Abstract
The Ste5 scaffold activates an associated mitogen-activated protein kinase cascade by binding through its RING-H2 domain to a Gβγ dimer (Ste4/Ste18) at the plasma membrane in a recruitment event that requires prior nuclear shuttling of Ste5. Genetic evidence suggests that Ste5 must oligomerize to function, but its impact on Ste5 function and localization is unknown. Herein, we show that oligomerization affects Ste5 activity and localization. The majority of Ste5 is monomeric, suggesting that oligomerization is tightly regulated. Increasing the pool of Ste5 oligomers increases association with Ste11. Remarkably, Ste5 oligomers are also more efficiently exported from the nucleus, retained in the cytoplasm by Ste11 and better recruited to the plasma membrane, resulting in constitutive activation of the mating mitogen-activated protein kinase cascade. Coprecipitation tests show that the RING-H2 domain is the key determinant of oligomerization. Mutational analysis suggests that the leucine-rich domain limits the accessibility of the RING-H2 domain and inhibits export and recruitment in addition to promoting Ste11 association and activation. Our results suggest that the major form of Ste5 is an inactive monomer with an inaccessible RING-H2 domain and Ste11 binding site, whereas the active form is an oligomer that is more efficiently exported and recruited and has a more accessible RING-H2 domain and Ste11 binding site.
INTRODUCTION
Many eukaryotic signal transduction pathways use cytoplasmic scaffolds to link membrane-associated signaling components such as receptors and G proteins to downstream enzymes such as kinases and phosphatases (Pawson and Scott, 1997; Burack and Shaw, 2000). The Ste5 scaffold of budding yeast is the prototype of a family of proteins that tether kinases of mitogen-activated protein kinase (MAPK) cascades in eukaryotes (Elion, 1995, 2001; Burack and Shaw, 2000; Nguyen et al., 2002; Roy et al., 2002). Ste5 assembles the mating pathway kinases Ste11 (MAPKKK), Ste7 (MAPKK), and Fus3 (MAPK) into an active signaling complex of high specific activity (Kranz et al., 1994; Choi et al., 1994; 1999) by binding the kinases through separate binding sites (Figure 1A; Choi et al., 1994; Inouye et al., 1997b). Ste5 is essential for the kinases to be activated in vivo, although they can phosphorylate one another in vitro (Errede et al., 1993; Neiman and Herskowitz, 1994; Wu et al., 1995).
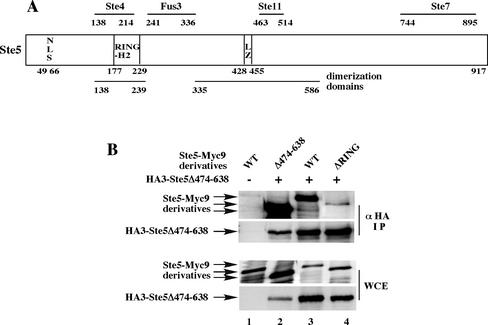
Figure 1.
The RING-H2 domain is essential for oligomerization of full-length Ste5. (A) Cartoon of Ste5 and its binding partners. LZ, leucinezipper. (B) Ste5 must have a RING-H2 domain to oligomerize efficiently. Coimmunoprecipitation of Myc- and HA-tagged derivatives of Ste5 by using extracts from ste5Δ (EY1775) cells expressing HA3-Ste5Δ474-638 (pYMW 68) with Ste5-Myc9 (pSKM19), Ste5ΔRING-Myc9 (pLS46), or Ste5Δ474-638-Myc9 (pYMW13).
A major function of Ste5 is to facilitate the activation of Ste11 by Ste20, a p21-activated kinase that is anchored to the plasma membrane through a Rho-type G protein Cdc42 (reviewed in Moskow et al., 2000; Lamson et al., 2002). Work from a variety of laboratories has led to the hypothesis that Ste5 activates Ste11 by binding to a heterotrimeric G protein at the plasma membrane and recruiting Ste11 to a pool of active Ste20 that is bound to Cdc42. On pheromone stimulation, the receptor activates a G protein by dissociating the Gα (Gpa1) subunit from the Gβγ dimer (Ste4/Ste18) (reviewed in Gustin et al., 1998). The Gβ subunit (Ste4) is then thought to bind to Ste5 (Whiteway et al., 1995; Inouye et al., 1997a, Feng et al., 1998) and to Ste20 (Leeuw et al., 1998). This recruitment event is thought to allow Ste20 to directly phosphorylate Ste11 in the Ste5 scaffold complex (Feng et al., 1998; Pryciak and Huntress, 1998; van Drogen et al., 2000).
The association between Ste5 and Ste4 is pheromone-dependent and tightly regulated. During vegetative growth, Ste5 shuttles continuously between the cytoplasm and nucleus, with a nuclear pool accumulating in G1 phase cells (Mahanty et al., 1999). In the presence of pheromone, Ste5 undergoes enhanced export from the nucleus to the cytoplasm, and the nuclear pool is recruited to plasma membrane. Ste5 rapidly accumulates at restricted cortical sites in G1 phase cells and at the projection tip at later times. A variety of experiments suggest that under physiological conditions, Ste5 must shuttle through the nucleus to be recruited to the plasma membrane and activate Fus3 (Mahanty et al., 1999). Fus3 is predominantly nuclear, whereas Ste7 and Ste11 are cytoplasmic (Mahanty et al., 1999; van Drogen et al., 2001), raising the possibility that nuclear shuttling helps Ste5 to assemble a signaling complex. Interestingly, the kinase suppressor of Ras scaffold also links MAPK cascade kinases (Raf, extracellular signal-regulated kinase kinase, extracellular signal-regulated kinase) to a membrane-linked anchor (Ras) (Nguyen et al., 2002; Roy et al., 2002) and shuttles through the nucleus (Brennan et al., 2002), suggesting that multiple aspects of Ste5 localization are conserved.
Ste5 forms dimers or higher order oligomers based on interallelic complementation, two-hybrid analysis, and coprecipitation (Yablonski et al., 1996; Feng et al., 1998). This property is shared by the JIP family of mammalian scaffolds (Yasuda et al., 1999) and may therefore have a conserved role in scaffold function. Genetic evidence argues that oligomerization is essential for Ste5 function and occurs through two domains, the RING-H2 domain and a central leucine-rich domain that spans a potential leucine zipper (Figure 1A; Yablonski et al., 1996). A demonstration of a direct role for either the RING-H2 domain or the leucine-rich domain in oligomerization is lacking. In addition, it is not known how oligomerization affects the ability of Ste5 to activate the MAPK cascade.
The relationship between oligomerization and recruitment is also unclear. It has been proposed that the association of Ste5 with Ste4 through the RING-H2 domain is a prerequisite for Ste5-Ste5 association and signaling (Inouye et al., 1997a). However, biochemical and two-hybrid analysis suggests that Ste5 oligomerization could occur constitutively (Feng et al., 1998). More recent analysis suggests that Ste5 undergoes an activating conformational change of interactions between the N- and C-terminal halves of the protein that regulates the associated kinases by allostery or alignment (Sette et al., 2000). Conformational changes could also regulate oligomerization and association with signaling components (Elion, 2001).
In this report, we analyzed how the oligomerization status of Ste5 affects nuclear shuttling, recruitment, association with signaling components, and ability to activate the MAPK cascade. Our results suggest that oligomerization of Ste5 positively regulates all of these events and that the activation of Ste5 involves a conformational switch from an inactive monomer to an active dimer.
MATERIALS AND METHODS
Strains and Plasmids
See Table 1 for a list of all plasmids and yeast strains used in this study. Yeast strains were grown in standard selective synthetic complete (SC) media. Cells with GAL1-driven genes were pregrown in medium containing 2% raffinose before induction in medium containing 2% galactose. Transformation of yeast was performed as described previously (Baker 1991) except that 20 μl of 1 M dithiothreitol was added after DNA was mixed with cells. Standard molecular biological techniques were used to construct all plasmids. All deletions and point mutations were made by site-directed polymerase chain reaction-based mutagenesis and were confirmed by sequencing at the Biopolymer Facility (Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA). Glutathione S-transferase (GST) from Schistosoma japonicum (Kaplan et al., 1997) was used for protein fusions.
Table 1.
