF-transfected B cells entering division was equal to ≈70% of the control. The effect on ttfd of transfected B cells stimulated with CD40L/anti-Ig was similar to CD40L-stimulated cells, with p21, PLZF, KLF4, and KLF9 increasing ttfd by ≈5, 9, 12, and 15 h, respectively, compared with the vector control.
Overexpression of KLFs Preferentially Delays the Entry into Division of Memory B Cells.
The outcome of overexpressing KLF4, KLF9, and PLZF on the proliferation of naïve versus memory B cells was next examined (Fig. 4 c–j). The combined stimulus of CD40L and anti-Ig was used because it mimics a T cell-dependent response in vivo. Overexpression of KLF4 dramatically reduced the amplitude of both subsets by ≈10-fold (Fig. 4 c, d, and l). Strikingly, overexpression of KLF4 affected the kinetics of the memory B cell response substantially more than that of naïve cells, with a ≈10% increase in ttfd of naïve B cells but ≈30% increase for memory B cells (Fig. 4k). Overexpression of KLF9 or PLZF also reduced the recruitment of naïve and memory B cells into division, albeit to a lesser extent than KLF4 (≈2- to 3-fold), indicating that modulation of this parameter was not subset-specific (Fig. 4 k and l). However, the difference in ttfd between B cells transfected to express KLF9 or PLZF and control-transfected cells was greater for memory than naïve cells (Fig. 4 e–h and k). Therefore, overexpression of KLF4, KLF9, and PLZF had a greater effect on ttfd of memory B cells compared with naïve B cells. Overexpression of p21 weakly modulated the kinetics of B cell responses. Thus, the KLF members may regulate entry of B cells into division via a p21-independent mechanism.
Discussion
Immunological memory is characterized by a rapid secondary response, which can produce up to 100-fold more Ag-specific B cells compared with the primary response (5). Various factors that may play a role in augmenting secondary responses have been investigated (4, 9, 10). However, despite the findings of these studies, the mechanisms that control this multifaceted response remain incompletely defined. Therefore, we set out to identify molecules that have critical roles in memory responses.
Human memory B cells enter division earlier than naïve B cells do when stimulated with CD40L in the absence or presence of different cytokines (13, 14). However, once in division, both subsets proliferate comparably (13), suggesting that earlier division entry is the key to why memory cells proliferate to a greater extent than naïve cells. To confirm that this result is attributable to intrinsic controls, combinations of CD40L, anti-Ig, and/or CpG were used to induce B cells into division (Fig. 1). The ttfd of naïve cells could be altered by combinations of different stimuli. In contrast, the ttfd of memory B cells was less susceptible to modulation, yet they always entered division earlier than naïve B cells did. Furthermore, when the B cell subsets were stimulated with titrations of CD40L (Fig. 1), memory cells retained their ability to enter division earlier than naïve B cells. Therefore, although extrinsic factors can direct certain aspects of B cell responses, and preferentially stimulate one subset over another (13, 14, 28–31), memory B cells fundamentally differ in physiology from naïve cells, which facilitates their earlier entry into division in response to the same stimuli.
A supposition of previous studies that investigated enhanced responses of murine memory versus naïve B cells was that naïve B cells are IgM+ and memory B cells are IgG+ (9, 10). Thus, the proposal that differences in responses of naïve and memory B cells result from differential signaling through IgM and IgG, respectively, is inconsistent with the observation that human IgM+ memory B cells proliferate with faster kinetics than IgM+ naïve B cells (Fig. 1) (13, 14). Furthermore, stimulation with CD40L and/or CpG was capable of inducing both memory B cell subsets to undergo a more robust response than naïve B cells, without the need for costimulation through the BcR (Fig. 1). Therefore, the enhanced proliferative ability of memory B cells is largely independent of Ig isotype (9, 10).