Yeast strains and plasmids used in this study
| Strains/plasmids | Genotype/description | Source | ||
|---|---|---|---|---|
| EY699 | ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 | R. Rothstein | ||
| EY957 | bar1Δ | Elion et al. (1993) | ||
| EY1775 | bar1Δ ste5Δ::TRP1 | Choi et al. (1994) | ||
| EYL357 | msn5::HIS3 | Mahanty et al. (1999) | ||
| EY2320 | ste11Δ | Lee and Elion (1999) | ||
| EYL1808 | ste4Δ::LEU2 ste5Δ::TRP1 | Feng et al. (1998) | ||
| EYL1809 | STE11-4 ste5Δ::TRP1 far1Δhis3Δ200 lys2::FUS1-HIS3 | Feng et al. (1998) | ||
| BY4741 | leu2 ura3 his3 met15 | Research Genetics | ||
| 2468 | ste4Δ::KANR leu2 ura3 his3 met15 | Research Genetics | ||
| PSY580 | ura3-52 trp1Δ63 leu2Δ1 | Seedorf and Silver (1997) | ||
| PSY1103 | rsl1-4 ura3-52 trp1Δ63 leu2Δ1 | Seedorf and Silver (1997) | ||
| LDY375 | nsp1ts::URA3 ade2 leu2 lys1 | Nehrbass et al. (1993) | ||
| pYMW4 | ste5Δ474-487-MYC9 | 2μ | URA3 | This study |
| pYMW7 | ste5Δ474-487 | CEN | URA3 | This study |
| pYMW13 | ste5Δ474-638-MYC9 | 2μ | URA3 | This study |
| pYMW27 | TAgNLS-GFP-GFP | 2μ | URA3 | This study |
| pYMW37 | ste5L482/485A-MYC9 | 2μ | URA3 | This study |
| pYMW39 | ste5L482/485A | CEN | URA3 | This study |
| pYMW49 | ste5Δ474-487-GST | 2μ | URA3 | This study |
| pYMW50 | STE5-GST | CEN | URA3 | This study |
| pYMW57 | ste5Δ474-487-GST | CEN | URA3 | This study |
| pYMW64 | ste5L482/485A-GST | 2μ | URA3 | This study |
| pYMW66 | HA3-ste5L482/485A | 2μ | LEU2 | This study |
| pYMW68 | HA3-ste5Δ474-638 | 2μ | LEU2 | This study |
| pYMW74 | HA3-STE5-GST | 2μ | LEU2 | This study |
| pYMW77 | STE5-GST | 2μ | URA3 | This study |
| pYMW81 | TAgNLS-STE5-GST | CEN | URA3 | This study |
| pYMW82 | TAgNLS-STE5-GST | 2μ | URA3 | This study |
| pYMW96 | GAL1-HA-STE4 | CEN | LEU2 | This study |
| pYMW97 | ste5C180A-GST | 2μ | URA3 | This study |
| pYMW98 | ste5C180A-GST | CEN | URA3 | This study |
| pYMW99 | ste5ΔNLS(Δ49-66)-GST | 2μ | URA3 | This study |
| pYMW100 | ste5ΔNLS(Δ49-69)-GST | CEN | URA3 | This study |
| pYMW102 | ste5L482/485A-GST | CEN | URA3 | This study |
| pYMW106 | HA3-ste5Δ474-487 | 2μ | LEU2 | This study |
| pYMW134 | MYC3-STE5-GST | 2μ | URA3 | This study |
| pYMW138 | MYC3-STE5 | 2μ | URA3 | This study |
| pSKM19 | STE5-MYC9 | 2μ | URA3 | Mahanty et al. (1999) |
| pSKM27 | GAL1-TAgNLS-STE5 | 2μ | URA3 | Mahanty et al. (1999) |
| pSKM31 | ste5C180A-MYC9 | 2μ | URA3 | Feng et al. (1998) |
| pSKM44 | TAgNLS-STE5 | CEN | URA3 | Mahanty et al. (1999) |
| pSKM47 | TAgNLS-STE5 | 2μ | URA3 | Mahanty et al. (1999) |
| pSKM87 | HA3-STE5 | 2μ | LEU2 | Mahanty et al. (1999) |
| pSKM90 | STE5-MYC9 | 2μ | HIS3 | Mahanty et al. (1999) |
| pSKM98 | TAgNLSK128T-STE5-MYC9 | 2μ | URA3 | Mahanty et al. (1999) |
| pLS46 | ste5ΔRING-MYC9 | 2μ | HIS3 | Feng et al. (1998) |
| pYBS138 | STE5 | CEN | URA3 | Kranz et al. (1994) |
| pYEE102 | FUS3-HA5 | CEN | HIS3 | Elion et al. (1993) |
| pKC55 | GAL1-STE7-MYC | CEN | HIS3 | Choi et al. (1994) |
| pNC245 | GAL1-STE11-MYC | CEN | TRP1 | Choi et al. (1994) |
| pJB207 | FUS1-LACZ | 2μ | LEU2 | Elion et al. (1990) |
All yeast strains are MATa. The EY and EYL strains are isogenic derivatives of W303a
Pheromone Assays
Halo assays were carried out as described previously (Elion et al., 1990) by using 5 μl of 100 μM α factor for bar1 strains and 5 μl of 2 mM α factor for BAR1 strains. α factor was synthesized by Dr. C. Dahl (Biopolymer Facility, Harvard Medical School). FUS1-lacZ expression was assayed as described previously (Farley et al., 1999) after inducing cells for 90 min in 50 nM α factor. Patch mating assays were done as described previously (Elion et al., 1990).
Indirect Immunofluorescence
Ste5 localization was monitored by indirect immunofluorescence by using derivatives tagged with either the Myc epitope or GST. A Ste5-Myc9 construct containing nine tandem copies of the Myc epitope at the C terminus of Ste5 was primarily used because previous work has established that it is nearly 100% functional when expressed at native levels from its own promoter (Feng et al., 1998; Mahanty et al., 1999). Ste5-Myc9 is more functional than N- and C-terminally tagged green fluorescent protein (GFP) derivatives of Ste5 and N-terminally tagged hemagglutinin (HA) and Myc derivatives of Ste5 and has been found to be a sensitive tool for the detection of plasma membrane recruitment after short exposure to α factor (Mahanty et al., 1999). Myc3-Ste5 and Myc3-Ste5-GST derivatives were also immunolocalized; however, both proteins were significantly more difficult to detect than Ste5-Myc9 because of fewer copies of the Myc epitope. Cells were grown to mid-logarithmic phase (A600 of ∼0.6) in selective SC medium with or without exposure to 50 nM α factor for 15 min and then fixed in 5% formaldehyde for 1 h at room temperature. Indirect immunofluorescence was performed essentially as described previously (Mahanty et al., 1999), except that the primary antibody was used at higher dilution (1:500–1000). This greatly improved the ability to detect Ste5 at the plasma membrane. Cells were incubated in primary antibody (mouse anti-Myc monoclonal 9E10 ascites or anti-GST polyclonal antibody) for 1.5 h at room temperature and then incubated in secondary antibody at a dilution of 1:300 (Cy3- or fluorescein isothiocyanate-conjugated antibody) for 1 h at room temperature.
Quantitation of Localization of Ste5
Nuclear accumulation was defined as the ability to detect a stronger signal in the nucleus compared with the cytoplasm. Nuclear exclusion was defined as a reduced amount of staining in the nucleus compared with the cytoplasm. Rim staining was defined as an enriched signal at the cortex of the cell. The percentage of total cells in the population that exhibited a particular localization pattern was determined by tallying 400–700 cells from two or more transformants in at least two experiments. Standard deviations were found to be an average of 2.4% for values in the range of ∼20–97% and 0.62% for values in the range of 1–17%. Minor variations in the numbers seem to reflect variations in strain backgrounds and growth conditions. For example, greater nuclear accumulation and rim staining of Ste5-Myc9 is detected in BAR1 cells compared with bar1Δ cells, in STE5 cells compared with ste5Δ cells, and in S288C strains compared with W303 strains. Greater nuclear accumulation was also detected in cells that only express the STE5-MYC9 plasmid without a second plasmid and when cells are grown in galactose medium. Note that the msn5Δ mutation decreased the intensity of rim staining relative to that of wild type, although it was more readily detected due to a reduced cytoplasmic pool. The rsl1-4 mutation did not completely block nuclear import of either Ste5-Myc9 or Ste5-GST, because individual 2μ transformants did not always exhibit nuclear exclusion of the Ste5 fusions. The block in recruitment of Ste5-GST was most evident in cells that also displayed an obvious block in nuclear import of Ste5-GST, as indicated by partial nuclear exclusion of Ste5-GST. Ste5-GST expression was heterogeneous in nsp1ts cells, so only the most brightly staining cells were tallied. Cells were visualized with an Axioskop 2 microscope (Carl Zeiss, Thornwood, NY) linked to a digital camera (C4742-95; Hamamatsu, Bridgewater, NJ).
Coimmunoprecipitation
Whole cell extracts were made as described previously (Elion et al., 1993) by using modified H buffer containing 200 mM NaCl. Coimmunoprecipitations and immunoblots were carried out as described previously (Elion et al., 1990; Feng et al., 1998) by using 250 μg to 2 mg of whole cell protein extract, 3–5 μg of 12CA5 (α-HA) or 1–2 μg of 9E10 (α-Myc) monoclonal antibody, and 30 μl of protein A agarose beads (Sigma-Aldrich, St. Louis, MO). Immunoblots were probed with 12CA5 and 9E10 or with α-Myc and α-HA polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoreactivity was detected with horseradish peroxidase-conjugated secondary antibody (enhanced chemiluminescence; Amersham Biosciences, Piscataway, NJ). Quantitation of enhanced chemiluminescence-detected bands was done using the Scion Image 1.62c densitometry program of the public domain software NIH image (available at//rsb.info.nih.gov/nih-image/). All immunoprecipitations were done a minimum of three times using two transformants and were reproducible.
RESULTS
The RING-H2 Domain Is the Major Determinant of Oligomerization
In previous coprecipitation tests, RING-H2 domain mutations (i.e., Ste5C180A, Ste5Δ143–313) did not block oligomerization of full-length Ste5, although they blocked oligomerization of a RING-H2 domain fragment. Surprisingly, the Ste5C180A mutation increased the ability of full-length Ste5 to oligomerize, suggesting that the RING-H2 domain inhibits the ability of the leucine-rich domain to oligomerize (Feng et al., 1998). These findings were consistent with genetic evidence that Ste5 oligomerizes through both the RING-H2 domain and the leucine-rich domain (Yablonski et al., 1996; Figure 1A) and suggested that the leucine-rich domain may be more critical than the RING-H2 domain for oligomerization of full-length Ste5.
A drawback of the prior biochemical analysis of oligomerization was the use of GST-tagged derivatives of Ste5, because GST forms stable dimers and can influence the oligomerization properties of a protein (McTigue et al., 1995; Maru et al., 1996; Tudyka and Skerra, 1997; Inouye et al., 2000). We therefore reexamined Ste5 oligomerization by using derivatives that had been tagged with epitopes. We first compared the ability of wild-type Ste5 and a Ste5ΔRING-H2 mutant (i.e., Ste5Δ177-229) to oligomerize with wild-type Ste5 and were unable to readily detect Ste5 × Ste5ΔRING hetero-oligomers in a coimmunoprecipitation assay, suggesting that the RING-H2 domain is essential for oligomerization (our unpublished data). We next used as a partner a deletion mutant (Ste5Δ474-638) that lacks almost half of the leucine-rich domain, accumulates to higher steady-state levels than wild-type Ste5, and homo-oligomerizes very efficiently (Figure 1B, lane 2). Ste5Δ474-638 could efficiently hetero-oligomerize with wild-type Ste5, but not with Ste5ΔRING (Figure 1B, compare lanes 3 and 4). Ste5ΔRING oligomerized equally poorly with both wild-type Ste5 and Ste5Δ474-638 (our unpublished data), indicating that the Δ474–638 deletion does not block the ability of the leucine-rich domain to oligomerize. Thus, the RING-H2 domain is the major determinant for oligomerization of full-length Ste5 and the leucine-rich domain plays a less critical role. Furthermore, only a portion of the previously defined leucine-rich domain may be directly involved in oligomerization.