Microarray analysis was used to determine the components of memory B cells that would allow more dynamic responses to stimulation compared with naïve cells. Molecules important for maintaining cell quiescence and controlling cell division (FOXO1A, PLZF, KLF4, and KLF9) were down-regulated in memory B cells, thus identifying them as possible candidates for the disparity between the proliferation of naïve and memory B cells (Fig. 2). Indeed, overexpression of KLF4, KLF9, or PLZF delayed entry of memory B cells into division such that their responses resembled those of naïve B cells transfected with the control vector. Furthermore, overexpression of KLF4, KLF9, and PLZF had a greater effect on ttfd of memory rather than naïve B cells, consistent with their reduced expression in memory B cells (Fig. 4). Therefore, we postulate that the accumulation of small modifications in the expression of a number of genes that maintain cellular quiescence allows memory B cells to enter division earlier than naïve B cells. A recent study demonstrated that, rather than causing an increase in a suite of genes involved in promoting B cell activation, engagement of an IgG BcR on murine B cells down-regulated a greater number of genes than observed after engagement of an IgM BcR (26). This finding led the authors to propose a “less-is-more” hypothesis, whereby IgG-expressing memory B cells are rapidly activated by virtue of the expression of fewer genes that may potentially prevent their differentiation into PC. Our finding that expression of genes that attenuate B cell proliferation is reduced in memory compared with naïve B cells is consistent with a less-is-more concept. However, an important difference is that many of the genes we observed to be decreased in switched memory B cells relative to naïve B cells also were reduced in IgM+ memory B cells, which suggests that signaling downstream of receptors other than the BcR (e.g., CD40, Toll-like receptors, etc.) contributes to enhanced responses of memory B cells and thus secondary Ab responses. This difference notwithstanding, decreased expression of KLFs and PLZF in memory B cells provides a mechanistic explanation for the hypothesis proposed by Goodnow and colleagues (26).
Differential expression of cell-cycle regulators has previously been documented during discrete stages of lymphocyte differentiation. By comparing a mixed population of germinal center and memory (i.e., IgDlo) B cells to naïve B cells, Wagner et al. (32) showed that p27 decreased and cyclin D3 increased more rapidly in activated germinal center/memory B cells compared with naïve B cells, correlating with greater proliferation of IgDlo cells in vitro. On the other hand, expression of the cell-cycle inhibitor p18 is important for differentiation of plasmablasts into nondividing PCs (33). Thus, PC and memory B cells differ in their patterns of expression of cell-cycle regulatory genes, with up-regulation of inhibitors during terminal differentiation, whereas memory cells down-regulate these genes to allow rapid proliferation in response to specific Ag. Furthermore, expression of KLF4 is rapidly down-regulated in murine B cells after stimulation through the BcR (22, 26), thus allowing Ag-primed cells to become activated more rapidly than unprimed B cells. Similarly, overexpression of KLF4 can induce cell-cycle arrest in murine leukemic pre-B cells (23). Additional studies have revealed roles for cell-cycle regulators during T cell differentiation. For instance, memory CD8+ T cells express higher levels of cyclins than naïve T cells do (34), whereas the level of p27 expression in CD4+ T cells establishes a threshold for inducing cell division after stimulation (35). Together with our results, it appears that the differential expression of cell-cycle regulators in naïve, and memory B cells set a threshold that extrinsic stimulation must overcome before cells can enter division. These studies of memory T and B cells demonstrate that regulation of the cell cycle is an important mechanism underlying their rapid responses to Ag.
It could be postulated that KLFs modulate the cell cycle by regulating the levels of p21. However, p21 did not alter proliferation to the same extent as KLFs or PLZF. Thus, although KLF4, KLF9, and PLZF are p21 transcriptional regulators (16, 17, 36), they may exert their effects on naïve B cells through a p21-independent mechanism. Consistent with this finding, KLF4 can inhibit transcription of cyclin D1 (37). Interestingly, activation of NF-κB induces cyclin D1 transcription and cell division (38). Furthermore, c-Rel and cyclin D1 are expressed at higher levels in memory B cells compared with naïve cells (data not shown). Thus, it is possible that KLF4 and KLF9 interact with the NF-κB pathway downstream of CD40 and BcR signaling to regulate B cell proliferation.
The memory B cell pool generated during a primary response is important for long-lasting humoral immunity. The dynamics of primary and secondary responses appear to result from reduced expression of cell-cycle regulators in memory B cells relative to naïve B cells. This allows memory cells, irrespective of Ig isotype, to enter the cell cycle more rapidly than naïve cells do, thereby providing an accelerated response to previously encountered Ag. This reprogramming of memory B cells occurs after activation and affinity selection to minimize the restraints on these cells with respect to their ability to enter division and receive stimulation from helper T cells. Collectively, these findings provide a framework for examining how dysregulation of cell-cycle-related mechanisms during B cell differentiation cause perturbed humoral immune responses, such as immunodeficiencies, autoimmunity, and B cell malignancies.
Materials and Methods
Reagents.
Ab are listed in SI Table 3. CpG 2006 and CpG C274 (12) were obtained from Sigma Genosys (Sydney, Australia). Recombinant CD40L has been described previously (13, 14).