Mutations in the Leucine-rich Domain Block Nuclear Accumulation and Recruitment
Our prior analysis showed that the recruitment of Ste5 to the plasma membrane was dependent on a putative nuclear localization signal (NLS; overlapping residues 49–66) and a RING-H2 domain (residues 177–229) (Figure 1A; Mahanty et al., 1999). To further understand how Ste5 localizes to the plasma membrane, we identified new mutations in Ste5 that block its ability to accumulate in nuclei during vegetative growth and be recruited to the plasma membrane in the presence of α factor mating pheromone. Two of these mutations, Ste5Δ474-487 and Ste5L482AL485A (L482AL485A is hereafter referred to as L482/485A), overlapped the leucine-rich domain and a region implicated in Ste11 binding. Indirect immunofluorescence showed that Ste5Δ474-487-Myc9 failed to accumulate in any nuclei, whereas Ste5L482/485A-Myc9 accumulated in only a few nuclei (Figure 2A and Table 2, lines 1–3). Ste5Δ474-487-Myc9 and Ste5L482/485A-Myc9 were also excluded from a much higher percentage of nuclei compared with Ste5-Myc9 (Figure 2A, % N.E.). Consistent with previous findings, wild-type Ste5-Myc9 accumulated in nuclei of 24% of the cells, of which ∼85% were unbudded (Figure 2A and Table 2, line 1) (Mahanty et al., 1999).
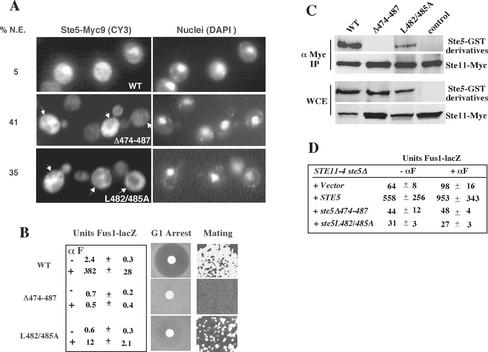
Figure 2.
Mutations in the leucine-rich domain prevent activation of Ste11 and block nuclear accumulation and recruitment of Ste5. (A) Ste5Δ474-487 and Ste5L482/485A are defective in nuclear accumulation and plasma membrane recruitment. Indirect immunofluorescence of Ste5 mutants tagged with nine copies of the Myc epitope as described in MATERIALS AND METHODS. Strains were ste5Δ (EY1775) expressing either Ste5-Myc9 (pSKM19), Ste5Δ474-487-Myc9 (pYMW4), or Ste5L482/485A-Myc9 (pYMW37) from 2μ plasmids. %N.E. is the percentage of cells in which Ste5-Myc9 derivatives seemed to be excluded from the nucleus. Nuclear exclusion (indicated by the arrows) was defined as a reduced immunofluorescence signal in the nucleus compared with the surrounding cytoplasm. (B) Quantitation of FUS1-lacZ expression, G1 arrest, and mating in STE5, ste5Δ474-487, and ste5L482/485A strains. For all assays a MATa bar1Δ ste5Δ (EY1775) strain was transformed with CEN plasmids expressing Ste5 (pYBS138), Ste5Δ474-487(pYMW7), or Ste5L482/485A (pYMW39). β-Galactosidase activity was assessed in strains that were cotransformed with a 2μ FUS1-lacZ plasmid (pJB207). (C) Ste5Δ474-487 and Ste5L482/485A do not efficiently associate with Ste11. Ste11-Myc (pNC245) was immunoprecipitated with 9E10 (anti-Myc), and immune complexes were tested for the presence of Ste5-GST (pYMW77), Ste5Δ474-487-GST (pYMW49), Ste5L482/485A-GST (pYMW64), or vector control by using anti-GST antibody. Strains were grown in 2% galactose medium for 5 h to induce expression of Ste11-Myc. (D) Ste5Δ474-487 and Ste5L482/485A fail to positively regulate STE11-4, a hyperactive version of Ste11. Quantitation of FUS1-lacZ expression in STE11-4 ste5Δ (EYL1809) cells cotransformed with CEN (YCplac22), STE5 CEN (pYBS138), ste5Δ474-487 CEN (pYMW7), or ste5L482/485A CEN (pYMW39) and FUS1-lacZ 2 μ (pJB207). For B and D, cells were induced with α factor (50 nM) for 90 min.
Table 2.
Localization of wild-type and mutant derivatives of Ste5
|
% Nuclear accumulation
|
% Rim staining
|
|||||
|---|---|---|---|---|---|---|
| Plasmids | Strainsa | Coexpressed with | Carbon sourceb | - α F | + α F | + α F |
| 1. Ste5-Myc9 | ste5Δ | Dex | 24±0.9 | 12±1.9 | ||
| 2. Ste5Δ474-487-Myc9 | ste5Δ | Dex | 0±0.6 | 0±0 | ||
| 3. Ste5L482/485-Myc9 | ste5Δ | Dex | 6±2.4 | 3±0.6 | ||
| 4. Ste5-Myc9 | STE5 | Dex | 29±4.8 | 16±2.7 | ||
| 5. Ste5Δ474-487-Myc9 | STE5 | Dex | 10±2.7 | 0±0.2 | ||
| 6. Ste5L482/485-Myc9 | STE5 | Dex | 14±2.4 | 9±2.2 | ||
| 7. Ste5-Myc9 | ste11Δ | Dex | 21±1.6 | |||
| 8. Ste5-Myc9 | ste5Δ | Vector | Dex | 18±6.4 | 8±3.1 | 17±5.0 |
| 9. Ste5-Myc9 | ste5Δ | ADH2-p-TAgNLS-(GFP)2 | Dex | 15±5.6 | 6±1.4 | 14±4.4 |
| 10. Ste5-Myc9 | ste5Δ | STE5-p-TAgNLS-STE5 | Dex | 28±7.9 | 14±2.8 | 41±5.0 |
| 11. Ste5-Myc9 | ste5Δ | ADH2-p-TAgNLS-(GFP)2 | Gal | 23±1.7 | 7±1.5 | 24±1.8 |
| 12. Ste5-Myc9 | ste5Δ | GAL1-p-TAgNLS-STE5 | Gal | 52±0.9 | 34±2.5 | 41±1.9 |
Strains were EY1775 (ste5Δ), EY957 (STE5) and EY2320 (ste11Δ)
Cells were grown in 2% of either dextrose (Dex) or galactose (Gal) medium
Ste5Δ474-487-Myc9 and Ste5L482/485A-Myc9 were also severely defective in their ability to be recruited to the plasma membrane. In the presence of α factor, Ste5Δ474-487-Myc9 failed to be recruited in any of the cells and Ste5L482/485A-Myc9 was weakly recruited in only a few cells (Table 2, lines 2–3). The correlation between the amount of nuclear accumulation and the amount of plasma membrane recruitment was in agreement with previous work (Mahanty et al., 1999). However, both localization defects were unexpected, because this region of Ste5 is distal to sequences involved in nuclear import or recruitment and did not support either nuclear import or export of heterologous proteins (our unpublished data).
Yeast strains expressing Ste5Δ474-487 and Ste5L482/485A in place of wild-type Ste5 were severely defective in mating pathway functions. Neither mutant could efficiently induce mating-specific transcription (as monitored with a FUS1-lacZ reporter gene), cell cycle arrest in G1 phase (as monitored in a growth inhibition halo assay), or form a significant number of diploids (as monitored in a patch mating assay) (Figure 2B). Ste5Δ474-487 was more defective than Ste5L482/485A in all of the assays, consistent with the relative severity of the two mutations.
Two lines of evidence indicated that the functional defects of Ste5Δ474-487 and Ste5L482/485A were linked to a reduced ability to activate Ste11. First, Ste5Δ474-487 and Ste5L482/485A were unable to efficiently associate with Ste11 in coprecipitation tests. In initial experiments, no interaction was detected between Ste11-Myc and HA3-tagged derivatives of either Ste5Δ474-487 or Ste5L482/485A (our unpublished data). Further analysis with GST-tagged derivatives of the mutants showed that Ste5Δ474-487 was more defective: Ste11-Myc failed to associate with Ste5Δ474-487-GST but could associate with a reduced amount of Ste5L482/485A-GST (Figure 2C). Second, neither Ste5Δ474-487 nor Ste5L482/485A could positively regulate a constitutively active form of Ste11, STE11-4 (Stevenson et al., 1992) that requires Ste5 for most of its basal activity (Figure 2D). Thus, residues 474–487 of Ste5 are critical for Ste11 association and activation.
The simplest interpretation of these findings was that the localization defects of Ste5Δ474-487 and Ste5L482/485A were a secondary consequence of their inability to bind to Ste11. However, two lines of evidence suggested that this was not the case. First, a ste11Δ null mutation did not block nuclear accumulation of wild-type Ste5 (Table 2, lines 7). Second, previous work suggests that Ste11 is not required for plasma membrane recruitment of Ste5 (Pryciak and Huntress, 1998). Collectively, these findings suggest that amino acids 474–487 define all or part of a novel domain that is required for nuclear accumulation and recruitment in addition to Ste11 binding.
Ste5Δ474-487 and Ste5L482/485A Have Enhanced Ability to Oligomerize and Associate with Ste4 and Ste7
Because the Δ474-487 and L482/485A mutations overlap the leucine-rich domain, we determined the ability of Ste5Δ474-487 and Ste5L482/485A to oligomerize. Myc- and HAtagged derivatives of each mutant were coexpressed in yeast and tested for their ability to coimmunoprecipitate from whole cell extracts. Both mutants formed abnormally high levels of homo-oligomers (Figure 3A), although they formed normal levels of hetero-oligomers with wild-type Ste5 (Figure 3B, only Ste5Δ474-487 is shown). Ste5Δ474-487 formed more homo-oligomers than Ste5L482/485 (Figure 3A, bottom two α-HA panels), suggesting that the more severe mutation had a stronger effect on oligomerization. The increase in oligomerization was unlikely to be the result of poor binding to Ste11, because equivalent levels of wild-type oligomers could be detected in wild-type and ste11Δ cells (our unpublished data). Thus, both mutants have an enhanced ability to oligomerize as long as both partners have the mutation.
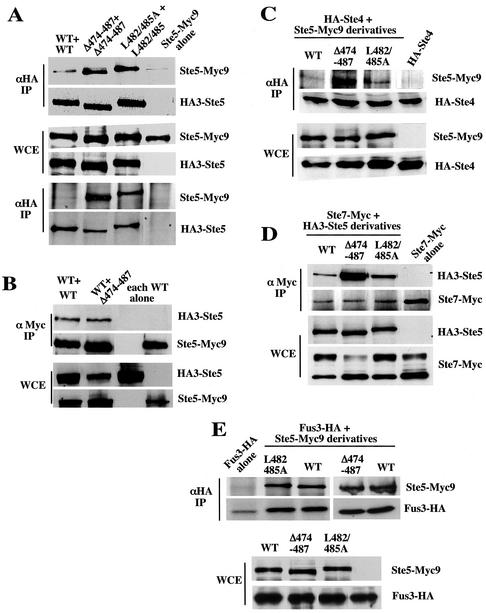
Figure 3.
Ste5Δ474-487 and Ste5L482/485A have an enhanced ability to oligomerize and associate with Ste4 (Gβ) and MKK Ste7. (A) Ste5Δ474-487 and Ste5L482/485A oligomerize more efficiently than wild-type Ste5. WCEs were made from EY1775 (ste5Δ) cells coexpressing Ste5-Myc9 (pSKM19), Ste5Δ474-487-Myc9 (pYMW4), or Ste5L482/485A-Myc9(pYMW37) with HA3-Ste5 (pSKM87), HA3-Ste5Δ474-487 (pYMW106), or HA3-Ste5L482/485A (pYMW66). (B) Ste5Δ474-487 hetero-oligomerizes with wild-type Ste5 at normal levels. WCEs were made from EY1775 coexpressing HA3-Ste5 (pSKM87) with either Ste5-Myc9 (pSKM19) or Ste5Δ474–487-Myc9 (pYMW4). (C–E) Ste5Δ474-487 and Ste5L482/485A associate with more Ste4 and Ste7 than wild-type Ste5. Coprecipitations were carried out by using WCEs from strains expressing HA3- or Myc9-tagged Ste5 with HA-Ste4 (pYMW96), Ste7-Myc (pKC55), or Fus3-HA5 (pYEE102).
The oligomerization results suggested that the Δ474–487 mutation defined a region in Ste5 that normally limited the ability of the RING-H2 domain to oligomerize. To determine whether the increase in oligomerization was dependent on the RING-H2 domain, we deleted the RING-H2 domain from Ste5Δ474-487. However, the double mutant was unstable. Next, we tested whether Ste5Δ474-487 and Ste5L482/485A could oligomerize with a partner that lacked the RING-H2 domain and found that they oligomerized at barely detectable levels, like wild-type Ste5 (our unpublished data, but see discussion of Figure 8C). This finding, taken together with the strong dependence on the RING-H2 domain for oligomerization of Ste5Δ474–638 (Figure 1B), strongly suggested that the enhanced oligomerization of Ste5Δ474-487 and Ste5L482/485A was the result of greater homo-oligomerization of the intact RING-H2 domain.
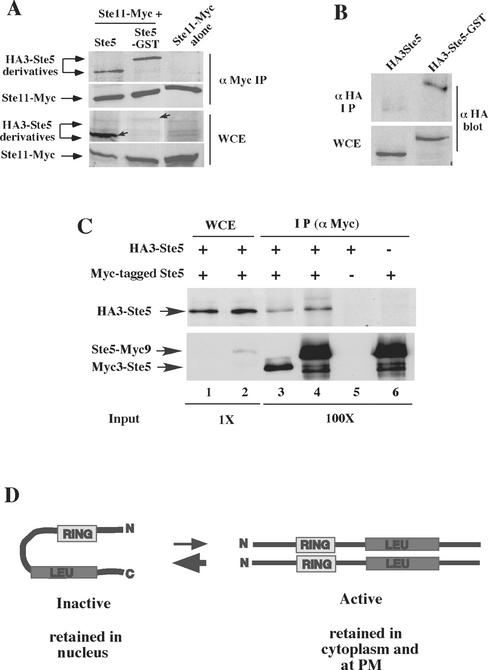
Figure 8.
Ste5-GST associates with more Ste11 and has a more accessible N terminus. (A) Ste5-GST associates with more Ste11. Ste11-Myc was immunoprecipitated from EY1775 cells expressing Ste11-Myc (pNC245) with either HA3-Ste5 (pSKM87) or HA3-Ste5-GST (pYMY74). Strains were grown in 2% galactose medium for 5 h to induce the expression of Ste11-Myc. (B) GST enhances the accessibility of the N terminus of Ste5 to antibody. Immunoblot of the relative level of HA3-Ste5 and HA3-Ste5-GST in whole cell extracts and α-HA (12CA5) immunoprecipitates. Strain EY1775 (ste5Δ) expressed either HA3-Ste5 (pSKM87) or HA3-Ste5-GST (pYMW74). (C) Ste5 oligomerizes at very low levels. Coimmunoprecipitation of Myc- and HA-tagged derivatives of Ste5 was done using extracts from ste5Δ (EY1775) cells cotransformed with HA3-Ste5 (pSKM87) and either Ste5-Myc9 (pSKM19) or Myc3-Ste5 (pYMW138). The left side (WCE) shows the amount of HA3-Ste5, Ste5-Myc9, and Myc3-Ste5 in 4.5 μg of whole cell extract. The right side (IP) shows the amount that is immunoprecipitated with 9E10 from 450 μg of WCE. At this exposure, Myc3-Ste5 is not detected in the WCE, whereas Ste5-Myc9 is detected due to six extra copies of Myc. (D) Model depicting Ste5 undergoing a conformational change from an inactive monomer to an active oligomer. The Ste5 monomer is shown in an autoinhibitory conformation that protects the RING-H2 domain and Ste11 binding site. The oligomer is shown in a more open conformation with more accessible RING-H2 domain and Ste11 binding site.
To further explore whether the RING-H2 domain was more accessible in Ste5Δ474-487 and Ste5L482/485A, we determined whether Ste5Δ474-487 and Ste5L482/485A had an increased ability to associate with Ste4. Strikingly, Ste5Δ474-487 and Ste5L482/485A both associated with more Ste4 than did wild-type Ste5, and Ste5Δ474-487 associated with more Ste4 than did Ste5L482/485A (Figure 3C).
Similar experiments were done with Fus3, which binds next to the RING-H2 domain, and with Ste7, which binds the C terminus of Ste5 (Figure 1A). Ste5Δ474-487 and Ste5L482/485A also had an increased ability to associate with Ste7, with Ste5Δ474-487 exhibiting the larger increase (Figure 3D). In contrast, both mutants associated with wild-type levels of Fus3 (Figure 3E), indicating that the mutations specifically affected interactions with the N- and C-terminal binding partners. Thus, Ste5Δ474-487 and Ste5L482/485A seem to have an altered conformation that makes the RING-H2 domain more accessible for oligomerization and binding to Ste4 and the C termini more accessible to Ste7.
A msn5Δ Mutation Restores Efficient Nuclear Accumulation to SteΔ474-487 and Ste5L482/485A
We next determined why Ste5Δ474-487 and Ste5L482/485A failed to accumulate in nuclei. Decreased nuclear accumulation can either be the result of a block in nuclear import or an increase in nuclear export. Two pieces of evidence suggested that Ste5Δ474-487 and Ste5L482/485A could be imported into nuclei. First, a Ste5(1–242) fragment that overlaps the NLS and the RING-H2 domain accumulated in as many nuclei as did full-length Ste5 (our unpublished data). Second, Ste5C180A-Myc9, which lacks a functional RING-H2 domain, accumulated in ∼10% more nuclei than Ste5-Myc9 in side-by-side comparisons. Thus, all of the information required for nuclear import of Ste5 resides in the first 242 amino acids of the protein and is not dependent on the status of either the leucine-rich domain or the RING-H2 domain.
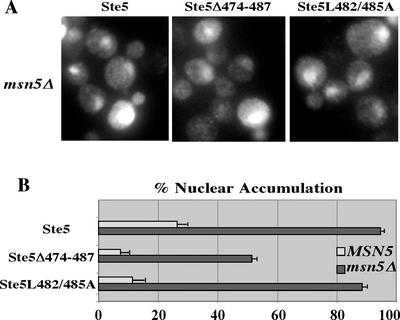
These findings suggested that Ste5Δ474-487 and Ste5L482/485A failed to accumulate in nuclei because they were more efficiently exported. To test this hypothesis, we looked at their localization in a msn5Δ strain that lacks the major exportin required for nuclear export of Ste5 (Mahanty et al., 1999). Lack of Msn5 causes Ste5 to accumulate in ∼90% of nuclei as long as it has a functional NLS. If Ste5Δ474-487 and Ste5L482/485A were defective in nuclear import, then they should not accumulate in msn5Δ nuclei. In contrast, if they were more efficiently exported, then they should accumulate in msn5Δ nuclei. Strikingly, Ste5Δ474-487 and Ste5L482/485A accumulated in a high percentage of msn5Δ nuclei (Figure 4, A and B), confirming our prediction. Thus, Ste5Δ474-487 and Ste5L482/485A are more efficiently exported than wild-type Ste5, and the Δ474–487 mutation defines a region in Ste5 that controls its accessibility to the nuclear export machinery in addition to Ste4 and Ste7.
Figure 4.
Ste5Δ474-487 and Ste5L482/485A are more efficiently exported than wild-type Ste5. (A) Mutation of the Msn5 exportin relocates Ste5Δ474-487 and Ste5L482/485A to the nucleus. Indirect immunofluoresence of vegetatively dividing wild-type (EY699) and msn5Δ (ELY357) cells expressing Ste5-Myc9 (pSKM19), Ste5Δ474-487-Myc9 (pYMW4), and Ste5L482/485A-Myc9 (pYMW37). (B) Quantitation of nuclear accumulation in MSN5 and msn5Δ cells.
Wild-Type Ste5 Suppresses the Nuclear Accumulation Defects of Ste5Δ474-487 and Ste5L482/485A
We wondered whether enhanced nuclear export was linked to the formation of homo-oligomers with an altered conformation. If this was the case, then Ste5Δ474-487 and Ste5L482/485A might be poorer substrates for export if they hetero-oligomerized with wild-type Ste5. This possibility was supported by the observation that the mutants formed fewer oligomers with a wild-type partner than they did with themselves (Figure 3, A and B). We tested whether formation of mutant × wild-type hetero-oligomers correlated with less nuclear export, by comparing the localization of Ste5Δ474-487-Myc9 and Ste5L482/485A-Myc9 in ste5Δ and STE5 strains. The presence of wild-type Ste5 partially suppressed the nuclear accumulation defect of both mutants, leading to more nuclear accumulation, in addition to better recruitment of Ste5L482/485A (Table 2, lines 2, 3, 5, and 6). Thus, the enhanced nuclear export of Ste5Δ474-487 and Ste5L482/485A could be linked to the formation of homo-oligomers that have an altered conformation.