Isolation of Human B Cells.
B cells were isolated from human spleens and fractionated into subsets of naïve, IgM memory, and isotype-switched memory cells by cell sorting (13, 14) (FACS Star+, Vantage, or Aria flow cytometer; Immunohistochemistry Systems from BD Biosciences, San Jose, CA). Institutional human ethics review committees approved all studies.
Immunofluorescence Staining.
Cells were labeled with mAbs as described in refs. 13 and 14. For intracellular staining, fixed cells were permeabilized in PBS/0.13% Tween-20 (13) and then stained with Abs. Data were acquired on a FACSCalibur and analyzed with FlowJo (Tree Star, Ashland, OR).
B Cell Proliferation and ttfd Assay.
B cell subsets were cultured with CD40L (13, 14), CpG (1 μg/ml), and/or anti-Ig (10 μg/ml). To determine ttfd, the mitotic inhibitor demecolcine (10 ng/ml; Sigma–Aldrich, Castle Hill, Australia) was added. Proliferation was measured by determining incorporation of [3H]thymidine [1 μCi/ml per well (1 Ci = 37 GBq); ICN Biomedicals, Irvine, CA]. Log normal distributions were fitted to the data by using Prism (GraphPad Software, San Diego, CA) (13, 14). The absolute number of cells was determined by adding CaliBRITE beads (BD Biosciences) to each well before harvest. BrdU incorporation was assessed as described in ref. 13.
Gene Expression Analysis.
RNA was isolated from each B cell subset (RNeasy kit; Qiagen, Doncaster, Australia), and complementary RNA was synthesized (14, 39). Biotin-labeled cRNA was hybridized to Human Genome U133 Set GeneChips (Affymetrix, Santa Clara, CA), and resulting data were analyzed by Affymetrix Microarray Suite software. Gene expression profiles were performed in duplicate from different donors.
sqPCR, Cloning, and Transfection.
Primer sequences are shown in SI Table 4 (Sigma Genosys). RNA was transcribed into cDNA, normalized for expression of GAPDH, and sqPCR analysis was performed. Primers for cloning included appropriate restriction enzyme sites. cDNA sequences were amplified with Pfu (1 unit; PerkinElmer, Melbourne, Australia) or Hi-Fidelity Taq (1 unit; Invitrogen, Carlsbad, CA) and subcloned into the pEGFP vector (BD) to generate fusion proteins with C-terminal GFP. Isolated B cells were transiently transfected (5–10 × 107 cells per ml) with 1–5 μg of plasmid by using a Nucleofector and the Human B Cell Nucleofector Kit (Amaxa Biosystems, Gaithersburg, MD). Cells were incubated in media for 10–12 h; CD20+CD27−GFP+ or CD20+CD27+GFP+ cells then were isolated by sorting. Efficiency of transfection ranged from 5% to 30% depending on the plasmid being used, and the transfection efficiency of naïve and memory B cells was comparable. Only transfected cells were sorted, thus obviating any effect that may be caused by residual nontransfected cells.
Supplementary Material
Acknowledgments
We thank Tony Basten for critical review of this manuscript, the Australian Red Cross Blood Service, the FACS facilities at Garvan and Centenary Institutes, and the Garvan Institute Microarray facility. This work was supported by the National Health and Medical Research Council (NHMRC) of Australia and Cancer Institute NSW. K.L.G. received a Postgraduate Research Award from the University of Sydney and a Cancer Institute NSW Research Scholar Award. S.G.T. is the recipient of a Career Development Award from NHMRC.
Abbreviations
- PC
plasma cell
- BcR
B cell receptor
- KLF
Krüppel-like factor
- PLZF
promyelocytic leukemia zinc finger
- ttfd
time to first division
- sq
semiquantitative
- Ag
antigen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703872104/DC1.