TAgNLS-Ste5 Drives Ste5-Myc9 to the Nucleus and Plasma Membrane
To further explore the possibility that Ste5 oligomers are recruited from a nuclear pool, we determined whether a nuclear localized form of Ste5, TAgNLS-Ste5, would stimulate nuclear accumulation and plasma membrane recruitment of wild-type Ste5, which is predominantly cytoplasmic (Mahanty et al., 1999). TAgNLS-Ste5 shuttles continuously through the nucleus but is predominantly nuclear in the absence and presence of mating pheromone due to greater reimport from the additional strong NLS. As a consequence, it is poorly recruited to the plasma membrane. We coexpressed untagged TAgNLS-Ste5 with Ste5-Myc9 and monitored nuclear accumulation and plasma membrane recruitment of Ste5-Myc9 by indirect immunofluorescence. TAgNLS-Ste5 was expressed at low levels from the STE5 promoter and at high levels from the GAL1 promoter (Table 2, lines 8–12). Remarkably, a low level of expression of TAgNLS-Ste5 was sufficient to increase the residency of Ste5-Myc9 in the nucleus during vegetative growth (-α factor) and cause a much larger increase in its recruitment in the presence of α factor. Greater expression of TAgNLS-Ste5 caused even greater nuclear accumulation of Ste5-Myc9 and a high level of recruitment. Moreover, more Ste5-Myc9 remained in the nucleus in the presence of α factor. Thus, the size of the nuclear pool of Ste5 is a rate-limiting factor in the amount that is recruited to the plasma membrane.
Three control experiments confirmed that the changes in Ste5-Myc9 localization were dependent on coexpression of nuclear-localized Ste5 and were not secondary consequences of sequestration of importins or exportins that normally regulate Ste5. First, the expression of another TAgNLS-tagged nuclear protein (TAgNLS-GFP-GFP) did not increase nuclear accumulation or recruitment of Ste5-Myc9 (Table 2, lines 9 and 11). Second, a msn5Δ mutation in the major exportin for Ste5 decreased Ste5-Myc9 recruitment (our unpublished data), suggesting that sequestration of Ste5 exportins is more likely to interfere with recruitment rather than enhance it. Third, cooverexpression of other Msn5 cargo (i.e., Far1 and Cdc24) did not increase nuclear accumulation or recruitment of Ste5-Myc9 (our unpublished data). Collectively, these findings support the possibility that TAgNLS-Ste5 × Ste5 hetero-oligomers are imported into the nucleus and subsequently recruited to the plasma membrane.
Fusing GST to Ste5 Greatly Increases the Pool of Oligomers and Stimulates Ste5 Activity and Recruitment
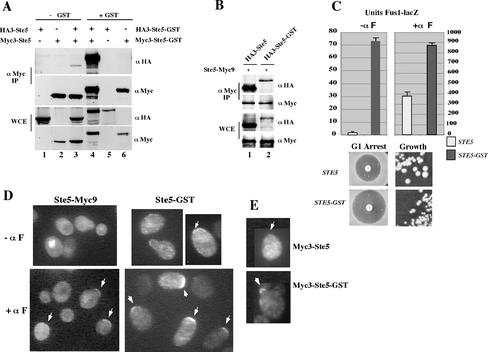
Our results raised the interesting possibility that Ste5 oligomers are more efficiently exported from the nucleus and recruited to the plasma membrane than are Ste5 monomers. We therefore tested whether artificially increasing the level of Ste5 oligomers would increase the pool of Ste5 that is exported and recruited. To make our analysis directly comparable with previous studies (Choi et al., 1994; Yablonski et al., 1996; Inouye et al., 1997a; Feng et al., 1998), we fused Ste5 to GST. We first determined whether GST enhances the formation of Ste5 oligomers in yeast whole cell extracts, by using HA- and Myc-tagged derivatives of Ste5 and Ste5-GST. Significantly more Myc3-Ste5-GST coprecipitated with HA3-Ste5-GST compared with the amount of Myc3-Ste5 that coprecipitated with HA3-Ste5 (Figure 5A, compare lanes 3 and 4). Densitometric analysis of duplicate samples revealed a 137-fold increase in the level of Ste5-GST oligomers compared with Ste5 oligomers. In contrast, HA3-Ste5-GST X Ste5-Myc9 hetero-oligomers were not more abundant than HA3-Ste5 × Ste5-Myc9 homo-oligomers (Figure 5B), demonstrating that the increase in Ste5-GST homo-oligomerization was due to interactions between the GST moieties. Therefore, the GST tag increases the level of Ste5 homo-oligomers to a point were they constitute most of the total pool.
Figure 5.
Increasing the pool of Ste5 oligomers by fusing it to GST stimulates Ste5 activity and recruitment to the plasma membrane. (A) Ste5-GST oligomerizes more efficiently. Myc- and HA-tagged derivatives of Ste5 were coimmunoprecipitated from extracts made from EY1775 (ste5Δ) cells coexpressing HA3-Ste5 (pSKM87) with Myc3-Ste5 (pYMW138), and HA3-Ste5-GST (pYMW74) with Myc3-Ste5-GST (pYMW134). (B) Enhanced oligomerization of Ste5-GST requires the presence of GST on both partners. Relative amount of Ste5-Myc9 that coimmunoprecipitates with HA3-Ste5 and HA3-Ste5-GST. (C) Ste5-GST hyperactivates the mating pathway. Quantitation of FUS1-lacZ activity, G1 arrest, and vegetative growth of EY1775 cells expressing Ste5 (pYBS138) and Ste5-GST (pYMW50) from CEN plasmids. For FUS1-lacZ assays, the cells also expressed FUS1-lacZ (pJB207). Strains were induced for 90 min with 50 nM α factor where indicated (+ α factor). (D) Ste5-GST is more readily detected at the plasma membrane than Ste5-Myc9. Indirect immunofluorescence of Ste5-GST and Ste5-Myc9. EY1775 (ste5Δ) cells expressing Ste5-Myc9 (pSKM19) or Ste5-GST (pYMW77) were grown at 30°C with (+αF) or without (-αF) a 15-min exposure to α factor. Ste5-Myc9 was detected with 9E10 antibody and Ste5-GST was detected with anti-GST antisera. (E) Myc3-Ste5-GST is recruited more efficiently than Myc3-Ste5. Indirect immunofluorecence was done on EY1775 (ste5Δ) cells expressing Myc3-Ste5 (pYMW138) or Myc3-Ste5-GST (pYMW134) after 30-min induction with α factor. Both proteins were detected with α-Myc (9E10) antibody.
The functional competency of Ste5-GST was determined by expressing it at native levels from its own promoter in a ste5Δ null strain and measuring various outputs of the pheromone response pathway. Ste5-GST was very hyperactive and constitutively activated the mating MAPK cascade, as shown by a 40-fold increase in β-galactosidase activity from the FUS1-lacZ reporter gene (Figure 5C, units Fus1-lacZ - αF) and slower vegetative growth compared with wild-type Ste5 (Figure 5C). Ste5-GST was also hyperactive in the presence of α factor and induced more FUS1-lacZ expression (Figure 5C, units Fus1-lacZ + αF), growth inhibition in a halo assay (Figure 5C) and diploid formation (Figure 6C). The enhanced pathway activation was not due to stabilization of Ste5 by the GST moiety, because the steady-state levels of HA3-Ste5-GST and Myc3-Ste5-GST were no greater than that of HA3-Ste5 and Myc3-Ste5 (Figure 5A, whole cell extract [WCE] panel compare lanes 3 and 4). Thus, Ste5-GST oligomers are much more active than wild-type Ste5 and do not require α factor to induce the mating pathway.
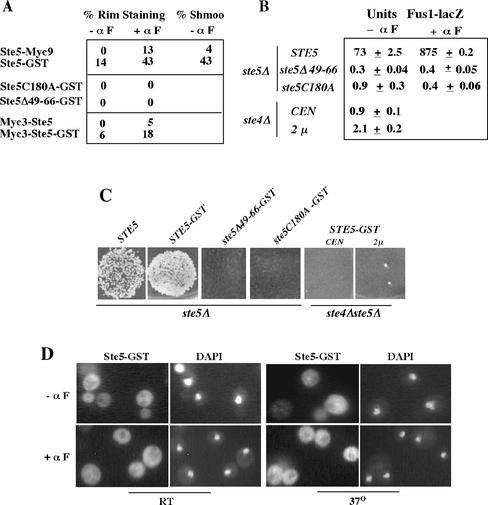
Figure 6.
Ste5-GST requires its RING-H2 domain, residues 49–66, and Ste4 to be recruited and activate the pathway. (A) Tally of plasma membrane recruitment of Ste5 constructs. EY1775 (ste5Δ) cells expressed Ste5-Myc9 (pSKM19), Ste5-GST (pYMW77), Myc3-Ste5 (pYMW138), Myc3-Ste5-GST (pYMW134), Ste5C180A-GST (pYMW97), or Ste5Δ49–66-GST (pYMW99). (B) Ste5-GST requires its RING-H2 domain, amino acids 47–66, and Ste4 to basally induce the mating pathway. Fus1-lacZ activity assessed in ste5Δ (EY1775) and ste4Δ (2468) cells harboring FUS1-lacZ (pJB207) with either CEN STE5-GST (pYMW50), CEN ste5Δ49-66-GST (pYMW100), CEN ste5C180A-GST (pYMW98), or 2μ STE5-GST (pYMW77). (C) Ste5-GST requires its RING-H2 domain, residues 47–66 and Ste4 to promote mating. Qualitative patch mating of a ste4Δ ste5Δ (EYL1808) strains harboring CEN STE5-GST (pYMY50) or 2μ STE5-GST (pYMY77) plasmids compared with a ste5Δ strain harboring CEN STE5 (pYBS138), CEN STE5-GST (pYMW50), and CEN ste5C180A-GST (pYMW98), and CEN ste5Δ49-66-GST (pYMW100). (D) Ste5-GST is poorly recruited in rsl1-4 cells. Ste5-GST was detected by indirect immunofluorescence as in Figure 5D. Cells grown at 25°C and then at 37°C for 1.5 h and compared with a wild-type control (FY23), which gave results similar to those in Figure 5D. Cells were incubated without (-αF) or with (+αF) 5 μM α factor for 15 min.
Ste5-GST was also more efficiently recruited to the plasma membrane compared with Ste5-Myc9, both during vegetative growth and in the presence of α factor (Figure 5D). Weak constitutive recruitment of Ste5-GST to the cell cortex could be detected in ∼14% of vegetatively dividing cells, compared with no detectable recruitment for Ste5-Myc9 (Figure 6A, % rim staining -αF). This basal recruitment was asymmetric and nearly always restricted to one side of the cell, as found for Ste5-Myc9 after brief α factor induction (Mahanty et al., 1999). Ste5-GST also underwent significantly enhanced recruitment to the cell cortex after brief (15-min) α factor induction, resulting in strongly detectable recruitment of Ste5-GST in 43% of cells. In contrast, Ste5-Myc9 could be detected at the plasma membrane of only 13% of cells. The Ste5-GST cells were also more enlarged and shmoo-like than the Ste5 cells (Figures 5D and 6A), suggesting that the enhanced recruitment induces more polarized growth.
To verify that the apparent increase in the recruitment of Ste5-GST compared with Ste5-Myc9 was not a secondary consequence of using different primary antibodies, we compared the ability of Myc3-Ste5 and Myc3-Ste5-GST to be recruited using the same α-Myc antibody. The Myc3-tagged proteins were much more difficult to detect than Ste5-Myc9 as a result of six fewer copies of the Myc epitope. Nevertheless, this direct comparison demonstrated that Myc3-Ste5-GST is more efficiently recruited to the plasma membrane than Myc3-Ste5 (Figures 5E and 6A). Thus, the GST tag simultaneously increases the pool of Ste5 that is oligomerized and stably recruited to the plasma membrane.
Ste5-GST Must Shuttle through the Nucleus and Be Recruited to Ste4 to Activate the Pathway
Prior genetic analysis led to the conclusion that oligomerization of Ste5 occurs as a consequence of binding to Gβγ dimers and that artificial oligomerization bypasses the need for binding to the Ste4 Gβ subunit (Inouye et al., 1997a). This conclusion was based on the ability of a Ste5-GST mutant derivative [Ste5(C177AC180A)-GST] to restore mating to a ste4Δ ste5Δ strain when it was overexpressed from the GAL1 promoter. However, nuclear shuttling and recruitment to Gβγ are critical for pathway activation when Ste5 is expressed at native levels (Mahanty et al., 1999). In addition, the GAL pathway is known to induce the expression of mating specific genes (Dolan and Gatlin, 1995; Ideker et al., 2001).
We determined whether the enhanced activity of Ste5-GST was dependent on recruitment, by testing for a functional dependence on the RING-H2 domain and Ste4. The C180A RING-H2 domain mutation, which blocks the association of Ste5 with Ste4 (Feng et al., 1998), completely abrogated the recruitment of Ste5-GST to the plasma membrane both in the absence and presence of mating pheromone (Figure 6A) and completely disrupted its function (Figure 6, B and C). In addition, a ste4Δ mutation in the Gβ subunit completely blocked the function of Ste5-GST, even when it was overexpressed from a multicopy 2μ plasmid (Figure 6, B and C). Thus, the RING-H2 domain and Ste4 are both absolutely required for Ste5-GST to activate the MAPK cascade, and oligomerization does not bypass the need for binding to Ste4. These results argue that pathway activation is dependent on recruitment of preformed Ste5 oligomers to Gβγ.
As a control, we also determined whether Ste5-GST must shuttle through the nucleus to be recruited to the plasma membrane, because this is the case for wild-type Ste5. The localization of Ste5-GST was dependent on amino acid residues required for nuclear localization of wild-type Ste5. A Ste5Δ49-66-GST derivative was unable to accumulate in nuclei (our unpublished data) or be recruited (Figure 6A) and was devoid of function (Figure 6, B and C). Furthermore, the recruitment of Ste5-GST to the plasma membrane was also dependent on the β-importin Rsl1/Kap95 and the nucleoporin Nsp1, which regulate nuclear import of wild-type Ste5 (Mahanty et al., 1999). Temperature sensitive rsl1-4 and nsp1ts mutations (Nehrbass et al., 1993; Seedorf and Silver, 1997) blocked the recruitment of Ste5-GST during vegetative growth at both permissive (25°C) and nonpermissive (37°C) temperatures, and greatly reduced recruitment at nonpermissive temperature in the presence of α factor (Figure 6D; shown is rsl1-4; see MATERIALS AND METHODS for details on nsp1ts). Consistent with this, Ste5-GST was also unable to induce morphological changes at either temperature in either mutant. Thus, recruitment of Ste5-GST is dependent on the same nuclear import machinery that regulates wild-type Ste5.
Fusing GST to TAgNLS-Ste5 Induces Its Export from the Nucleus
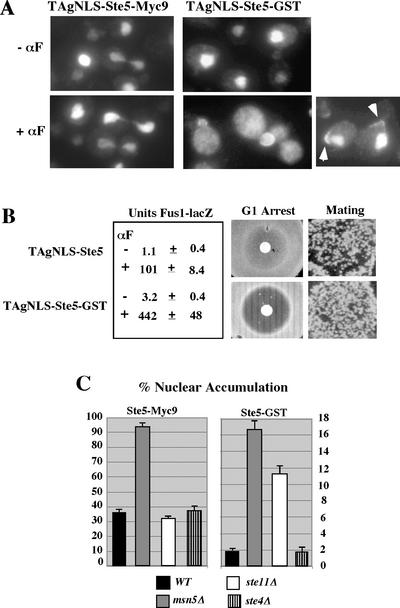
We next determined whether increasing the pool of Ste5 oligomers increases the pool of Ste5 that is exported from the nucleus. We used the predominantly nuclear TAgNLS-Ste5 derivative to determine whether fusion of GST to Ste5 increases its export from the nucleus. TAgNLS-Ste5 was predominantly nuclear both in the absence and presence of α factor (Figure 7A) and unable to efficiently activate the mating MAPK cascade (Figure 7B), presumably as a result of its reimport into the nucleus. Similarly, TAgNLS-Ste5-GST was also predominantly nuclear during vegetative growth (Figure 7A) and did not constitutively activate the pathway (Figure 7B), indicating that it is readily imported into the nucleus. In contrast, during α factor stimulation, a much greater pool of TAgNLS-Ste5-GST was in the cytoplasm of budded cells and at the plasma membrane of unbudded cells compared with TAgNLS-Ste5 (Figure 7A). The better recruitment of TAgNLS-Ste5-GST correlated with much greater pathway activation and projection formation (Figure 7, A and B). Thus, fusion of GST to TAgNLS-Ste5 promotes its export from the nucleus and recruitment to the plasma membrane in the presence of α factor, suggesting that Ste5 oligomers are more efficiently exported in addition to being more efficiently recruited.
Figure 7.
Ste5-GST is more efficiently exported from the nucleus. (A) Fusion of GST to TAgNLS-Ste5 induces its export and recruitment. Indirect immunofluorescence of TAgNLS-Ste5-Myc9 and TAgNLS-Ste5-GST expressed from 2μ plasmids pSKM47 and pYMW82 in strain EY1775. TAgNLS-Ste5-Myc9 was detected with 9E10 antibody and GST was detected with anti-GST antisera. (B) TAgNLS-Ste5-GST has an enhanced ability to activate the mating pathway. TAgNLS-Ste5-Myc9 and TAgNLS-Ste5-GST were expressed from CEN plasmids pSKM44 and pYMW81 in strain EY1775. Pheromone responses were assayed as described in MATERIALS AND METHODS. (C) Ste5-GST shuttles but is retained in the cytoplasm by Ste11. Nuclear accumulation of Ste5-GST and Ste5-Myc9 quantitated in WT, msn5Δ, ste11Δ, and ste4Δ cells that had been subjected to indirect immunofluorescence to detect either Ste5-GST or Ste5-Myc9 and stained with 4,6-diamidino-2-phenylindole to detect nuclear DNA.
Ste5-GST Is More Efficiently Exported and Is Retained by Ste11 in the Cytoplasm
The enhanced nuclear export of TAgNLS-Ste5-GST strongly suggested that Ste5-GST was also more efficiently exported from the nucleus. Interestingly, Ste5-GST accumulated in only 2% of total nuclei in ste5Δ cells, suggesting that it may be more efficiently exported, as found for Ste5Δ474-487 and Ste5L482/485A. We therefore compared the ability of Ste5-GST to accumulate in the nuclei of MSN5 and msn5Δ cells (Figure 7C). The msn5Δ mutation increased nuclear accumulation of Ste5-GST by more than eightfold compared with a less than threefold increase for Ste5-Myc9. The greater fold-increase in nuclear accumulation for Ste5-GST compared with Ste5 indicates that Ste5-GST is more efficiently exported than Ste5. Ste5-GST accumulated in fewer nuclei than wild-type Ste5 in the msn5Δ strain, suggesting that it is also more efficiently exported by Msn5-independent export pathways that operate in the absence of Msn5 (Mahanty et al., 1999) and involve multiple exportins (Wang and Elion, unpublished data). These findings suggest that Ste5-GST is more efficiently exported to the cytoplasm than wild-type Ste5 both in the absence and presence of mating pheromone.
The fact that Ste5-GST constitutively activates the mating MAPK cascade raised the possibility that it mimics the effects of α factor and induces its own export. We therefore determined whether mutations that block MAPK cascade activation would increase nuclear accumulation of Ste5-GST. A ste4Δ mutation did not increase nuclear accumulation of Ste5-GST, indicating that Ste5-GST does not induce its own export in the absence of α factor (Figure 7C). Interestingly, however, a ste11Δ mutation caused a sixfold increase in nuclear accumulation of Ste5-GST, with no effect on nuclear accumulation of Ste5-Myc9 (Figure 7C). Ste11 is cytoplasmic and largely excluded from nuclei (Mahanty et al., 1999), suggesting that it retains Ste5-GST in the cytoplasm. Thus, the low level of nuclear accumulation of Ste5-GST compared with Ste5-Myc9 is the combined effect of more efficient export from the nucleus and better retention in the cytoplasm by Ste11.