References
- 1.Lanzavecchia A. Nature. 1985;314:537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 2.Liu YJ, Banchereau J. Immunologist. 1996;4:55–66. [Google Scholar]
- 3.McHeyzer-Williams LJ, McHeyzer-Williams MG. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 4.Liu YJ, Barthelemy C, de Bouteiller O, Arpin C, Banchereau J. Immunity. 1995;2:239–248. doi: 10.1016/1074-7613(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 5.McHeyzer-Williams LJ, Cool M, McHeyzer-Williams MG. J Exp Med. 2000;191:1149–1166. doi: 10.1084/jem.191.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maruyama M, Lam KP, Rajewsky K. Nature. 2000;407:636–642. doi: 10.1038/35036600. [DOI] [PubMed] [Google Scholar]
- 7.Liu YJ, Oldfield S, MacLennan IC. Eur J Immunol. 1988;18:355–362. doi: 10.1002/eji.1830180306. [DOI] [PubMed] [Google Scholar]
- 8.Tangye SG, Liu YJ, Aversa G, Phillips JH, de Vries JE. J Exp Med. 1998;188:1691–1703. doi: 10.1084/jem.188.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin SW, Goodnow CC. Nat Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 10.Wakabayashi C, Adachi T, Wienands J, Tsubata T. Science. 2002;298:2392–2395. doi: 10.1126/science.1076963. [DOI] [PubMed] [Google Scholar]
- 11.Jelinek DF, Splawski JB, Lipsky PE. J Immunol. 1986;136:83–92. [PubMed] [Google Scholar]
- 12.Bernasconi NL, Traggiai E, Lanzavecchia A. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 13.Tangye SG, Avery DT, Deenick EK, Hodgkin PD. J Immunol. 2003;170:686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- 14.Good KL, Bryant VL, Tangye SG. J Immunol. 2006;177:5236–5247. doi: 10.4049/jimmunol.177.8.5236. [DOI] [PubMed] [Google Scholar]
- 15.McConnell MJ, Chevallier N, Berkofsky-Fessler W, Giltnane JM, Malani RB, Staudt LM, Licht JD. Mol Cell Biol. 2003;23:9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, Buettner R. Mol Cell Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Yusuf I, Andersen HM, Fruman DA. J Immunol. 2006;176:2711–2721. doi: 10.4049/jimmunol.176.5.2711. [DOI] [PubMed] [Google Scholar]
- 20.Dallas PB, Gottardo NG, Firth MJ, Beesley AH, Hoffmann K, Terry PA, Freitas JR, Boag JM, Cummings AJ, Kees UR. BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 22.Fruman DA, Ferl GZ, An SS, Donahue AC, Satterthwaite AB, Witte ON. Proc Natl Acad Sci USA. 2002;99:359–364. doi: 10.1073/pnas.012605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA, Huettner CS, Fruman DA. Blood. 2007;109:747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Proc Natl Acad Sci USA. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker RJ, Boon RA, Rondaij MG, Kragt A, Volger OL, Elderkamp YW, Meijers JC, Voorberg J, Pannekoek H, Horrevoets AJ. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 26.Horikawa K, Martin SW, Pogue SL, Silver K, Peng K, Takatsu K, Goodnow CC. J Exp Med. 2007;204:759–769. doi: 10.1084/jem.20061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bovia F, Salmon P, Matthes T, Kvell K, Nguyen TH, Werner-Favre C, Barnet M, Nagy M, Leuba F, Arrighi JF, et al. Blood. 2003;101:1727–1733. doi: 10.1182/blood-2001-12-0249. [DOI] [PubMed] [Google Scholar]
- 28.Arpin C, Banchereau J, Liu YJ. J Exp Med. 1997;186:931–940. doi: 10.1084/jem.186.6.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tangye SG, Avery DT, Hodgkin PD. J Immunol. 2003;170:261–269. doi: 10.4049/jimmunol.170.1.261. [DOI] [PubMed] [Google Scholar]
- 30.Wagner EF, Hanna N, Fast LD, Kouttab N, Shank PR, Vazquez A, Sharma S. J Immunol. 2000;165:5573–5579. doi: 10.4049/jimmunol.165.10.5573. [DOI] [PubMed] [Google Scholar]
- 31.Tangye SG, Hodgkin PD. Immunology. 2004;112:509–520. doi: 10.1111/j.1365-2567.2004.01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner EF, Hleb M, Hanna N, Sharma S. J Immunol. 1998;161:1123–1131. [PubMed] [Google Scholar]
- 33.Tourigny MR, Ursini-Siegel J, Lee H, Toellner KM, Cunningham AF, Franklin DS, Ely S, Chen M, Qin XF, Xiong Y, et al. Immunity. 2002;17:179–189. doi: 10.1016/s1074-7613(02)00364-3. [DOI] [PubMed] [Google Scholar]
- 34.Kaech SM, Hemby S, Kersh E, Ahmed R. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 35.Rowell EA, Walsh MC, Wells AD. J Immunol. 2005;174:3359–3368. doi: 10.4049/jimmunol.174.6.3359. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttridge DC, Albanese C, Reuther JY, Baldwin AS., Jr Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baugh LR, Hill AA, Brown EL, Hunter CP. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.