Ste5-GST Associates with More Ste11 and Has a More Accessible N Terminus
A prediction from the localization results is that Ste5-GST should associate with more Ste11 than wild-type Ste5. We compared the ability of Ste11-Myc to associate with HA3-Ste5 and HA3-Ste5-GST in coprecipitation tests. Ste11-Myc was expressed from the GAL1 promoter whereas the Ste5 derivatives were constitutively expressed from the STE5 promoter, allowing formation of a steady-state pool of Ste5 oligomers before the expression of Ste11. It was not possible to express HA3-Ste5-GST to as high a level as HA3-Ste5 in cells that also expressed Ste11-Myc, because of hyperactivation of the MAPK cascade. Therefore, the abundance of HA3-Ste5-GST in the whole cell extracts was much lower than that of HA3-Ste5 (Figure 8A, WCE). In sharp contrast, equivalent amounts of HA3-Ste5-GST and HA3-Ste5 coprecipitated with Ste11-Myc (Figure 8A, α Myc IP), indicating that a much greater percentage of the total pool of HA3-Ste5-GST was associated with Ste11 compared with that of HA3-Ste5. Thus, Ste5-GST oligomers have an enhanced ability to associate with Ste11.
The large increase in the association of Ste5-GST with Ste11 suggested that the Ste11 binding domain was more accessible, raising the possibility that the Ste5-GST oligomers have a more open conformation as suggested for Ste5Δ474-487 and Ste5L482/485A. To test this possibility, we determined whether the N terminus of the Ste5-GST fusion was more accessible than that of wild-type Ste5, by comparing the ability of the 12CA5 antibody to immunoprecipitate HA3-Ste5 and HA3-Ste5-GST. Remarkably, HA3-Ste5-GST was more efficiently immunoprecipitated than HA3-Ste5 (Figure 8B). Similar results were found with Myc3-Ste5 and Myc3-Ste5-GST when they were expressed individually (our unpublished data) or together (Figure 5A, α-Myc panels, lanes 3 and 4). Thus, the N terminus of Ste5-GST is more accessible to the antibody, suggesting that it has a more open conformation.
Only a Minor Fraction of the Total Pool of Ste5 Is Oligomerized in Diluted Whole Cell Extracts
Our analysis argued that the formation of a Ste5 oligomer is a key rate-limiting step in determining the ability of Ste5 to be recruited to the plasma membrane and activate the pathway. We therefore estimated the fraction of total Ste5 that is oligomerized in yeast whole cell extracts, by assessing the relative ability of functional epitope-tagged derivatives of Ste5 to associate in the coimmunoprecipitation assay. Figure 8C shows that roughly <1% of the total input of HA3-Ste5 coprecipitated with either Myc3-Ste5 or Ste5-Myc9, suggesting that the majority of HA3-Ste5 is monomeric. Similar results were obtained when the immunoprecipitation was performed in the opposite direction, and α factor treatment did not affect the total amount of oligomerization detected (our unpublished data). Thus, the majority of Ste5 is likely to be monomeric, suggesting that oligomerization is a tightly regulated event.
DISCUSSION
Oligomerization Constitutively Activates Ste5
Although indirect genetic evidence suggested that oligomerization of Ste5 is required for its function, it was not known whether Ste5 oligomers transduce the signal through the mating MAPK cascade. Herein, we show that increasing the pool of Ste5 oligomers with a GST fusion induces strong constitutive activation of the mating MAPK cascade, suggesting that Ste5 oligomers are the active form of Ste5 during signal transmission. Several lines of evidence argue that the increase in Ste5 activity is dependent on the dimerization property of GST rather than a steric effect that alters the structure of Ste5. First, the effect of GST on oligomerization requires that both partners be fused to GST. Second, GST increases the level of Ste5 oligomers to the point where they constitute the majority of the pool. Third, Ste5 is also hyperactivated when it is fused to a small leucine-zipper that forms stable parallel dimers (our unpublished data) but not when it is fused to GFP, which is the same size as GST but does not efficiently dimerize (Mahanty et al., 1999). These findings, together with previous glycerol gradient sedimentation analysis of GST-Ste5 multikinase complexes (Choi et al., 1994; 1999), are most consistent with the active form of Ste5 being a dimer. The fact that Ste5-GST has multiple enhancements at the level of association with signaling components and localization suggests that oligomerization is a key rate-limiting step in the activation of Ste5.
The Major Form of Ste5 in Cells Is an Inactive Monomer
We found that only a very minor fraction of the total pool of Ste5 forms oligomers in coimmunoprecipitation experiments (Figure 8C). Thus, the majority of Ste5 is monomeric under our coimmunoprecipitation conditions. The low level of Ste5 oligomers suggests that oligomerization is tightly regulated and could require stabilizing factors that are diluted in our extracts. Two lines of evidence lead us to favor the possibility that the major form of Ste5 is an autoinhibited monomer in which contacts between the N- and C-terminal halves of the protein decrease the accessibility of the RING-H2 domain and Ste11 binding site (Figure 8D). First, previous work indicates that the N- and C-terminal halves of Ste5 can associate (Sette et al., 2000). Second, we find that the major pool of Ste5 does not efficiently associate with Ste4 and Ste11 unless Ste5 has an increased capacity to oligomerize. The potential existence of a monomer that protects the RING-H2 domain from oligomerization makes biological sense given the propensity of these domains to form higher order oligomers (Borden, 2000; Kentsis et al., 2002) and the potential for inappropriate recruitment and pathway activation. The fact that Ste5-GST constitutively hyperactivates the MAPK cascade (Figure 5) suggests that too high a pool of oligomers would cause inappropriate pathway activation and provides a rationale for why the steady state level of oligomers needs to be kept low.
The Availability of the RING-H2 Domain Determines the Amount of Oligomerization
We find that oligomerization of full-length Ste5 is largely controlled by the RING-H2 domain, with the leucine-rich domain playing a less critical role (Figure 1B). Interestingly, a good correlation was found between oligomerization and accessibility of Ste5 to proteins that bind to different regions of the protein, including the N terminus (epitope antibody), RING-H2 domain (Ste4), leucine-rich domain (Ste11), and C termini (Ste7) (Figures 3 and 8, A and B). Thus, the oligomer may have a more accessible RING-H2 domain and Ste11 and Ste7 binding sites compared with the monomer (Figure 8D). This possibility is supported by preliminary results suggesting that Ste5-GST has an enhanced ability to bind Ste4 (our unpublished data).
Ste5 may oligomerize before associating with either Ste11 or Ste4, because Ste5 does not require either Ste4 or Ste11 to oligomerize (Yablonski et al., 1996; our unpublished data), associates with more Ste11 and is better recruited as a Ste5-GST fusion (Figure 8A). This interpretation is consistent with the observation that the same RING-H2 domain mutation blocks association with Ste4 as well as oligomerization of the RING-H2 domain (Feng et al., 1998). It is tempting to speculate that Ste5 first undergoes a conformational change to allow oligomerization of the RING-H2 domain before binding to Ste4 and Ste11.
Our interpretation contrasts the view of Sette et al. (2000) who have proposed that the active form of Ste5 undergoes stronger interactions between the N and C halves of the protein, either through intramolecular contacts in a closed monomer or intermolecular contacts in an antiparallel dimer. In their study, hyperactivating mutations in either the N terminus (Ste5 P44L) or C termini (Ste5 S770K) caused better coprecipitation of the N-terminal and C-terminal halves of the protein with no effect on oligomerization. However, the use of GST fusions in this analysis could have interfered with the detection of potential effects of the mutations on oligomerization. In light of our findings, the P44L and S770K mutations would be predicted to increase the accessibility of nearby RING-H2 and leucine-rich domains for oligomerization. This prediction is supported by findings in Sette et al. (2000): 1) GST-Ste5P44L(1–518) associates more readily with Ste5P44L(1–518) than it does with wild-type Ste5(1–518); and 2) GST-Ste5 (1–518) and GST-Ste5P44L(1–518) both associate more readily with full-length Ste5P44L than with full-length Ste5. Furthermore, our model does not rule out the possibility that interactions between the N and C termini of the protein could be involved in activation of Ste5; for example, N to C interactions could occur between dimers or within a dimer (see Elion, 2001 for a discussion).
The Leucine-rich Domain Negatively Regulates the Accessibility of the RING-H2 Domain and Vice Versa
The finding that mutations in the leucine-rich domain increase the ability of Ste5 to oligomerize and to associate with Ste4 suggests that the leucine-rich domain restricts the availability of the RING-H2 domain (Figure 3). Conversely, the fact that a C180A RING-H2 domain mutant oligomerizes more efficiently and has an enhanced capacity to be suppressed by overexpression of Ste11 (Feng et al., 1998), suggests that the RING-H2 domain restricts the availability of the leucine-rich domain and the Ste11 binding site. Although multiple interpretations are possible, the simplest is that the Ste5Δ474-487 and Ste5L482/485 mutations define a domain that makes intramolecular contacts with the RING-H2 domain or a part of the protein that influences its accessibility. An attractive possibility is that these intramolecular interactions maintain Ste5 in an autoinhibitory conformation that limits the accessibility of both the RING-H2 domain and the Ste11 binding site (Figure 8D).
Oligomerization Does Not Bypass a Requirement for Recruitment of Ste5 to Ste4
Previous work led to the conclusion that oligomerization of Ste5 bypasses the need for Ste4 for pathway activation (Inouye et al., 1997a). This conclusion was based on the ability of a Ste5(C177AC180A)-GST RING-H2 domain mutant to suppress the mating defect of a ste4Δ mutant when it was overexpressed from the GAL1 promoter. However, we find that when Ste5-GST or Ste5C180A-GST are expressed from the STE5 promoter at native or overexpressed levels, the RING-H2 domain and Ste4 are both essential for basal and induced signaling. Thus, Ste5 signaling capacity is tightly linked to its recruitment to Gβγ, presumably as a preformed dimer. The previous results can be reconciled by postulating that the need for regulated recruitment to the plasma membrane is bypassed if the amount of Ste5 in the cytoplasm is high enough to allow interactions with Ste20 at the cell cortex. This interpretation is supported by the fact that overexpressed Ste5(C177AC180A)-GST still requires Ste20 to activate the pathway (Sette et al., 2000). It is also supported by the observation that signal transduction can be restored to a Ste5Δ49-66 mutant that fails to shuttle through the nucleus when it is overexpressed (Elion, 2002).
Conformationally Distinct Forms of Ste5 May Localize Differently in the Cell
Our analysis reveals a strong link between oligomerization and nuclear export and recruitment. The major monomeric form of Ste5 shuttles through the nucleus, but is efficiently retained in G1 phase nuclei. In contrast, Ste5 derivatives that oligomerize more readily are more efficiently exported from the nucleus, better retained in the cytoplasm by Ste11 and more efficiently recruited to the plasma membrane. These observations raise the intriguing possibility that oligomerization could be regulated at the level of localization. For example, Ste5 oligomers might accumulate in subcellular compartments with a higher concentration of Ste5 or other proteins that stabilize the oligomer. Negative regulatory events could also prevent the accumulation of oligomers; for example, an inhibitor could bind monomers and block oligomerization or degrade oligomers at the end of a cycle of signaling. A rate-limiting step in recruitment is the amount of Ste5 that is in the nucleus. Thus, it is possible that oligomerization occurs in the nucleus or during nuclear shuttling. Interestingly, key binding partners of Ste5 are differentially distributed in the nucleus (Msn5, Fus3), cytoplasm (Ste11, Ste7), and at the plasma membrane (Ste4), suggesting that Ste5 conformation could be regulated by sequential binding to these proteins. The most obvious potential regulator is Fus3, which phosphorylates a domain near the RING-H2 domain (Kranz, Satterberg, and Elion, unpublished data). However, the available evidence suggests that α factor does not increase oligomerization and Fus3 is not required for nuclear accumulation or recruitment (Wang and Elion, unpublished data), arguing against this possibility.
A second interesting possibility is that Ste5 oligomers form as a result of binding of the nuclear export machinery. This is strongly supported by the tight link between oligomerization, nuclear export and recruitment revealed by comparative analysis of Ste5 mutants (i.e., wild-type Ste5, Ste5Δ474-487, Ste5L282/485A, Ste5-GST, and TAgNLS-Ste5-GST). Further support comes from the observation that amino acid residues in Ste5 that mediate nuclear export of a heterologous protein also influence the accessibility of the RING-H2 domain (Wang and Elion, unpublished data). For example, nuclear exportins might bind to inactive monomers in the nucleus, induce a conformational change that increases the accessibility of the RING-H2 domain and promotes the formation of oligomers, which are exported and subsequently stabilized by binding to Ste11 in the cytoplasm and Ste4 at the plasma membrane. Because Ste11 seems to preferentially bind to Ste5 oligomers, it has the potential to increase the cytoplasmic pool of oligomers by preventing intermolecular interactions between the N- and C-terminal halves of the protein. The binding of Ste4 to the RING-H2 domain has a similar potential. A prediction of this model is that Ste11 should be required for efficient recruitment of Ste5. Remarkably, the behavior of Ste5Δ474-487 fulfills this prediction because it is selectively defective in binding to Ste11 and cannot be recruited even when it's nuclear accumulation defect is suppressed (Table 2).
Acknowledgments
We thank John Pringle, Dan Finley, Johannes Walters, and Maosong Qi for helpful comments on the manuscript. This research was supported by National Institutes of Health grant GM-46962 (to E.A.E.).
References
- Baker, R. (1991). Rapid colony transformation of Saccharomyces cerevisiae. Nucleic Acids Res. 19, 1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden, K.L.B. (2000). RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295, 1103-1112. [DOI] [PubMed] [Google Scholar]
- Brennan, J.A., Volle, D.J., Chaika, O.V., and Lewis, R.E. (2002). Phosphorylation regulates the nucleocytoplasmic distribution of kinase suppressor of Ras. J. Biol. Chem. 277, 5369-5377. [DOI] [PubMed] [Google Scholar]
- Burack, W.R., and Shaw, A.S. (2000). Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 12, 211-216. [DOI] [PubMed] [Google Scholar]
- Choi, K.-Y., Kranz, J.A., Mahanty, S.K., and Elion, E.A. (1999). Characterization of Fus3 localization, Active Fus3 localizes in complexes of varying size and specific activity. Mol. Biol. Cell 10, 1553-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.-Y., Satterberg, B., Lyons, D.M., and Elion, E.A. (1994). Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78, 499-512. [DOI] [PubMed] [Google Scholar]
- Dolan, J.W., and Gatlin, J.E. (1995). A role for the Gal11 protein in pheromone-induced transcription in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 212, 854-860. [DOI] [PubMed] [Google Scholar]
- Elion, E.A. (1995). Ste5, a meeting place for MAP kinases and their associates. Trends Cell Biol. 5, 322-327. [DOI] [PubMed] [Google Scholar]
- Elion, E.A. (2001). The Ste5 Scaffold. J. Cell Sci. 114, 3967-3978. [DOI] [PubMed] [Google Scholar]
- Elion, E.A. (2002). How to monitor nuclear shuttling. In: Methods in Enzymology, Guide to Yeast Genetics and Molecular Biology, ed. G. Fink and C. Guthrie, New York: Academic Press.
- Elion, E.A., Grisafi, P.L., and Fink, G.R. (1990). FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell 60, 649-664. [DOI] [PubMed] [Google Scholar]
- Elion, E.A., Satterberg, B., and Kranz, J.E. (1993). FUS3 phosphorylates multiple components of the mating signal transduction cascade. Evidence for STE12 and FAR1. Mol. Biol. Cell 4, 495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede, B., Gartner, A., Zhou, Z., Nasmyth, K., and Ammerer, G. (1993). MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature 362, 261-264. [DOI] [PubMed] [Google Scholar]
- Farley, F.W., Satterberg, B., Goldsmith, E.J., and Elion, E.A. (1999). Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics 151, 1425-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y., Song, L., Kincaid, E., Mahanty, S.K., and Elion, E.A. (1998). Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 8, 267-278. [DOI] [PubMed] [Google Scholar]
- Gustin, M.C., Albertyn, J., Alexander, M.R., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 62, 1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker, T., Thorsson, V., Ranish, J.A., Christmas, R., Buhler, J., Eng, J.K., Bumgarner, R., Goodlett, D.R., Aebersold, R., and Hood, L. (2001). Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292, 929-934. [DOI] [PubMed] [Google Scholar]
- Inouye, C., Dhillon, N., and Thorner, J. (1997b). Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics 147, 479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye, C., Dhillon, N., and Thorner, J. (1997a). Ste5 RING-H2 domain-Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278, 103-106. [DOI] [PubMed] [Google Scholar]
- Inouye, K., Mizutani, S., Koide, H., and Kaziro, Y. (2000). Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 275, 3737-3740. [DOI] [PubMed] [Google Scholar]
- Kaplan, W., Husler, P., Klump, H., Erhardt, J., Sluiscremer, N., and Dirr, H. (1997). Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase, a detoxification enzyme and fusion protein affinity tag. Protein Sci. 6, 399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis, A., Gordon, R.E., and Borden, K.L.B. (2002). Self-assembly properties of a model RING domain. Proc. Natl. Acad. Sci. USA 99, 667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, J., Satterberg, B., and Elion, E.A. (1994). The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 8, 313-327. [DOI] [PubMed] [Google Scholar]
- Lamson, R.E., Winters, M.J., and Pryciak, P.M. (2002). Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 22, 2932-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B., and Elion, E.A. (1999). The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc. Natl. Acad. Sci. USA 96, 12679-12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw, T., Wu, C.L., Schrag, J.D., Whiteway, M., Thomas, D.Y., and Leberer, E. (1998). Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391, 191-195. [DOI] [PubMed] [Google Scholar]
- Mahanty, S.K., Wang, Y., Farley, F.W., and Elion, E.A. (1999). Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98, 501-512. [DOI] [PubMed] [Google Scholar]
- Maru, Y., Afar, D.E.., Witte, O.N., and Shibuya, M. (1996). The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem. 271, 15353-15357. [DOI] [PubMed] [Google Scholar]
- McTigue, M.A., Williams, D.R., and Tainer, J.A. (1995). Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonicum and its complex with the leading antischistosomal drug praziquantel. J. Mol. Biol. 246, 21-27. [DOI] [PubMed] [Google Scholar]
- Moskow, J.J., Gladfelter, A.S., Lamson, R.E., Pryciak, P.M., and Lew, D.J. (2000). Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 7559-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass, U., Fabre, E., Dihlmann, S., Herth, W., and Hurt, E.C. (1993). Analysis of nucleocytoplasmic transport in a thermosensitive mutant of nuclear pore protein NSP1. Eur. J. Cell Biol. 62, 1-12. [PubMed] [Google Scholar]
- Neiman, A.M., and Herskowitz, I. (1994). Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc. Natl. Acad. Sci. USA 91, 3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A., et al. (2002). Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 22, 3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson, T., and Scott, J.D. (1997). Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075-2080. [DOI] [PubMed] [Google Scholar]
- Pryciak, P.M., and Huntress, F.A. (1998). Membrane recruitment of the kinase cascade scaffold protein Ste5 by the G-βγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12, 2684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, F., Laberge, G., Douziech, M., Ferland-McCollough, D., and Therrien, M. (2002). KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16, 427-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf, M., and Silver, P.A. (1997). Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA 94, 8590-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette, C., Inouye, C.J., Stroschein, S.L., Iaquinta, P.J., and Thorner, J. (2000). Mutational analysis suggests that activation of the yeast pheromone response MAPK pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 11, 4033-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, B.J., Rhodes, N., Errede, B., and Sprague, G.F., Jr. (1992). Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 6, 1293-1304. [DOI] [PubMed] [Google Scholar]
- Tudyka, T., and Skerra, A. (1997). Glutathione S-transferase can be used as an enzymatically active dimerization module for a recombinant protease inhibitor, and functionally secreted into the periplasm of Escherichia coli. Protein Sci. 6, 2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen, F., O'Rourke, S.M., Stucke, V.M., Jaquenoud, M., Neiman, A.M., and Peter, M. (2000). Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10, 630-639. [DOI] [PubMed] [Google Scholar]
- van Drogen, F., Stucke, V.M., Jorritsma, G., and Peter, M. (2001). MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 3, 1051-1059. [DOI] [PubMed] [Google Scholar]
- Whiteway, M.S., Wu, C., Leeuw, T., Clark, K., Fourest-Lieuvin, A., Thomas, D.Y., and Leberer, E. (1995). Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science 269, 1572-1575. [DOI] [PubMed] [Google Scholar]
- Wu, C., Whiteway, M., Thomas, D.Y., and Leberer, E. (1995). Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270, 15984-15992. [DOI] [PubMed] [Google Scholar]
- Yablonski, D., Marbach, I., and Levitzki, A. (1996). Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc. Natl. Acad. Sci. USA 93, 13864-13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, J., Whitmarsh, A.J., Cavanagh, J., Sharma, M., and Davis, R.J. (1999). The JIP group of mitogen-activated protein kinase scaffold proteins, Mol. Cell. Biol. 19, 7245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]